ABSTRACT
We recently demonstrated that the virion host shutoff (vhs) protein, an mRNA-specific endonuclease, is required for efficient herpes simplex virus 1 (HSV-1) replication and translation of viral true-late mRNAs, but not other viral and cellular mRNAs, in many cell types (B. Dauber, J. Pelletier, and J. R. Smiley, J. Virol. 85:5363–5373, 2011, http://dx.doi.org/10.1128/JVI.00115-11). Here, we evaluated whether the structure of true-late mRNAs or the timing of their transcription is responsible for the poor translation efficiency in the absence of vhs. To test whether the highly structured 5′ untranslated region (5′UTR) of the true-late gC mRNA is the primary obstacle for translation initiation, we replaced it with the less structured 5′UTR of the γ-actin mRNA. However, this mutation did not restore translation in the context of a vhs-deficient virus. We then examined whether the timing of transcription affects translation efficiency at late times. To this end, we engineered a vhs-deficient virus mutant that transcribes the true-late gene US11 with immediate-early kinetics (IEUS11-ΔSma). Interestingly, IEUS11-ΔSma showed increased translational activity on the US11 transcript at late times postinfection, and US11 protein levels were restored to wild-type levels. These results suggest that mRNAs can maintain translational activity throughout the late stage of infection if they are present before translation factors and/or ribosomes become limiting. Taken together, these results provide evidence that in the absence of the mRNA-destabilizing function of vhs, accumulation of viral mRNAs overwhelms the capacity of the host translational machinery, leading to functional exclusion of the last mRNAs that are made during infection.
IMPORTANCE The process of mRNA translation accounts for a significant portion of a cell's energy consumption. To ensure efficient use of cellular resources, transcription, translation, and mRNA decay are tightly linked and highly regulated. However, during virus infection, the overall amount of mRNA may increase drastically, possibly overloading the capacity of the translation apparatus. Our results suggest that the HSV-1 vhs protein, an mRNA-specific endoribonuclease, prevents mRNA overload during infection, thereby allowing translation of late viral mRNAs. The requirement for vhs varies between cell types. Further studies of the basis for this difference likely will offer insights into how cells regulate overall mRNA levels and access to the translational apparatus.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a large enveloped nuclear DNA virus. Like all viruses, HSV-1 critically depends on the translation machinery of the host cell to synthesize its more than 70 encoded proteins. Therefore, it is not surprising that HSV-1 employs several strategies to stimulate viral protein synthesis, including directly enhancing translation initiation, preventing global shutdown of translation by host stress kinases, and reducing competition from cellular mRNAs for the translation machinery (reviewed in references 1 and 2).
Translation of most transcripts is cap dependent and initiated by the cap-binding complex eIF4F (composed of eIF4E, eIF4G, eIF4A, eIF4B, and eIF4H) binding to the mRNA 5′ m7G-cap via its eIF4E subunit. Bound eIF4G recruits the 43S preinitiation complex (containing the 40S ribosomal subunit and additional eIFs), which scans the mRNA 5′ untranslated region (UTR) to locate the start codon. The 60S ribosomal subunit then joins to form a translation-competent 80S ribosome. Scanning is facilitated by the RNA helicase activity of eIF4A, which unwinds secondary structures in the 5′UTR in collaboration with the helicase cofactors eIF4B and eIF4H. Actively translated mRNAs are thought to form a closed-loop structure bridged by interactions between eIF4G and PABP bound to the 3′ poly(A) tail of the mRNA (reviewed in reference 3).
Three HSV-1 proteins have been shown to enhance assembly of the cap-binding complex. The viral Ser/Thr kinase US3 activates mTORC1 to hyperphosphorylate eIF4E-BP, which then releases eIF4E (4). The cap-binding capacity of eIF4E can be increased further through p38-mnk signaling triggered by ICP27 (5, 6). Also, the viral protein ICP6 enhances the interaction of eIF4E and eIF4G (7).
Cellular stress kinases, such as the double-stranded RNA (dsRNA)-dependent kinase PKR or the endoplasmic reticulum (ER) stress kinase PERK, block translation initiation by phosphorylating translation initiation factor eIF2α in an attempt to curb viral gene expression (reviewed in reference 8). However, the HSV-1 proteins ICP34.5, US11, and gB counteract this global shutdown of protein synthesis by directing the cellular phosphatase PP1a to dephosphorylate eIF2α, inhibiting the activation of PKR and preventing the activation of PERK, respectively (9–11).
Lastly, HSV-1 significantly alters the cellular translation profile by crippling cellular gene expression and flooding the cell with viral transcripts. The virion host shutoff (vhs) protein, encoded by the gene UL41, plays a crucial role in this viral take-over. vhs is an RNase that enters the cell as part of the tegument and destabilizes most cellular and viral mRNAs (reviewed in reference 12). It targets actively translating mRNAs through its interaction with the helicase isoforms eIF4AI and eIF4AII, as well as the helicase cofactors eIF4H and eIF4B (13–17). The cellular RNase XrnI contributes to the vhs-mediated mRNA decay (18). This attack on cellular transcripts has two main effects: stunting the antiviral immune response (reviewed in references 19 and 20) and reducing competition for the translation machinery to enhance access for viral mRNAs. vhs also destabilizes viral mRNAs. Although seemingly counterintuitive, this degradation it is thought to sharpen the transitions between immediate-early (IE), early (E), leaky-late (L1), and true-late (L2) gene expression, thereby further reducing the amount of mRNAs that compete for translation initiation. Late during infection the RNase activity of vhs is dampened through interaction with the tegument proteins VP22, VP16, and possibly UL47 (21–26). These interactions may also facilitate incorporation of vhs into the tegument of progeny virions.
Although the mRNA destabilizing activity of vhs and its role in immune evasion are well documented, until recently the importance of vhs for viral gene expression was unclear. In addressing this issue, we have previously provided two lines of evidence that vhs is able to stimulate mRNA translation. First, we showed that vhs increases translation of the second cistron driven by several internal ribosome entry site (IRES) elements or viral 5′UTRs in a bicistronic reporter gene assay (27). Second, we established that vhs is crucial for translation of viral true-late (L2) mRNAs and for efficient virus replication in several cell types (28). In the latter study, we found that levels of true-late proteins, such as gC, US11, and UL47, were reduced in HeLa cells and several other cell types infected with vhs-deficient HSV-1 due to impaired translation of the respective mRNAs independent of eIF2α phosphorylation. The concomitant accumulation of inactive translation initiation complexes in restrictive cells in the absence of vhs triggered formation of stress granules. Strikingly, translation of earlier viral transcripts and cellular RNAs, such as γ-actin mRNA, was not affected, even at late times postinfection, when translation of viral true-late mRNA was severely impaired.
In contrast to the situation in HeLa cells and other restrictive cell lines, vhs is not required for translation of viral true-late mRNAs or efficient virus replication in Vero cells (28). Although the molecular basis for this striking cell type difference remains unclear, it is possible that HeLa and Vero cells differ in the resilience of their translational apparatus and/or their ability to control overall mRNA levels during HSV infection. Consistent with the former possibility, we found that viral protein synthesis is substantially more sensitive to eIF4A inhibition by hippuristanol in HeLa cells than in Vero cells (28). This result suggests that eIF4A activity is limiting in HeLa cells.
