ABSTRACT
The structural pattern of infectivity matrices, which contains infection data resulting from inoculations of a set of hosts by a set of parasites, is a key parameter for our understanding of biological interactions and their evolution. This pattern determines the evolution of parasite pathogenicity and host resistance, the spatiotemporal distribution of host and parasite genotypes, and the efficiency of disease control strategies. Two major patterns have been proposed for plant-virus genotype infectivity matrices. In the gene-for-gene model, infectivity matrices show a nested pattern, where the host ranges of specialist virus genotypes are subsets of the host ranges of less specialized viruses. In contrast, in the matching-allele (MA) model, each virus genotype is specialized to infect one (or a small set of) host genotype(s). The corresponding infectivity matrix shows a modular pattern where infection is frequent for plants and viruses belonging to the same module but rare for those belonging to different modules. We analyzed the structure of infectivity matrices between Potato virus Y (PVY) and plant genotypes in the family Solanaceae carrying different eukaryotic initiation factor 4E (eIF4E)-coding alleles conferring recessive resistance. Whereas this system corresponds mechanistically to an MA model, the expected modular pattern was rejected based on our experimental data. This was mostly because PVY mutations involved in adaptation to a particular plant genotype displayed frequent pleiotropic effects, conferring simultaneously an adaptation to additional plant genotypes with different eIF4E alleles. Such effects should be taken into account for the design of strategies of sustainable control of PVY through plant varietal mixtures or rotations.
IMPORTANCE The interaction pattern between host and virus genotypes has important consequences on their respective evolution and on issues regarding the application of disease control strategies. We found that the structure of the interaction between Potato virus Y (PVY) variants and host plants in the family Solanaceae departs significantly from the current model of interaction considered for these organisms because of frequent pleiotropic effects of virus mutations. These mutational effects allow the virus to expand rapidly its range of host plant genotypes, make it very difficult to predict the effects of mutations in PVY infectivity factors, and raise concerns about strategies of sustainable management of plant genetic resistance to viruses.
INTRODUCTION
The interaction pattern between host and parasite genotypes has important consequences on their respective evolution and on issues regarding the application of disease control strategies. This pattern determines to a large extent the maintenance of genetic diversity in host and parasite populations (1), the structure of these populations in space and time (2, 3), and the evolution of parasite pathogenicity and host resistance (4).
Different models of host-parasite interaction and coevolution have been proposed (3, 5) (Fig. 1a to c). The gene-for-gene (GFG) model is the genetic system of interaction which was most frequently postulated for plant-pathogen interactions (4). In this system, a pathogen elicitor interacts with a host factor and triggers a specific defense reaction in the host which leads to the inhibition of infection. In contrast, the matching-allele (MA) model, initially proposed for an invertebrate immune system (5), describes a system where infection of a host by a parasite requires a specific match between some of their interacting factors. A pure MA model is an extreme form of biological specificity, where a parasite with a given genotype is only able to infect hosts belonging to a single genotype, and, reciprocally, hosts of a given genotype can only be infected by parasites of a single genotype (5). Compared to this one-to-one interaction pattern, a more relaxed model supposes that a parasite with a given genotype is able to infect hosts belonging to a few genetically related genotypes (and vice versa). In that situation, the host-parasite interaction matrix is organized into interaction modules, where host and parasite genotypes belonging to the same module are more preferentially compatible with each other (i.e., hosts are infected and parasites are infectious) than with members of other modules.
FIG 1.
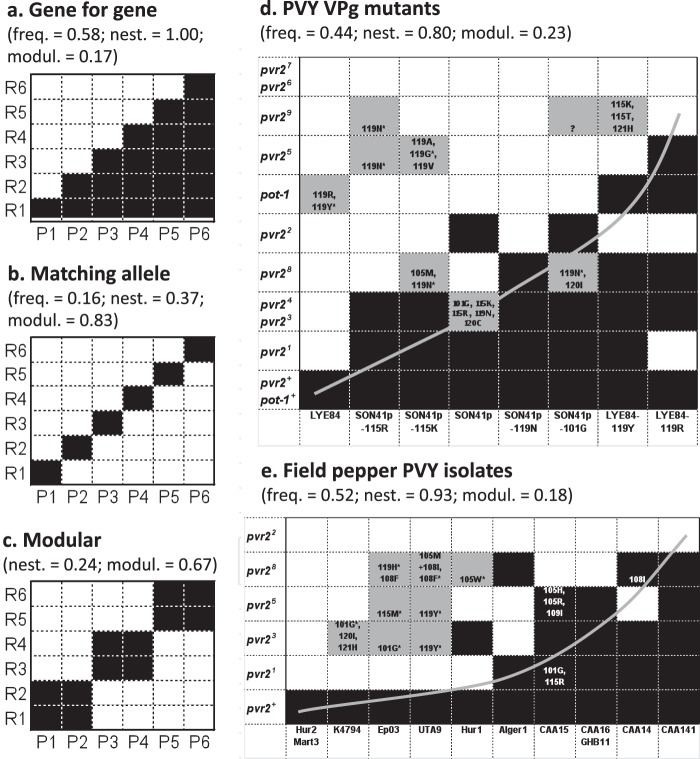
Interaction matrices between host and parasite genotypes. (a to c) Three theoretical interaction matrices between host genotypes carrying different resistance genes or alleles (R1 to R6) and parasite pathotypes (P1 to P6). Black boxes indicate infection of the host with the indicated genotype by the parasite with the indicated genotype, and white boxes indicate lack of infection. (d and e) Experimental interaction matrices between PVY variants (columns) and plants of Capsicum annuum or Solanum habrochaites with genotypes (rows) carrying different alleles at the pvr2 or pot-1 locus, respectively. Black boxes indicate infection of 100% of inoculated plants, whereas white boxes indicate no infection. Gray boxes indicate <100% infection and occurrence of additional mutations in the VPg pathogenicity factor. Amino acid substitutions observed in the VPg of the viral progeny are indicated within boxes. The question mark indicates that a single plant was infected, and no sequence was obtained for the VPg-coding region. Asterisks indicate PVY populations, almost all of which show secondary mutations in the VPg-coding region, that were chosen for back-inoculation in the same plant genotype. For all of them, all (25 of 25) back-inoculated plants were infected 15 days after inoculation. These matrices differ by the frequency of compatibility (infection) cases (freq) and the nestedness (nest; estimated with the algorithm of Rodríguez-Gironés and Santamaría [19]) and modularity (modul; estimated with an exhaustive search algorithm) of these compatibility cases. The rows and columns of matrices shown in panels d and e were ordered to evidence the significant nested pattern (gray curve) (Table 1). No frequency of compatibility cases is indicated for the modular model because it depends greatly on the size of modules.
