Abstract
Sulfolobus mutants resistant to archaeal lytic virus Sulfolobus islandicus rod-shaped virus 2 (SIRV2) were isolated, and mutations were identified in two gene clusters, cluster sso3138 to sso3141 and cluster sso2386 and sso2387, encoding cell surface and type IV secretion proteins, respectively. The involvement of the mutations in the resistance was confirmed by genetic complementation. Blocking of virus entry into the mutants was demonstrated by the lack of early gene transcription, strongly supporting the idea of a role of the proteins in SIRV2 entry.
TEXT
To date, relatively few archaeal viruses have been characterized, and most of those that have been characterized infect acidothermophilic members of the order Sulfolobales. Despite their limited number of around 50 species, they exhibit considerably greater morphological diversity than the more extensively characterized bacteriophages, about 95% of which show head-tail morphologies. Archaeal viruses, in contrast, exhibit fusiform shapes, often with one or two tails, bottle shapes, bearded-globular forms, and a wide variety of rod-like and filamentous morphotypes which often carry small terminal appendages (1–3). This morphological diversity suggests that the archaeal viruses may employ a variety of mechanisms to enter their hosts, but current insights into entry mechanisms are limited to an OppA transporter protein, Sso1273, possibly providing a receptor site for the Acidianus two-tailed virus (ATV) in Sulfolobus solfataricus P2 (4). And very recently, microscopic studies suggested that Sulfolobus islandicus rod-shaped virus 2 (SIRV2) enters the host cell by attaching and moving through a pilus-like filament; however, the nature of the structure and the identity of the involved proteins remain elusive (5).
Sulfolobus solfataricus P2 is an acidothermophilic crenarchaeon that can host a wide range of archaeal viruses, many of which are propagated stably (1, 3, 6). Moreover, few of the viruses appear to induce cell lysis, possibly reflecting a need to minimize contact with the harsh hot acidic environment. However, recent studies have identified a few viruses that can enter a lytic phase, including the Sulfolobus turreted icosahedral virus (STIV), the two-tailed fusiform (ATV), and, more recently, the rudivirus SIRV2 (7–9).
SIRV2 is classified in the family Rudiviridae together with other well-characterized viruses, including SIRV1 (10, 11), ARV1 (12) and SRV1, (13), all of which are rod shaped and lack an envelope, and their genomes consist of linear double-stranded DNA with covalently closed ends (10, 14, 15). In a recent microarray analysis of S. solfataricus infected with SIRV2, we demonstrated that the viral genes were activated at different times and that mainly stress-response host genes and those implicated in vesicle formation were downregulated (16). The results also illustrated that SIRV2 infection at a multiplicity of infection (MOI) of 30 resulted in growth inhibition of S. solfataricus 5E6 (16). In the present experiment, the culture was infected at a lower MOI (<1) which also led to a growth retardation, but the infected culture could enter the exponential-growth phase at 80 h postinfection (p.i.) (Fig. 1A). The surviving cells (named 5E6R) appeared to be resistant to SIRV2 because, in contrast to the sensitive 5E6 strain, no growth retardation was observed when 5E6R was diluted and infected with SIRV2 at the same MOI (Fig. 1).
FIG 1.
(A) Growth retardation of S. solfataricus 5E6 upon SIRV2 infection. (B) Resistance of S. solfataricus 5E6R to SIRV2. OD600, optical density at 600 nm.
