ABSTRACT
The capacity of influenza A viruses to cross species barriers presents a continual threat to human and animal health. Knowledge of the human-swine interface is particularly important for understanding how viruses with pandemic potential evolve in swine hosts. We sequenced the genomes of 141 influenza viruses collected from North American swine during 2002 to 2011 and identified a swine virus that possessed all eight genome segments of human seasonal A/H3N2 virus origin. A molecular clock analysis indicates that this virus—A/sw/Saskatchewan/02903/2009(H3N2)—has likely circulated undetected in swine for at least 7 years. For historical context, we performed a comprehensive phylogenetic analysis of an additional 1,404 whole-genome sequences from swine influenza A viruses collected globally during 1931 to 2013. Human-to-swine transmission occurred frequently over this time period, with 20 discrete introductions of human seasonal influenza A viruses showing sustained onward transmission in swine for at least 1 year since 1965. Notably, human-origin hemagglutinin (H1 and H3) and neuraminidase (particularly N2) segments were detected in swine at a much higher rate than the six internal gene segments, suggesting an association between the acquisition of swine-origin internal genes via reassortment and the adaptation of human influenza viruses to new swine hosts. Further understanding of the fitness constraints on the adaptation of human viruses to swine, and vice versa, at a genomic level is central to understanding the complex multihost ecology of influenza and the disease threats that swine and humans pose to each other.
IMPORTANCE The swine origin of the 2009 A/H1N1 pandemic virus underscored the importance of understanding how influenza A virus evolves in these animals hosts. While the importance of reassortment in generating genetically diverse influenza viruses in swine is well documented, the role of human-to-swine transmission has not been as intensively studied. Through a large-scale sequencing effort, we identified a novel influenza virus of wholly human origin that has been circulating undetected in swine for at least 7 years. In addition, we demonstrate that human-to-swine transmission has occurred frequently on a global scale over the past decades but that there is little persistence of human virus internal gene segments in swine.
INTRODUCTION
Influenza A viruses (IAVs) circulating in domestic swine populations present important economic concerns for the swine industry and a pandemic threat for humans. The H1N1 influenza pandemic of 2009 highlighted the risk that genetically diverse IAVs in swine (swIAVs) present for humans and the importance of understanding the evolutionary processes that generate their diversity (1, 2). Swine infected with IAVs of both avian and human origin have the capacity to generate novel viruses with genome segments of multiple host origins through reassortment and have therefore been referred to as “mixing vessels” (3). The influenza A virus genome is comprised of eight discrete genome segments: PB2, PB1, PA, HA, NP, NA, MP, and NS. Of these, those encoding hemagglutinin (HA) and neuraminidase (NA) are the most antigenically important and define the three subtypes that circulate in swine: H1N1, H1N2, and H3N2. These swine subtypes are similar to those presently found in humans (H1N1 and H3N2) but represent only a subset of those found in wild birds (16 HA subtypes and 9 NA subtypes), which are thought to be the natural reservoir for IAVs (4). Spillover events of IAVs occur frequently between host species, as exemplified by the isolation of avian H5N1 (5), avian H7N9 (6), and swine H3N2 “variant” (H3N2v) (7) viruses from humans in recent years. To date, however, none of these viruses have sustained onward transmission in humans, and the various barriers to successful adaptation to a new mammalian host species represent one of the most important outstanding questions in influenza virus biology (8, 9).
A characteristic that distinguishes swine from other mammalian IAV hosts, including humans, horses, canines, and seals, is the number of IAVs from a different host (in this case, mainly humans) that have successfully adapted to onward transmission in swine (10–15). The global frequency of human-to-swine transmission of the 2009 H1N1 pandemic virus (pH1N1) in recent years reinforces the importance of “reverse zoonosis” of human viruses as a major source of IAV diversity in swine (16–19). In North American swine, these recently introduced pH1N1 viruses cocirculate with several major IAV lineages in swine, including triple-reassortant H3N2 swine viruses, human-origin H1N2 (δ-1) swine viruses, and classical H1N1 (γ) swine viruses (20), with frequent reassortment between lineages (17, 21). Since the 1970s, swine in Europe have been infected with Eurasian H1N1 viruses of avian origin and H3N2 viruses of human origin (22), and pH1N1 viruses now circulate as well (18). Both North American and European swIAV lineages have been introduced into Asian swine populations, with notable proliferation of the Eurasian avian-like H1N1 lineage in the last decade (23). Surveillance for IAVs in swine is limited in South America, Africa, and Australia, but viruses of human origin, including pH1N1, have been detected on all three continents in recent years (24–26).
To further understand the evolutionary mechanisms that generate the extensive genetic diversity of swIAVs in North America, particularly the role of human-to-swine transmission, we performed whole-genome sequencing (coding regions) of 141 IAVs collected from North American swine during 2003 to 2011. We characterize a new swine virus of wholly human seasonal H3N2 origin that has circulated undetected in swine for at least 7 years. To provide a broader historical context for this unusual swine virus, we also estimated the global frequency of introductions of human seasonal IAVs into swine since the 1960s. Our time-scaled phylogenetic analysis of globally available whole-genome swIAV sequence data reveals that human-to-swine introductions occur frequently but that transmission of human internal gene segments in swine is a comparatively rare event.
MATERIALS AND METHODS
Data collection and sample preparation.
For the years 2002 to 2011, influenza A virus samples collected from swine from routine diagnostic submissions were randomly selected from the influenza virus archive at the University of Minnesota Veterinary Diagnostic Laboratory (UMVDL). These samples were selected to best represent the full time period and the three geographical regions of U.S. hog production: the southeast region (U.S. states of Alabama, Georgia, Kentucky, North Carolina, Tennessee, and Virginia), the south-central/west region (Arkansas, Colorado, New Mexico, Oklahoma, and Texas), and the midwest region (Iowa, Illinois, Indiana, Kansas, Minnesota, Missouri, and Nebraska). Samples from swine in Canada (2005 to 2011) and Mexico (2010 to 2011) also were selected from UMVDL, as available. Original specimen material (nasal swab supernatant or lung tissue homogenate stored at −80°C) was aliquoted and sent to the J. Craig Venter Institute (JCVI) in Rockville, MD, for sequencing.