In the present communication, we evaluated two possible models to explain why only viral true-late mRNAs depend on vhs for efficient translation during infection of restrictive HeLa cells. Both are based on the assumption that access to the translation machinery is limited at late times during infection with vhs-deficient HSV-1 due to accumulation of higher-than-normal amounts of mRNA. One model proposes that the highly structured 5′UTRs of HSV true-late mRNAs present an obstacle to translation initiation in the absence of vhs. The second model instead proposes that mRNA structure plays little, if any, role and that late mRNAs are functionally excluded from polysomes, because they accumulate after the cellular translational machinery has been overwhelmed by the accumulation of excess mRNA earlier during infection. We present evidence supporting the latter hypothesis.
MATERIALS AND METHODS
Cells.
HeLa (cervical carcinoma) and Vero (African green monkey kidney) cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% or 5% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Medium for Cre-Vero cells, stably expressing Cre recombinase, contained additional hygromycin B (400 μg/ml) to maintain the transgene.
Viruses.
The wild-type (wt) HSV-1 strain KOS37 was derived from the bacterial artificial chromosome (BAC) containing the genome of KOS37 (29), obtained from David Leib. Recombinant viruses designed for this study were created using the KOS37 BAC and a kanamycin/streptomycin dual selection marker (30) or en passant mutagenesis (31). The new BACs then were transfected into Cre-Vero cells to generate infectious viral particles. Viruses were passaged on Cre-Vero cells to remove the BAC sequence from the viral genome.
To generate the gC 5′UTR exchange mutants 1-140, 21-140, and 41-140, the kanamycin/streptomycin selection marker cassette was PCR amplified from the plasmid pSK+ KanaRpsL using the oligonucleotides JRS673 and JRS681 (Table 1). This PCR fragment was used to replace the coding sequence for the gC 5′UTR from positions +41 to +140 of the KOS37 BAC using the recombineering protocol described previously (30). The selection marker cassette and the remaining portion of the gC 5′UTR then was replaced by PCR fragments coding for the γ-actin 5′UTR using the same protocol. The DNA fragments were generated by PCR amplification using the overlapping oligonucleotide pairs JRS675/JRS676 (41-140), JRS706/JRS676 (21-140), and JRS707/JRS676 (1-140) (Table 1).
TABLE 1.
Primers used for generating the viruses ΔSma, 1-140, 1-140-ΔSma, 21-140, 21-140-ΔSma, 41-140, 41-140-ΔSma, IEUS11, and IEUS11-ΔSma
| Name | Sequence 5′–3′ |
|---|---|
| JRS781 | TGCCTTTTCCAACATGCGCCGGCGCGGCACCTCTCTGGCCTCGGGGACCCAAGCTTCACGCTGCCGCAAG |
| JRS782 | GGGGATCCAGGGGGAGGTCCTCGTCGTCTTCGTATCCGCCGGCGATCTGTGAACTAGTGGATCCCCCCGA |
| JRS783 | TGCCTTTTCCAACATGCGCCGGCGCGGCACCTCTCTGGCCTCGGGGACCCGGGACGTTACCGGGGGCCACCCCGGCCCCAGGTCGTCCTCCTCGGAGATA |
| JRS784 | TATCTCCGAGGAGGACGACCTGGGGCCGGGGTGGCCCCCGGTAACGTCCCGGGTCCCCGAGGCCAGAGAGGTGCCGCGCCGGCGCATGTTGGAAAAGGCA |
| JRS673 | GGGCTACCCTCACTACCGAGGGCGCTTGGTCGGGAGGCCGCATCGAACGCAAGCTTCACGCTGCCGCAAG |
| JRS681 | CACAACAGGCTCCAGAGGACCACGGCAAGGCCCACCCGCCCCGGGGCCATGAACTAGTGGATCCCCCCGA |
| JRS675 | GGGCTACCCTCACTACCGAGGGCGCTTGGTCGGGAGGCCGCATCGAACGCGTCTCAGTCGCCGCTGCCAGCTCTCGCACTCTGTTCTTCCGCCGCTCCGC |
| JRS676 | CACAACAGGCTCCAGAGGACCACGGCAAGGCCCACCCGCCCCGGGGCCATTGCGACCGGCAGAGAAACGCGACGGCGGAGCGGCGGAAGAACAG |
| JRS706 | TAAATTCCGGAAGGGGACACGGGCTACCCTCACTACCGAGGGCGCTTGGTGTCTCAGTCGCCGCTGCCAGCTCTCGCACTCTGTTCTTCCGCCGCTCCGC |
| JRS707 | CCCCGGATGGGGCCCGGGTATAAATTCCGGAAGGGGACACGGGCTACCCTGTCTCAGTCGCCGCTGCCAGCTCTCGCACTCTGTTCTTCCGCCGCTCCGC |
| JRS842 | ACCCGGGCGGCGGGGGGCGGGTCTCTCCGGCGCACATAAAAGGGGGCGTGAGGACCGGGATAGGGATAACAGGGTAATCGATTT |
| JRS843 | GCACGGCGGTTCTGGCCGCCTCCCGGTCCTCACGCCCCCTTTTATGTGCGCCGGAGAGACGCCAGTGTTACAACCAATTAACC |
The virus mutant IEUS11 was created using en passant mutagenesis (31). The selection marker cassette I-SceI-AphAI was PCR amplified from pEP-KanS using oligonucleotides JRS842 and JRS843 (Table 1) and introduced into the KOS37 BAC, relocating the coding region from downstream of the US12 TATA box to downstream of the US11 TATA box. The marker cassette was seamlessly removed by in vivo I-SceI cleavage as described in reference 31.
A 588-nucleotide deletion in the UL41 open reading frame (ORF), exactly as described in the original KOS-derived ΔSma virus (32), was introduced into the KOS37 BAC and the BACs of 1-140, 21-140, 41-140, and IEUS11 to generate the KOS37 derivatives ΔSma, 1-140-ΔSma, 21-140-ΔSma, 41-140-ΔSma, and IEUS11-ΔSma. First, the kanamycin/streptomycin selection marker cassette (30) was PCR amplified from the plasmid pSK+ KanaRpsL using the oligonucleotides JRS781 and JRS782 and introduced into the respective BACs, creating a deletion in the UL41 ORF. The cassette then was seamlessly replaced by a DNA fragment created by annealing the oligonucleotides JRS783 and JRS784 (Table 1).
Viruses were propagated and the titers were determined in Vero cells. Virus absorption, infection, and mock infections were carried out at 37°C at a multiplicity of 10 PFU/cell.
Drug treatments.
Phosphonoacetic acid (PAA; Sigma) was used at 300 μg/ml starting at 1 h postinfection (hpi) to inhibit viral DNA replication. Cycloheximide (CHX) was used at 100 μg/ml. Cells were pretreated for 30 min prior to infection and treated over the course of the infection.
Western blot analysis.
Mock-infected and infected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (1% IGEPAL CA-630, 150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.1% SDS, 0.25% sodium deoxycholate, 1× Roche protease inhibitor, 10 mM Na-β-glycerophosphate, 2 mM Na3VO4, 1 mM NaF). Cleared lysates were analyzed by 8% or 11% SDS-PAGE, transferred to a nitrocellulose membrane (Hybond ECL; Amersham), and immunoblotted with primary antibodies specific for the HSV-1 proteins gC (mouse; P1104; Virusys Corporation), gB (mouse; P1123; Virusys Corporation), ICP4 (mouse, P1101; Virusys Corporation), ICP27 (mouse; P1113; Virusys Corporation), ICP34.5 (rabbit; gift from Ian Mohr), US11 (mouse; gift from Ian Mohr), UL47 (rabbit; gift from Gillian Elliott), thymidine kinase (TK; rabbit; gift of William C. Summers), and VP16 (mouse; LP1; gift from Tony Minson) and the cellular proteins β-actin (mouse; A5441; Sigma), phospho-eIF2α (rabbit; 9721; Cell Signaling), and eIF2α (rabbit; 9722; Cell Signaling). Primary antibodies were detected by suitable secondary antibodies coupled to Alexa Flour 680 (Invitrogen) or IRDye800 (Rockland) using the Odyssey infrared imaging system (LICOR).