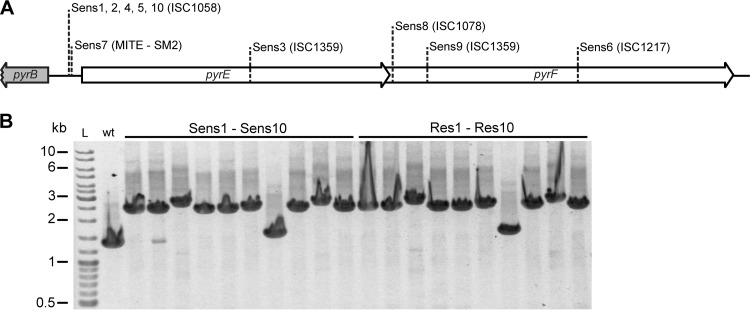
In order to manipulate the SIRV2-sensitive S. solfataricus 5E6 strain genetically (17), 10 pyrEF mutants, labeled Sens1 to Sens10, were isolated from Gelrite plates containing 5-fluoroorotic acid (5′-FOA). Their mutation sites in the pyrEF gene region were identified by a combination of PCR amplification, restriction digest analysis, and sequencing (17). All the mutations were shown to result from transposon insertions, either IS elements or miniature inverted terminal repeat elements (MITEs), and the insertions occurred in the coding sequences or within the single promoter (Fig. 2A). These results are consistent with the previous reports demonstrating high transposition activity in S. solfataricus and its contribution to chromosomal plasticity (18–20). Following the procedure described above, SIRV2-resistant cultures were generated for each of the pyrEF mutants. Single colonies were then produced from the cultures by streaking onto Gelrite plates to yield the purified resistant strains Res1 to Res10. The stability of the transposon insertions in the pyrEF genes was tested for each of the 10 pyrEF mutants (Sens1 to Sens10) and their corresponding SIRV2-resistant colonies (Res1 to Res10) by growing them in rich media containing uracil (17) for 3 days without transfer, prior to total DNA extraction and PCR amplification of the pyrEF regions. Each transposon insertion appeared to be stable, because no wild-type PCR bands were observed, except a weak wild-type band produced in Sens2, consistent with the undetectable reversion rates for Sulfolobus transposons recorded earlier (19, 20). Since Res2 did not generate the wild-type band, the extra PCR product in Sens2 was probably due to a minor contamination of the colony by wild-type cells (Fig. 2B).
FIG 2.
Analysis of the pyrEF mutants derived from S. solfataricus 5E6. (A) Types and insertion sites of transposons inserted in the pyrEF gene region of different pyrEF mutants. (B) PCR amplification of the pyrEF region from Sens1 to Sens10 and from Res1 to Res10. wt, wild type.
Sens1, Sens3, Sens7, and Sens8 were selected for a transformation test because they carried different transposons located at different insertion sites (Fig. 2A). Shuttle vector pEXA was used for transformation (21), and water was used in the negative control. While Sens7 and Sens8 appeared unstable after electroporation, Sens1 and Sens3 yielded transformants without colony formation in the negative control. Thus, we focused on Sens1 and its resistant mutants for further studies of SIRV2 susceptibility.
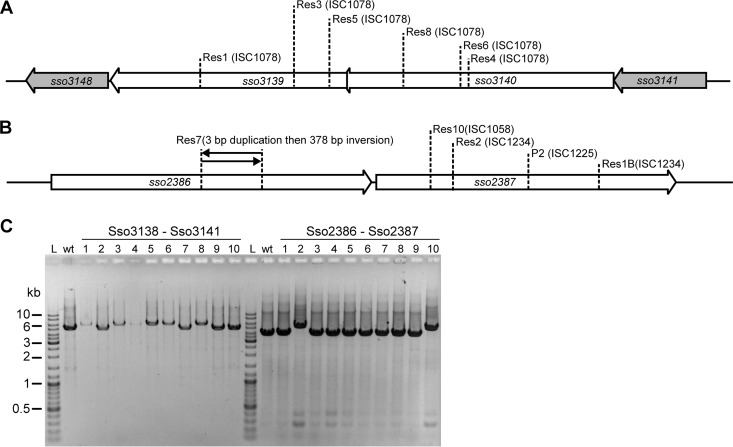
The SIRV2-resistant cells were enriched directly from the SIRV2-sensitive culture; therefore, the only selective pressure appeared to occur either upon SIRV2 infection or during virus-induced cell lysis. Moreover, since the active clustered regularly interspaced short palindromic repeat (CRISPR) loci A, B, C, and D were all lost from the 5E6 host strain (16), the residual CRISPR loci E and F, which lack the spacer acquisition cas genes, were unlikely to be responsible for the resistance (21, 22). Therefore, we inferred that resistance arose as a result of mutated host genes that are important for the SIRV2 life cycle. To identify such mutations, the genomes of strains Sens1 and Res1 were resequenced by the use of a Hiseq 2000 sequencer, yielding about 200-fold coverage. The sequencing reads of both strains were aligned with the genome sequence of S. solfataricus P2 (23) using the R2R program (24) to identify mismatches as well as insertions and deletions. Mutations to the P2 genome in the resequenced genomes of Sens1 and Res1 were then compared manually. Only one mutation was detected and constituted a single insertion of ISC1078 into Res1 but not Sens1. The insertion was localized in sso3139, a gene encoding a conserved hypothetical protein lying within an operon (Fig. 3A).