The GFG, MA, and modular models correspond to interaction matrices that differ in the frequency and structure of compatibility cases (Fig. 1a to c). In a dynamic GFG coevolution, parasites adapt to a new resistance gene or allele in the host without losing their adaptations to older forms of host resistance. Accordingly, cross-infectivity, where a parasite with a given genotype is able to infect hosts belonging to different genotypes, is frequent. A symmetrical situation occurs with hosts, where new resistance genes or alleles protect against recently evolved parasite genotypes and also against older parasite genotypes. As a result, the interaction matrix shows a nested, stair-like compatibility pattern (Fig. 1a). In contrast, under the MA model and, to a lesser extent, under the modular model, parasite mutations conferring infectivity to hosts with newly evolved resistance genes simultaneously abolish the capacity to infect older host genotypes. This model is characterized by rare cross-infectivity for parasites. The interaction matrix shows a modular pattern, where compatibility cases are concentrated along the diagonal. As a consequence, the interaction matrices corresponding to these different models can be distinguished by the frequency of compatibility cases, their nestedness (i.e., the maximal degree of concentration of the compatibility cases in the lower right portion of the matrix that can be achieved after row and column permutation), and their modularity (i.e., the maximal degree of concentration of the compatibility cases into modules that group host and parasite genotypes). It is important to keep in mind that host-parasite interaction matrices where rows and/or columns are permuted are equivalent and that the total number of equivalent matrices obtained through row and/or column permutations becomes rapidly very large as the number of rows and columns increases, causing computational issues.
In plant-virus interactions, both the GFG and MA models have been postulated (4). The interaction matrix between pepper (Capsicum spp.) plants representing genotypes with different alleles at the L locus, conferring dominant resistance, and tobamoviruses (genus Tobamovirus) shows a nested pattern which fits with the GFG model. Furthermore, the tobamovirus genotypes with the largest spectrum of infectivity show fitness costs in terms of accumulation in susceptible plant genotypes (6), a prediction of the GFG model. In contrast, interaction between plants with genotypes carrying eukaryotic initiation factor 4E or 4G (eIF4E or eIF4G, respectively)-mediated recessive resistance and different groups of viruses was assumed to correspond to an MA model. Indeed, in these pathosystems, infectivity depends on a direct physical interaction between a host factor (eIF4E or eIF4G) and a virus factor (most frequently, the genome-linked viral protein, or VPg) (7, 8). The most exhaustive study is that of the interaction between rice recessive resistance alleles at the rymv1 locus (mainly alleles rymv1–2 and rymv1–3) and Rice yellow mottle virus (RYMV; genus Sobemovirus) (9–11). As in the MA model, cross-infectivity was rare since among the RYMV isolates that could adapt to plants carrying the resistance alleles, 89% (34/38) were able to infect only genotypes with rymv1–2 or only genotypes with rymv1–3 (11). This “converse genetic barrier” for adaptation to rymv1–2 and rymv1–3 was later shown to be conferred by a particular mutation in the VPg of RYMV (9). However, given the small set of resistance alleles examined, it is presently not possible to obtain statistical evidence about the structure of this interaction matrix and to assign it to a GFG or MA model.
In this study, we examined the interaction pattern between 12 plant genotypes in the family Solanaceae carrying different eIF4E-mediated resistance/susceptibility factors and Potato virus Y (PVY; genus Potyvirus) genotypes. Although the mechanistic bases of virus infectivity and plant resistance in this system fit with the assumptions of the MA or modular model, these were rejected after the structure of the interaction matrix was analyzed. One reason was a particularly high frequency of pleiotropic cross-infectivity mutations in PVY conferring simultaneously an adaptation to the resistance controlled by several genes or alleles and contrasting with the rice-RYMV system. Such interaction patterns and the causative mutations in plants and viruses have important consequences in terms of resistance management.
MATERIALS AND METHODS
Plant material.
Ten pepper (Capsicum annuum) and two Solanum habrochaites (a tomato wild relative) inbred lines were used for infectivity tests. The pepper accessions had different alleles at the pvr2 locus encoding eIF4E: Yolo Wonder was the susceptible reference carrying the pvr2+ allele, whereas Yolo Y, Florida VR2, HD285, PI322719, SC81, Maroc1, Serrano Vera Cruz, PI195301, and Chile de Arbol carried alleles pvr21 to pvr29, respectively (7). These 10 alleles correspond to highly similar copies of an eIF4E which differ by a small number of amino acid substitutions (7, 12). The S. habrochaites accessions carried different alleles at the pot-1 locus orthologous to the pepper pvr2 locus: PI247087 carried the pot-1 recessive resistance allele, whereas PI134417 was the susceptible reference (pot-1+) (13). Among all these resistance genes or alleles, only pvr21 and pvr22 are used extensively in breeding programs. They are present in 14% of 364 pepper cultivars registered in the European varietal catalogue between 1980 and 2010 (22% between 1990 and 2000) (A. Palloix, unpublished data).
Potato virus Y variants.
Two sets of PVY variants were used: (i) viral populations derived from the two cDNA clones SON41p and LYE84 and from several of their VPg mutants (14, 15) and (ii) representative isolates collected from pepper crops corresponding to different haplotypes according to the VPg sequence. Four mutants of SON41p, named S101G, T115K, T115R, and D119N according to the position and nature of the amino acid substitution in the VPg, were obtained after experimental evolution in HD285 carrying the pvr23 resistance allele (14). A fifth mutant, S120C, was excluded because of its lack of stability and difficulty in obtaining a homogeneous inoculum. Each mutation was introduced into the SON41p clone by site-directed mutagenesis and shown to be sufficient for pathogenicity against pvr23. Similarly, two VPg mutants of LYE84, named H119R and H119Y, were obtained after experimental evolution in PI247087 carrying the pot-1 resistance allele, and each mutation was introduced into the LYE84 clone by site-directed mutagenesis (15; also the present study). Mutations H119R (15) and H119Y (this study) were shown to be sufficient for pathogenicity against pot-1.
The sequences of the VPg-coding regions of 57 PVY isolates collected from pepper crops were determined (references 12, 16, and 17 and the present study), and 12 haplotypes were observed based on the amino acid diversity in the region spanning positions 101 to 123, shown to be critical for pathogenicity toward eIF4E-mediated resistance (14, 16). A single isolate was chosen to represent each haplotype for infectivity tests (Fig. 2).
FIG 2.

Characteristics and VPg sequence of pepper PVY isolates used in the present study. The sequence alignment of the central part of the VPg (amino acid positions 101 to 123) is shown, with dots indicating the presence of the same amino acid as in the first sequence. Year of collection, place of collection, and accession number of the VPg-coding sequence are indicated, respectively, in parentheses.
Analysis of the infectivity properties of PVY variants.