IAV genome sequencing.
The complete genomes of 141 influenza viruses collected from North American swine were sequenced at JCVI. Viral RNA was isolated using the ZR 96 Viral RNA kit (Zymo Research Corporation, Irvine, CA, USA). The influenza A genomic RNA segments were simultaneously amplified from 3 μl of purified RNA using a multisegment RT-PCR strategy (27, 28). The multisegment reverse transcription-PCR (M-RT-PCR) amplicons were sequenced using Nextera Library construction fusing the MiSeq platform (Illumina, Inc., San Diego, CA, USA). Additionally, M-RT-PCR amplicons sheared for 7 min and Ion Torrent-compatible barcoded adapters were ligated to create 200-bp libraries that were purified and sequenced using Ion Torrent (Life Technologies, Grand Island, NY, USA).
Phylogenetic analysis.
In addition to the 141 viral genomes sequenced for this study, 1,404 whole-genome sequences from swIAVs collected globally during 1931 to 2012 were downloaded from the Influenza Virus Resource (29) at GenBank. Sequence alignments for each of the six internal gene segments (PB2, PB1, PA, NP, MP, and NS), as well as H1, H3, N1, and N2 separately, were generated using MUSCLE v3.8.31 (30), with manual correction using Se-Al v2.0 (31). Neighbor-joining trees were inferred using PAUP (32) for each of the eight viral genome segments to identify which lineage each swine virus segment was most closely related to: (i) North American classical swine viruses, (ii) North American triple reassortant swine viruses, (iii) Eurasian avian-like swine viruses, (iv) human pandemic 2009 H1N1 viruses, (v) human seasonal influenza viruses (H3N2 or H1N1), or (vi) avian influenza viruses (data available upon request). We then focused our analysis on the evolutionary relationships between human seasonal influenza A viruses and the set of swine viruses found to be closely related to human seasonal viruses of the H1N1, H1N2, and H3N2 subtypes (see Table S2 in the supplemental material). Due to the relatively high level of sampling of IAVs from U.S. swine during 2009 to 2013 compared to other geographical regions and time periods, 100 of these viruses were randomly selected for inclusion in the study after it was determined that none of the excluded viruses belonged to an unrepresented swIAV lineage of human origin. The program Path-O-Gen v1.4.0 (available at http://tree.bio.ed.ac.uk/software/pathogen/) was used to identify potential contaminants that substantially deviated from the linear regression of root-to-tip genetic distance against time, which were subsequently removed from the study.
The final data set of swIAV sequences that are closely related to seasonal human influenza viruses and used in this study included the following: (i) 84 swine H1 sequences, with 108 human seasonal H1 sequences sampled globally from 1978 to 2009 included as background; (ii) 208 swine H3 sequences, with 251 human seasonal H3 sequences sampled globally from 1968 to 2013 as background; (iii) six swine N1 sequences, with 116 human seasonal N1 sequences sampled globally from 1977 to 2009 included as background; (iv) 350 swine N2 sequences, with 325 human seasonal N2 sequences sampled globally from 1957 to 2013 as background; (v) 305 swine PB1 sequences, with 126 human seasonal PB1 sequences sampled globally from 1968 to 2013 as background; and (vi) four swine sequences each for the PB2, PA, NP, MP, and NS segments, with 126 human seasonal PB2, PA, NP, MP, NS sequences sampled globally from 1968 to 2013 as background (see Table S2 in the supplemental material). The phylogenetic relationships of each of the 10 data sets were inferred using the maximum likelihood (ML) method available in the program RAxML v7.2.6 (33), incorporating a general time-reversible (GTR) model of nucleotide substitution with a gamma-distributed (Γ) rate variation among sites. To assess the robustness of each node, a bootstrap resampling process was performed (500 replicates), again using the ML method available in RAxML v7.2.6. Due to the computational complexity of these processes, we utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Divergence times.
A time-scaled Bayesian approach was employed to estimate the timing of each human-to-swine transmission event observed across the 10 phylogenies (i.e., PB2, PB1, PA, H1, H3, NP, N1, N2, MP, and NS). To that end, we employed a relaxed uncorrelated lognormal (UCLN) molecular clock, a flexible Bayesian skyline plot (BSP) demographic model (10 piece-wise constant groups), and a general time-reversible (GTR) model of nucleotide substitution with a gamma-distributed (Γ) rate variation among sites. The analysis was repeated for the H1, H3, and N2 segments implementing a host-specific local clock (HSLC) model (as performed by Worobey et al. [34]) to demonstrate that observed differences in the evolutionary rate of IAVs in humans and swine hosts did not compromise our inferred phylogeny or time scale. Host-specific clades were specified for all introductions listed in Table 1, except in the case of singleton viruses, which were ignored. The MCMC chain was run separately three times for each of the data sets for at least 100 million iterations, with subsampling every 10,000 iterations. The analysis utilized the BEAST package v1.8.0 (35) and BEAGLE (36) to improve computational performance. For viruses for which only the year of viral collection was available, the lack of tip date precision was accommodated by sampling across a 1-year window from 1 January to 31 December. All parameters reached convergence, as assessed visually using Tracer v.1.5, with statistical uncertainty reflected in values of the 95% highest posterior density (HPD). The initial 10% of the chain was removed as burn-in, runs were combined using LogCombiner v1.8.0, and maximum clade credibility (MCC) trees were summarized using TreeAnnotator v1.8.0. The estimate of each human-to-swine transmission event is provided by the interval between two nodes on the phylogeny, including the 95% HPD: the node where a swine clade associated with an introduction coalesces with the background human diversity (i.e., the human-swine node) and the node representing the time to the most recent common ancestor (tMRCAs) for each swine clade (the swine node).