RNA extraction and Northern blot analysis.
Total RNA was isolated from cells in 35-mm culture dishes using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples (10 μg) were subjected to 1.2% agarose-formaldehyde gel electrophoresis, visualized by SYBR Gold staining, and transferred to a Genescreen membrane (NEN). Hybridizations of gC, US11, and TK probes, radiolabeled with 32P by random priming, were performed using ExpressHyb (Clontech) according to the user's manual. The gC and US11 probes were generated by PCR amplification of viral DNA using the primers 5′ CCCAAACCCAAGAACAACAC and 5′-TGTTCGTCAGGACCTCCTCT (gC), 5′-GGCGACCCAGATGGTTACTT and 5′-GCGAGCCGTACGTGGTTC (US11), or 5′-GCTTCGTACCCCTGCCATCA and 5′-GGTCATGCTGCCCATAAGGTATC (TK). Signals were analyzed with the Fujifilm FLA-5100 phosphorimager and Image Gauge, version 4.22.
Polysome analysis.
HeLa or Vero cells growing in two 100-mm culture dishes were infected with KOS37, ΔSma, IEUS11, or IEUS11-ΔSma virus at a multiplicity of infection (MOI) of 10. Cells were lysed at 12 hpi in lysis buffer (250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 50 mM Tris-HCl, pH 7.4, 0.5% Triton X-100, 100 μg/ml cycloheximide, RNaseOUT). Cleared lysates were layered onto gradients of 10 to 50% sucrose solution in 50 mM Tris-HCl, pH 7.4, 25 mM KCl, 1.5 mM MgCl2, 100 μg/ml cycloheximide, 2 mM dithiothreitol (DTT), and RNaseOUT and centrifuged in an SW40 rotor for 105 min at 38,000 rpm and 4°C. Twenty fractions were collected from the top of the gradient. Samples were Proteinase K treated and RNA was phenol-chloroform extracted. Half of each RNA sample was subjected to Northern blot analysis as described above to determine the distribution of selected mRNA species across the gradient.
RESULTS
Experimental rationale and design of test viruses.
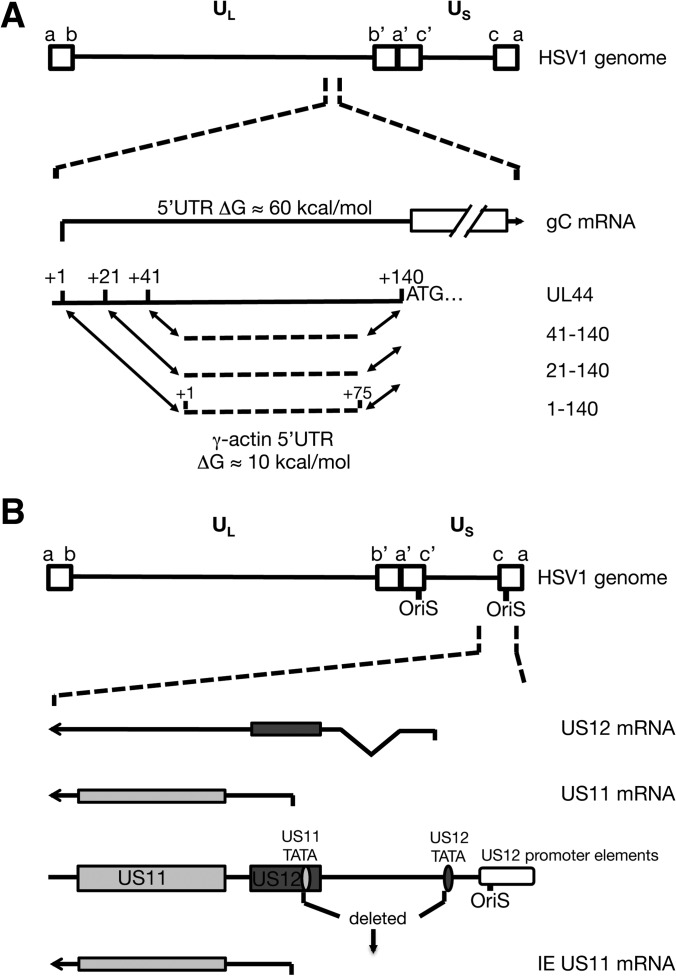
We previously demonstrated that the mRNAs encoding the true-late (L2) proteins gC and US11 are poorly translated in HeLa cells infected with vhs-deficient HSV-1 (ΔSma), while translation of earlier viral mRNA species as well as cellular mRNAs is not affected (28). It is well established that infection with a vhs-deficient virus leads to accumulation of higher levels of viral and cellular mRNAs than occurs during wild-type infection (33–35). This accumulation of mRNA could lead to increased competition between mRNAs for access to the translation machinery. As outlined in the introduction, we considered two possible models to account for the poor translation of HSV true-late mRNAs in the absence of vhs. The first suggests that the highly structured 5′UTRs of HSV true-late mRNAs imposes a barrier to initiation under conditions of mRNA competition, whereas the second suggests instead that true-late mRNAs are disadvantaged because they accumulate after the cellular translational apparatus has been overwhelmed by mRNAs produced earlier during infection. We tested the first hypothesis by monitoring the effects of reducing the secondary structure of the gC mRNA 5′UTR (Fig. 1A) and evaluated the second by scoring the effects of expressing the L2 US11 mRNA at earlier times postinfection by placing it under the control of an IE promoter (Fig. 1B).
FIG 1.
Schematic diagram of the viral genes mutated to study the effect of UTR structure (A) and expression kinetics (B) on translational activity in wild-type or vhs-null HSV-1. (A) The DNA coding for the 5′UTR of the UL44 mRNA was replaced completely (1-140) or partially (21-140 and 41-140) with DNA coding for the γ-actin 5′UTR. (B) The region downstream of the US12 TATA box was deleted, including the US11 TATA box, to put the transcription of the US11 gene under the control of the IE US12 promoter.
HSV DNA is GC rich (68%); therefore, HSV 5′UTRs tend to be relatively structured. For example, the gC and US11 mRNA 5′UTRs display predicted free energies (ΔGs) of −63 and −81 kcal/mol, respectively, whereas the cellular γ-actin mRNA contains a 5′UTR with little secondary structure (ΔG ≈ −10 kcal/mol by Mfold). Translation of mRNAs with highly structured 5′UTRs is more dependent on eIF4A helicase function than translation of mRNAs with relatively unstructured 5′UTRs (36–38), and we previously found that accumulation of L2 proteins is highly susceptible to inhibition by the eIF4A inhibitor hippuristanol (28). Therefore, we reasoned that HSV-1 L2 mRNAs are at a relative disadvantage due to their structured 5′UTRs when competition for translation factors is high, such as during the late phases of vhs-null infection of HeLa cells. To test this hypothesis, we created viral mutants in which the gC 5′UTR is replaced by the 75-nucleotide-long γ-actin 5′UTR (Fig. 1A). The replacement was either complete (from the transcription start site at +1 up to the start codon at +140, ΔG = −10 kcal/mol) or partial (for positions +21 to +140 or +41 to +140, ΔGs of −22 and −32 kcal/mol, respectively) (Fig. 1A). The partial replacements were introduced because of earlier reports describing regulatory elements in the 5′ noncoding region of the gC/UL44 gene that affect transcription (39, 40). All virus mutants were created using a bacterial artificial chromosome (BAC) containing the genome of the KOS37 strain (wt) or vhs-deficient KOS37 (ΔSma) and are named according to their respective mutations: 1-140, 1-140-ΔSma, 21-140, 21-140-ΔSma, 41-140, and 41-140-ΔSma.