FIG 3.
Different mutations in the SIRV2-resistant strains and their stability. (A) Transposon insertions in sso3139 and sso3140. (B) Mutations in sso2386 and sso2387. (C) PCR amplification of the mutation region from different resistant strains.
Next we tested whether other resistant strains also carried mutations in sso3139 or in other genes of the same operon by employing a primer pair (5′-GCTACGCTTCTAACAAACCTAATCTG and 5′-CGAAACTTGCGAAACAACTACCT) designed to amplify the whole operon region. After PCR amplification, restriction digestion, and sequencing, another 5 strains were shown to contain mutations at different locations within sso3139 or the adjacent sso3140 (Fig. 3A). Interestingly, all the 6 mutations were produced by ISC1078 insertion (Fig. 3A) and appeared to be stably maintained (Fig. 3C).
To identify possible mutations in the other 4 resistant strains, genome resequencing followed by PCR analyses of relevant genes was performed (using primers 5′-GAGTCTGGGGAAAATCGGTAAAGTT and 5′-TGGCATTGTAACCCTAATTGCTTCT). These revealed IS element insertions in sso2387 of Res2 and Res10 (Fig. 3B). Sso2387 in Sens2 and Sens10 contains 577 amino acids (aa) but only 283 aa in the sequenced S. solfataricus P2 genome (23). An analysis of the sequences around sso2387 in S. solfataricus P2 revealed that it is a partial gene that resulted from an ISC1225 insertion (Fig. 3B), which could explain the resistance of the wild-type P2 strain to SIRV2 (13 and this work). Interestingly, an inversion in sso2386 was detected in Res7 whereas no mutations were identified in Res9 that could be linked to SIRV2 resistance (Fig. 3B).
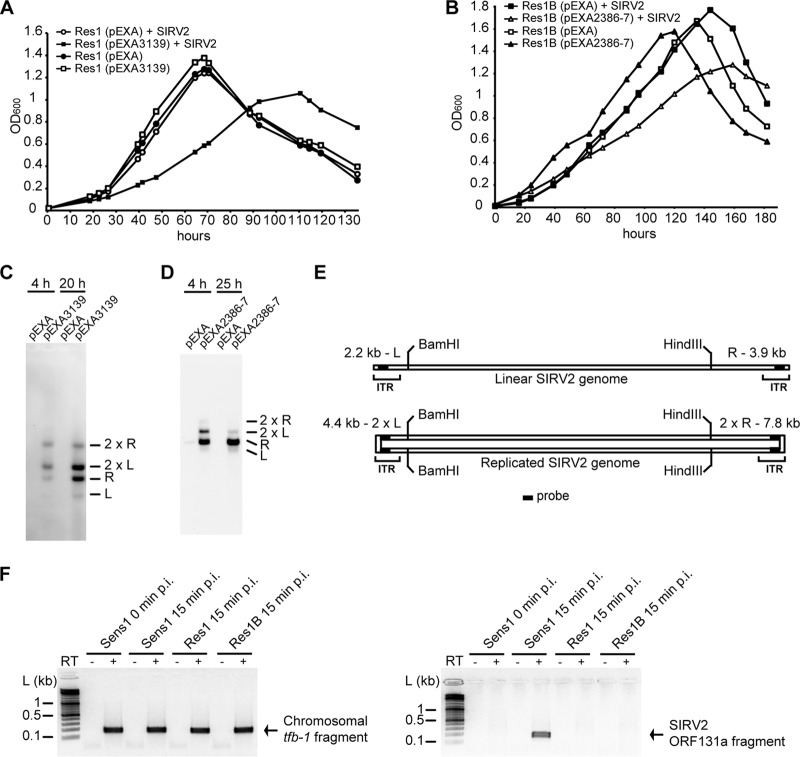
The frequently observed mutations in cluster sso3139 and sso3140 and cluster sso2386 and sso2387 in the resistant strains strongly suggest that the two gene clusters are important for the SIRV2 life cycle. To confirm the implication of the mutations in the gained resistance, genetic complementation was performed for the mutated genes. As described above, Sens1 appeared stable during genetic manipulation, and we thus selected Res1 for complementation of sso3139 mutation. For complementation of mutations in the other gene cluster, Res1B, carrying an ISC1234 insertion in sso2387 (Fig. 3B), was isolated from SIRV2-infected Sens1. Res1 cells were transformed with vector pEXA2 containing sso3139, and Res1B cells were transformed with vector pEXA2 containing sso2386 and sso2387. After SIRV2 was added into the cultures, growth retardation occurred in the complemented cells, while the noncomplemented culture, transformed with the empty vector, showed a growth rate similar to that of the uninfected culture (Fig. 4A and B). Further, Southern hybridization (17) using a probe derived from the SIRV2 inverted terminal repeats (ITR) detected signals only from the complemented cells (Fig. 4C and D) and the multiple hybridized bands were consistent with ongoing replication (Fig. 4E) (10, 25). The absence of SIRV2 signal in the resistant strains indicates a defect in the virus life cycle.