Because direct bombardment of pepper or S. habrochaites plants with PVY cDNA was unsuccessful, we used Nicotiana clevelandii as a first host for cDNA bombardment. Then, the virus populations obtained from cDNA clones of SON41p, LYE84, and their six VPg mutants were inoculated mechanically onto plants representing 10 pepper and 2 S. habrochaites genotypes with different eIF4Es, as previously described (15). For SON41p and LYE84, inoculation of plants carrying recessive resistance genes or alleles was also performed after an additional passage of the virus population in plants of the susceptible pepper and S. habrochaites reference genotypes, respectively. The 12 representative PVY field isolates chosen as described above were inoculated mechanically onto the six pepper plants with genotypes featuring the pvr21, pvr22, pvr23, pvr25, or pvr28 allele. The pvr24 allele was not tested to avoid redundancy with pvr23 (see the Results section), and the pvr26, pvr27, and pvr29 alleles were not retained because plant accessions carrying these alleles were not (or rarely) infected by SON41p, LYE84, or their mutants (see the Results section). Symptoms were recorded from 2 to 5 weeks after inoculation, and PVY was detected by a double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) and reverse transcription-PCR (RT-PCR) at 5 weeks after inoculation in apical leaves. At least two independent experiments, each comprising at least 20 plants per virus-plant genotype combination, were performed.
The nucleotide sequences of the VPg-coding regions of PVY populations that infected plants carrying resistance genes or of susceptible control plants were determined as described previously (12, 14) from a total of four (when available) infected plants per virus-plant genotype combination.
The H119Y mutation observed in the VPg-coding region of the PVY population infecting the PI247087 accession of S. habrochaites carrying the pot-1 resistance gene after inoculation by LYE84 was introduced into the LYE84 cDNA clone by site-directed mutagenesis and homologous recombination in Saccharomyces cerevisiae as described by Ayme et al. (14).
Statistical analysis of the structure of virus-plant infectivity matrices.
Three criteria were chosen to describe virus-plant infectivity matrices and to compare them to expectations of the GFG, MA, and modular models: (i) the total number of compatibility cases in the matrix, (ii) their nestedness, and (iii) their modularity. Methods to estimate nestedness and modularity are described in Weitz et al. (18). Nestedness varies usually from 0 (low nestedness) to 1 (high nestedness) and was estimated by three different algorithms (19–21) with the package bipartite of the R software program (http://cran.r-project.org/). Modularity reflects the concentration of compatibility cases within modules compared with random distribution regardless of modules (18, 22) and varies from −1 (antimodular matrix) to +1 (high modularity matrix). Values close to zero correspond to random partitions into modules of randomly distributed compatibility cases. Given the contrasts in statistical power to detect modules between methods (23), we used five different algorithms, implemented in the package igraph of the R software program to estimate modularity (22, 24–28) (Table 1). For statistical significance assessment, the nestedness and modularity of the plant-virus interaction matrices obtained experimentally (in brief experimental matrices) were compared to two different null models as described by Weitz et al. (18). In the first one (Bernoulli random null model), the same total number of compatibility cases as in the experimental matrices was randomly distributed in matrices containing the same number of rows and columns as the experimental matrices. In the second one (probabilistic degree null model), each plant virus combination in the matrix was assigned a probability of being a compatible interaction which was equal to the mean of the frequencies of compatibility cases in the same column and in the same row in the experimental matrix. One thousand (for modularity) or 10,000 (for nestedness) simulations were performed for both null models. For all analyses, redundant or empty rows or columns (i.e., rows or columns sharing the same compatibility cases or containing no compatibility cases, respectively) were withdrawn.
TABLE 1.
Nestedness and modularity of PVY-plant infectivity matrices
| Parameter and algorithm | Value for the parameter in:a |
|||
|---|---|---|---|---|
| PVY mutants |
PVY isolates |
|||
| Including susceptible plant genotypes | Excluding susceptible plant genotypes | Including susceptible plant genotypes | Excluding susceptible plant genotypes | |
| Nestedness | ||||
| binmatnest2b | 0.80 (0.032*, 0.193) | 0.71 (0.301, 0.541) | 0.93 (0.021*, 0.106) | 0.77 (0.340, 0.579) |
| NODF2c | 0.71 (0.011*, 0.172) | 0.56 (0.231, 0.397) | 0.85 (0.009**, 0.028*) | 0.73 (0.128, 0.267) |
| WINEd | 0.61 (0.011*, 0.117) | 0.53 (0.059, 0.332) | 0.64 (0.015*, 0.070) | 0.47 (0.289, 0.400) |
| Modularity | ||||
| leading.eigenvector.communitye | 0.22 (0.371, 0.326) | 0.25 (0.316, 0.292) | 0.18 (0.374, 0.343) | 0.19 (0.077, 0.255) |
| spinglass.communityf | 0.20 (NA, NA) | 0.22 (NA, NA) | 0.18 (NA, NA) | 0.22 (NA, NA) |
| optimal.communityg | 0.23 (0.765, 0.516) | 0.27 (0.433, 0.328) | 0.18 (0.734, 0.468) | 0.19 (0.107, 0.327) |
| edge.betweenness.communityh | 0.11 (0.527, 0.373) | 0.25 (0.191, 0.178) | 0 | 0 |
| infomap.communityi | 0 | 0 | 0 | 0 |
Nestedness and modularity were estimated with different algorithms of the R software package, and estimation values are indicated separately for PVY VPg mutants or field isolates and including or excluding the Capsicum annuum and Solanum habrochaites susceptible reference genotype plants that were infected by all PVY variants (Fig. 1d and e). Values in parentheses are the frequencies of matrices simulated under the Bernoulli null model and the probabilistic degree null model, respectively, showing higher nestedness or modularity estimates than the experimental infectivity matrices, except when no module was detected (modularity indicated as 0). Totals of 10,000 (nestedness) or 1,000 (modularity) matrices were simulated independently under the two null models. Nestedness was significant at the 5% (*) and 1% (**) type I error threshold (in bold). NA, not available (the method cannot work with unconnected graphs, many of which were obtained in the matrices simulated under the null models).
R function according to Rodríguez-Gironés and Santamaría (19).
R function according to Almeida-Neto et al. (20).
R function according to Galeano et al. (21).
R function according to Newman (22).
R function according to Newman and Girvan (24), Reichardt and Bornholdt (25), and Traag and Bruggeman (26).
R function according to Brandes et al. (27).
R function according to Newman and Girvan (24).
R function according to Rosvall and Bergstrom (28).
RESULTS
Infectivity of PVY variants in Capsicum annuum and Solanum habrochaites.
