ABSTRACT
Rotaviruses (RVs) are 11-segmented, double-stranded RNA viruses that cause severe gastroenteritis in children. In addition to an error-prone genome replication mechanism, RVs can increase their genetic diversity by reassorting genes during host coinfection. Such exchanges allow RVs to acquire advantageous genes and adapt in the face of selective pressures. However, reassortment may also impose fitness costs if it unlinks genes/proteins that have accumulated compensatory, coadaptive mutations and that operate best when kept together. To better understand human RV evolutionary dynamics, we analyzed the genome sequences of 135 strains (genotype G1/G3/G4-P[8]-I1-C1-R1-A1-N1-T1-E1-H1) that were collected at a single location in Washington, DC, during the years 1974 to 1991. Intragenotypic phylogenetic trees were constructed for each viral gene using the nucleotide sequences, thereby defining novel allele level gene constellations (GCs) and illuminating putative reassortment events. The results showed that RVs with distinct GCs cocirculated during the vast majority of the collection years and that some of these GCs persisted in the community unchanged by reassortment. To investigate the influence of protein coadaptation on GC maintenance, we performed a mutual information-based analysis of the concatenated amino acid sequences and identified an extensive covariance network. Unexpectedly, amino acid covariation was highest between VP4 and VP2, which are structural components of the RV virion that are not thought to directly interact. These results suggest that GCs may be influenced by the selective constraints placed on functionally coadapted, albeit noninteracting, viral proteins. This work raises important questions about mutation-reassortment interplay and its impact on human RV evolution.
IMPORTANCE Rotaviruses are devastating human pathogens that cause severe diarrhea and kill >450,000 children each year. The virus can evolve by accumulating mutations and by acquiring new genes from other strains via a process called reassortment. However, little is known about the relationship between mutation accumulation and gene reassortment for rotaviruses and how it impacts viral evolution. In this study, we analyzed the genome sequences of human strains found in clinical fecal specimens that were collected at a single hospital over an 18-year time span. We found that many rotaviruses did not reassort their genes but instead maintained them as specific sets (i.e., constellations). By analyzing the encoded proteins, we discovered concurrent amino acid changes among them, which suggests that they are functionally coadapted to operate best when kept together. This study increases our understanding of how rotaviruses evolve over time in the human population.
INTRODUCTION
Rotaviruses (RVs) are ubiquitous human pathogens that cause severe watery diarrhea in young children (1). The 23-kb viral genome is comprised of 11 distinct double-stranded RNA segments (i.e., genes) and codes for 6 structural proteins (VP1 to VP4, VP6, and VP7) and 6 nonstructural proteins (NSP1 to NSP6) (1). Like other segmented RNA viruses, RVs are able to increase their genetic diversity (i) by accumulating point mutations due to error-prone genome replication and (ii) by reassorting genes during host coinfection (2, 3). Compared to evolution by mutation, which occurs more slowly, at a rate of 10−3 to 10−5 changes per site per round of copying, reassortment can have immediate phenotypic consequences because it creates chimeric progeny with genes derived from multiple parental strains (4, 5). In some cases, reassortment can confer selective advantages on the virus, for example, by increasing its capacity to replicate within the host or allowing it to infect a new host (4). However, reassortment might also impose fitness costs if it unlinks genes/proteins that have accumulated compensatory, coadaptive mutations and therefore operate best when kept together (6, 7). However, little is known about the dynamic interplay between mutation accumulation and gene reassortment that drives RV evolution. Given the sustained burden of RV disease and reduced vaccine efficacy in many parts of the world, studies elucidating the evolutionary mechanisms of locally and globally circulating human strains are warranted (8). This information is expected to enhance our ability to predict and prepare for emerging RVs, thereby ensuring future protection of children against severe diarrheal disease.