FIG 4.
Cluster sso3138 to sso3141 and cluster sso2386 and sso2387 are involved in SIRV2 entry. (A) Growth retardation of sso3139-complemented Res1 upon SIRV2 infection. Res1 (pEXA), Res1 transformed with expression vector pEXA; Res1 (pEXA3139), Res1 transformed with expression vector pEXA containing sso3139. (B) Growth retardation of sso2386-and-sso2387-complemented Res1B upon SIRV2 infection. Res1B (pEXA), Res1B transformed with expression vector pEXA; Res1B (pEXA2386–2387), Res1B transformed with expression vector pEXA containing sso2386 and sso2387. (C and D) Visualization of SIRV2 DNA replication in Res1 (C) and Res1B (D) transformants infected with SIRV2. Plasmid constructs contained in the transformants are labeled on top of each lane, and the sampling time p.i. is indicated as hours. L and R designate the left and right terminal fragments, respectively, after a double digestion with BamHI and HindIII (see panel E). (E) Schematic presentation of SIRV2 genomic map and the formation of terminal duplex replicative intermediates (2L and 2R), as described previously (12). The locations of the probe (filled rectangle) in the termini are indicated. ITR, inverted terminal repeat. (F) RT-PCR amplification of sso0446 (tfb-1) (left panel) and SIRV2 ORF131a transcript fragments (right panel). “+” and “−” indicate the presence and absence of reverse transcriptase (RT), respectively.
To gain insights into the functions of the two gene clusters, the protein sequences of the genes were firstly analyzed by the use of program TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) for the possible presence of transmembrane helices. Sso3138, Sso3139, and Sso3140 were predicted to be primarily located extracellularly (see Fig. S1 in the supplemental material), correlating with a previous prediction of the presence of class III signal peptides at their N termini (26). Among these, Sso3140 was confirmed to be a membrane-associated protein in a proteomics study (27). In contrast to the other 3 proteins, Sso3141 was predicted to contain two transmembrane helices, one at the N terminus and the other at the C terminus, while the sequence between them was presumed to be located intracellularly. Therefore, it appears that the proteins encoded in the operon form a membrane-associated cell surface structure and may function as a receptor for SIRV2. Moreover, it was demonstrated recently that Sso2386 carries multiple transmembrane helices and that Sso2387 constitutes an ATPase associated with a type IV secretion system, and they were designated AapF and AapE, respectively (28). Further, homologs of both are essential for the formation of the adhesive type IV pilus of S. acidocaldarius (28). The association with the cell membrane of proteins encoded by both gene clusters strongly indicates their involvement in the entry process of SIRV2.
