ABSTRACT
The helper component proteinase (HCPro) is an indispensable, multifunctional protein of members of the genus Potyvirus and other viruses of the family Potyviridae. This viral factor is directly involved in diverse steps of viral infection, such as aphid transmission, polyprotein processing, and suppression of host antiviral RNA silencing. In this paper, we show that although a chimeric virus based on the potyvirus Plum pox virus lacking HCPro, which was replaced by a heterologous silencing suppressor, caused an efficient infection in Nicotiana benthamiana plants, its viral progeny had very reduced infectivity. Making use of different approaches, here, we provide direct evidence of a previously unknown function of HCPro in which the viral factor enhances the stability of its cognate capsid protein (CP), positively affecting the yield of virions and consequently improving the infectivity of the viral progeny. Site-directed mutagenesis revealed that the ability of HCPro to stabilize CP and enhance the yield of infectious viral particles is not linked to any of its previously known activities and helped us to delimit the region of HCPro involved in this function in the central region of the protein. Moreover, the function is highly specific and cannot be fulfilled by the HCPro of a heterologous potyvirus. The importance of this novel requirement in regulating the sorting of the viral genome to be subjected to replication, translation, and encapsidation, thus contributing to the synchronization of these viral processes, is discussed.
IMPORTANCE Potyviruses form one of the most numerous groups of plant viruses and are a major cause of crop loss worldwide. It is well known that these pathogens make use of virus-derived multitasking proteins, as well as dedicated host factors, to successfully infect their hosts. Here, we describe a novel requirement for the proper yield and infectivity of potyviral progeny. In this case, such a function is performed by the extensively studied viral factor HCPro, which seems to use an unknown mechanism that is not linked to its previously described activities. To our knowledge, this is the first time that a factor different from capsid protein (CP) has been shown to be directly involved in the yield of potyviral particles. Based on the data presented here, we hypothesize that this capacity of HCPro might be involved in the coordination of mutually exclusive activities of the viral genome by controlling correct assembly of CP in stable virions.
INTRODUCTION
The family Potyviridae is one of the largest groups of plant viruses, with 173 definite members so far, which are distributed in seven different genera (1). The genomes of members of this family consist of a single-stranded, positive-sense RNA molecule, with the only exception being the viruses belonging to the genus Bymovirus, which have a bipartite genome. Members of the genera Potyvirus, Ipomovirus, Macluravirus, Tritimovirus, Rymovirus, and Brambyvirus have genomes ranging from 8.2 to 11 kb in size, which are encapsidated in flexuous rods 11 to 14 nm in diameter by 680 to 950 nm in length (2). These viral particles (or virions) comprise around 2,000 subunits of the capsid protein (CP) arranged all around the RNA molecule (3, 4). Once inside the cell and after uncoating, the genomic RNA is translated to produce a large polyprotein and a truncated frameshift product, comprising in most cases the following factors (from N to C termini): P1, the helper component proteinase (HCPro), P3 (and P3N-PIPO), 6K1, CI, 6K2, NIa (VPg plus Pro), NIb, and CP. The polyprotein is then processed by three virus-encoded proteases in order to release the mature factors needed for the infection process (5, 6).
As viruses have small, compact genomes, all viral functions must be performed by a limited number of encoded proteins; therefore, it is usual to find viral factors participating in different processes of the infection cycle. Such is the case for the multitasking potyviral HCPro, which was initially identified as a helper required for aphid-mediated plant-to-plant transmission (7), likely working as a bridge between insect mouthparts and virions (8). Supporting this idea, two conserved motifs were identified in HCPro as critical for insect-mediated transmission: the KITC motif, which is located in the N-terminal part of the protein and seems to be crucial for the retention of viral particles in aphid stylets, and the PTK motif, located near the C terminus and likely implicated in HCPro-CP interaction (9–13). Interestingly, HCPro was found to be associated with virus particles, being located in a protruding tip at one end of the virion, but the actual roles of these particular HCPro units are unknown (14, 15).
HCPro has a protease activity that autocatalytically cleaves its C terminus to release itself from the rest of the viral polyprotein (16). Furthermore, the cysteine and histidine catalytic residues were mapped by site-directed mutagenesis to the C-terminal half of the protein, supporting the hypothesis that HCPro closely resembles members of the papain-type family of cysteine proteases (17). The first high-resolution crystal structure of an HCPro protease domain was recently solved, giving new and important insights into its unique protein folding, self-cleavage mechanism, and substrate specificity (18).
At the end of the 1990s, three independent laboratories found that the potyviral HCPro is also a potent inhibitor of the RNA-silencing pathway (19–21), supporting the idea that the RNA-silencing mechanism is, among other functions, a host defensive barrier against viruses. Several reviews of RNA silencing and its role in antiviral defense have been recently published (22–25). The mechanism by which HCPro acts as an RNA-silencing suppressor (RSS) appears to rely on its capacity to specifically interact with/sequester virus-derived 21-nucleotide (nt) small RNAs (26–28) and to interact with the small RNA methyltransferase HEN1 (29) and other host factors (30, 31). Interestingly, since its first characterization, HCPro has also been related to several events during viral infection, such as genome amplification, both cell-to-cell and long-distance movements, pathogenicity, and viral synergism (reviewed in references 32 and 33). These capacities were attributed to the role of HCPro as an RSS (34); however, more recently, other functions have been proposed for this factor that might not be directly related to RNA-silencing suppression (35–37).
Despite the multiple key roles of HCPro during viral infection, this protein can be replaced by unrelated RSSs from different viruses in the genome of Plum pox virus (PPV) (genus Potyvirus) (38, 39). Here, we report that, although the replacement of HCPro does not have a significant impact on the ability of PPV to infect Nicotiana benthamiana plants, as long as the RSS efficiently inhibited the host silencing-based antiviral defense, a massive reduction in the initiation of a new infection in plant-to-plant manual passages was perceived for the progeny of PPV chimeras expressing heterologous RSSs. Based on this observation, we show direct evidence of a crucial species-specific role for HCPro in enhancing the yield of intact viral particles that, as a consequence, strongly affects virus infectivity. Site-directed mutagenesis revealed that this ability is not linked to the previously described HCPro activities and helped to map the protein region involved in this novel function. Altogether, these data not only indicate that HCPro, besides its well-defined skills, is directly involved in the yield of infectious particles, but also highlight the existence of a novel requirement for the efficient infectivity of a potyvirus.
MATERIALS AND METHODS
Plants.
Agroinfiltration assays were performed in N. benthamiana plants. Viral infectivity assays were performed in N. benthamiana and Nicotiana clevelandii. The plants were grown in a greenhouse maintained at 16 h light with supplementary illumination and at 19 to 23°C.
Plasmids.
Gateway technology (Invitrogen) was applied to construct binary plasmids expressing PPV 5′ untranslated region (UTR)-P1-HCPro by following the instructions of the manufacturer. Site-directed mutagenesis of HCPro was carried out by two PCR steps as described by Herlitze and Koenen (40). The primers and templates used for PCR amplification and site-directed mutagenesis of HCPro are available upon request. BP Clonase (Invitrogen) reactions were used to introduce the PCR fragments into the donor vector pDONR-207 (Invitrogen) and to generate the entry clones.
Expression vectors producing wild-type and mutant versions of PPV 5′ UTR-P1-HCPro (pMDC32-P1HCPro and derivatives) were constructed by LR Clonase (Invitrogen) reactions between the corresponding pDONR-P1HCPro entry clones and the destination vector, pMDC32 (provided by Mark Curtis, University of Zurich) (41). Binary plasmids expressing PPV HCPro (p35S-HCPPV) and PPV 5′ UTR-P1-HCPro (p35S-P1HCPPV) (42), PPV P3-6K1-CI-6K2-NIa-NIb-green fluorescent protein (GFP)-CP (pLONG-GFP) (43), TEV 5′ UTR-P1-HCPro-P3-6K1 (p35S-P1/HC-Pro) (44), and the Tomato bushy stunt virus (TBSV) P19 silencing suppressor (35S-p19) (45) have been previously described.