TABLE 1.
Characteristics of the 20 introductions of human seasonal influenza A viruses into swine, 1965 to 2013
| Introduction | Regiona | Lineage | Related human virus | Representative swine virus | Segment(s) |
|---|---|---|---|---|---|
| 1 | N. America | Triple reassortant | A/New York/641/1996 H3N2 | A/sw/Texas/4199/2/1998 H3N2 | PB1, H3, N2 |
| 2 | N. America | A/New York/564/1997 H3N2 | A/sw/North Carolina/2003 H3N2 | H3 | |
| 3 | N. America | A/Auckland/588/2000 H3N2 | A/sw/Nebraska/00188/2003 H3N2 | H3, N2 | |
| 4 | N. America | A/Auckland/602/2001 H3N2 | A/sw/Iowa/H03LDH5/2003 H3N2 | H3, N2 | |
| 5 | N America | N2-2002 | A/Denmark/5/2002 H3N2 | A/swine/North Carolina/SG1464/2004 H3N2 | N2 |
| 6 | N. America | δ-1 | A/Michigan/2/2003 H1N2 | A/sw/Oklahoma/0174/2007 H1N2 | H1 |
| 7 | N. America | δ-2 | A/Michigan/1/2003 H1N1 | A/sw/North Carolina/01550/2007 H1N1 | H1, N1 |
| 8 | N. America | A/Morioka/52/2002 H3N2 | A/sw/Saskatchewan/02903/2009 H3N2 | Whole genomeb | |
| 9 | Europe | A/Bilthoven/2813/1975 H3N2 | A/sw/England/87842/1990 H3N2 | Whole genomeb | |
| 10 | Europe | A/Bilthoven/5931/1974 H3N2 | A/sw/Spain/82108/2007 H3N2 | H3, N2 | |
| 11 | Europe | A/Christs Hospital/157/1983 H1N1 | A/sw/England/1382/2010 H1N2 | H1 | |
| 12 | Europe | A/Michigan/3/2003 H1N2 | A/sw/England/00003/2009 H1N2 | H1 | |
| 13 | Asia | A/Bilthoven/17938/1969 H3N2 | A/sw/Miyazaki/1/2006 H1N2 | N2 | |
| 14 | Asia | A/Oslo/244/1997 H3N2 | A/sw/Guangdong/606/2010 H1N2 | N2 | |
| 15 | Asia | A/New York/562/1996 H3N2 | A/sw/Chonburi/NIAH106952-026/2011 H3N2 | H3, N2 | |
| 16 | Asia | A/Christchurch/10/2004 H3N2 | A/sw/Hong Kong/2503/2011 H3N2 | H3, N2 | |
| 17 | Asia | A/Malaysia/34291/2006 H1N1 | A/sw/Binh Doung/02-16/2010 H1N2 | H1 | |
| 18 | S. America | A/Auckland/602/2001 H3N2 | A/sw/Argentina/CIP051-A2/2008 H3N2 | Whole genomeb | |
| 19 | S. America | A/Michigan/2/2003 H1N2 | A/sw/Argentina/CIP051-StaFeN2/2010 H1N2 | H1, N2 | |
| 20 | S. America | A/Michigan/1/2003 H1N1 | A/sw/Argentina/CIP051-BsAs76/2009 H1N1 | H1, N1 |
Region where the introduction is thought to have originated in swine. N. America, North America; S. America, South America.
All eight genome segments were inferred to be of human origin.
HA/NA analysis.
As more swIAV sequence data are available for the HA and NA segments than for the entire genome, a more complete analysis of human-to-swine transmission of these antigens was obtained by using the same methods of Bayesian phylogenetic inference described above for all available swIAV sequence data for the H1 (n = 761), H3 (n = 341), N1 (n = 15), and N2 (n = 936) segments (see Tables S3 and S4 in the supplemental material). Although it was impossible to evaluate reassortment with internal gene segments, it was useful to assess the number of human-to-swine introductions involving the antigenic segments that could not be detected using whole-genome sequences.
Nucleotide sequence accession numbers.
All data sequenced for this study were submitted to the Influenza Virus Resource at the National Center for Biotechnology Information's GenBank (29), and accession numbers are available in Table S1 in the supplemental material.
RESULTS
Identification of a novel unreassorted human influenza virus in Canadian swine.