The failure of viral entry into Res1 and Res1B cells was further confirmed by reverse transcription-PCR analysis of one of the early genes, ORF131a (17). RNA extracted from cells taken at 15 min p.i. was DNase I treated and reverse transcribed (SuperScript II reverse transcriptase; Invitrogen). PCR performed on the cDNAs detected ORF131a only from Sens1 cells, while the positive-control sso0446 (tfb-1) gene was detected in all the 3 strains (Fig. 4F). This strongly supports the conclusion that the proteins encoded by the two gene clusters are involved in SIRV2 entry. A likely scenario is that gene cluster sso3138 to sso3141 encodes a surface receptor for SIRV2 and that gene cluster sso2386 and sso2387 is involved in the secretion of the receptor components.
Except in Escherichia coli, very few virus receptors are known in the domains of Bacteria and Archaea (29). The primary receptors for E. coli filamentous phages are pili which retract toward the cell surface, bringing the phages to the secondary receptor located in the periplasm (30). Linear archaeal viruses, including rudiviruses, have been observed to attach to pili (5, 31, 32). Future work is needed to determine the association of the two identified gene clusters with the structure of pili. To our knowledge, this is the first work providing genetic and biochemical evidence for a possible receptor system in archaeal virus entry.
Supplementary Material
ACKNOWLEDGMENTS
We thank Roger A. Garrett for critically reading the manuscript.
This work is supported by a European Union FP7 grant (265933).
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01495-14.
REFERENCES
- 1.Prangishvili D, Forterre P, Garrett RA. 2006. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 4:837–848. 10.1038/nrmicro1527 [DOI] [PubMed] [Google Scholar]
- 2.Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 35:1035–1054. 10.1111/j.1574-6976.2011.00280.x [DOI] [PubMed] [Google Scholar]
- 3.Peng X, Garrett RA, She Q. 2012. Archaeal viruses–novel, diverse and enigmatic. Sci. China Life Sci. 55:422–433. 10.1007/s11427-012-4325-8 [DOI] [PubMed] [Google Scholar]
- 4.Erdmann S, Scheele U, Garrett RA. 2011. AAA ATPase p529 of Acidianus two-tailed virus ATV and host receptor recognition. Virology 421:61–66. 10.1016/j.virol.2011.08.029 [DOI] [PubMed] [Google Scholar]
- 5.Quemin ER, Lucas S, Daum B, Quax TE, Kuhlbrandt W, Forterre P, Albers SV, Prangishvili D, Krupovic M. 2013. First insights into the entry process of hyperthermophilic archaeal viruses. J. Virol. 87:13379–13385. 10.1128/JVI.02742-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zillig W, Arnold HP, Holz I, Prangishvili D, Schweier A, Stedman K, She Q, Phan H, Garrett R, Kristjansson JK. 1998. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 2:131–140. 10.1007/s007920050052 [DOI] [PubMed] [Google Scholar]
- 7.Fu CY, Johnson JE. 2012. Structure and cell biology of archaeal virus STIV. Curr. Opin. Virol. 2:122–127. 10.1016/j.coviro.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Häring M, Vestergaard G, Rachel R, Chen L, Garrett RA, Prangishvili D. 2005. Virology: independent virus development outside a host. Nature 436:1101–1102. 10.1038/4361101a [DOI] [PubMed] [Google Scholar]
- 9.Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, Forterre P, Tenaillon O, Bernander R, Prangishvili D. 2009. A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. U. S. A. 106:11306–11311. 10.1073/pnas.0901238106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng X, Blum H, She Q, Mallok S, Brugger K, Garrett RA, Zillig W, Prangishvili D. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226–234. 10.1006/viro.2001.1190 [DOI] [PubMed] [Google Scholar]
- 11.Prangishvili D, Arnold HP, Gotz D, Ziese U, Holz I, Kristjansson JK, Zillig W. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vestergaard G, Haring M, Peng X, Rachel R, Garrett RA, Prangishvili D. 2005. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336:83–92. 10.1016/j.virol.2005.02.025 [DOI] [PubMed] [Google Scholar]