A previously described full-length infectious cDNA clone of PPV (pIC-PPV-NKGFP) (46) was used as the backbone to generate the HCPro mutant variants of the virus. Hence, the RsrII/SexAI fragment from PPV, which encodes a region including the C-terminal part of P1 and almost the complete HCPro, was replaced with analogous fragments from the corresponding pDONR-P1HCPro entry vectors. Chimeric PPV viruses carrying either the P1b or P19 coding sequence from Cucumber vein yellowing virus (CVYV) or TBSV, respectively, instead of that from PPV HCPro were previously described (38, 39).
Viral infection.
For biolistic inoculation, the Helios Gene Gun System (Bio-Rad) was used as previously described (47). For manual inoculation of plants using plasmids, 6 μg of DNA from the indicated constructs was spread on two leaves of plants that had previously been dusted with Carborundum. For manual inoculation of plants using sap extracts as the source of virus, infected leaves were ground with sodium phosphate buffer (5 mM; pH 7.5) with an ice-cold pestle (2 ml/mg), and the sap produced was used to inoculate plants as described above.
Agroinfiltration and fluorescence imaging.
Either healthy or infected N. benthamiana plants were infiltrated with Agrobacterium tumefaciens strain C58C1 carrying the indicated plasmids, as previously described (42). GFP fluorescence was observed with an epifluorescence stereomicroscope using excitation and barrier filters at 450/90 nm and 500/550 nm, respectively, and photographed with an Olympus DP70 digital camera.
Western blot analysis.
Infected/agroinfiltrated leaves were ground to fine powder under liquid nitrogen in a mortar and stored at −80°C until use. For preparation of protein samples under denaturing conditions, the powders were thawed in SDS buffer (150 mM Tris-HCl, pH 7.5, 6 M urea, 2% SDS, and 5% β-mercaptoethanol) (2 ml/mg). Protein samples under native conditions were prepared by thawing the powder in sodium phosphate buffer (5 mM; pH 7.5; 2 ml/mg). The elapsed time between native extraction and addition of denaturing/loading buffer to stop the incubation is indicated below. Samples were boiled for 10 min, and cell debris was removed by centrifugation at 18,000 × g at 4°C for 10 min. The supernatants were resolved on SDS-PAGE (12% acrylamide), electroblotted to a nitrocellulose membrane, and subjected to Western blot analysis. Specific viral proteins were detected using anti-HCPro, anti-P1b, or anti-CP rabbit serum as the primary antibody and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) as the secondary reagent. GFP was detected using a mixture of two anti-GFP monoclonal antibodies (Roche Applied Science) as primary detection reagents and HRP-conjugated sheep anti-mouse IgG (GE Healthcare Life Science) as the secondary antibody. The immunostained proteins were visualized by enhanced chemiluminescence detection with a LifeABlot kit (Euroclone). Ponceau red staining of the membranes was used to check the global protein content of the samples.
Electron microscopy.
The presence of virion-like particles in extracts prepared from infected leaves was determined by immunosorbent electron microscopy. Hence, samples were incubated for 5 min with collodion-coated carbon-stabilized copper grids, which had previously been treated with anti-CP serum. Negative staining was done with 2% uranyl acetate, and the grids were then observed using a JEM 1011 electron microscope (Jeol). Images were taken with an ES1000W Erlangshen charge-coupled-device (CCD) camera (Gatan), and ImageJ software (48) was used to measure the lengths of viral particles.
RESULTS
The supply of PPV HCPro in trans complements the defect in infectivity of PPV-P1P1b viral progeny.
PPV chimeras expressing unrelated RSSs instead of HCPro had been previously built and characterized (38). Preliminary observations using these modified viruses showed that although they could cause efficient infection in Nicotiana plants, the viral progeny seemed to have reduced infectivity in plant-to-plant manual passages (data not shown). Because of these observations, we hypothesized that the weak infectivity of these chimeric viruses was due to the lack of an HCPro-supplied function. To test this idea, systemically infected leaves of N. benthamiana plants that had been inoculated biolistically with PPV-P1P1b, a chimera in which the HCPro coding sequence had been replaced by that of CVYV P1b (Fig. 1A) (38), were infiltrated at 21 days after inoculation with diverse A. tumefaciens strains carrying different binary plasmids, thus supplying specific proteins to the systemically infected tissue in trans (for simplicity, we refer to each A. tumefaciens strain by the plasmid it carries). Subsequently, these agroinfiltrated leaf patches were used to prepare crude leaf extracts under native conditions for the inoculation of N. clevelandii leaves, and the infectivity of the chimera was estimated from the total number of GFP foci that it produced (Fig. 1B). Using this approach, we found that the progeny of PPV-P1P1b that had been supplemented with either empty vector or a plasmid expressing the strong RSS P19 from TBSV produced just a few GFP foci (Fig. 1C). In contrast, when the chimera had been supplemented in trans with an HCPro-expressing plasmid (either p35S-P1HCPPV or p35S-HCPPV), the quantity of GFP infection foci in the inoculated leaves was extraordinarily boosted (Fig. 1C). The number of foci produced by viral progeny derived from 8 independent HCPro-supplied N. benthamiana plants showed reproducible and clear enhancement of PPV-P1P1b infectivity (data not shown).
FIG 1.
PPV HCPro expressed in trans complements the infectivity deficiency of PPV-P1P1b progeny. (A) Schematic representation of the viral cDNA of the PPV-P1P1b clone. (B) Experimental approach followed for testing the effect of HCPro added in trans on PPV-P1P1b infectivity. (1) N. benthamiana plants were infected with PPV-P1P1b by biolistic inoculation; (2) 2 weeks later, systemically infected leaves were infiltrated with cultures of A. tumefaciens strains carrying different binary vectors (the expression of the indicated proteins supplemented in trans by agroinfiltration was confirmed by carrying out an RNA-silencing suppression test of these constructs in parallel); (3) agroinfiltrated patches were harvested 5 days after infiltration; and (4) extracts prepared from these patches were used for manual inoculation of N. clevelandii plants. (C) Representative photographs taken under an epifluorescence microscope of N. clevelandii leaves from two plants inoculated with extracts from N. benthamiana plants infected with PPV-P1P1b and supplemented by agroinfiltration with the indicated proteins. —, empty vector. (D) Western blot analysis of the native extracts from the infected and agroinoculated N. benthamiana plants (two plants per sample) used to inoculate the N. clevelandii plants shown in panel C. A polyclonal serum specific for PPV CP and a monoclonal antibody raised against GFP were utilized for viral accumulation assessment. Protein sizes indicated on the right (in kilodaltons) were estimated from the positions of prestained molecular mass markers (New England BioLabs) run in the same gel.
Moreover, an immunoblot analysis of samples from the first inoculated plants used as the inoculum in the infectivity assay showed that levels of viral CP in those biolistically inoculated N. benthamiana plants that had been supplemented with HCPro were much higher than those in plants agroinfiltrated with either the empty vector or p35s-p19 (Fig. 1D). In addition, the appearance of a smaller band of around 15 kDa that reacted with the anti-CP serum correlated with lower levels of intact CP, whereas the accumulation of GFP was not affected by the expression of different proteins in trans (Fig. 1D). Altogether, these results indicate that CP is specifically degraded in the absence of HCPro and that this degradation has a negative impact on the infectivity of the viral progeny.
PPV HCPro specifically affects the stability of CP.