In a first set of experiments, plants of C. annuum and S. habrochaites representing genotypes with different alleles at the pvr2 and pot-1 locus, respectively, encoding highly similar eIF4E copies (7) were inoculated mechanically with virus populations produced from cDNA clones of isolates SON41p and LYE84 and of their VPg mutants (Fig. 1d). Mutants S101G, T115K, T115R, and D119N of SON41p had been selected by C. annuum accession HD285 plants carrying the resistance allele pvr23, and mutants H119R and H119Y of LYE84 had been selected by S. habrochaites accession PI247087 plants carrying the resistance allele pot-1. Three categories of reactions were observed in plants at 5 weeks after inoculation. In 44% of plant-PVY combinations (42/96), 100% of plants were infected at the systemic level, and no additional mutation was observed in the VPg-coding region of the PVY populations. In 46% (44/96) of cases, no plant was infected at the systemic level. Finally, in 10% of cases (10/96), the infection frequency was below 100% (from 2.2 to 82.6%), and secondary nonsynonymous substitutions were always observed in the VPg-coding region of the PVY progeny (Fig. 1d). These secondary mutations were at codon positions 101, 105, 115, 119, 120, and 121 of the VPg, which were shown to determine pathogenicity toward the pvr2 and/or pot-1 gene (14–16). Concerning the mutations observed in the progeny of SON41p and LYE84 VPg mutants, the responsibility of the identified secondary mutations in the VPg-coding region in determining infection was not formally established (this would have required introducing these mutations by site-directed mutagenesis into the cDNA clones of the PVY mutants). However, six of these PVY mutants carrying secondary mutations were randomly chosen for back-inoculation to the same plant genotype (Fig. 1d). For all of them, 100% (25 of 25) of plants were infected 15 days after inoculation, and no additional mutation was observed in the VPg-coding region of the viral progeny. This shows clearly that the infected plants of the latter category correspond to resistance breakdown (RB) events that occurred during the test and do not represent the initial infectivity properties of the PVY mutant. For this reason, plant-PVY combinations corresponding to this third category were considered incompatibility cases in analyses of the interaction patterns of infectivity matrices. Importantly, for all plant genotypes corresponding to this third category for the VPg mutants, no infection was observed after inoculation by the PVY populations derived from the initial cDNA clones (SON41p or LYE84), even after an additional passage in a pepper or S. habrochaites plant with the reference susceptibility genotype to produce the inocula, evidencing an evolutionary springboard effect conferred by the first acquired RB mutation (see below).
As a whole, plants with the reference susceptibility genotypes of C. annuum and S. habrochaites were 100% infected by all PVY variants. In contrast, none of the plants carrying pvr26 or pvr27 were infected. The C. annuum plants with genotype pvr23 or pvr24 showed the same behaviors toward all PVY variants. This is in accordance with the fact that they possess very similar eIF4E sequences, differing by a single amino acid substitution (7, 12), and suggests that they are redundant in terms of interaction specificity with PVY. In total, the PVY variants were able to infect plants belonging to 1 to 5 plant genotypes (3.25 on average) carrying resistance genes or alleles (i.e., excluding the reference susceptibility plant genotypes), of a total of 10, if we consider only cases where 100% of plants were infected.
In a second set of experiments, the six pepper plants with the pvr2+, pvr21, pvr22, pvr23, pvr25, or pvr28 genotype were inoculated mechanically with 12 representative PVY field isolates corresponding to different haplotypes based on the amino acid diversity of the central part of the VPg, which determines pathogenicity toward recessive resistance genes in the Solanaceae (Fig. 1e). Results were quite similar to those obtained with SON41p, LYE84, and their VPg mutants. We observed the same three categories of reactions as previously for SON41p, LYE84, and their mutants. Again, the category where the infection frequency was below 100% and where secondary nonsynonymous substitutions were observed in the VPg-coding region of the PVY progeny corresponded to RB events that occurred during the test. Indeed, for eight randomly chosen PVY mutants carrying secondary mutations, 100% (25 of 25) of plants were infected 15 days after back-inoculation to the same plant genotype, and no additional mutation was observed in the VPg-coding region of the viral progeny (Fig. 1e). Compared to the previous experiment, a fourth category of reaction was observed in three PVY isolate-pvr2 resistance allele combinations, with 100% infection and occurrence of amino acid substitutions in the VPg compared to the sequence of the virus from the inoculum or from susceptible reference plants. This category includes isolate CAA14 versus a pvr28 plant and isolate CAA15 versus a pvr21 or pvr25 plant (Fig. 1e). It is possible that these isolates were only partly adapted to the resistance alleles, and their fitness was increased by the observed mutations. Alternatively, minor variants present in the PVY inoculum could have been selected during the experiment. All plants of the susceptible reference genotype were infected by all isolates. In contrast, none of the plants of the pvr22 genotype were infected. The isolates were able to infect plants belonging to 0 to 4 plant genotypes (1.58 on average) carrying pvr2 resistance alleles (excluding the reference susceptibility plant genotype), of a total of 5, if we consider only cases where 100% plants were infected. This corresponds roughly to the same infectivity probability as with the PVY mutants, with a probability of 0.316 for PVY field isolates to infect plants carrying a pvr2 resistance allele and of 0.325 for SON41p, LYE84, and their mutants.
Analysis of the interaction pattern between PVY variants and Capsicum annuum and Solanum habrochaites.
The three proposed host-parasite interaction models (GFG, MA, and modular) are characterized by different frequencies of compatibility cases in infectivity matrices as well as different structures of these compatibility cases in terms of nestedness and modularity (Fig. 1a to c). The frequency of compatibility cases observed in our experimental matrices was much higher than that expected under the MA model (P < 0.004; Fisher's exact tests) but similar to that expected under the GFG model (P > 0.34; Fisher's exact tests). Results were similar for the matrices obtained with field isolates or VPg mutants of SON41p and LYE84 and keeping or excluding plants with susceptible C. annuum and S. habrochaites reference genotypes. Results were also highly consistent between the three nestedness estimation algorithms and between the five modularity estimation algorithms (Table 1). The experimental matrices obtained for the VPg mutants or the field isolates showed low modularity values (<0.23) but high nestedness values (0.61 to 0.93) (Table 1; Fig. 1d and e). In addition, the experimental matrices were not more modular than matrices generated under the null models (at least 32.6% of the simulated matrices had higher modularity values than the experimental matrices). In contrast, the experimental matrices were significantly more nested than matrices generated under the Bernoulli null model (see Materials and Methods) (18) (P = 0.009 to 0.032, depending on the matrix and the algorithm). However, they were not, except in one case, significantly more nested than matrices generated under the probabilistic degree null model at the 5% error threshold. The rather marginal nestedness observed in the experimental matrices was also influenced by the presence of plants with the reference susceptibility genotypes, which were infected by all PVY variants, and neither nestedness nor modularity was significant if we considered only plant genotypes with resistance alleles.