We also considered the possibility that the timing of L2 transcription rather than mRNA structure was responsible for the poor translation of true-late mRNAs in the absence of vhs. According to this model, without the mRNA-destabilizing activity of vhs, one or more limiting translation factors are sequestered by cellular and viral IE and E mRNAs, hindering translation of mRNAs made during the later stages of infection. To test this hypothesis, we examined the effects of driving accumulation of an L2 mRNA at earlier times postinfection on its translation in the absence of vhs. We first sought to create an IE-gC virus by fusing the IE ICP27 promoter to gC coding sequences. However, this virus failed to accumulate appreciable levels of gC mRNA prior to the onset of viral DNA replication (data not shown), perhaps due to the presence of negatively acting posttranscriptional control sequences within gC coding sequences (41, 42); therefore, it was not suitable for our study. As an alternative strategy, we created IEUS11 viral mutants that contain the L2 US11 ORF under the control of the immediate-early promoter of the US12 gene (Fig. 1B). The IEUS11 and IEUS11-ΔSma mutants were constructed by deleting the viral DNA sequences extending downstream of the US12 TATA box to downstream of the US11 TATA box, similar to a deletion found in a virus mutant described earlier (10). These mutants lack most of the US12 ORF coding for the ICP47 protein. However, as ICP47 is involved in immune evasion by downregulating antigen presentation by major histocompatibility complex class 1 (MHC-I), it is dispensable for virus replication in cell culture (43).
Replacing the 5′UTR does not change the expression kinetics of the gC transcript.
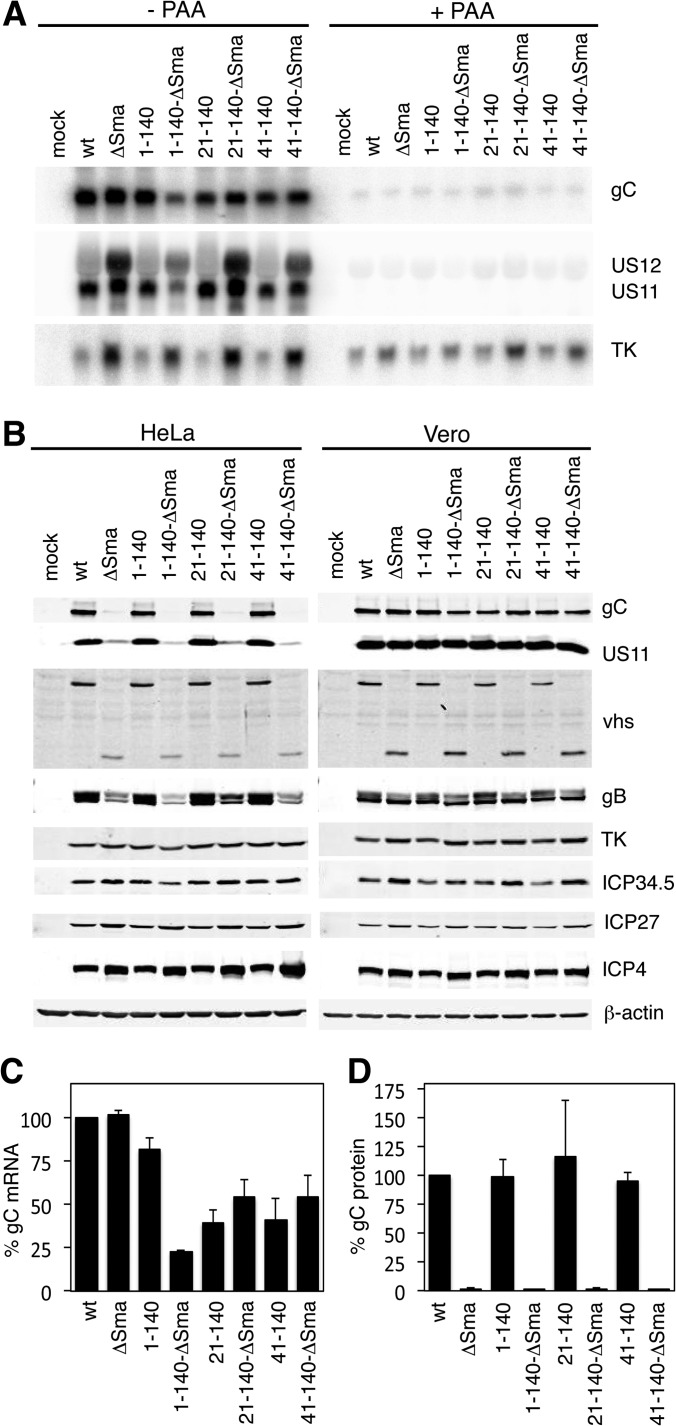
Earlier reports have shown that the 5′ noncoding region of the gC gene contains regulatory sequences that are necessary for optimal expression from the gC promoter (39, 40). Therefore, we performed Northern blot analysis to test whether the UTR exchange mutants still were able to transcribe gC mRNA and whether gC mRNA accumulated with true-late kinetics. HeLa cells were infected with KOS37 (wt) and the vhs-deficient mutant ΔSma, as well as with the vhs-competent and vhs-deficient versions of the UTR mutants 1-140, 21-140, and 41-140, at an MOI of 10 for 12 h. All viruses accumulated gC mRNA of the predicted size (albeit to varied levels), and RNA levels were severely reduced by treating infected cells with phosphonoacetic acid (PAA), which inhibits viral DNA replication and transcription of true-late genes (Fig. 2A). These data confirmed that the UTR replacement did not change the expression kinetics of gC. All of the gC UTR mutants displayed lower levels of gC mRNA than the wild-type virus (quantified in Fig. 2C). In contrast to the 21-140 and 41-140 substitution mutants, which accumulated approximately equivalent levels of gC mRNA in the presence and absence of vhs (ca. 40 to 55% of wild-type levels), the 1-140-ΔSma isolate displayed markedly reduced gC mRNA levels relative to its vhs+ counterpart (23% versus 82% of wild-type levels). This reduction likely does not stem from either the UTR exchange or the vhs mutation, as this isolate also displayed lower levels of US11 and TK mRNA than any of the other vhs-deficient viruses. These findings suggest that the 1-140-ΔSma virus acquired one or more additional mutations that reduce viral gene expression during BAC recombineering or plaque passaging. Consistent with previous work (33–35), ΔSma and the vhs-deficient 21-140 and 41-140 exchange mutants displayed markedly elevated levels of the IE US12 and E TK mRNAs, as well as somewhat elevated levels of the L2 gC and US11 mRNAs, relative to their wild-type counterparts (Fig. 2B). This finding likely reflects the well-established ability of vhs to destabilize viral mRNAs belonging to all three kinetic classes (33–35).
FIG 2.