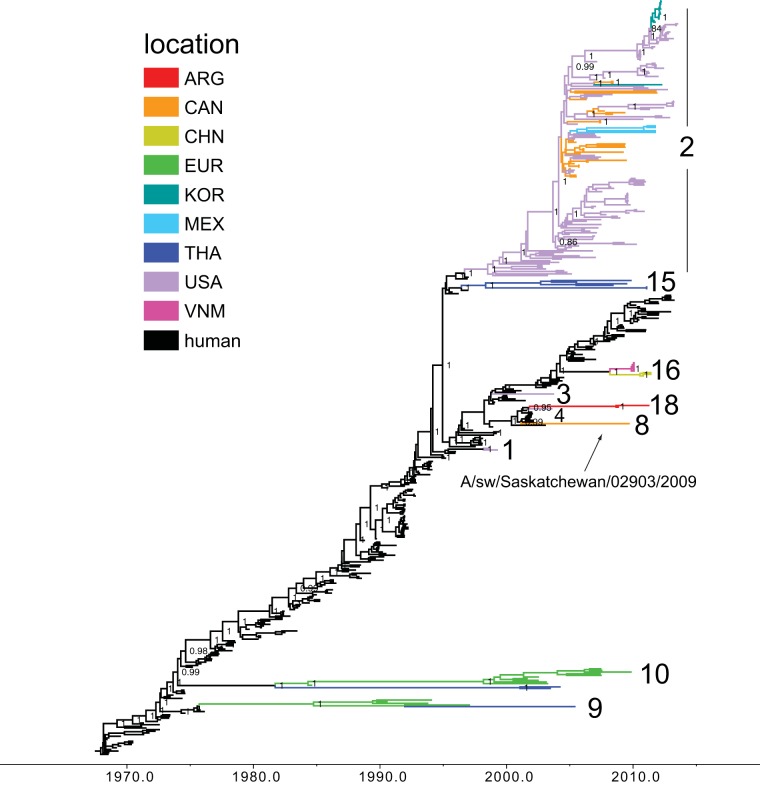
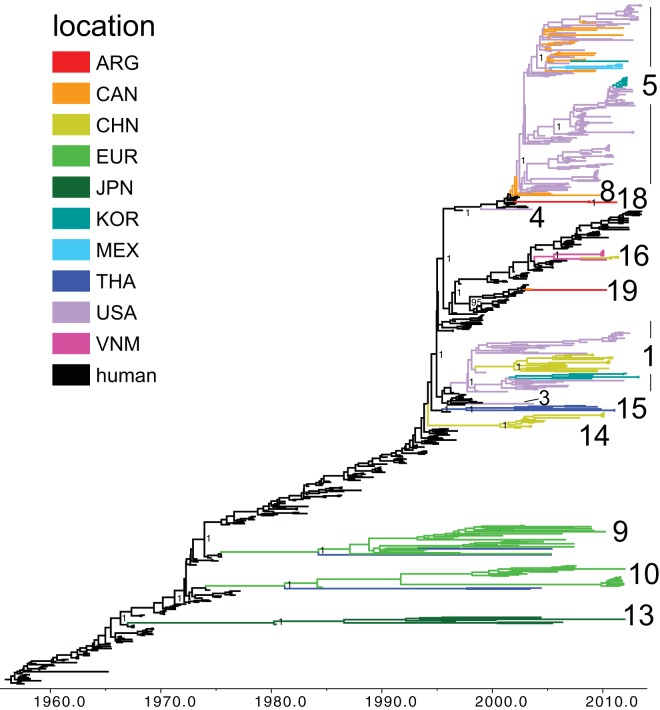
Among the newly sequenced IAVs from North American swine, we identified a previously undetected introduction of a human seasonal H3N2 virus into a population of Canadian swine, represented by the isolate A/sw/Saskatchewan/02903/2009(H3N2). Notably, all eight of the viral genome segments are closely related to human seasonal A/H3N2 viruses collected during 2001 to 2002 [e.g., A/New York/74/2002(H3N2)], indicating that this virus has not reassorted with other swine viruses since entering that swine population (introduction 8) (Table 1 and Fig. 1, 2, and 3; phylogenies for other segments are available in Fig. S1 to S7 in the supplemental material). The long branch lengths that separate A/sw/Saskatchewan/02903/2009(H3N2) from the most closely related human viruses on the maximum likelihood (ML) phylogenetic trees indicate that this swine virus lineage has circulated undetected for many years (e.g., the representative ML tree for the PB2 is available in Fig. S8 in the supplemental material). From the time-scaled MCC tree we estimate that the time to the most recent common ancestor (tMRCA) for A/sw/Saskatchewan/02903/2009(H3N2) and the most closely related human viruses falls between 2000.77 and 2002.74 (95% HPD) across the phylogenies inferred for each of the eight genome segments, representing an estimated 7 to 9 years of undetected circulation of this virus before its collection on 24 September 2009. As the intensity of sampling and sequencing of human influenza viruses in North America greatly exceeds that conducted in swine, the long phylogenetic branch adjoining A/sw/Saskatchewan/02903/2009(H3N2) and human seasonal viruses is more likely to arise from lack of sampling of swine viruses than unsampled human viruses.
FIG 1.
Phylogenetic relationships between human and swine H3 segments. A time-scaled Bayesian MCC tree inferred for the HA (H3) sequences of 208 swine IAVs identified as of human seasonal virus origin and 251 human seasonal H3N2 influenza viruses, collected 1968 to 2013, is shown. Branches of human seasonal H3N2 influenza virus origin are in black, while branches associated with viruses from swine are shaded by country or continent of origin: ARG, Argentina; CAN, Canada; CHN, China (including Hong Kong SAR and Taiwan); EUR, Europe; KOR, South Korea; MEX, Mexico; THA, Thailand; VNM, Vietnam. Posterior probabilities of >0.9 are included for key nodes, and the 10 discrete introductions of the human H3 segment into swine that are supported by high posterior probabilities and long branch lengths are labeled according to Table 1.
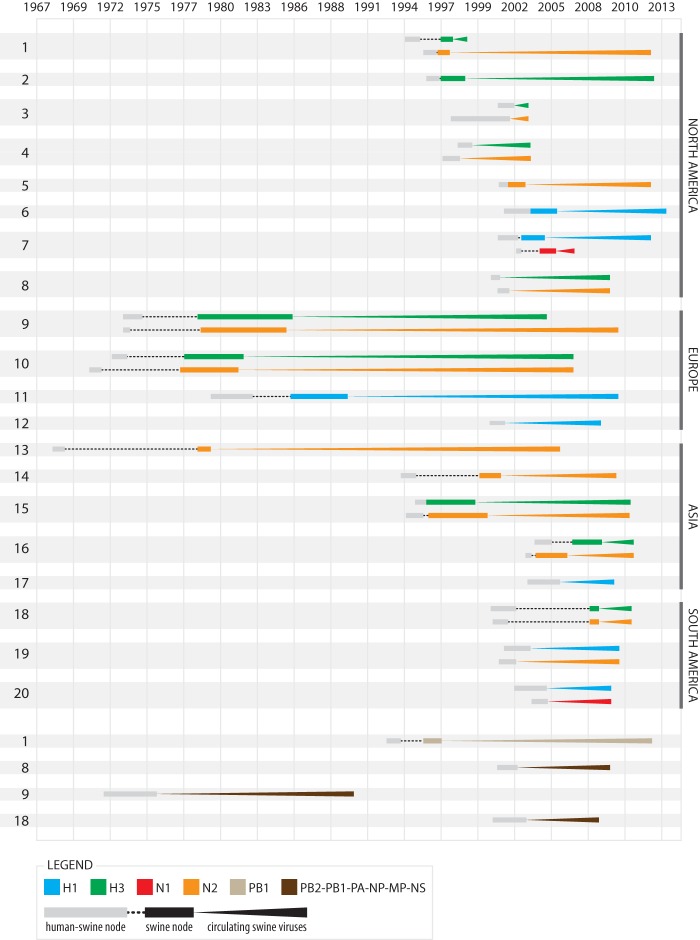
FIG 2.