Our current understanding of RV genetic diversity is consistent with the notion that most human strains belong to one of two cocirculating genogroups: (i) Wa-like genogroup 1 or (ii) DS-1-like genogroup 2 (9). These two genogroups were first described in the mid-1980s on the basis of RNA hybridization assays using laboratory prototype strains (10). Today, human RVs are classified as either genogroup 1 or 2 as a result of their overall genotype level gene constellation (GC) (9, 11). In particular, using a classification scheme that assigns a genotype to the VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/6 genes based upon the percent nucleotide identity cutoff values, genogroup 1 strains exhibit the genotype level GC G1/3/4/9/12-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1. In contrast, genogroup 2 RVs exhibit the genotype level GC G2-P[4]/P[6]-I2-R2-C2-M2-A2-N2-T2-E2-H2. Intergenogroup gene reassortment can occur, but such strains have only rarely caused disease in otherwise healthy children (12–17). In fact, in most geographical locations, genogroup 1 strains tend to outnumber both genogroup 2 strains and intergenogroup reassortant strains (9). This observation suggests that there may be selective pressures acting to promote the maintenance of human RVs with certain GCs, thereby restricting the prevalence of gene reassortment among cocirculating strains.
To better understand the limitations of gene reassortment among human RVs, our laboratories have been systematically analyzing the sequences of genogroup 1 strains found in fecal specimens that were collected from children with RV gastroenteritis at Children's Hospital National Medical Center in Washington, DC (18–21). This specimen collection is unique in that it includes samples from as early as 1974—just a few years after the discovery of RVs—to as late as 1991 (22) (Table 1). Thus, an unprecedented 18-year surveillance time span is captured, providing a unique opportunity to analyze RV evolution over time at a single geographical location. We have previously reported on the genome sequencing and analysis of 62 genogroup 1 strains (51 G3P[8] and 11 G4P[8] RVs) from this collection via a semiautomated reverse transcription (RT)-PCR/Sanger nucleotide-sequencing pipeline at the J. Craig Venter Institute (JCVI). In the current study, we worked with JCVI to deduce the genome sequences of an additional 73 genogroup 1 RVs (72 G1P[8] RVs and 1 G3P[8] RV) from this archival Washington, DC, fecal-specimen collection using next-generation techniques (Ion PGM [Life Technologies] and Illumina HiSeq platforms). Using maximum-likelihood phylogenetic methods, the individual genes of the 73 newly sequenced RVs were compared to each other, to those of the 62 previously sequenced RVs from the collection, and to those of ∼200 human and animal strains from around the world. The results of this study identified new alleles in the viral population and provide additional evidence that genogroup 1 RVs with the same genotype level GC but distinct subgenotype, allele level GCs cocirculated in Washington, DC, during each of the collection years. RVs with some allele level GCs were transient, while others were found to persist in the community for >12 years, unchanged by gene reassortment. Using a mutual information-based approach, we directly investigated the influence of compensatory, coadaptive mutations in the viral proteins on GC maintenance. An extensive genome-wide amino acid covariance network was identified that included all 12 DC RV proteins. Unexpectedly, two viral structural proteins that are not thought to directly interact (VP4 and VP2) showed the highest number of intermolecular connections. These results suggest that selection pressures acting on coadapted interacting and noninteracting viral proteins tempered gene reassortment among the genogroup 1 DC RVs and influenced the persistence of viruses with specific allele level GCs in the community.
TABLE 1.
Preliminary G/P types of RVs in Washington, DC, archival fecal specimens
| Yr | No. of specimens for G/P genotype: |
|||
|---|---|---|---|---|
| G1P[8] | G2P[4] | G3P[8] | G4P[8] | |
| 1974 | 20 | 0 | 1 | 1 |
| 1975 | 31 | 1 | 8 | 0 |
| 1976 | 8 | 2 | 49 | 2 |
| 1977 | 20 | 6 | 4 | 8 |
| 1978 | 55 | 1 | 11 | 1 |
| 1979 | 44 | 8 | 6 | 1 |
| 1980 | 37 | 3 | 10 | 18 |
| 1981 | 5 | 0 | 0 | 0 |
| 1987 | 6 | 0 | 0 | 0 |
| 1988 | 112 | 0 | 7 | 1 |
| 1989 | 60 | 0 | 8 | 0 |
| 1991 | 30 | 6 | 46 | 0 |
| Total no. (%) | 428 (56) | 27 (4) | 150 (20) | 32 (4) |
MATERIALS AND METHODS
Sample collection, RNA extraction, and G/P typing.