To confirm the previous observation that PPV CP becomes susceptible to proteolytic degradation in the absence of PPV HCPro, an approach similar to that described above (Fig. 1B) was used in the next experiment. To gain insight into the specificity of the observed effect, a plasmid expressing the HCPro protein from another potyvirus, Tobacco etch virus (TEV), was also used to agroinfiltrate N. benthamiana leaves that had been systemically infected with the PPV-P1P1b chimera. Protein extracts from the infiltrated patches were prepared under both denaturing and native conditions, and CP accumulation was assessed by Western blot analysis. In agreement with the previous result, CP was clearly detected in protein extracts from infected tissues supplemented with two different plasmids expressing PPV HCPro (Fig. 2A, Phosphate). In contrast, levels of viral CP extracted under native conditions were barely detected, not only in samples from the infected plants that had been supplemented in trans with either the empty vector or TBSV P19, but also in those from plants expressing the homologous RSS HCPro from TEV, thus revealing the high specificity of the protective action of HCPro on its cognate CP. High intensity of CP-specific degradation bands of around 15 kDa correlated again with low accumulation of the intact protein, whereas the accumulation of GFP was not affected by the different agroinfiltration treatments (Fig. 2A, Phosphate).
FIG 2.
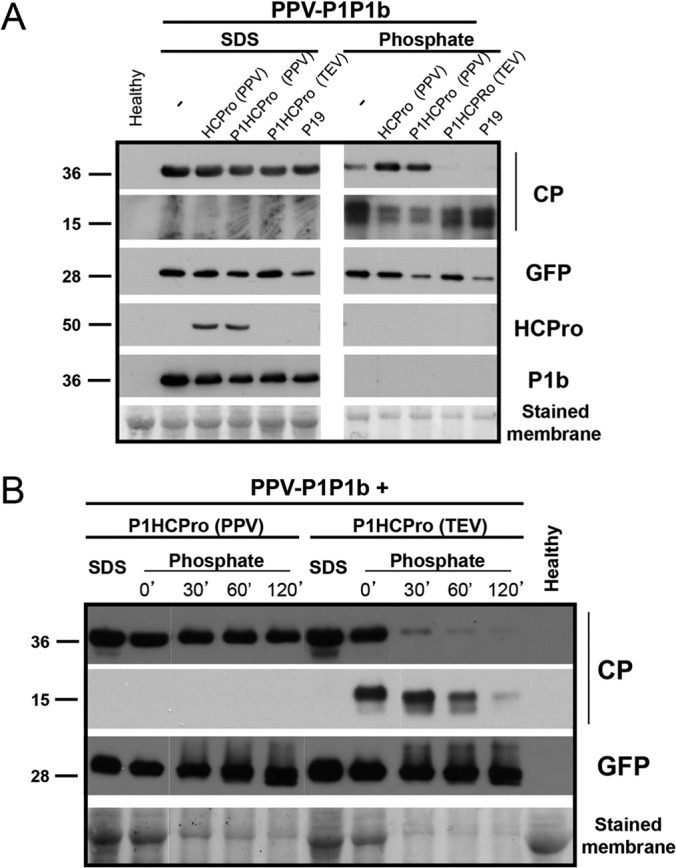
Stabilization of PPV CP by HCPro is species specific. (A) Analysis of CP stability in native and denatured extracts of N. benthamiana plants infected with PPV-P1P1b and supplemented by agroinfiltration with the indicated proteins. (B) Time course of CP degradation in extracts of N. benthamiana plants infected with PPV-P1P1b and supplemented with either PPV or TEV P1HCPro factors. Extracts prepared in SDS buffer (SDS) and the time elapsed after the extraction of total proteins with phosphate buffer (0 min [0′], 30 min, 60 min, and 120 min) are indicated. Protein accumulation and stability were assessed by Western blot analysis with polyclonal sera specific for PPV CP, PPV HCPro, CVYV P1b, and a monoclonal antibody raised against GFP. Protein sizes indicated on the left (in kilodaltons) were estimated from the positions of prestained molecular mass markers (New England BioLabs) run in the same gels. Membranes stained with Ponceau red showing the RubisCO large subunit were included as loading controls. The expression of the indicated proteins supplemented in trans by agroinfiltration was confirmed by carrying out an RNA-silencing suppression test of the constructs in parallel.
Surprisingly, the above-described differences in the accumulation of CP were not observed in protein extracts prepared under denaturing conditions (Fig. 2A, SDS), supporting the idea that the specific instability of PPV CP in the absence of PPV HCPro occurs ex vivo. It is worth mentioning that PPV HCPro and CVYV P1b could not be efficiently extracted from leaf tissues by using phosphate buffer (native conditions) due to the absence of a reducing agent in the buffer (Fig. 2 and data not shown).
To gain new insight into this particular HCPro-mediated effect, a time course analysis of CP accumulation levels after native extraction was also carried out (Fig. 2B). The test showed that not only under denaturing conditions (Fig. 2B, SDS), but also a short time after native extraction (Fig. 2B, Phosphate, lanes 0′), levels of PPV CP were similar independently of either PPV or TEV HCPro expression in trans. However, after incubation for 30 min at room temperature, PPV CP produced in the presence of TEV HCPro dropped to hardly detectable levels, whereas the levels of CP produced in the presence of PPV HCPro remained unaltered for at least 2 h (Fig. 2B). In agreement with this observation, degradation bands of CP appeared only in the samples supplemented with TEV HCPro (Fig. 2B). On the other hand, virus-derived GFP was highly stable regardless of whether it was produced in the presence of PPV or TEV HCPro (Fig. 2B). These results confirm that the effect of HCPro on CP stability is species specific and can be detected exclusively after protein extraction under nondenaturing conditions.
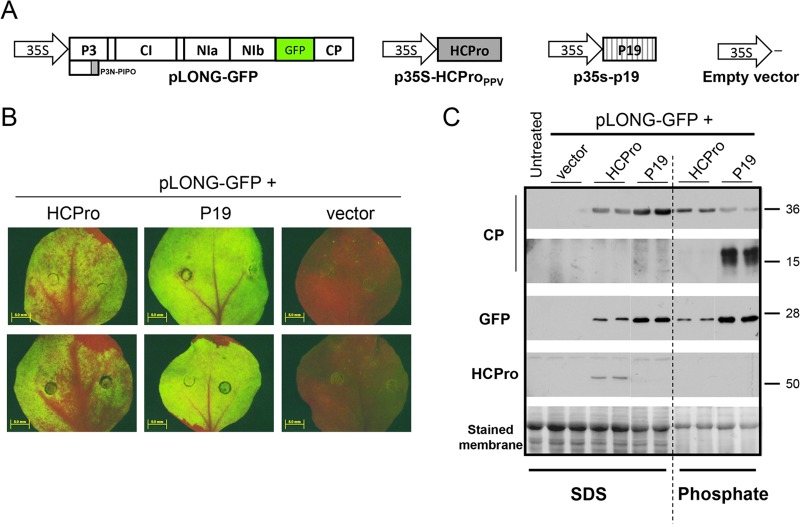
PPV HCPro also has an effect on CP stability in a transient-expression assay.
To assess the effect of HCPro on CP stability in a different context than a viral infection, the proteins were expressed by agroinfiltration in N. benthamiana: HCPro from p35S-HCPPV, and CP as part of a truncated PPV polyprotein starting in P3 from pLONG-GFP. Either p35s-p19 or the empty vector was agroinfiltrated with pLONG-GFP as a control (Fig. 3A). Since the long mRNA produced from pLONG-GFP is a highly sensitive substrate for the RNA-silencing machinery, GFP fluorescence can be detected when only this construct is coinfiltrated with either p35S-HCPPV or p35s-p19, as they express PPV HCPro and TBSV P19 RSSs, respectively (Fig. 3B). Accumulation levels of GFP and PPV CP, extracted under either denaturing or native conditions from the agroinfiltrated leaf patches, were tested by Western blot analysis. The absence of GFP (and CP) in patches infiltrated with pLONG-GFP plus the empty vector confirmed fluorescence observations under UV light (Fig. 3C). Similar to the previously observed results, whereas the accumulation of CP under denaturing conditions in patches expressing pLONG-GFP plus p35s-p19 was elevated and was even higher than in those patches expressing pLONG-GFP plus p35S-HCPPV, CP levels sharply declined when the extraction was carried out under native conditions. Additionally, CP-specific degradation bands were also detected in these samples as an indication of CP instability (Fig. 3C). In contrast, the amounts of CP extracted from patches expressing pLONG-GFP plus p35S-HCPPV under native and denaturing conditions were similar, and no CP degradation bands were detected (Fig. 3C). As expected, the accumulation of GFP was not affected by the extraction conditions. Together, these results indicate that the specific HCPro-mediated effect on CP stability initially observed in viral infections is mimicked in an agroinfiltration test.