Analyses of viral mRNA and protein levels. (A) The UTR exchange mutants transcribe gC mRNA with true-late kinetics. HeLa cells were mock treated or infected with wt, ΔSma, 1-140, 1-140-ΔSma, 21-140, 21-140-ΔSma, 41-140, and 41-140-ΔSma viruses at an MOI of 10 and were left untreated or treated with PAA, respectively. RNA was extracted at 12 hpi and analyzed by Northern blotting with probes specific for US11, TK, and gC. (B) UTR mutants do not show increased translation of gC. HeLa or Vero cells were mock treated or infected with wt, ΔSma, 1-140, 1-140-ΔSma, 21-140, 21-140-ΔSma, 41-140, and 41-140-ΔSma viruses at an MOI of 10. Cells were lysed at 12 hpi and analyzed by Western blotting with the indicated antibodies. (C and D) Quantification of the relative levels of gC mRNA (C) and protein (D) in infected HeLa cells at 12 hpi. Values represent averages from 3 independent experiments.
Secondary structure of the gC mRNA 5′UTR is not responsible for reduced translation in the absence of vhs.
We next analyzed viral protein accumulation to determine if replacing the gC mRNA 5′UTR with that of γ-actin mRNA rescued gC protein expression of HeLa cells in the absence of vhs. HeLa and Vero cells were infected with wt and ΔSma viruses and the vhs-competent and vhs-deficient pairs of the UTR mutants 1-140, 21-140, and 41-140 at an MOI of 10. Cells were lysed 12 hpi and analyzed by Western blotting with antibodies for representative proteins of all kinetic classes (Fig. 2B). Accumulation of the viral proteins ICP4 and ICP27 (IE), TK (E), and ICP34.5 (L1) was comparable between the wt and mutants in both HeLa and Vero cells, while glycoprotein B (L1) was reduced. As previously reported, US11 and gC showed severely reduced accumulation in HeLa cells infected with ΔSma, while accumulation was comparable between infections with vhs-competent and vhs-deficient viruses in Vero cells. All of the vhs-competent gC UTR substitution mutants accumulated essentially wild-type levels of gC protein in HeLa cells, and these levels were severely reduced in the corresponding vhs-null derivatives. In Vero cells, gC protein reached comparable levels in all infections. Although the amount of gC mRNA in HeLa cells infected with 1-140-ΔSma was only 28% of that in 1-140-infected cells (Fig. 2C), Western blotting detected only about 2% of the levels of gC protein obtained in 1-140-infected HeLa cells (Fig. 2D). Thus, translation of the 1-140 mRNA is severely impaired in HeLa cells in the absence of vhs. It is interesting that although the gC mRNA levels in HeLa cells infected with vhs-competent 21-140 and 41-140 were only about 40% of that in wt-infected cells, the amount of gC protein detected was comparable to that of the wt (Fig. 2D). This observation suggests that the translation machinery is working at close to capacity in HeLa cells infected with wild-type HSV. Overall, our characterization of the UTR exchange mutants suggests that the structure of the 5′UTR of the gC mRNA is not the reason for the low translational activity in HeLa cells infected with vhs-deficient HSV-1. We assume that the reduced translation of other L2 gene products observed in the absence of vhs also is independent of their 5′UTR structures.
IEUS11 mutants express US11 with immediate-early kinetics.
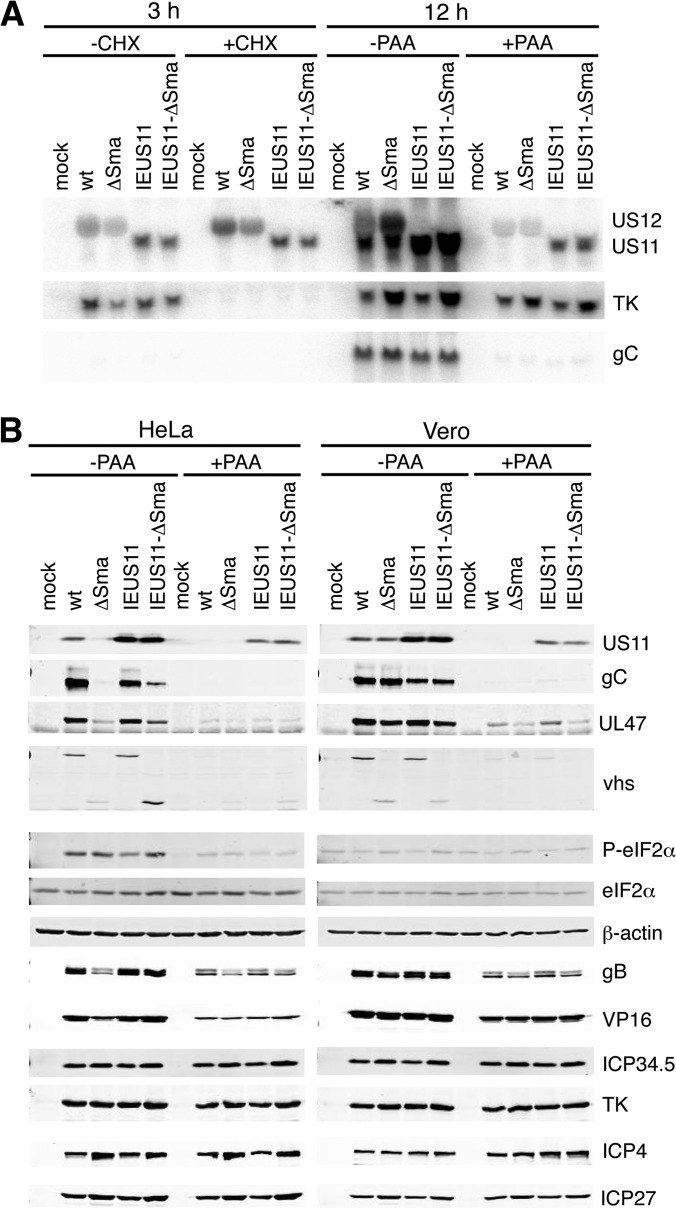
As described above, the IEUS11 mutants contain a deletion that places the US11 ORF in juxtaposition to the IE US12 promoter (Fig. 1B). Based on previous work (10), this mutation should lead to IE expression of US11 and abrogate US12 mRNA. To determine the expression kinetics of the US11 ORF in wt-, ΔSma-, IEUS11-, and IEUS11-ΔSma-infected HeLa cells, we treated the cells with cycloheximide (CHX) and PAA (Fig. 3A). While transcription of early genes depends on newly transcribed IE gene products, IE genes are transcribed even when translation is blocked by CHX. Expression of L2 genes is blocked by PAA treatment. Northern blot analysis of RNA extracted at 3 hpi from infected HeLa cells left untreated or treated with CHX demonstrated that IEUS11 and IEUS11-ΔSma express US11 mRNA with IE kinetics (Fig. 3A). PAA treatment of cells infected for 12 h confirmed that wt and ΔSma viruses expressed US11 with true-late kinetics. The US11-specific probe also hybridizes to the US12 mRNA, and we confirmed that, as expected, the IEUS11 mutants do not accumulate US12 mRNA. Accumulation of the E TK mRNA was inhibited by CHX, confirming that CHX was active. The Northern blot also shows that US11 mRNA is transcribed throughout the whole replication cycle in IEUS11-infected cells, as the mRNA signal was strongest in infections that were allowed to proceed for 12 h without PAA treatment.
FIG 3.