Introductions of human seasonal influenza viruses into swine, 1965 to 2013. A summary of the 20 introductions of seasonal human IAVs into swine resulting in sustained transmission in swine (for at least 1 year) by segment and region. Introductions involving the HA and NA segments are depicted in the upper portion of the figure, and the subset involving internal gene segments are presented in the lower portion. Each colored line represents a human-to-swine transmission event of a segment. Introductions are numbered 1 to 20 in accordance with Table 1. The timing of each human-to-swine transmission event is estimated from the tMRCAs inferred from the MCC trees, with gray boxes indicating the 95% HPD interval between the swine clade and most closely related human viruses and the black box indicating the 95% HPD interval for the swine clade only. Each line extends forward in time up to the most recently sampled swine virus of that lineage.
FIG 3.
Phylogenetic relationships between human and swine N2 segments. A time-scaled Bayesian MCC tree inferred for the NA (N2) sequences of 350 swine influenza viruses identified as of human seasonal virus origin and 325 human H3N2 and H1N2 seasonal influenza viruses, collected from 1957 to 2013, is shown. Labeling and shading are similar to those in Fig. 1.
Frequency of introduction of human seasonal viruses into swine, 1965 to 2013.
To interpret the evolution of the A/sw/Saskatchewan/02903/2009(H3N2) virus within a broader historical context, we determined the global frequency of human-to-swine transmission events of seasonal IAVs at a whole genome level (summarized in Fig. 2). We focused only on IAV introductions that sustained transmission in swine for more than 1 year, as this provides evidence of successful adaptation to a new host species.
During the period between 1965 to 2013 we identified 20 stable introductions of seasonal human influenza A viruses in part or in entirety (Fig. 2 and Table 1). The majority (n = 13) of introductions involved human seasonal viruses of the H3N2 subtype. Four introductions involved human seasonal H1N1 viruses, and three involved human seasonal H1N2 viruses, despite the fact that this reassortant subtype circulated in humans only from 2001 to 2003 (Fig. 3 and 4 and Table 1; also, see Table S5 in the supplemental material). Our estimate of 20 human-to-swine introductions over this time period is conservative and excludes (i) swine viruses of human origin that lacked evidence of sustained transmission for at least 1 year in swine (n = 19) (see Table S5 in the supplemental material); (ii) any human-to-swine transmission events for which whole-genome sequence data were not available; (iii) any human-to-swine transmission events involving pandemic influenza viruses of the past century, including the North American classical swine influenza virus lineage, which may also have been transmitted by humans to swine in North America during the 1918 pandemic (34), and (iv) the global introductions of human pH1N1 viruses that have occurred in swine since 2009, which have been described in detail elsewhere (16).
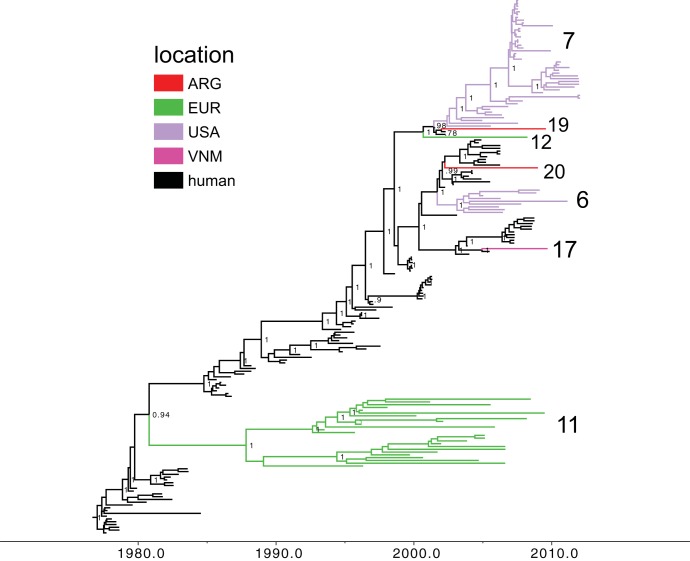
FIG 4.
Phylogenetic relationships between seasonal human and swine H1 segments. A time-scaled Bayesian MCC tree inferred for HA (H1) sequences of 84 swine influenza viruses identified as of human seasonal virus origin and 108 human seasonal H1N1 and H1N2 influenza viruses, collected from 1977 to 2009, is shown. Labeling and shading are similar to those in Fig. 1.
Human-to-swine introductions were observed across a wide range of countries and continents, despite substantial differences in the intensity of IAV surveillance conducted in swine populations globally over the time period (Table 1 and Fig. 2). The frequency of introductions was generally associated with the intensity of sampling of swIAVs in a given region, with the highest number (n = 8) of viral introductions being identified in North American swine, where the availability of sequence data was highest. Five introductions were observed in Asian swine, four in European swine, and three in South American swine.
Reassortment patterns of human seasonal viruses introduced into swine, 1918 to 2013.