Fecal specimens were collected from infants and young children who were hospitalized with diarrhea at Children's Hospital National Medical Center, Washington, DC, as described previously (18–21). The fecal specimens were tested for evidence of RV using electron microscopy and for viral antigens using an enzyme-linked immunosorbent assay (ELISA) (23). RNA was extracted from RV-positive samples using TRIzol (Invitrogen), and samples were classified into preliminary G/P types based on the results of a microtiter plate hybridization-based PCR-ELISA (Table 1) (23). No demographic or clinical information is available for these samples. This study was evaluated by the National Institutes of Health Office for Human Subjects Protection and was deemed exempt from review by the Institutional Review Board.
RT-PCR and nucleotide sequencing.
RNAs from a representative subset of G1P[8]-positive fecal specimens (230 samples in total) were sent to JCVI (Rockville, MD) for RT-PCR and next-generation nucleotide sequencing (Table 2). Long-range RT-PCR amplicons were generated using the Qiagen One-Step RT-PCR kit (Qiagen). The reaction mixtures were incubated in a thermocycler at 45°C for 30 min, 95°C for 15 min, and then 50 cycles of 94°C for 10 s, 55°C for 3 min, and 68°C for 10 min. A final extension step occurred at 68°C for 10 min. The primer sequences used for RT-PCR are included in the GenBank accession pages. The amplicons were treated with shrimp alkaline phosphatase and exonuclease I (New England BioLabs), quantitated with SYBR green (Life Technologies), and pooled in equal-nanogram amounts.
TABLE 2.
Summary of analysis statuses for G1P[8]-positive archival fecal specimens
| Yr | No. of G1P[8] specimens collected | No. of specimens sent to JCVI | No. of single-genome sequences | No. of mixed-genome sequences |
|---|---|---|---|---|
| 1974 | 20 | 20 | 7 | 0 |
| 1975 | 31 | 19 | 11 | 0 |
| 1976 | 8 | 8 | 3 | 0 |
| 1977 | 20a | 9a | 5 | 2a |
| 1978 | 55b | 12b | 4b | 0 |
| 1979 | 44 | 31 | 7 | 2 |
| 1980 | 37a | 14a | 3 | 3a |
| 1981 | 5 | 5 | 0 | 0 |
| 1987 | 6 | 6 | 0 | 0 |
| 1988 | 112 | 65 | 20 | 8 |
| 1989 | 60 | 29 | 9 | 3 |
| 1991 | 30 | 12 | 4 | 3 |
| Total | 428 | 230 | 73 | 21 |
Two samples (DC1127 and DC1230) were determined to each contain a G4 RV by sequence analysis.
One sample (DC799) was determined to contain a G3 RV by sequence analysis.
For sequencing using the Ion PGM system (Life Technologies), pooled amplicons were sheared for 15 min, and compatible barcoded adapters were ligated to the sheared DNA using the Ion Xpress Plus Fragment Library Kit (Life Technologies) to create 200-bp libraries. The libraries were pooled in equal volumes and cleaned with AMPure XP Reagent (Agencourt). Quantitative PCR was performed on the pooled libraries to assess the quality of the pool and to determine the template dilution factor for emulsion PCR. The pool was diluted appropriately and amplified on Ion Sphere particles (Life Technologies) during emulsion PCR on the Ion OneTouch 2 system (Life Technologies). The emulsion was broken, and the pool was cleaned and enriched for template-positive Ion Sphere particles on the Ion OneTouch ES instrument (Life Technologies). Sequencing was performed on the Ion PGM system using a 316 chip.