Kinetic analyses of viral mRNA and protein levels. (A) The IEUS11 virus transcribes US11 with IE kinetics. HeLa cells were mock treated or were infected with wt, ΔSma, IEUS11, and IEUS11-ΔSma viruses at an MOI of 10 and were left untreated or were treated with CHX or PAA, respectively. RNA was extracted at 12 hpi and analyzed by Northern blotting with probes specific for US11, TK, and gC. (B) IE transcription of US11 mRNA enhances its translation at late times during infection. HeLa or Vero cells were mock treated or infected with wt, ΔSma, IEUS11, and IEUS11-ΔSma virus at an MOI of 10 and were left untreated or were treated with PAA. Cells were lysed at 12 hpi and analyzed by Western blotting with the indicated antibodies.
Immediate-early US11 transcripts are translated late during infection without dependency on vhs.
We next performed Western blot analyses of HeLa and Vero cells infected with wt, ΔSma, IEUS11, and IEUS11-ΔSma viruses. Accumulation of the viral proteins ICP4 and ICP27 (IE), TK (E), and ICP34.5 (L1) at 12 hpi was comparable between all viruses in both HeLa and Vero cells (Fig. 3B). Interestingly, ICP34.5 levels were not reduced in PAA-treated cells, indicating that its expression is regulated more like an early gene than a late gene. As expected, levels of VP16 and gB (L1) as well as UL47, vhs, gC, and US11 (L2) were comparable in wt- and ΔSma-infected Vero cells. Expression of leaky-late proteins was significantly reduced, while that of true-late proteins was severely impaired in ΔSma-infected HeLa cells compared to wt-infected HeLa cells (Fig. 3B). PAA treatment confirmed the leaky-late and true-late expression kinetics of the respective proteins (Fig. 3B). In accordance with the IE expression profile of US11 mRNA in cells infected with IEUS11 mutants (Fig. 3A), we also detected US11 protein in PAA-treated HeLa and Vero cells infected with IEUS11 and IEUS11-ΔSma (Fig. 3B). This observation demonstrates that translation of the US11 mRNA prior to the start of replication does not depend on vhs activity. Furthermore, infection with vhs-competent and vhs-deficient IEUS11 mutants led to accumulation of high levels of US11 protein in untreated HeLa and Vero cells. Comparing PAA-treated and untreated cells, we saw an equal increase in US11 protein levels for vhs-competent and vhs-deficient IEUS11 mutants. This increase most likely is due to translation during the late stage of infection, supporting our hypothesis that earlier transcription of US11 mRNA can reverse the translation inhibition at late times in ΔSma virus infection.
US11 has been shown to inhibit the eIF2α kinase PKR, and IE expression of US11 has been shown to functionally compensate for the deletion of ICP34.5 during HSV-1 infection (10, 44). However, we found that phosphorylation of eIF2α was comparable between ΔSma- and IEUS11-ΔSma-infected HeLa cells (Fig. 3B). Moreover, the levels were comparable to those observed during wild-type infection. These observations are in accordance with our previous conclusion that reduced translation of late mRNAs in the absence of vhs is not due to enhanced eIF2α phosphorylation (28). These findings differ from previous reports of increased eIF2α and PKR phosphorylation in the absence of vhs (35, 45). It is possible that these differences reflect the different cell types used (mouse embryonic fibroblasts and a human fibrosarcoma cell line, whereas we used HeLa cells in the present study). Furthermore, other stress kinases, such as PERK, HRI, and GCN2, could contribute to the phosphorylation of eIF2α. The contribution of these kinases to the phosphorylation status of eIF2α will be addressed in future studies.
Interestingly, the levels of the L2 UL47 and gC proteins were moderately increased in IEUS11-ΔSma-infected HeLa cells compared to those of ΔSma-infected cells, although not to the levels observed during infection with the vhs-competent virus (Fig. 3B). At least for gC, this increase does not appear to be due to higher mRNA levels (Fig. 3A, bottom).
Taken in combination, these results indicate that early expression or overexpression of US11 restored US11 protein levels in IEUS11-ΔSma-infected HeLa cells to wild-type levels and also boosted accumulation of other late gene products, albeit to a much lesser degree.
US11 mRNA from the IEUS11-ΔSma mutant is translationally active at late times postinfection.
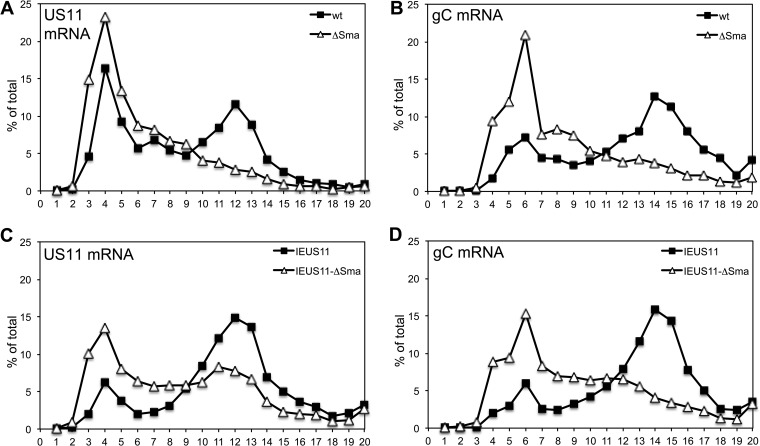
To directly assess the translational activity of US11 and gC mRNAs, we performed polysome analyses. We infected HeLa cells with wt, ΔSma, IEUS11, or IEUS11-ΔSma virus, prepared lysates at 12 hpi, and analyzed polysome profiles by sucrose density centrifugation. RNA was extracted from fractions 1 to 20 (top to bottom) and subjected to agarose gel electrophoresis. Staining of rRNA with SYBR Gold showed that 40S ribosomal subunits are found in fractions 3 and 4, 60S-80S complexes are found in fractions 5 to 7, and higher-numbered fractions represent polysomes (data not shown). The RNA was further analyzed by Northern blotting with probes specific for gC and US11 mRNA and quantified by scanning with a phosphorimager (data not shown). The distribution of the respective mRNA signal over the gradients is graphically represented in Fig. 4 as a percentage of the total mRNA signal present in each fraction. As shown previously (28), US11 and gC mRNAs from wt-infected cells were found mainly in polysome fractions, while US11 and gC mRNA from ΔSma-infected cells were shifted toward fractions representing monosomes or preinitiation complexes (Fig. 4A and B). This finding indicated that translation initiation on these transcripts is strongly compromised in the absence of vhs. In cells infected with IEUS11 mutants, we found a strong increase in the percentage of US11 mRNA in polysome fractions for both vhs-competent and vhs-deficient virus mutants (Fig. 4C). This result suggests that earlier accumulation of US11 mRNA yields an increase in translationally active US11 mRNA at late times; thus, it alleviates the restricted phenotype of the ΔSma virus for this mRNA. In contrast, we found only a slight increase in the percentage of gC mRNA in polysome fractions in IEUS11 and IEUS11-ΔSma viruses compared to wt and ΔSma viruses, respectively (Fig. 4D). The results from this polysome analysis correlate well with the Western blot data. Taken together, the effect of immediate-early US11 expression seems to be 2-fold. First, IEUS11 mRNA is translated more efficiently at late times during infection in the absence of vhs, possibly because it initially accesses translation factors before they become limiting. Second, earlier expression and/or overexpression of US11 also may aid translation of other late mRNAs, albeit to a much lesser degree. We have not yet addressed which of the multiple functions of US11 are responsible for the latter effect, which does not appear to involve changes in the levels of eIF2α phosphorylation.
FIG 4.