Comparisons across the phylogenies inferred for the eight genome segments indicate that, upon introduction into the swine population, human-origin IAVs rapidly acquired internal gene segments from other IAVs circulating in swine via reassortment. For example, the vast majority of human PB2 segments (96.9%) were replaced in North American swine by triple reassortant internal gene (TRIG) segments, were replaced in European swine by Eurasian avian-origin PB2 segments (73.1%), and were replaced in Asian swine by a combination of PB2 segments of classical swine virus lineage (20.5%), TRIGs (48.2%), and the Eurasian avian-origin lineage (18.1%) (see Fig. S9 in the supplemental material). Notably, 80% (16/20) of the human-to-swine introductions identified in this study were associated with the onward transmission of only the human-origin HA and/or NA antigenic segments and no internal genes in swine (Table 1). Evidence of onward transmission of the N1 (see Fig. S5 in the supplemental material) or the six internal gene segments (PB2, PB1, PA, NP, MP, or NS) (see Fig. S1 to S4, S6, and S7 in the supplemental material) of human origin was limited. The pattern of reassortment appears to be bimodal: either the full human virus genome constellation is conserved, or all six internal genes are replaced in swine by reassortment. Only in the case of the triple-reassortant virus that emerged in the late 1990s in North American swine was onward transmission of a single human-origin internal gene segment gene segment (in this case PB1; introduction 1) (Table 1; also, see Fig. S2 in the supplemental material) maintained outside a full human genome background (37).
Three singleton unreassorted viruses were identified in swine that contained all eight segments of human seasonal virus origin as well as evidence of transmission in swine for at least 1 year, based on long phylogenetic branch lengths and divergence times: A/sw/Saskatchewan/02903/2009(H3N2), described above; A/sw/England/87842/1990(H3N2); and A/sw/Argentina/CIP051-A2/2008(H3N2) (introductions 8, 9, and 18, respectively) (Table 1 and Fig. 1 and 2; also, see Fig. S1 to S7 in the supplemental material). The virus A/sw/Ontario/52156/2003(H1N2) also had all eight genome segments of human seasonal virus origin but did not meet the criteria for transmission in swine for at least 1 year (see Fig. S1 to S7 and Table S5 in the supplemental material).
Evolutionary rates.
Rates of nucleotide substitution for the H1, H3, and N2 segments were consistently lower in human seasonal viruses than in closely related viruses in swine (Table 2; also, see Fig. S10 to S12 in the supplemental material). Importantly, despite this rate variation, using a host-specific local clock (HSLC) model (see Fig. S10 to S12 in the supplemental material) did not change the topology of the phylogenies that were inferred using a relaxed molecular clock.
TABLE 2.
Evolutionary rates of IAVs in humans in swinea
| Segment | Host | Introduction | Evolutionary rate (95% HPD)b |
|---|---|---|---|
| H1 | Human | NA | 2.71 (2.44–3.03) |
| Swine | 6 | 3.74 (3.21–4.33) | |
| Swine | 7 | 4.52 (3.75–5.28) | |
| Swine | 11 | 4.24 (3.85–4.66) | |
| H3 | Human | NA | 3.14 (2.89–3.39) |
| Swine | 2 | 4.15 (3.81–4.46) | |
| Swine | 9 | 6.15 (4.99–7.30) | |
| Swine | 10 | 4.11 (3.62–4.62) | |
| Swine | 15 | 4.60 (3.80–5.41) | |
| Swine | 16 | 4.07 (3.07–5.09) | |
| Swine | 18 | 3.95 (2.91–4.96) | |
| N2 | Human | NA | 2.33 (2.14–2.51) |
| Swine | 1 | 3.35 (3.01–3.67) | |
| Swine | 5 | 2.87 (2.62–3.11) | |
| Swine | 9 | 3.62 (3.28–3.95) | |
| Swine | 10 | 3.24 (2.89–3.59) | |
| Swine | 13 | 3.34 (2.96–3.71) | |
| Swine | 14 | 2.85 (2.31–3.44) | |
| Swine | 15 | 3.82 (3.19–4.45) | |
| Swine | 16 | 3.75 (2.83–4.73) | |
| Swine | 18 | 2.85 (2.00–3.73) |
Comparison of the rates of evolution for the H1, H3, and N2 segments in humans and major clades of human-origin viruses in swine associated with the introductions presented in Table 1. NA, not applicable.
Values are 103 nucleotide substitutions per site per year.
DISCUSSION
We have identified a fully human-origin influenza virus—A/sw/Saskatchewan/02903/2009(H3N2)—which may have circulated undetected in swine for 7 to 9 years. More broadly, we have identified 20 discrete introductions of human seasonal influenza A viruses into swine in the Americas, Europe, and Asia since 1965 that successfully sustained onward transmission in swine for at least 1 year. Our phylogenetic analysis at a whole-genome level indicates that human-origin internal genes frequently are replaced in swine via reassortment, while human-origin HA and NA segments are maintained in the swine virus population, increasing antigenic diversity and complicating the development of effective cross-protective influenza vaccines (38). These findings have important implications for expanding swIAV surveillance to fully capture worldwide viral diversity as well as directions for future research aimed at understanding the human-swine interface.
The patterns of reassortment for human-origin viruses in swine were striking and suggest that there may be evolutionary constraints on human internal gene segments in swine, although experimental studies are required to test this hypothesis. The identification of A/sw/Saskatchewan/02903/2009(H3N2) was therefore notable and suggests that viruses of wholly human origin can persist in swine for substantial periods of time, at least in certain ecological niches. All swine viruses of wholly human origin are singletons on the tree (introductions 8, 9, and 18), and it is therefore difficult to assess how widely wholly human origin-lineages disseminated in local swine populations, particularly as all three viruses were detected in swine populations where surveillance is not systematically conducted or reported to public databases (Argentina, Canada, and England). Here, the inference of onward transmission in swine is based on the overwhelmingly higher level of influenza virus surveillance in humans than in swine and the low probability that long branch lengths arise from unsampled human viruses. The 1918 pandemic H1N1 virus potentially provides another example of a human virus for which all eight genome segments successfully transmitted onward in swine. However, the scarcity of sequence data from the 1918 pandemic in humans and swine confounds attempts to infer whether the 1918 virus was first transmitted from humans to swine, or vice versa.