DISCUSSION
The interaction patterns between PVY and pepper and S. habrochaites differ significantly from the matching allele or modular models.
Interactions between plant and virus genotypes were proposed to correspond to the GFG or MA model on the basis of the structure of infectivity matrices and of the molecular mechanisms determining infectivity of the virus and resistance of the plant (4). However, plant-virus infectivity matrices have been only rarely determined and usually comprise only a small number of plant and/or virus genotypes, hampering any statistical analysis of their structure. Interaction between plants with genotypes harboring various alleles at eIF4E (or eIF4G)-encoding loci controlling susceptibility or recessive resistance and different groups of viruses were considered emblematic of the MA model (4). Indeed, in these systems, infection was shown to depend on a specific match and a direct physical interaction between the plant eIF4E (or eIF4G) and a virus pathogenicity factor, usually the VPg (7, 8). Mutations in the plant factor that abolish interaction with the virus VPg confer resistance to the plant, and mutations in the virus VPg that restore interaction with the mutated plant factor are responsible for infectivity of the virus in plants carrying resistance alleles. However, again, little data were available to support this model on the basis of the structure of the interaction matrix between plant and virus genotypes.
The infectivity matrices that we obtained with PVY clones and mutants or with field isolates and genotypes of C. annuum and S. habrochaites plants did not show any evidence of modularity, as would have been the case for the MA model or for a more relaxed modular model. This was mainly due to a high frequency of cross-infectivity, each PVY variant usually being able to infect plants representing several genotypes with different resistance alleles. However, even taking into account this high frequency of cross-infectivity, our experimental matrices were not more modular than matrices generated at random. The lack of modularity indicates that there is no tendency for PVY variants with similar VPgs to infect plants with similar eIF4Es (and vice versa). As a consequence, it is not possible to predict the infectivity properties of a given PVY isolate from those of its closest VPg sequence variants.
In contrast, significant nestedness was detected for some of our experimental matrices, which could be reminiscent of the GFG model of interaction. However, this effect was rather marginal, and the molecular mechanism of interaction between PVY and plants carrying recessive resistance alleles does not correspond to an elicitor-receptor interaction triggering specific plant defenses, as usually considered in the GFG model (3). It should be noted, however, that nested, but not modular, patterns of interaction were frequently detected in phage-bacteria interactions (29) and could be a rather general pattern of interaction.
The PVY-plant interaction considered here is consequently intermediate between the GFG and MA models, sharing the mechanistic bases of the MA model and the extensive cross-infectivity of the GFG model. One explanation could be that potyvirus VPgs possess intrinsically disordered domains, especially in the central part which corresponds to the pathogenicity determinant against recessive resistance genes (30–32), which can confer the ability to bind different ligands (33, 34) and/or to bind a large set of allelic forms of a given ligand like eIF4E. However, this structural flexibility has some limits, and, in contrast with the GFG model, we did not observe any PVY variant with universal infectivity (Fig. 1d and e).
In addition to this static view of the virus-plant interaction pattern at a given evolutionary time, it is also important to consider their genetic bases in a more dynamic view to unravel their causes and consequences. Remarkably, highly similar structural patterns of infectivity matrices were observed for SON41p, LYE84, and their mutants, on one hand (hence representing a very low virus genetic diversity), and for PVY isolates collected from pepper fields worldwide, on the other hand (hence, comprising a much larger genetic diversity). This suggests that the same genetic mechanisms could be involved in determining the observed interaction patterns.
Widespread cross-infectivity and evolutionary springboard effects of PVY mutations in solanaceous crops.
The PVY VPg mutants used in the present study were the results of experimental evolution of populations derived from the SON41p and LYE84 clones. Initially, SON41p was infectious only in pepper plants carrying pvr21 or pvr22 in addition to plants with the susceptibility alleles, and LYE84 was infectious only in plants with the susceptibility alleles. After a first set of inoculations, SON41p gained infectivity toward the pvr23 resistance allele in pepper, and LYE84 gained infectivity toward the pot-1 resistance allele in S. habrochaites (14, 15). These RBs were due to precise amino acid substitutions in the VPg (Fig. 1d). When the VPg mutants of SON41p and LYE84 were inoculated onto the set of plants carrying different eIF4E alleles, different kinds of pleiotropic effects were observed.
The first kind of pleiotropic effect can be named cross-infectivity by analogy to cross-resistance of microbes, insects, or weeds to different (bio)chemical compounds in a medical or agricultural context (35–39). It can be defined as the effect of a single mutational event which leads to the breakdown of at least two plant resistance genes or alleles, an initial one, which exerts selection pressure on the pathogen population leading to the fixation of an RB mutation, and a second one, which does not play any role in the fixation of the RB mutation. As best examples of cross-infectivity, the breakdown mutations selected by pot-1 resistance in S. habrochaites resulted also in the breakdown of four distinct pvr2 resistance alleles in pepper (Fig. 1d and 3). Similar cross-infectivity effects are also expected between tobacco (Nicotiana tabacum) and pepper resistance genes. Indeed, it was shown previously that the VPg of PVY was also the pathogenicity factor corresponding to the va gene in tobacco (40, 41). Sequence comparisons indicated that the S101G and D119G substitutions in the VPg of PVY SON41p allowed the breakdown of the va2 resistance allele in tobacco. These two substitutions also allowed the breakdown of the pvr23 allele (14, 42), displaying consequently a cross-infectivity effect between the va2 and pvr23 alleles in tobacco and pepper cultivars, respectively (Fig. 3). Obviously, this definition is only meaningful if the two resistance genes considered have different specificities, i.e., different spectra of action toward the pathogen diversity. Since the pvr23 and pvr24 resistance alleles showed the same specificities of action toward the eight PVY clones and mutants tested (Fig. 1d), which is consistent with their sequence similarity (one amino acid difference only) (7), we would not define as cross-infectivity the effect of VPg mutations involved in the simultaneous breakdown of pvr23 and pvr24.
FIG 3.
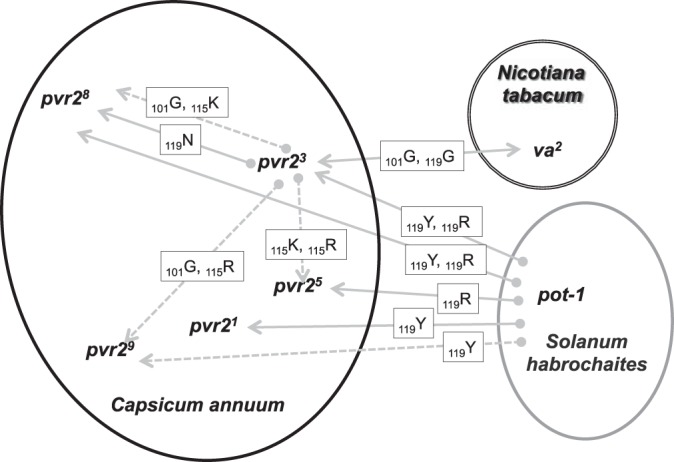
Cross-infectivity and springboard effects of PVY mutations involved in resistance breakdown (RB) in three solanaceous species. Arrows with solid lines correspond to cross-infectivity effects, and arrows with broken lines correspond to evolutionary springboard effects of RB mutations. Mutations in boxes correspond to amino acid substitutions in PVY VPg conferring RB. Arrows point toward the second resistance gene or allele for which cross-infectivity or springboard effects are observed after fixation of the mutation involved in the breakdown of a first resistance gene. The double-headed arrow indicates that the two considered resistance genes can select the mutation (symmetrical cross-infectivity).