Polysome analyses. HeLa cells were infected with wild-type, ΔSma, IEUS11, or IEUS11-ΔSma virus at an MOI of 10. Cells were lysed 12 hpi and analyzed by sedimentation through a 10 to 50% sucrose gradient. Samples were taken from top to bottom, and extracted RNA was analyzed by Northern blotting for the viral mRNAs for gC and US11. Bands were quantified by a phosphorimager, and the distribution of the mRNA across the gradient is shown as a percentage of the total signal of the respective mRNA. The graph represents one of two comparable experiments.
DISCUSSION
We previously reported that vhs is necessary for efficient translation of HSV true-late gene products in certain cell types, such as HeLa cells, but not in Vero cells (28). In this study, we sought to determine why the translation of true-late transcripts is particularly sensitive to the restrictive conditions created by infection with a vhs-deficient virus while translation of earlier transcripts and cellular mRNAs is not affected.
Our findings demonstrate that the secondary structure of the 5′UTRs of true-late mRNAs is not the primary culprit for poor translation initiation on these transcripts. Replacing the 5′UTR of gC with the less structured γ-actin 5′UTR did not rescue translation of gC in cells infected with the vhs-deficient virus mutants. However, transcribing the true-late gene US11 with IE kinetics clearly increased its translational activity at late times postinfection. This result argues that the translational defect observed for late mRNAs in the absence of vhs does not stem from one or more structural features of the affected mRNAs but instead arises because these transcripts accumulate late during infection. The simplest model is that when conditions become limiting at late times during infection in ΔSma-infected HeLa cells, the translation rate of viral proteins depends on whether the respective mRNAs were transcribed early enough to access the translation machinery. This model proposes that mRNAs that have already been assembled into polysomes are at an advantage for translation initiation compared to newly transcribed mRNAs when translation factors are limiting. This advantage could be due to the proposed closed-loop structure of polysomes that might allow recycling of ribosomes or the tethering of eIF4F to the mRNA through its interaction with PABP, keeping it separate from the pool of free translation initiation factors (3).
Characterization of the IEUS11 mutants also revealed a smaller positive effect of earlier expression and/or increased accumulation of the US11 protein on translation of other late proteins. US11 is an RNA-binding protein that binds to and inhibits activation of the dsRNA-dependent kinase PKR (10, 44, 46). As a result, US11 helps prevent inhibition of translation initiation by phosphorylated eIF2α and consequent global shutdown of protein synthesis. However, the effects of US11 on late gene expression observed in our study seem to be independent of eIF2α phosphorylation, as the level of eIF2α phosphorylation was comparable in ΔSma- and IEUS11-ΔSma-infected cells. US11 has multiple other functions, such as inhibition of 2′-5′oligoadenylate synthetase (47), inhibition of alpha/beta interferon induction by binding to mda-5, RIG-I, and PACT (48–50), recruitment of nucleolin to promote nuclear egress of virions (51), inhibition of staurosporine and heat shock-induced apoptosis (52), prevention of PKR-dependent autophagy (53), and prevention of HIPK2-induced growth arrest (54). Which of these functions is responsible for the improved translation of late gene products is a subject for further studies.
vhs-null mutants display a striking host range phenotype, with translation of late mRNAs being severely impaired in HeLa cells but not Vero cells. Given the results described in this communication, we suggest that the levels of mRNA that accumulate during vhs-null infection in HeLa cells overload the translation apparatus. Vero cells might be better at regulating overall (cellular and viral) mRNA levels and/or have a higher translational capacity. Recent studies in yeast stress that maintaining proper mRNA levels is vital for a cell by demonstrating that regulation of transcription, translation, and mRNA decay are linked (55, 56). Furthermore, computational modeling in yeast suggests that ribosomes are the limiting component of the translation apparatus (57–59). This configuration is thought to combine efficient use of resources with flexibility in terms of changes in expression levels. To the best of our knowledge, there have been no studies published that examine how much total cellular mRNA levels can increase before the capacity of the mammalian translational machinery is exceeded.
In conclusion, our results support our earlier hypothesis that vhs is necessary to prevent mRNA overload in HeLa cells but not in Vero cells. The unrestricted accumulation of mRNA leads to reduced translation of true-late gene products, most likely because they are transcribed when the pool of free translation factors and ribosomes has become severely diminished. Determining the mechanisms that enable tighter control of mRNA levels in Vero cells compared to HeLa cells should provide interesting insights into the regulation of mRNA decay pathways and/or transcriptional control.
ACKNOWLEDGMENTS
We thank Ian Mohr for the US11 antibody.
This work was funded by an operating grant from the Canadian Institutes for Health Research (FRN MOP 37995). B.D. was supported by a postdoctoral fellowship from Alberta Innovates-Health Solutions, and J.R.S. is the Canada Research Chair in Molecular Virology (Tier I).
Footnotes
Published ahead of print 11 June 2014
REFERENCES
- 1.Smith RW, Graham SV, Gray NK. 2008. Regulation of translation initiation by herpesviruses. Biochem. Soc. Trans. 36:701–707. 10.1042/BST0360701 [DOI] [PubMed] [Google Scholar]
- 2.Walsh D, Mohr I. 2011. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 9:860–875. 10.1038/nrmicro2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 11:113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, Mohr I. 2010. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 24:2627–2639. 10.1101/gad.1978310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hargett D, McLean T, Bachenheimer SL. 2005. Herpes simplex virus ICP27 activation of stress kinases JNK and p38. J. Virol. 79:8348–8360. 10.1128/JVI.79.13.8348-8360.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18:660–672. 10.1101/gad.1185304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh D, Mohr I. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 20:461–472. 10.1101/gad.1375006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samuel CE. 1993. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603–7606 [PubMed] [Google Scholar]
- 9.He B, Gross M, Roizman B. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 94:843–848. 10.1073/pnas.94.3.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohr I, Gluzman Y. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759–4766 [PMC free article] [PubMed] [Google Scholar]
- 11.Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J. Virol. 81:3377–3390. 10.1128/JVI.02191-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Read GS. 2013. Virus-encoded endonucleases: expected and novel functions. Wiley Interdiscip. Rev. RNA 4:693–708. 10.1002/wrna.1188 [DOI] [PubMed] [Google Scholar]
- 13.Doepker RC, Hsu W-L, Saffran HA, Smiley JR. 2004. Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J. Virol. 78:4684–4699. 10.1128/JVI.78.9.4684-4699.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng P, Everly DN, Read GS. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 79:9651–9664. 10.1128/JVI.79.15.9651-9664.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page HG, Read GS. 2010. The virion host shutoff endonuclease (UL41) of herpes simplex virus interacts with the cellular cap-binding complex eIF4F. J. Virol. 84:6886–6890. 10.1128/JVI.00166-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarma N, Agarwal D, Shiflett LA, Read GS. 2008. Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus. J. Virol. 82:6600–6609. 10.1128/JVI.00137-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiflett LA, Read GS. 2013. mRNA decay during herpes simplex virus (HSV) infections: mutations that affect translation of an mRNA influence the sites at which it is cleaved by the HSV virion host shutoff (Vhs) protein. J. Virol. 87:94–109. 10.1128/JVI.01557-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. 2012. A common strategy for host RNA degradation by divergent viruses. J. Virol. 86:9527–9530. 10.1128/JVI.01230-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paladino P, Mossman KL. 2009. Mechanisms employed by herpes simplex virus 1 to inhibit the interferon response. J. Interferon Cytokine Res. 29:599–607. 10.1089/jir.2009.0074 [DOI] [PubMed] [Google Scholar]