FIG 3.
Specific degradation of PPV CP in the absence of PPV HCPro in a transient-expression assay. (A) Schematic representation of constructs used for the transient-expression test. (B) GFP fluorescence of leaves agroinfiltrated with the indicated constructs. Pictures of two independent N. benthamiana plants taken under an epifluorescence microscope at 6 days postinfiltration are shown. (C) Western blot analysis of N. benthamiana leaf patches that were agroinfiltrated with pLONG-GFP plus constructs expressing PPV HCPro, TBSV P19, or empty vector. Total protein extracts prepared in either denaturing (SDS) or nondenaturing (phosphate) buffer are indicated. Polyclonal sera specific for PPV CP and PPV HCPro and a monoclonal antibody raised against GFP were utilized for protein accumulation assessments. Protein sizes indicated on the right (in kilodaltons) were estimated from the positions of prestained molecular mass markers (New England BioLabs) run in the same gel. The membrane stained with Ponceau red showing the RubisCO large subunit was included as a loading control.
Positively charged amino acids located in the central region of HCPro, but no others required for aphid transmission, RNA-silencing suppression,or self-cleavage, are involved in CP stabilization.
HCPro is a multifunctional protein involved in polyprotein processing, aphid transmission, and suppression of antiviral RNA silencing (33). A site-directed mutagenic approach was carried out in order to find a putative link between these roles and the effect of HCPro on CP stability observed here. The amino acids that were changed to alter specific functions of the protein (protease activity, aphid transmission, and RNA-silencing suppression) are listed in Table 1.
TABLE 1.
Point mutations introduced in PPV HCPro
| Activity/domain | Mutation | Expected effect | Tested inb: | Reference |
|---|---|---|---|---|
| Transmission (HCPro connects virions and aphids) | K52E, K52Aa | Disrupt interaction between HCPro and aphids | TVMV, TEV | 9, 10, 13 |
| P310A, T311A | Disrupt interaction between HCPro and CP | ZYMV | 11 | |
| RNA-silencing suppression (HCPro sequesters virus-derived small RNAs) | L134H | Abolish RNA-silencing suppression | PPV | 49 |
| RK182,4AA | ZYMV, TEV, TuMV, PVY, ClYVV | 27, 34, 72 | ||
| RK246,7AA | TEV | 34, 72 | ||
| Positively charged motif | RRH234,5,6AAAa | |||
| Cysteine protease (HCPro cleaves its C terminus) | C344A, H417A | Abolish self-cleavage | TEV | 34, 72 |
Not previously tested, but included in this study.
TVMV, Tobacco vein mottling virus; ZYMV, Zucchini yellowing mosaic virus; TuMV, Turnip mosaic virus; PVY, Potato virus Y; ClYVV, Clover yellowing vein virus.
The capacities of the HCPro variants to allow the expression of pLONG-GFP were analyzed in a coagroinfiltration test (data not shown). GFP fluorescence derived from pLONG-GFP, revealing the presence of an active RSS, was observed in plants expressing wild-type HCPro; the mutants likely affected in aphid transmission; and the protein with the mutation C344A, which affects its protease active center. The previously reported PPV HCPro L134H null anti-silencing mutant (49); 2 out of 3 mutants affected in positively charged amino acids of the central region of the protein (RK182,4AA and RK246,7AA); and the H427A mutant, expected to disturb protease activity, were unable to prevent the silencing of pLONG-GFP (data not shown).
The abilities of the different HCPro mutants to prevent the degradation of CP were also assessed using the coagroinfiltration assay. In addition to pLONG-GFP and the indicated plasmid expressing either the wild-type or a mutant version of HCPro, p35s-p19 was also included in order to ensure both high and homogeneous levels of expression regardless of the RNA-silencing suppression activity of the HCPro variant (Fig. 4A). Protein extracts were prepared from agroinfiltrated leaf patches under denaturing or native conditions, and CP accumulation levels were assessed by Western blot analysis at the extraction time or after in vitro incubation (Fig. 4B). As expected, the expression of pLONG-GFP with p35s-p19 and the empty vector produced high levels of CP, as detected after denaturing extraction (lanes S) and immediately after native extraction (lanes 0′), but CP accumulation dropped until it was hardly detected after 45 min of incubation at room temperature (Fig. 4B). In contrast, when pLONG-GFP and p35s-p19 were coinfiltrated with pMDC32-P1HCPro-derived plasmids expressing either wild-type HCPro or some of the HCPro mutants, CP was still detected at a high level even after incubation in the nondenaturing buffer (Fig. 4B). Mutations that did not disrupt the capacity of HCPro to prevent CP degradation included K52A, K52E, P310A, and T311E, which are predicted to affect aphid transmission; C344A, which is expected to disturb protease activity; and L134H, which was shown to abolish RNA-silencing suppression.
FIG 4.
Effects of diverse mutations in PPV HCPro on its capacity to prevent CP degradation. (A) Schematic representation of virus-derived constructs used in the assay. (B) Western blot analysis of N. benthamiana leaf patches that were agroinfiltrated with p35s-p19 and pLONG-GFP plus constructs expressing the indicated P1-HCPro variants or empty vector. Extracts prepared in SDS buffer (lanes S) and the time in minutes elapsed after the extraction of total proteins with phosphate buffer (0 min and 45 min) are indicated. Polyclonal sera specific for PPV CP and PPV HCPro and a monoclonal antibody raised against GFP were utilized for protein accumulation assessments. Protein sizes indicated on the left (in kilodaltons) were estimated from the positions of prestained molecular mass markers (New England BioLabs) run in the same gels. Membranes stained with Ponceau red showing the RubisCO large subunit were included as loading controls.
A different result was observed when HCPro variants carrying mutations in positively charged amino acids located in the central region of the protein (RRH234,5,6AAA, RK246,7AA, or RK182,4AA) were expressed. Hence, in these cases, levels of CP were nearly undetectable by Western blot analysis after 45 min of incubation (Fig. 4B), suggesting that these HCPro amino acids participate in preventing CP degradation. Interestingly, whereas mutations in amino acids RRH234,5,6 did not disturb the capacity of HCPro to suppress the reporter silencing (data not shown), they strongly diminished the ability of HCPro to prevent CP degradation (Fig. 4B), indicating a very specific effect of these amino acids in this novel activity of HCPro. It is worth mentioning that infiltration of the mixture that carried pMDC32-P1HCPro H427A, which was built to produce an HCPro protein with a defect in its protease activity, gave rise to a very low accumulation of HCPro, which was therefore unable both to suppress RNA silencing (data not shown) and to avoid CP degradation (Fig. 4B). In this case, the introduced mutation likely disrupts HCPro folding or stability.
HCPro is an enhancer of viral particle yield.