The second kind of pleiotropic effect, which we named the evolutionary springboard effect, occurs when a first plant resistance gene (or allele) leads to the fixation of a first RB mutational event in the virus population which further favors the breakdown of a second resistance gene (or allele) through an additional mutational event(s). In that case, the direct inoculation of the initial virus population (SON41p or LYE84) to plants carrying the second resistance gene did not lead to infection, even after a supplementary passage in susceptible reference plants before inoculation, evidencing the evolutionary springboard effect. As best examples of the springboard effect, mutations T115K and T115R that were selected by the pvr23 resistance allele favored the breakdown of pvr25 and pvr28 and, respectively, of pvr25 and pvr29 (Fig. 1d and 3).
Cross-infectivity and springboard effects correspond to positive or synergistic pleiotropy effects, where a single mutation has two favorable effects for the pathogen, allowing infection of, or acquisition of, RB properties toward plant genotypes representing two different resistance genes or alleles. In contrast, the third case of pleiotropic effect identified in this study corresponds to antagonistic pleiotropy, where a mutation allows the breakdown of a first resistance gene and abolishes simultaneously the capacity of breakdown of a second one. Antagonistic pleiotropy was observed among PVY VPg mutations involved in the breakdown of alleles pvr22 and pvr23 in pepper, such as mutations T115K, T115R, and D119N (Fig. 1d).
The ability of many field PVY isolates to infect pepper plants with different pvr2 resistance alleles is likely the result of cross-infectivity effects of mutations. Only the pvr21 and pvr22 recessive resistance genes have been largely deployed worldwide. Whereas none of the isolates was able to infect pvr22 plants, six of them were breaking the pvr21 allele without the requirement of additional mutations in the VPg (Fig. 1e). Four of these isolates (Alger1, GHB11, CAA16, and CAA141) (15; also our unpublished data) were collected in plants homozygous for pvr21 (no data are available for the plant origin of the other two isolates). The selective cause of the pvr21-breaking capacity of these isolates was probably the pvr21 allele itself, and their capacity to infect plants having genotypes with other pvr2 resistance alleles is the by-product of the fixation of the pvr21-breaking mutation. Supporting this assumption, the six isolates infecting pepper plants carrying the pvr21 genotype had a significantly higher capacity to infect pepper plants with additional genotypes than the six isolates that were not infecting the pvr21 pepper (P = 0.005; Fisher exact test). Unfortunately, it was impossible to reconstruct the mutational pathways leading to pvr21 breakdown for the former six isolates because of the large number of mutations at the amino acid positions critical for pathogenicity toward pvr2 plants compared to isolates that did not break pvr21 (Fig. 2).
Such synergistic pleiotropic effects of infectivity mutations are rare in plant-pathogen interactions, and only a few cross-infectivity effects have been described (11, 43–45). To our knowledge, no evolutionary springboard effect among RB mutations has been described so far. For example, in the most exhaustive study, only one of eight (12.5%) RB mutations in the VPg of RYMV conferred simultaneously the capacity to infect rice plants with the rymv1–2 and rymv1–3 resistance alleles, and 4 of 38 RYMV isolates (11%) were infectious in both kinds of rice genotypes (11), showing the rarity of cross-infectivity in this system. However, such effects could have been underestimated because of the small size of the plant-pathogen interaction matrices usually analyzed. In comparison, in our system, we observed seven occurrences of cross-infectivity effects and seven of evolutionary springboard effects (Fig. 3), which represents 17.5% for each if we include the pvr26 and pvr27 alleles (7 of 40 combinations between VPg mutations and pvr2 alleles that could reveal pleiotropic effects).
Consequences on resistance management strategies.
Evaluating whether mutations involved in the breakdown of different plant resistance genes or alleles are independent or not is crucial since it determines the risk of emergence of multivirulent pathogens (i.e., pathogens breaking down simultaneously the resistance controlled by several genes or alleles) and the sustainability of disease control strategies based on genetic resistance (46, 47). In this respect, the evolutionary pathways leading to multivirulence and the different cases of pleiotropic effects of RB mutations (cross-infectivity, evolutionary springboards, and antagonistic pleiotropy) determine the probability of emergence of RB populations (47, 48).
Cross-infectivity and springboard effects are likely to decrease the efficiency of resistance management strategies such as varietal rotations or mixtures. The fact that these effects occur also between different plant species and even genera like Nicotiana, Solanum, and Capsicum (Fig. 3) indicates that the different crop species in the agricultural landscape should be considered simultaneously in this regard. Wolfe (49) reviewed four mechanisms by which growing mixtures of plant cultivars carrying different resistance genes or alleles in the same fields increased resistance durability: (i) the decrease of host density for the pathogen, compared to situations with 100% susceptible plants (or plants in which resistance is broken down), (ii) the barrier effect reducing transmission efficiency due to nonhosts, (iii) the counterselection of RB mutations through the fitness costs of these mutations in hosts lacking the corresponding resistance, and (iv) the decrease of selection pressure for RB variants compared to situations with 100% of plants carrying the resistance gene. The same four mechanisms are also in play during rotation strategies if we take into account several consecutive cropping seasons. Cross-infectivity and springboard effects will suppress or decrease the action of these four mechanisms and are therefore expected to reduce drastically the efficiency of the mixture and rotation strategies. Indeed, a pathogen isolate carrying cross-infectivity mutations (or showing evolutionary springboard effects) will be able to infect (or to evolve RB capacity toward) a larger panel of cultivars in the mixture, hence increasing its host density (mechanism i) and reducing the barrier effects (mechanism ii). Also, several cultivars in the mixture will contribute to select and maintain the same RB mutations in the pathogen population, in the case of both cross-infectivity and evolutionary springboard effects, which will reduce the effects of counterselection (mechanism iii) and of decreased selection pressure (mechanism iv).
ACKNOWLEDGMENTS
We thank Joël Chadœuf (INRA PACA) for helpful discussions about estimations of nestedness and modularity and Josselin Montarry (INRA Rennes) for improving the manuscript.