The detection of independent introductions of human seasonal IAVs in swine in at least eight different countries highlights the potential for IAV diversity to emerge in any part of the world. Although swIAVs have the capacity to rapidly disseminate within regions (39), the identification of A/sw/Saskatchewan/02903/2009(H3N2) indicates that novel viruses also may persist on more local scales outside major swine production centers, where they are less likely to be detected by surveillance. Human-to-swine transmission events also are likely to go undetected in cases where human viruses are transmitted onward in swine for only short time periods, or in earlier decades, for which only serological data, partial gene sequences, or historical anecdotal records are available (40–42). In fact, a broader analysis of all swIAV sequence data available for the full-length HA and NA sequences revealed an additional 21 human-to-swine transmission events that met our criteria for evidence of onward transmission in swine but lacked internal gene sequence data (see Fig. S13 to S15 and Table S6 in the supplemental material). An additional 19 putative human-to-swine introductions lacked evidence of onward transmission in swine for more than 1 year (see Table S5 in the supplemental material). Finally, our study did not consider pH1N1 viruses because prior studies provide global estimates of the frequency of human-to-swine transmission of these viruses since 2009 (16). Taken together, these data make it clear that our estimate of human-to-swine transmission represents a conservative, lower-bound estimate, and increased swIAV sequencing on a global scale is required to capture the full scale of human-to-swine transmission (43–46).
Although swine viruses may pose a threat to humans, it is important to note that the human-swine interface appears to be strongly asymmetrical for influenza, and to date, the 2009 H1N1 pandemic virus represents the lone documented influenza virus of certain swine origin to sustain transmission in humans (data available from the 1918, 1957, and 1968 pandemics are insufficient at this time to determine whether a swine “intermediary” mammalian host was involved in the transmission of avian H1N1, H2N2, or H3N2 viruses to humans). Further understanding of the ecological and epidemiological circumstances under which human-to-swine transmission events occur is therefore vital for the health of both humans and swine and merits further research.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sagar Goyal and Devi Patnayak, along with Wendy Wiese, Becca Wheeldon, and Lotus Smasal at UMVDL, for their virology work and Rebecca Halpin, Nadia Fedorova, and Timothy B. Stockwell for their assistance in viral sequencing.
Partial support for this work was provided by a grant from the Saskatchewan Ministry of Agriculture, contract number C70077. This work was supported in part by the Multinational Influenza Seasonal Mortality Study (MISMS), an on-going international collaborative effort to understand influenza epidemiology and evolution, led by the Fogarty International Center, National Institutes of Health, with funding from the Office of Global Affairs at the Department of Health and Human Services [MIN, CV]. This project has also been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN272200900007C. E.C.H. is supported by an NHMRC Australia Fellowship.
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01080-14.
REFERENCES
- 1.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier R a M, Pappas C, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JSM, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- 3.Scholtissek C. 1990. Pigs as the “mixing vessel” for the creation of new pandemic influenza A viruses. Med. Princ. Pract. 2:65–71 [Google Scholar]
- 4.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taubenberger J, Morens D. 2009. Pandemic influenza–including a risk assessment of H5N1. Rev. Sci. Tech. 28:187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowling BJ, Jin L, Lau EHY, Liao Q, Wu P, Jiang H, Tsang TK, Zheng J, Fang VJ, Chang Z, Ni MY, Zhang Q, Ip DKM, Yu J, Li Y, Wang L, Tu W, Meng L, Wu JT, Luo H, Li Q, Shu Y, Li Z, Feng Z, Yang W, Wang Y, Leung GM, Yu H. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382:129–137. 10.1016/S0140-6736(13)61171-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, Boulton R, Haddy L, Biggerstaff M, Brammer L, Trock S, Burns E, Gomez T, Wong KK, Katz J, Lindstrom S, Klimov A, Bresee JS, Jernigan DB, Cox N, Finelli L. 2013. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin. Infect. Dis. 57:S4–S11. 10.1093/cid/cit272 [DOI] [PubMed] [Google Scholar]
- 8.Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen CW. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199–210. 10.1016/S0168-1702(02)00027-8 [DOI] [PubMed] [Google Scholar]
- 11.Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. 2004. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. 103:67–73. 10.1016/j.virusres.2004.02.015 [DOI] [PubMed] [Google Scholar]
- 12.Karasin A, Carman S, Olsen C. 2006. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005). J. Clin. Microbiol. 44:1123–1126. 10.1128/JCM.44.3.1123-1126.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon KJ, Krauss S, Webster RG. 2000. Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet. Microbiol. 74:47–58. 10.1016/S0378-1135(00)00165-6 [DOI] [PubMed] [Google Scholar]
- 14.Campitelli L, Donatelli I, Foni E, Castrucci MR, Fabiani C, Kawaoka Y, Krauss S, Webster RG. 1997. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology 232:310–318. 10.1006/viro.1997.8514 [DOI] [PubMed] [Google Scholar]