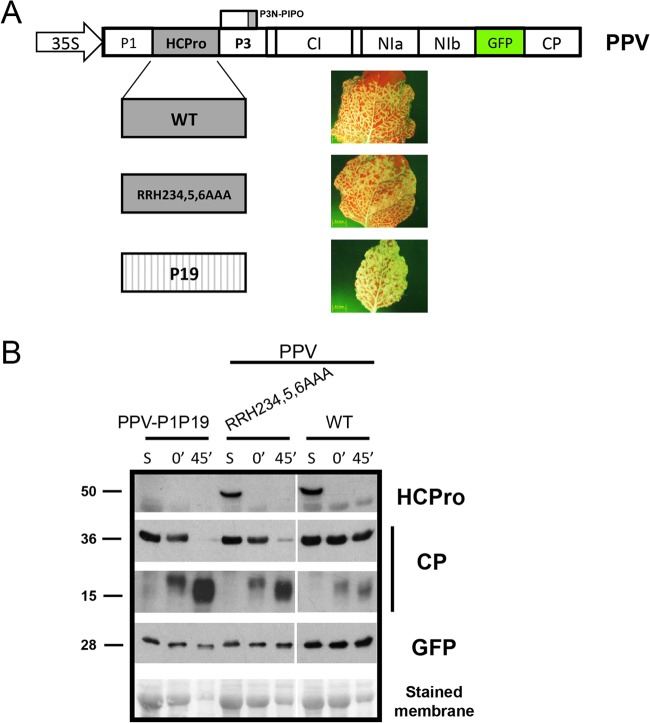
Just one of the HCPro mutated variants (RRH234,5,6AAA) affected the ability of the protein to prevent CP degradation without altering its RNA-silencing suppression activity. In order to test the biological significance of CP instability in the context of the viral infection, the HCProRRH234,5,6AAA coding sequence was engineered into the PPV genome. Additionally, the previously described PPV-P1P19 chimera (39), in which the HCPro coding sequence had been replaced by that of the TBSV P19 RSS plus a corresponding PPV NIa cleavage site to allow P19 release, was also used (Fig. 5A). In this case, the PPV-P1P19 variant was expected to produce an unstable CP, similar to that of the PPV-P1P1b chimera, due to the absence of HCPro. All these constructs were used to infect N. benthamiana plants. Interestingly, there were no outstanding visual differences between the phenotypes caused by these viruses; hence, systemic symptoms and GFP fluorescence in both inoculated and systemically infected leaves appeared at similar times after inoculation, displaying comparable intensities (data not shown and Fig. 5A). Both the accumulation and the stability of viral CP were determined by Western blot assay, as described above, from infected noninoculated upper leaves. Whereas extractions with a denaturing buffer showed that these viruses accumulated similar levels of PPV CP (Fig. 5B, lane S), the native extraction verified that CP produced in plants infected with HCProRRH234,5,6AAA- and P19-expressing PPV variants were highly susceptible to proteolytic degradation after in vitro incubation (Fig. 5B, lanes 45′). As expected, the appearance of specific CP degradation bands was much more prominent in these samples (Fig. 5B). It is worth mentioning that a slightly lower accumulation of the HCProRRH234,5,6AAA mutant virus CP than of the wild-type PPV CP was observed by extraction under denaturing conditions in additional independent experiments (data not shown).
FIG 5.
Degradation of CP derived from a PPV that carries the RRH234,5,6AAA mutation in HCPro. (A) Schematic representation of full-length cDNA of PPV and its derivatives, in which wild-type (WT) HCPro was replaced by either the HCPro RRH234,5,6AAA mutant or TBSV P19 RSS. Representative photographs taken at 14 days postinoculation under an epifluorescence microscope of equivalent N. benthamiana leaves systemically infected with the corresponding viruses are also shown. (B) Western blot analysis of noninoculated upper leaves of N. benthamiana plants infected with the indicated viruses. Extracts prepared in SDS buffer (lanes S) and the time in minutes elapsed after the extraction of total proteins with phosphate buffer (0 min and 45 min) are indicated. Polyclonal sera specific for PPV CP and PPV HCPro and a monoclonal antibody raised against GFP were utilized for protein accumulation assessments. Protein sizes indicated on the left (in kilodaltons) were estimated from the positions of prestained molecular mass markers (New England BioLabs) run in the same gel. The membrane stained with Ponceau red showing the RubisCO large subunit was included as a loading control.
Since virion purification by traditional methods failed several times to obtain viral particles from PPV-HCProRRH234,5,6AAA (data not shown), an alternative approach was used in order to determine whether the CP degradation was due to defects in the assembly of viral particles. Thus, crude extracts from plants infected with both the wild-type and the mutated viruses were directly observed by electron microscopy. The time elapsed between protein extraction and the incubation of extracts with grids was strictly limited to minimize CP degradation. Moreover, the electron microscopy analyses were done on 1/10-diluted and nondiluted extracts of plants infected with wild-type and HCProRRH234,5,6AAA mutant viruses, respectively, to ensure that the amount of CP was much higher in the sample corresponding to the mutated variant (Fig. 6A). Typical flexuous rod-like virions were easily detected in wild-type-PPV-containing extracts (Fig. 6B, left). In contrast, viral particles were barely detected in the sap derived from those plants infected with HCProRRH234,5,6AAA-expressing PPV, even when the extract contained around 10 times more CP that the wild-type-containing protein extract (Fig. 6A). Most importantly, the mutant-derived rods were much shorter than their wild-type-derived counterparts (Fig. 6B, right). Fifty particles of each virus type were selected at random and measured in order to estimate their size distribution. Hence, whereas the majority of wild-type-derived virions (56%) were 850 to 950 nm in length, almost all particles derived from the mutant virus (94%) were less than 450 nm long (Fig. 6C). It is important to mention that a few full-length viral particles were found in the mutant-derived sample, which suggests that the ability of PPV-HCProRRH234,5,6AAA to form virions was not completely abolished. This result was additionally supported by the detection of a small amount of CP sedimented at the position of entire virions after centrifugation in a sucrose density gradient (data not shown).
FIG 6.
The PPV mutant that expresses HCProRRH234,5,6AAA has a defect in the yield of viral particles. (A) Western blot analysis of extracts from noninoculated upper leaves of N. benthamiana infected with the indicated viruses. Dilutions of the extract of leaves infected with wild-type PPV are shown. Polyclonal serum specific for PPV CP was utilized for protein accumulation assessment. The asterisk indicates the dilution of wild-type PPV used for electron microscopy observation. (B) Electron microscopy images of PPV particles trapped with anti-CP serum from extracts of N. benthamiana plants infected with the indicated viruses. (C) Size distribution of 50 randomly selected particles from the indicated viruses. Note that the particle lengths are only an approximate estimation based on the calibration of the electron microscope.
Altogether, these data show that the incapacity of HCProRRH234,5,6AAA to prevent CP degradation correlates directly with a defect in the accumulation of full-length virions, suggesting that HCPro plays a key role as an enhancer in the proper yield of viral particles.
DISCUSSION
In this paper, we report a previously unknown function of HCPro: enhancement of viral particle yield. This capacity seems to be independent of its previously described activities, as shown by a mutagenic approach (Fig. 4). Hence, the well-described interaction between the PTK motif of HCPro and CP, which is involved in aphid transmission, does not appear to be necessary for the novel function (see below). In addition, the proteinase activity is not required, since while the HCPro protease domain works only in cis (18), the ability of HCPro to stabilize CP can be supplied in trans (Fig. 1 to 4), and it is not affected by a point mutation in the crucial cysteine of the proteinase catalytic triad (C344A) (Fig. 4B). Finally, it does not seem to be linked to RNA-silencing suppression, either, as a mutant in which this activity is abolished (L134H) protects CP from degradation (Fig. 4B), whereas another mutant that efficiently suppresses RNA silencing (RRH234,5,6AAA) is unable to stabilize CP (Fig. 4 to 6). Interestingly, this novel function is highly specific (Fig. 3), which suggests that HCPro acts only upon its cognate CP. Species-specific interactions of the potyviral CP with other viral factors, such as the cylindrical inclusions, have been previously reported (50). Given that coinfections by several viruses are not unusual in nature (51), the prevention of functional heterologous interactions might be an evolutionary strategy to avoid promoting the propagation of competitor viruses.