This work was supported by the SYSTERRA program of the Agence Nationale de la Recherche, a French-Tunisian PHC Utique bilateral project (10G0905), and a project from the Maladies Infectieuses Emergentes interdisciplinary program of the Centre National de la Recherche Scientifique.
Footnotes
Published ahead of print 18 June 2014
REFERENCES
- 1.Luijckx P, Fienberg H, Duneau D, Ebert D. 2013. A matching-allele model explains host resistance to parasites. Curr. Biol. 23:1085–1088. 10.1016/j.cub.2013.04.064 [DOI] [PubMed] [Google Scholar]
- 2.Gandon S, Capowiez Y, Dubois Y, Michalakis Y, Olivieri I. 1996. Local adaptation and gene-for-gene coevolution in a metapopulation model. Proc. Biol. Sci. 263:1003–1009. 10.1098/rspb.1996.0148 [DOI] [Google Scholar]
- 3.Dybdahl MF, Storfer A. 2003. Parasite local adaptation: Red Queen versus Suicide King. Trends Ecol. Evol. 18:523–530. 10.1016/S0169-5347(03)00223-4 [DOI] [Google Scholar]
- 4.Sacristán S, García-Arenal F. 2008. The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9:369–384. 10.1111/j.1364-3703.2007.00460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal A, Lively CM. 2002. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol. Ecol. Res. 4:79–90 [Google Scholar]
- 6.Fraile A, Pagan I, German A, Saez E, García-Arenal F. 2011. Rapid genetic diversification and high fitness penalties associated with pathogenicity evolution in a plant virus. Mol. Biol. Evol. 28:1425–1437. 10.1093/molbev/msq327 [DOI] [PubMed] [Google Scholar]
- 7.Charron C, Nicolaï M, Gallois JL, Robaglia C, Moury B, Palloix A, Caranta C. 2008. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 54:56–68. 10.1111/j.1365-313X.2008.03407.x [DOI] [PubMed] [Google Scholar]
- 8.Hébrard E, Poulicard N, Gérard C, Traore O, Wu HC, Albar L, Fargette D, Bessin Y, Vignols F. 2010. Direct interaction between the Rice yellow mottle virus (RYMV) VPg and the central domain of the rice eIF(iso)4G1 factor correlates with rice susceptibility and RYMV virulence. Mol. Plant Microbe Interact. 23:1506–1513. 10.1094/MPMI-03-10-0073 [DOI] [PubMed] [Google Scholar]
- 9.Poulicard N, Pinel-Galzi A, Traoré O, Vignols F, Ghesquière A, Konate G, Hébrard E, Fargette D. 2012. Historical contingencies modulate the adaptability of Rice yellow mottle virus. PLoS Pathog. 8:e1002482. 10.1371/journal.ppat.1002482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinel-Galzi A, Rakotomalala M, Sangu E, Sorho F, Kanyeka Z, Traoré O, Sérémé D, Poulicard N, Rabenantoandro Y, Séré Y, Konaté G, Ghesquière A Hébrard E, Fargette D. 2007. Theme and variations in the evolutionary pathways to virulence of an RNA plant virus species. PLoS Pathog. 3:e180. 10.1371/journal.ppat.0030180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traoré O, Pinel-Galzi A, Issaka S, Poulicard N, Aribi J, Ake S, Ghesquière A, Séré Y, Konate G, Hébrard E, Fargette D. 2010. The adaptation of Rice yellow mottle virus to the eIF(iso)4G-mediated rice resistance. Virology 408:103–108. 10.1016/j.virol.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 12.Moury B, Charron C, Janzac B, Simon V, Gallois J-L, Palloix A, Caranta C. 3 December 2013. Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome-linked viral protein (VPg): a game of mirrors impacting resistance spectrum and durability. Infect. Genet. Evol. 10.1016/j.meegid.2013.11.024 [DOI] [PubMed] [Google Scholar]
- 13.Ruffel S, Gallois JL, Lesage ML, Caranta C. 2005. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 274:346–353. 10.1007/s00438-005-0003-x [DOI] [PubMed] [Google Scholar]
- 14.Ayme V, Souche S, Caranta C, Jacquemond M, Chadoeuf J, Palloix A, Moury B. 2006. Different mutations in the genome-linked protein VPg of Potato virus Y confer virulence on the pvr23 resistance in pepper. Mol. Plant Microbe Interact. 19:557–563. 10.1094/MPMI-19-0557 [DOI] [PubMed] [Google Scholar]
- 15.Moury B, Morel C, Johansen E, Guilbaud L, Souche S, Ayme V, Caranta C, Palloix A, Jacquemond M. 2004. Mutations in Potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant Microbe Interact. 17:322–329. 10.1094/MPMI.2004.17.3.322 [DOI] [PubMed] [Google Scholar]
- 16.Ayme V, Petit-Pierre J, Souche S, Palloix A, Moury B. 2007. Molecular dissection of the Potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88:1594–1601. 10.1099/vir.0.82702-0 [DOI] [PubMed] [Google Scholar]
- 17.Ben Khalifa M, Simon V, Fakhfakh H, Moury B. 2012. Tunisian Potato virus Y isolates with unnecessary pathogenicity towards pepper: support for the matching allele model in eIF4E resistance-potyvirus interactions. Plant Pathol. 61:441–447. 10.1111/j.1365-3059.2011.02540.x [DOI] [Google Scholar]
- 18.Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB, Hochberg ME. 2013. Phage-bacteria infection networks. Trends Microbiol. 21:82–91. 10.1016/j.tim.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez-Gironés MA, Santamaría L. 2006. A new algorithm to calculate the nestedness temperature of presence-absence matrices. J. Biogeogr. 33:924–935. 10.1111/j.1365-2699.2006.01444.x [DOI] [Google Scholar]
- 20.Almeida-Neto M, Guimaraes P, Guimaraes PR, Loyola RD, Ulrich W. 2008. A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos 117:1227–1239. 10.1111/j.0030-1299.2008.16644.x [DOI] [Google Scholar]
- 21.Galeano J, Pastor JM, Iriondo JM. 2008. Weighted-interaction nestedness estimator (WINE): a new estimator to calculate over frequency matrices. Environ. Model. Softw. 24:1342–1346. 10.1016/j.envsoft.2009.05.014 [DOI] [Google Scholar]
- 22.Newman ME. 2006. Modularity and community structure in networks. Proc. Natl. Acad. Sci. U. S. A. 103:8577–8582. 10.1073/pnas.0601602103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldecoa R, Marín I. 2013. Exploring the limits of community detection strategies in complex networks. Sci. Rep. 3:2216. 10.1038/srep02216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman ME, Girvan M. 2004. Finding and evaluating community structure in networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 69:026113. 10.1103/PhysRevE.69.026113 [DOI] [PubMed] [Google Scholar]