- 13.Vestergaard G, Shah SA, Bize A, Reitberger W, Reuter M, Phan H, Briegel A, Rachel R, Garrett RA, Prangishvili D. 2008. Stygiolobus rod-shaped virus and the interplay of crenarchaeal rudiviruses with the CRISPR antiviral system. J. Bacteriol. 190:6837–6845. 10.1128/JB.00795-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blum H, Zillig W, Mallok S, Domdey H, Prangishvili D. 2001. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281:6–9. 10.1006/viro.2000.0776 [DOI] [PubMed] [Google Scholar]
- 15.Prangishvili D, Koonin EV, Krupovic M. 2013. Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem. Soc. Trans. 41:443–450. 10.1042/BST20120313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okutan E, Deng L, Mirlashari S, Uldahl K, Halim M, Liu C, Garrett RA, She Q, Peng X. 2013. Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells. RNA Biol. 10:875–885. 10.4161/rna.24537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng L, Zhu H, Chen Z, Liang YX, She Q. 2009. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13:735–746. 10.1007/s00792-009-0254-2 [DOI] [PubMed] [Google Scholar]
- 18.Martusewitsch E, Sensen CW, Schleper C. 2000. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J. Bacteriol. 182:2574–2581. 10.1128/JB.182.9.2574-2581.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redder P, Garrett RA. 2006. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 188:4198–4206. 10.1128/JB.00061-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blount ZD, Grogan DW. 2005. New insertion sequences of Sulfolobus: functional properties and implications for genome evolution in hyperthermophilic archaea. Mol. Microbiol. 55:312–325. 10.1111/j.1365-2958.2004.04391.x [DOI] [PubMed] [Google Scholar]
- 21.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 79:35–49. 10.1111/j.1365-2958.2010.07452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erdmann S, Garrett RA. 2012. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 85:1044–1056. 10.1111/j.1365-2958.2012.08171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PM, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U. S. A. 98:7835–7840. 10.1073/pnas.141222098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skovgaard O, Bak M, Lobner-Olesen A, Tommerup N. 2011. Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 21:1388–1393. 10.1101/gr.117416.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oke M, Kerou M, Liu H, Peng X, Garrett RA, Prangishvili D, Naismith JH, White MF. 2011. A dimeric Rep protein initiates replication of a linear archaeal virus genome: implications for the Rep mechanism and viral replication. J. Virol. 85:925–931. 10.1128/JVI.01467-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szabó Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschröder M. 2007. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189:772–778. 10.1128/JB.01547-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pham TK, Sierocinski P, van der Oost J, Wright PC. 2010. Quantitative proteomic analysis of Sulfolobus solfataricus membrane proteins. J. Proteome Res. 9:1165–1172. 10.1021/pr9007688 [DOI] [PubMed] [Google Scholar]
- 28.Henche AL, Ghosh A, Yu X, Jeske T, Egelman E, Albers SV. 2012. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ. Microbiol. 14:3188–3202. 10.1111/j.1462-2920.2012.02898.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 30.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. 2011. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 13:51–76 http://www.horizonpress.com/cimb/v/v13/51.pdf [PubMed] [Google Scholar]
- 31.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendörfer M, Kristjansson JK. 1993. Screening for Sulfolobales, their plasmids and their viruses in Icelandic Solfataras. Syst. Appl. Microbiol. 16:609–628. 10.1016/S0723-2020(11)80333-4 [DOI] [Google Scholar]
- 32.Bettstetter M, Peng X, Garrett RA, Prangishvili D. 2003. AFV1, a novel virus infecting hyperthermophilic archaea of the genus acidianus. Virology 315:68–79. 10.1016/S0042-6822(03)00481-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.