Our results show that an HCPro function is required to keep CP stable in native leaf extracts (Fig. 2). Since HCPro was not detectable in these extracts, it is unlikely that the protein physically protects CP (e.g., by inhibiting a protease that degrades CP). Hence, we speculate that the ex vivo degradation of the CP that is produced in the absence of HCPro occurs because most CP subunits are not assembled in viral particles or are improperly assembled, and under these conditions, the CP is extremely sensitive to proteases. Furthermore, the detected CP products of around 15 kDa would be the result of that proteolytic process. The appearance of high-molecular-weight forms of CP correlates with a defect in the novel function of HCPro (Fig. 6A), which is in good agreement with published data showing that potyviral CP is ubiquitinated and degraded under certain conditions (52). CP degradation was not observed when extracts were prepared under denaturing conditions (Fig. 2 to 5), supporting the assumption that this viral protein is specifically and actively degraded by the action of host factors once cells are disrupted under native conditions.
What is the molecular mechanism by which HCPro enhances the yield of intact viral particles? Although the answer to this question is unknown at this time, we envisage two not mutually exclusive scenarios of action for the protein: (i) HCPro is involved in initial steps of the assembly of CP subunits in virus particles; (ii) HCPro either directly or indirectly stabilizes virions once they are formed. In any case, the absence of this HCPro function would affect the yield of full-length viral particles, as observed in this study. Intriguingly, despite the enormous amounts of data that have been published about potyviruses, very little is known about their genome encapsidation process (4, 53, 54). Besides the essential role of CP in viral particle assembly, there is no information about other viral or host factors required for this event. In addition to the expected detection of VPg (55), HCPro was found associated with viral particles (14) in a protrusion located at one of the ends (15). Further experiments also detected the presence of CI in these protruding tips (56). Although the presence of HCPro in virions might be attributed to its role in viral transmission, as previously suggested (15), it could also have an alternative function in the infection process. Moreover, given the capacities of HCPro and CI to interact in vivo (57, 58), coordinated recruiting of both viral factors toward the virion tip seems conceivable.
Potyviral CP is subjected to posttranslational modifications that likely influence its functions during viral infection. On one hand, CP is phosphorylated by the protein kinase CK2, a modification that seems to regulate the RNA binding function of CP and therefore likely controls the formation and/or stability of viral ribonucleoproteins (59–61). On the other hand, O-GlcNAcylation modifications carried out by the host protein SECRET AGENT were also found in PPV CP (59, 62). Interestingly, a mutagenic approach showed that O-GlcNAcylation might have a role in virion assembly and/or stability (63). Hence, the involvement of HCPro in the regulation of virion assembly by modulation of posttranslational modifications of CP is an attractive possibility that is worthy of exploration in the future.
Even though in vivo interaction between HCPro and CP from PPV has not been observed by bimolecular fluorescence complementation and yeast two-hybrid assays (reference 58 and our unpublished results), there are numerous reports showing that potyviral HCPro and CP from several viruses are able to interact in diverse systems (8, 11, 57, 64–67). Thus, it is reasonable to think that direct interaction between HCPro and CP is required for the enhancement of viral particle yield. However, such interaction should be a type of contact different from that involved in aphid transmission, since mutations in either P310 or T311 from HCPro, which have been previously associated with CP-HCPro interaction and aphid transmission (9–13), seem to have no or very little effect on CP stability (Fig. 4). In agreement with this assumption, binding between HCPro and CP from non-aphid-transmissible viruses was previously observed (14, 67), which suggests that the interaction between these two viral factors might have a biological role besides aphid-mediated plant-to-plant transmission, such as the one proposed here.
The need for an additional viral factor(s) for the correct yield of full-length viral particles has also been described for closteroviruses. The filamentous virions from members of this family possess a long body formed by CP and a short tail formed mainly by the minor capsid protein (CPm), as well as another diverged copy of CP and a virus-derived HSP70 chaperone (68), resulting in a peculiar “rattlesnake-like” structure. In the absence of any of the above-mentioned proteins, shorter particles are mostly produced (69, 70). Whereas in the case of closteroviruses, it is well established that the viral particle, apart from genome protection, works as a movement-specialized device (68), this does not seem to be the case for PPV, as variants yielding short viral particles efficiently move through the whole plant (Fig. 5). Our studies then open another question: why is this novel function of HCPro required for potyviruses?
The steps of the viral infection process must be perfectly coordinated in space and time to avoid overlapping of potentially exclusionary events. Hence, strict control over CP expression is required to separate early viral translation and replication from late genome encapsidation. In many plant viruses, this is achieved by the late expression of CP from independently controlled subgenomic RNAs. Unlike that of other RNA viruses, the genome expression of potyviruses is mainly produced through the proteolytic processing of a single polyprotein, producing equimolar amounts of most of the viral factors at the same time. Because of this, these viruses must use a different strategy to avoid having premature encapsidation interfere with the required translation and replication of the genomic RNA. What is known at present about potyviral RNA trafficking within an infected cell, including the control of viral RNA encapsidation, has been recently reviewed (71). To explain how the appropriate synchronization can be achieved, a proposed model suggests that the interaction of CP with the host factors HSP70 and CPIP controls the cellular levels of CP and, consequently, genome encapsidation (52). Based on the data presented here, we hypothesize that the spatiotemporal availability of HCPro in a given infected cell could also have a role in the intricate coordination of translation, replication, and encapsidation. Hence, whereas low cellular HCPro levels at early stages of infection would negatively affect the yield of virions, higher accumulation of the protein at later times would enhance the encapsidation process at the expense of translation/replication. On the other hand, if an enhancer of viral particle yield were a strict requirement for potyviruses in general, it would be expected that potyviruses encoding no HCPro (e.g., some ipomoviruses) manage to supply that enhancer in an alternative way. Further research to elucidate the method by which HCPro (or another viral protein) affects CP stability and virion yield might help to reveal the unknown mechanism used by potyviruses to coordinate such diverse processes as uncoating, translation, replication, escape from antiviral defenses, encapsidation, and movement together in a single cell.
ACKNOWLEDGMENTS
This work was supported by grants BIO2010-18541 and BIO2013-49053-R from the Spanish MICINN.
We thank Elvira Domínguez and Beatriz García for technical assistance, Cristina Patiño for support in the electron microscopy experiments, Juan José López-Moya for critical readings of the manuscript, and Jake B. B. Harris for English proofreading.