- 20.Smiley JR. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063–1068. 10.1128/JVI.78.3.1063-1068.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott G, Mouzakitis G, O'Hare P. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam Q, Smibert CA, Koop KE, Lavery C, Capone JP, Weinheimer SP, Smiley JR. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575–2581 [PMC free article] [PubMed] [Google Scholar]
- 23.Mbong EF, Woodley L, Dunkerley E, Schrimpf JE, Morrison LA, Duffy C. 2012. Deletion of the herpes simplex virus 1 UL49 gene results in mRNA and protein translation defects that are complemented by secondary mutations in UL41. J. Virol. 86:12351–12361. 10.1128/JVI.01975-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sciortino MT, Taddeo B, Giuffrè-Cuculletto M, Medici MA, Mastino A, Roizman B. 2007. Replication-competent herpes simplex virus 1 isolates selected from cells transfected with a bacterial artificial chromosome DNA lacking only the UL49 gene vary with respect to the defect in the UL41 gene encoding host shutoff RNase. J. Virol. 81:10924–10932. 10.1128/JVI.01239-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu M, Taddeo B, Zhang W, Roizman B. 2013. Selective degradation of mRNAs by the HSV host shutoff RNase is regulated by the UL47 tegument protein. Proc. Natl. Acad. Sci. U. S. A. 110:E1669–E1675. 10.1073/pnas.1305475110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smibert CA, Popova B, Xiao P, Capone JP, Smiley JR. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saffran HA, Read GS, Smiley JR. 2010. Evidence for translational regulation by the herpes simplex virus virion host shutoff protein. J. Virol. 84:6041–6049. 10.1128/JVI.01819-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dauber B, Pelletier J, Smiley JR. 2011. The herpes simplex virus 1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J. Virol. 85:5363–5373. 10.1128/JVI.00115-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gierasch WW, Zimmerman DL, Ward SL, Vanheyningen TK, Romine JD, Leib DA. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206. 10.1016/j.jviromet.2006.03.014 [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Zhao Y, Leiby M, Zhu J. 2009. A new positive/negative selection scheme for precise BAC recombineering. Mol. Biotechnol. 42:110–116. 10.1007/s12033-009-9142-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430. 10.1007/978-1-60761-652-8_30 [DOI] [PubMed] [Google Scholar]
- 32.Read GS, Karr BM, Knight K. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwong AD, Frenkel N. 1987. Herpes simplex virus-infected cells contain a function (s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. U. S. A. 84:1926–1930. 10.1073/pnas.84.7.1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oroskar AA, Read GS. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasieka TJ, Lu B, Crosby SD, Wylie KM, Morrison LA, Alexander DE, Menachery VD, Leib DA. 2008. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J. Virol. 82:5527–5535. 10.1128/JVI.02047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelletier J, Sonenberg N. 1985. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40:515–526. 10.1016/0092-8674(85)90200-4 [DOI] [PubMed] [Google Scholar]
- 37.Shahbazian D, Parsyan A, Petroulakis E, Topisirovic I, Martineau Y, Gibbs BF, Svitkin Y, Sonenberg N. 2010. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol. Cell. Biol. 30:1478–1485. 10.1128/MCB.01218-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382–394. 10.1017/S135583820100108X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Homa FL, Otal TM, Glorioso JC, Levine M. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 6:3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weir JP, Narayanan PR. 1990. Expression of the herpes simplex virus type 1 glycoprotein C gene requires sequences in the 5′ noncoding region of the gene. J. Virol. 64:445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins KD, Gregonis J, Borge S, Rice SA. 2003. Transactivation of a viral target gene by herpes simplex virus ICP27 is posttranscriptional and does not require the endogenous promoter or polyadenylation site. J. Virol. 77:9872–9884. 10.1128/JVI.77.18.9872-9884.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedlackova L, Perkins KD, Meyer J, Strain AK, Goldman O, Rice SA. 2010. Identification of an ICP27-responsive element in the coding region of a herpes simplex virus type 1 late gene J. Virol. 84:2707–2718. 10.1128/JVI.02005-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411–415. 10.1038/375411a0 [DOI] [PubMed] [Google Scholar]
- 44.Cassady KA, Gross M, Roizman B. 1998. The herpes simplex virus US11 protein effectively compensates for the gamma1(34.5) gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sciortino MT, Parisi T, Siracusano G, Mastino A, Taddeo B, Roizman B. 2013. The virion host shutoff RNase plays a key role in blocking the activation of protein kinase R in cells infected with herpes simplex virus 1. J. Virol. 87:3271–3276. 10.1128/JVI.03049-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khoo D, Perez C, Mohr I. 2002. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J. Virol. 76:11971–11981. 10.1128/JVI.76.23.11971-11981.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez R, Mohr I. 2007. Inhibition of cellular 2′-5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J. Virol. 81:3455–3464. 10.1128/JVI.02520-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kew C, Lui PY, Chan CP, Liu X, Au SW, Mohr I, Jin DY, Kok KH. 2013. Suppression of PACT-induced type I interferon production by herpes simplex virus 1 Us11 protein. J. Virol. 87:13141–13149. 10.1128/JVI.02564-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulvey M, Camarena V, Mohr I. 2004. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon requires both the Us11 and gamma(1)34.5 gene products. J. Virol. 78:10193–10196. 10.1128/JVI.78.18.10193-10196.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J. Virol. 86:3528–3540. 10.1128/JVI.06713-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greco A, Arata L, Soler E, Gaume X, Coute Y, Hacot S, Calle A, Monier K, Epstein AL, Sanchez JC, Bouvet P, Diaz JJ. 2012. Nucleolin interacts with US11 protein of herpes simplex virus 1 and is involved in its trafficking. J. Virol. 86:1449–1457. 10.1128/JVI.06194-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Javouhey E, Gibert B, Arrigo AP, Diaz JJ, Diaz-Latoud C. 2008. Protection against heat and staurosporine mediated apoptosis by the HSV-1 US11 protein. Virology 376:31–41. 10.1016/j.virol.2008.02.031 [DOI] [PubMed] [Google Scholar]
- 53.Lussignol M, Queval C, Bernet-Camard MF, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. 2013. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 87:859–871. 10.1128/JVI.01158-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giraud S, Diaz-Latoud C, Hacot S, Textoris J, Bourette RP, Diaz JJ. 2004. US11 of herpes simplex virus type 1 interacts with HIPK2 and antagonizes HIPK2-induced cell growth arrest. J. Virol. 78:2984–2993. 10.1128/JVI.78.6.2984-2993.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haimovich G, Daniel Medina A, Sebastien Causse Z, Garber M, Millán-Zambrano G, Barkai O, Chávez S, José Pérez-Ortín E, Darzacq X, Choder M. 2013. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153:1000–1011. 10.1016/j.cell.2013.05.012 [DOI] [PubMed] [Google Scholar]
- 56.Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. 2010. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143:552–563. 10.1016/j.cell.2010.10.033 [DOI] [PubMed] [Google Scholar]
- 57.Chu D, Barnes DJ, von der Haar T. 2011. The role of tRNA and ribosome competition in coupling the expression of different mRNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 39:6705–6714. 10.1093/nar/gkr300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu D, von der Haar T. 2012. The architecture of eukaryotic translation. Nucleic Acids Res. 40:10098–10106. 10.1093/nar/gks825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. 2013. Rate-limiting steps in yeast protein translation. Cell 153:1589–1601. 10.1016/j.cell.2013.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]