- 15.de Jong JC, Smith DJ, Lapedes AS, Donatelli I, Campitelli L, Barigazzi G, Van Reeth K, Jones TC, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. 2007. Antigenic and genetic evolution of swine influenza A (H3N2) viruses in Europe. J. Virol. 81:4315–4322. 10.1128/JVI.02458-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson MI, Gramer MR, Vincent AL, Holmes EC. 2012. Global transmission of influenza viruses from humans to swine. J. Gen. Virol. 93:2195–2203. 10.1099/vir.0.044974-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton J-C, Lowe J, Gramer M, Webby RJ. 2011. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis. 17:1624–1629. 10.3201/eid1709.110338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starick E, Lange E, Fereidouni S, Bunzenthal C, Höveler R, Kuczka A, grosse Beilage E, Hamann H-P, Klingelhöfer I, Steinhauer D, Vahlenkamp T, Beer M, Harder T. 2011. Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J. Gen. Virol. 92:1184–1188. 10.1099/vir.0.028662-0 [DOI] [PubMed] [Google Scholar]
- 19.Vijaykrishna D, Poon LLM, Zhu HC, Ma SK, Li OTW, Cheung CL, Smith GJD, Peiris JSM, Guan Y. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529. 10.1126/science.1189132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir. Viruses 7(Suppl 4):S42–S51. 10.1111/irv.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson MI, Vincent AL, Kitikoon P, Holmes EC, Gramer MR. 2012. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011. J. Virol. 86:8872–8878. 10.1128/JVI.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown IH. 2013. History and epidemiology of swine influenza in Europe. Curr. Top. Microbiol. Immunol. 370:133–146. 10.1007/82_2011_194 [DOI] [PubMed] [Google Scholar]
- 23.Vijaykrishna D, Smith GJD, Pybus OG, Zhu H, Bhatt S, Poon LLM, Riley S, Bahl J, Ma SK, Cheung CL, Perera RAPM, Chen H, Shortridge KF, Webby RJ, Webster RG, Guan Y, Peiris JSM. 2011. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473:519–522. 10.1038/nature10004 [DOI] [PubMed] [Google Scholar]
- 24.Holyoake PK, Kirkland PD, Davis RJ, Arzey KE, Watson J, Lunt RA, Wang J, Wong F, Moloney BJ, Dunn SE. 2011. The first identified case of pandemic H1N1 influenza in pigs in Australia. Aust. Vet. J. 89:427–431. 10.1111/j.1751-0813.2011.00844.x [DOI] [PubMed] [Google Scholar]
- 25.Njabo KY, Fuller TL, Chasar A, Pollinger JP, Cattoli G, Terregino C, Monne I, Reynes J-M, Njouom R, Smith TB. 2012. Pandemic A/H1N1/2009 influenza virus in swine, Cameroon, 2010. Vet. Microbiol. 156:189–192. 10.1016/j.vetmic.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereda A, Cappuccio J, Quiroga MA, Baumeister E, Insarralde L, Ibar M, Sanguinetti R, Cannilla ML, Franzese D, Escobar Cabrera OE, Craig MI, Rimondi A, Machuca M, Debenedetti RT, Zenobi C, Barral L, Balzano R, Capalbo S, Risso A, Perfumo CJ. 2010. Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg. Infect. Dis. 16:304–307. 10.3201/eid1602.091230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza a viruses. J. Virol. 83:10309–10313. 10.1128/JVI.01109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou B, Wentworth DE. 2012. Influenza A virus molecular virology techniques. Methods Mol. Biol. 865:175–192. 10.1007/978-1-61779-621-0_11 [DOI] [PubMed] [Google Scholar]
- 29.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601. 10.1128/JVI.02005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambaut A. 2002. Sequence alignment editor, version 2.0. http://tree.bio.ed.ac.uk/software/seal/. Accessed 15 December 2013 [Google Scholar]
- 32.Swofford DL. 2003. Phylogenetic analysis using parsimony (and other methods). 4.0. Sinauer Associates, Sunderland, MA [Google Scholar]
- 33.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 34.Worobey M, Han G-Z, Rambaut A. 2014. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature 508:254–257. 10.1038/nature13016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suchard MA, Rambaut A. 2009. Many-core algorithms for statistical phylogenetics. Bioinformatics 25:1370–1376. 10.1093/bioinformatics/btp244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon KJ, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q, Madson D, Miller CL, Harris DLH. 2012. Vaccine development for protecting swine against influenza virus. Anim. Health Res. Rev. 13:181–195. 10.1017/S1466252312000175 [DOI] [PubMed] [Google Scholar]
- 39.Nelson MI, Lemey P, Tan Y, Vincent A, Lam TT-Y, Detmer S, Viboud C, Suchard MA, Rambaut A, Holmes EC, Gramer M. 2011. Spatial dynamics of human-origin H1 influenza A virus in North American swine. PLoS Pathog. 7:e1002077. 10.1371/journal.ppat.1002077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundin WD. 1970. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature 228:857. [DOI] [PubMed] [Google Scholar]
- 41.Brown IH, Harris PA, Alexander DJ. 1995. Serological studies of influenza viruses in pigs in Great Britain 1991–2. Epidemiol. Infect. 114:511–520. 10.1017/S0950268800052225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morens DM, Taubenberger JK. 2014. A possible outbreak of swine influenza, 1892. Lancet Infect. Dis. 14:169–172. 10.1016/S1473-3099(13)70227-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takemae N, Nguyen T, Ngo LT, Hiromoto Y, Uchida Y, Pham VP, Kageyama T, Kasuo S, Shimada S, Yamashita Y, Goto K, Kubo H, Le VT, Van Vo H, Do HT, Nguyen DH, Hayashi T, Matsuu A, Saito T. 2013. Antigenic variation of H1N1, H1N2 and H3N2 swine influenza viruses in Japan and Vietnam. Arch. Virol. 158:859–876. 10.1007/s00705-013-1616-8 [DOI] [PubMed] [Google Scholar]
- 44.Cappuccio JA, Pena L, Dibárbora M, Rimondi A, Piñeyro P, Insarralde L, Quiroga MA, Machuca M, Craig MI, Olivera V, Chockalingam A, Perfumo CJ, Perez DR, Pereda A. 2011. Outbreak of swine influenza in Argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J. Gen. Virol. 92:2871–2878. 10.1099/vir.0.036590-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poonsuk S, Sangthong P, Petcharat N, Lekcharoensuk P. 2013. Genesis and genetic constellations of swine influenza viruses in Thailand. Vet. Microbiol. 167:314–326. 10.1016/j.vetmic.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 46.Takemae N, Parchariyanon S, Damrongwatanapokin S, Uchida Y, Ruttanapumma R, Watanabe C, Yamaguchi S, Saito T. 2008. Genetic diversity of swine influenza viruses isolated from pigs during 2000 to 2005 in Thailand. Influenza Other Respir. Viruses 2:181–189. 10.1111/j.1750-2659.2008.00062.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.