Footnotes
Published ahead of print 18 June 2014
REFERENCES
- 1.Adams MJ, Zerbini FM, French R, Rabenstein F, Stenger DC, Valkonen JPT. 2012. Family Potyviridae, p 1069–1090 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Elsevier, Oxford, United Kingdom [Google Scholar]
- 2.López-Moya JJ, Valli A, García JA. 15 March 2009. Potyviridae. Encyclopedia of Life Sciences (ELS). John Wiley & Sons, Ltd.,Chichester, United Kingdom [Google Scholar]
- 3.Shukla DD, Ward CW. 1989. Structure of potyvirus coat proteins and its application in the taxonomy of the potyvirus group. Adv. Virus Res. 36:273–314. 10.1016/S0065-3527(08)60588-6 [DOI] [PubMed] [Google Scholar]
- 4.Kendall A, McDonald M, Bian W, Bowles T, Baumgarten SC, Shi J, Stewart PL, Bullitt E, Gore D, Irving TC, Havens WM, Ghabrial SA, Wall JS, Stubbs G. 2008. Structure of flexible filamentous plant viruses. J. Virol. 82:9546–9554. 10.1128/JVI.00895-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Moya JJ, García JA. 2008. Potyviruses, p 313–322 In Mahy BWJ, Van RegenmortelMHV. (ed), Encyclopedia of virology, 3rd ed. Elsevier, Oxford, United Kingdom [Google Scholar]
- 6.Ivanov KI, Eskelin K, Lõhmus A, Mäkinen K. 10 April 2014. Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 10.1099/vir.0.064220-0 [DOI] [PubMed] [Google Scholar]
- 7.Govier DA, Kassanis B. 1974. A virus-induced component of plant sap needed when aphids acquire potato virus Y from purified preparations. Virology 61:420–426. 10.1016/0042-6822(74)90278-5 [DOI] [PubMed] [Google Scholar]
- 8.Blanc S, López-Moya JJ, Wang RY, García-Lampasona S, Thornbury DW, Pirone TP. 1997. A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology 231:141–147. 10.1006/viro.1997.8521 [DOI] [PubMed] [Google Scholar]
- 9.Atreya CD, Pirone TP. 1993. Mutational analysis of the helper component-proteinase gene of a potyvirus: effects of amino acid substitutions, deletions, and gene replacement on virulence and aphid transmissibility. Proc. Natl. Acad. Sci. U. S. A. 90:11919–11923. 10.1073/pnas.90.24.11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanc S, Ammar ED, Garcia-Lampasona S, Dolja VV, Llave C, Baker J, Pirone TP. 1998. Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J. Gen. Virol. 79:3119–3122 [DOI] [PubMed] [Google Scholar]
- 11.Peng YH, Kadoury D, Gal-On A, Wang Y, Raccah B. 1998. Mutations in the HC-Pro gene of zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J. Gen. Virol. 79:897–904 [DOI] [PubMed] [Google Scholar]
- 12.Dombrovsky A, Gollop N, Chen SB, Chejanovsky N, Raccah B. 2007. In vitro association between the helper component-proteinase of zucchini yellow mosaic virus and cuticle proteins of Myzus persicae. J. Gen. Virol. 88:1602–1610. 10.1099/vir.0.82769-0 [DOI] [PubMed] [Google Scholar]
- 13.Fernández-Calvino L, Goytia E, López-Abella D, Giner A, Urizarna M, Vilaplana L, López-Moya JJ. 2010. The helper-component protease transmission factor of tobacco etch potyvirus binds specifically to an aphid ribosomal protein homologous to the laminin receptor precursor. J. Gen. Virol. 91:2862–2873. 10.1099/vir.0.022335-0 [DOI] [PubMed] [Google Scholar]
- 14.Manoussopoulos IN, Maiss E, Tsagris M. 2000. Native electrophoresis and Western blot analysis (NEWeB): a method for characterization of different forms of potyvirus particles and similar nucleoprotein complexes in extracts of infected plant tissues. J. Gen. Virol. 81:2295–2298 [DOI] [PubMed] [Google Scholar]
- 15.Torrance L, Andreev IA, Gabrenaite-Verhovskaya R, Cowan G, Mäkinen K, Taliansky ME. 2006. An unusual structure at one end of potato potyvirus particles. J. Mol. Biol. 357:1–8. 10.1016/j.jmb.2005.12.021 [DOI] [PubMed] [Google Scholar]
- 16.Carrington JC, Cary SM, Parks TD, Dougherty WG. 1989. A second proteinase encoded by a plant potyvirus genome. EMBO J. 8:365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh CS, Carrington JC. 1989. Identification of essential residues in potyvirus proteinase HC-Pro by site-directed mutagenesis. Virology 173:692–699. 10.1016/0042-6822(89)90582-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo B, Lin J, Ye K. 2011. Structure of the autocatalytic cysteine protease domain of potyvirus helper-component proteinase. J. Biol. Chem. 286:21937–21943. 10.1074/jbc.M111.230706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasschau KD, Carrington JC. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470. 10.1016/S0092-8674(00)81614-1 [DOI] [PubMed] [Google Scholar]
- 20.Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739–6746. 10.1093/emboj/17.22.6739 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. U. S. A. 95:13079–13084. 10.1073/pnas.95.22.13079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding SW. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10:632–644. 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- 23.Aliyari R, Ding SW. 2009. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 227:176–188. 10.1111/j.1600-065X.2008.00722.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding SW, Lu R. 2011. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr. Opin. Virol. 1:533–544. 10.1016/j.coviro.2011.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Incarbone M, Dunoyer P. 2013. RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18:382–392. 10.1016/j.tplants.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 26.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25:2768–2780. 10.1038/sj.emboj.7601164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiboleth YM, Haronsky E, Leibman D, Arazi T, Wassenegger M, Whitham SA, Gaba V, Gal-On A. 2007. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 81:13135–13148. 10.1128/JVI.01031-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valli A, Oliveros JC, Molnar A, Baulcombe D, García JA. 2011. The specific binding to 21-nt double-stranded RNAs is crucial for the anti-silencing activity of Cucumber vein yellowing virus P1b and perturbs endogenous small RNA populations. RNA 17:1148–1158. 10.1261/rna.2510611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamous RM, Boonrod K, Fuellgrabe MW, Ali-Shtayeh MS, Krczal G, Wassenegger M. 2011. The helper component-proteinase of the Zucchini yellow mosaic virus inhibits the Hua Enhancer 1 methyltransferase activity in vitro. J. Gen. Virol. 92:2222–2226. 10.1099/vir.0.031534-0 [DOI] [PubMed] [Google Scholar]
- 30.Anandalakshmi R, Marathe R, Ge X, Herr JM, Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance VB. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142–144. 10.1126/science.290.5489.142 [DOI] [PubMed] [Google Scholar]
- 31.Endres MW, Gregory BD, Gao Z, Foreman AW, Mlotshwa S, Ge X, Pruss GJ, Ecker JR, Bowman LH, Vance V. 2010. Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog. 6:e1000729. 10.1371/journal.ppat.1000729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maia IG, Haenni A-L, Bernardi F. 1996. Potyviral HC-Pro: a multifunctional protein. J. Gen. Virol. 77:1335–1341. 10.1099/0022-1317-77-7-1335 [DOI] [PubMed] [Google Scholar]
- 33.Syller J. 2005. The roles and mechanisms of helper component proteins encoded by potyviruses and caulimoviruses. Physiol. Mol. Plant Pathol. 67:119–130. 10.1016/j.pmpp.2005.12.005 [DOI] [Google Scholar]
- 34.Kasschau KD, Carrington JC. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71–81. 10.1006/viro.2001.0901 [DOI] [PubMed] [Google Scholar]
- 35.Ala-Poikela M, Goytia E, Haikonen T, Rajamäki ML, Valkonen JP. 2011. Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85:6784–6794. 10.1128/JVI.00485-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haikonen T, Rajamäki ML, Valkonen JP. 2013. Interaction of the microtubule-associated host protein HIP2 with viral helper component proteinase is important in infection with Potato virus A. Mol. Plant Microbe Interact. 26:734–744. 10.1094/MPMI-01-13-0023-R [DOI] [PubMed] [Google Scholar]