- 25.Reichardt J, Bornholdt S. 2006. Statistical mechanics of community detection. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 74:016110. 10.1103/PhysRevE.74.016110 [DOI] [PubMed] [Google Scholar]
- 26.Traag VA, Bruggeman J. 2009. Community detection in networks with positive and negative links. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 80:036115.27. 10.1103/PhysRevE.80.036115 [DOI] [PubMed] [Google Scholar]
- 27.Brandes U, Delling D, Gaertler M, Gorke R, Hoefer M, Nikoloski Z, Wagner D. 2008. On modularity clustering. IEEE Trans. Knowl. Data Eng. 20:172–188. 10.1109/TKDE.2007.190689 [DOI] [Google Scholar]
- 28.Rosvall M, Bergstrom CT. 2008. Maps of information flow reveal community structure in complex networks. Proc. Natl. Acad. Sci. U. S. A. 105:1118–1123. 10.1073/pnas.0706851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. 2011. Statistical structure of host-phage interactions. Proc. Natl. Acad. Sci. U. S. A. 108:E288–E297. 10.1073/pnas.1101595108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grzela R, Szolajska E, Ebel C, Madern D, Favier A, Wojtal I, Zagorski W, Chroboczek J. 2008. Virulence factor of Potato virus Y genome-attached terminal protein VPg is a highly disordered protein. J. Biol. Chem. 283:213–221. 10.1074/jbc.M705666200 [DOI] [PubMed] [Google Scholar]
- 31.Hébrard E, Bessin Y, Michon T, Longhi S, Uversky VN, Delalande F, Van Dorsselaer A, Romero P, Walter J, Declerk N, Fargette D. 2009. Intrinsic disorder in viral proteins genome-linked: experimental and predictive analyses. Virol. J. 6:23. 10.1186/1743-422X-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rantalainen KI, Uversky VN, Permi P, Kalkkinen N, Dunker AK, Makinen K. 2008. Potato virus A genome-linked protein VPg is an intrinsically disordered molten globule-like protein with a hydrophobic core. Virology 377:280–288. 10.1016/j.virol.2008.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elena SF, Rodrigo G. 2012. Towards an integrated molecular model of plant-virus interactions. Curr. Opin. Virol. 2:719–724. 10.1016/j.coviro.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 34.Jiang J, Laliberté J-F. 2011. The genome-linked protein VPg of plant viruses—a protein with many partners. Curr. Opin. Virol. 1:347–354. 10.1016/j.coviro.2011.09.010 [DOI] [PubMed] [Google Scholar]
- 35.George AM. 1996. Multidrug resistance in enteric and other Gram-negative bacteria. FEMS Microbiol. Lett. 139:1–10. 10.1111/j.1574-6968.1996.tb08172.x [DOI] [PubMed] [Google Scholar]
- 36.Han HP, Yu Q, Purba E, Li M, Walsh M, Friesen S, Powles SB. 2012. A novel amino acid substitution Ala-122-Tyr in ALS confers high-level and broad resistance across ALS-inhibiting herbicides. Pest Manag. Sci. 68:1164–1170. 10.1002/ps.3278 [DOI] [PubMed] [Google Scholar]
- 37.Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 32:386–408. 10.1111/j.1574-6976.2007.00097.x [DOI] [PubMed] [Google Scholar]
- 38.Race E. 2001. Cross-resistance within the protease inhibitor class. Antivir. Ther. 6(Suppl 2):29–36 [PubMed] [Google Scholar]
- 39.Tabashnik BE, Unnithan GC, Masson L, Crowder DW, Li X, Carriere Y. 2009. Asymmetrical cross-resistance between Bacillus thuringiensis toxins Cry1Ac and Cry2Ab in pink bollworm. Proc. Natl. Acad. Sci. U. S. A. 106:11889–11894. 10.1073/pnas.0901351106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuta C, Nishimura M, Morishita H, Hataya T. 1999. A single amino acid change in viral genome-associated protein of Potato virus Y correlates with resistance breaking in “virgin A mutant” tobacco. Phytopathology 89:118–123. 10.1094/PHYTO.1999.89.2.118 [DOI] [PubMed] [Google Scholar]
- 41.Lacroix C, Glais L, Verrier JL, Jacquot E. 2011. Effect of passage of a Potato virus Y isolate on a line of tobacco containing the recessive resistance gene va2 on the development of isolates capable of overcoming alleles 0 and 2. Eur. J. Plant Pathol. 130:259–269. 10.1007/s10658-011-9751-0 [DOI] [Google Scholar]
- 42.Montarry J, Doumayrou J, Simon V, Moury B. 2011. Genetic background matters: a plant-virus gene-for-gene interaction is strongly influenced by genetic contexts. Mol. Plant Pathol. 12:911–920. 10.1111/j.1364-3703.2011.00724.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdul-Razzak A, Guiraud T, Peypelut M, Walter J, Houvenaghel MC, Candresse T, Le Gall O, German-Retana S. 2009. Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E-mediated resistance against Lettuce mosaic potyvirus. Mol. Plant Pathol. 10:109–113. 10.1111/j.1364-3703.2008.00513.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houterman PM, Cornelissen BJ, Rep M. 2008. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 4:e1000061. 10.1371/journal.ppat.1000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parlange F, Daverdin G, Fudal I, Kuhn ML, Balesdent MH, Blaise F, Grezes-Besset B, Rouxel T. 2009. Leptosphaeria maculans avirulence gene AvrLm4–7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape and circumvents Rlm4-mediated recognition through a single amino acid change. Mol. Microbiol. 71:851–863. 10.1111/j.1365-2958.2008.06547.x [DOI] [PubMed] [Google Scholar]
- 46.Mundt CC. 1990. Probability of mutation to multiple virulence and durability of resistance gene pyramids. Phytopathology 80:221–223. 10.1094/Phyto-80-221 [DOI] [Google Scholar]
- 47.Fabre F, Bruchou C, Palloix A, Moury B. 2009. Key determinants of resistance durability to plant viruses: Insights from a model linking within- and between-host dynamics. Virus Res. 141:140–149. 10.1016/j.virusres.2008.11.021 [DOI] [PubMed] [Google Scholar]
- 48.Fabre F, Montarry J, Coville J, Senoussi R, Simon V, Moury B. 2012. Modelling the evolutionary dynamics of viruses within their hosts: a case study using high-throughput sequencing. PLoS Pathog. 8:e1002654. 10.1371/journal.ppat.1002654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe MS. 1985. The current status and prospects of multiline cultivars and variety mixtures for disease resistance. Annu. Rev. Phytopathol. 23:251–273. 10.1146/annurev.py.23.090185.001343 [DOI] [Google Scholar]