- 37.Haikonen T, Rajamäki ML, Tian YP, Valkonen JP. 2013. Mutation of a short variable region in HCpro protein of Potato virus A affects interactions with a microtubule-associated protein and induces necrotic responses in tobacco. Mol. Plant Microbe Interact. 26:721–733. 10.1094/MPMI-01-13-0024-R [DOI] [PubMed] [Google Scholar]
- 38.Carbonell A, Dujovny G, García JA, Valli A. 2012. The Cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in Plum pox virus infection in a host-specific manner. Mol. Plant Microbe Interact. 25:151–164. 10.1094/MPMI-08-11-0216 [DOI] [PubMed] [Google Scholar]
- 39.Maliogka VI, Calvo M, Carbonell A, García JA, Valli A. 2012. Heterologous RNA-silencing suppressors from both plant- and animal-infecting viruses support plum pox virus infection. J. Gen. Virol. 93:1601–1611. 10.1099/vir.0.042168-0 [DOI] [PubMed] [Google Scholar]
- 40.Herlitze S, Koenen M. 1990. A general and rapid mutagenesis method using polymerase chain reaction. Gene 91:143–147. 10.1016/0378-1119(90)90177-S [DOI] [PubMed] [Google Scholar]
- 41.Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133:462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valli A, Martín-Hernández AM, López-Moya JJ, García JA. 2006. RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus (CVYV), a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80:10055–10063. 10.1128/JVI.00985-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucini C. 2004. Expresión de proteínas heterólogas en plantas por medio del virus de la sharka (PPV). Universidad Politécnica de Madrid, Madrid, Spain [Google Scholar]
- 44.Delgadillo MO, Sáenz P, Salvador B, García JA, Simón-Mateo C. 2004. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 85:993–999. 10.1099/vir.0.19735-0 [DOI] [PubMed] [Google Scholar]
- 45.Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949–956. 10.1046/j.1365-313X.2003.01676.x [DOI] [PubMed] [Google Scholar]
- 46.Fernández-Fernández MR, Mouriño M, Rivera J, Rodríguez F, Plana-Durán J, García JA. 2001. Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus-based vector. Virology 280:283–291. 10.1006/viro.2000.0762 [DOI] [PubMed] [Google Scholar]
- 47.Salvador B, Delgadillo MO, Saénz P, García JA, Simón-Mateo C. 2008. Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol. Plant Microbe Interact. 21:20–29. 10.1094/MPMI-21-1-0020 [DOI] [PubMed] [Google Scholar]
- 48.Abramoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 49.González-Jara P, Atencio FA, Martínez-García B, Barajas D, Tenllado F, Díaz-Ruíz JR. 2005. A single amino acid mutation in the plum pox virus helper component-proteinase gene abolishes both synergistic and RNA silencing suppression activities. Phytopathology 95:894–901. 10.1094/PHYTO-95-0894 [DOI] [PubMed] [Google Scholar]
- 50.Langenberg WG. 1993. Structural proteins of three viruses in the potyviridae adhere only to their homologous cylindrical inclusions in mixed infections. J. Struct. Biol. 110:188–195. 10.1006/jsbi.1993.1021 [DOI] [PubMed] [Google Scholar]
- 51.Syller J. 2012. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13:204–216. 10.1111/j.1364-3703.2011.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hafren A, Hofius D, Ronnholm G, Sonnewald U, Mäkinen K. 2010. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22:523–535. 10.1105/tpc.109.072413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu XJ, Shaw JG. 1998. Evidence that assembly of a potyvirus begins near the 5′ terminus of the viral RNA. J. Gen. Virol. 79:1525–1529 [DOI] [PubMed] [Google Scholar]
- 54.Anindya R, Savithri HS. 2003. Surface-exposed amino- and carboxy-terminal residues are crucial for the initiation of assembly in Pepper vein banding virus: a flexuous rod-shaped virus. Virology 316:325–336. 10.1016/S0042-6822(03)00593-2 [DOI] [PubMed] [Google Scholar]
- 55.Puustinen P, Rajamaki ML, Ivanov KI, Mäkinen K. 2002. Detection of the potyviral genome-linked protein VPg in virions and its phosphorylation by host kinases. J. Virol. 76:12703–12711. 10.1128/JVI.76.24.12703-12711.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabrenaite-Verkhovskaya R, Andreev IA, Kalinina NO, Torrance L, Taliansky ME, Mäkinen K. 2008. Cylindrical inclusion protein of potato virus A is associated with a subpopulation of particles isolated from infected plants. J. Gen. Virol. 89:829–838. 10.1099/vir.0.83406-0 [DOI] [PubMed] [Google Scholar]
- 57.Guo DY, Rajamäki ML, Saarma M, Valkonen JPT. 2001. Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 82:935–939 [DOI] [PubMed] [Google Scholar]
- 58.Zilian E, Maiss E. 2011. Detection of plum pox potyviral protein-protein interactions in planta using an optimized mRFP-based bimolecular fluorescence complementation system. J. Gen. Virol. 92:2711–2723. 10.1099/vir.0.033811-0 [DOI] [PubMed] [Google Scholar]
- 59.Fernández-Fernández MR, Camafeita E, Bonay P, Méndez E, Albar JP, García JA. 2002. The capsid protein of a plant single-stranded RNA virus is modified by O-linked N-acetylglucosamine. J. Biol. Chem. 277:135–140. 10.1074/jbc.M106883200 [DOI] [PubMed] [Google Scholar]
- 60.Ivanov KI, Puustinen P, Merits A, Saarma M, Mäkinen K. 2001. Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 276:13530–13540. 10.1074/jbc.M009551200 [DOI] [PubMed] [Google Scholar]
- 61.Ivanov KI, Puustinen P, Gabrenaite R, Vihinen H, Rönnstrand L, Valmu L, Kalkkinen N, Mäkinen K. 2003. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell 15:2124–2139. 10.1105/tpc.012567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen D, Juárez S, Hartweck L, Alamillo JM, Simón-Mateo C, Pérez JJ, Fernández-Fernández MR, Olszewski NE, García JA. 2005. Identification of secret agent as the O-GlcNAc transferase that participates in Plum pox virus infection. J. Virol. 79:9381–9387. 10.1128/JVI.79.15.9381-9387.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez JJ, Udeshi ND, Shabanowitz J, Ciordia S, Juárez S, Scott CL, Olszewski NE, Hunt DF, García JA. 2013. O-GlcNAc modification of the coat protein of the potyvirus Plum pox virus enhances viral infection. Virology 442:122–131. 10.1016/j.virol.2013.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang SH, Lim WS, Kim KH. 2004. A protein interaction map of soybean mosaic virus strain G7H based on the yeast two-hybrid system. Mol. Cells 18:122–126 [PubMed] [Google Scholar]
- 65.Seo JK, Kang SH, Seo BY, Jung JK, Kim KH. 2010. Mutational analysis of interaction between coat protein and helper component-proteinase of Soybean mosaic virus involved in aphid transmission. Mol. Plant Pathol. 11:265–276. 10.1111/j.1364-3703.2009.00603.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin L, Shi Y, Luo Z, Lu Y, Zheng H, Yan F, Chen J, Adams MJ, Wu Y. 2009. Protein-protein interactions in two potyviruses using the yeast two-hybrid system. Virus Res. 142:36–40. 10.1016/j.virusres.2009.01.006 [DOI] [PubMed] [Google Scholar]
- 67.Roudet-Tavert G, German-Retana S, Delaunay T, Delécolle B, Candresse T, Le Gall O. 2002. Interaction between potyvirus helper component-proteinase and capsid protein in infected plants. J. Gen. Virol. 83:1765–1770 [DOI] [PubMed] [Google Scholar]
- 68.Dolja VV, Kreuze JF, Valkonen JPT. 2006. Comparative and functional genomics of the closteroviruses. Virus Res. 117:38–51. 10.1016/j.virusres.2006.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satyanarayana T, Gowda S, Ayllon MA, Dawson WO. 2004. Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5 ′ region. Proc. Natl. Acad. Sci. U. S. A. 101:799–804. 10.1073/pnas.0307747100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tatineni S, Gowda S, Dawson WO. 2010. Heterologous minor coat proteins of Citrus tristeza virus strains affect encapsidation, but the coexpression of HSP70h and p61 restores encapsidation to wild-type levels. Virology 402:262–270. 10.1016/j.virol.2010.03.042 [DOI] [PubMed] [Google Scholar]
- 71.Mäkinen K, Hafren A. 2014. Intracellular coordination of potyviral RNA functions in infection. Front. Plant Sci. 5:110. 10.3389/fpls.2014.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kasschau KD, Cronin S, Carrington JC. 1997. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228:251–262. 10.1006/viro.1996.8368 [DOI] [PubMed] [Google Scholar]