For sequencing using the Illumina HiSeq (Illumina), pooled amplicons were randomly amplified and prepared for sequencing using a sequence-independent single-primer amplification (SISPA) method described previously (24). Briefly, 50 to 200 ng DNA was combined with dimethyl sulfoxide and a chimeric oligonucleotide containing a known 22-nucleotide (nt) barcode sequence followed by a 3′ random hexamer. The mixture was incubated at 95°C for 5 min and immediately placed on ice. The denatured DNA template was then incubated with the Klenow fragment 3′-5′-exo (New England BioLabs) at 37°C for 60 min, followed by 75°C for 10 min. The resulting DNA was amplified by PCR using AmpliTaq Gold (Life Technologies) for 35 cycles (94°C for 30 s, 55°C for 30 s, and 68°C for 45 s). PCR mixtures contained primers corresponding to the known 22-nt barcode sequence. The resulting DNA was then treated with exonuclease I (New England BioLabs) at 37°C for 60 min, followed by incubation at 72°C for 15 min. The SISPA products were normalized and pooled into a single reaction mixture that was purified using a QIAquick PCR purification kit (Qiagen). Samples were gel purified to select for SISPA products 300 to 500 bp in size, which were used for Illumina HiSeq sequencing (100-bp paired-end reads).
The sequence reads from the Ion PGM and Illumina HiSeq were sorted by barcode, trimmed, and assembled using the clc_novo_assemble program (CLC bio). The resulting contigs were searched against custom full-length RV segment nucleotide databases to find the closest reference sequence for each segment. All Ion PGM and Illumina HiSeq sequence reads were then mapped to the selected reference segments using the clc_ref_assemble_long program (CLC bio). Gaps and some of the ambiguous bases in the assemblies were resolved by Sanger sequencing, as described previously (25, 26). The nucleotide sequences determined in the current study (1,124 total gene sequences) were deposited in GenBank (see “Nucleotide sequence accession numbers” below and Tables S1 and S2 in the supplemental material). Genotypes were determined for each gene using the RotaC Web server (http://rotac.regatools.be/; 11, 27).
Nucleotide sequence alignments, phylogenetic analyses, and calculation of codon dN/dS values.
Nucleotide sequence alignments and phylogenies were generated with Geneious Pro v5.6.5 (Biomatters) using the ClustalW and PhyML plug-ins (28, 29). The general time reversal substitution model with gamma-distributed rate variation among sites was chosen based upon the results of Akaike information criterion ranking of each alignment as implemented in MEGA 5.0 (30, 31). Trees were constructed with the nucleotide sequences of >97% of each viral gene (see the figure legends for exact numbers), and 1,000 pseudoreplicates were generated for bootstrapping analyses. Intragenotypic alleles were defined based upon the results of McDonald et al. (25, 26) and based upon the clustering of gene sequences into monophyletic branches of the individual trees (25, 26). Monophyletic groupings were collapsed using FigTree v1.4, and the figures were colorized using Adobe Illustrator CS5 (Adobe Systems). The accession numbers and genotypes of previously sequenced RV genes used in the phylogenetic analyses are provided in Tables S3 to S7 in the supplemental material. The dN/dS values for each codon were determined using SNAP v.1.1.1 and the codon-aligned nucleotide sequences of the complete open reading frames for genes VP7, VP4, VP6, VP1, and NSP2 to NSP6 (32–34). For VP2, VP3, and NSP1, the open reading frame alignments were trimmed to match the length of the shortest sequence prior to calculating dN/dS values: VP2 (nucleotides 1 to 141 and 2653 to 2691 were removed), VP3 (nucleotides 1 to 9 were removed), and NSP1 nucleotides 1 to 134 were removed).
Identification of allele-specific amino acid changes and covariation analyses.
To identify allele-specific amino acid changes, alignments were constructed for each of the 12 RV proteins in Geneious Pro v5.5.2 using the ClustalW plug-in with the BLOSUM cost matrix. Individual alignments were created for each viral protein using the sequences of all DC RVs (i.e., derived from both mixed and nonmixed fecal specimens) in all possible pairwise allele combinations. Amino acid changes that showed both complete intra-allelic conservation and complete interallelic variation were quantitated. For the covariation analyses, the amino acid sequences of the DC RV proteins from the nonmixed fecal specimens only were concatenated in the order VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5-NSP6 and aligned. The complete concatenated alignments were then trimmed down to protein “pairs” in all 66 possible combinations (i.e., VP7-VP4, VP7-VP6, VP7-VP1, etc.). Gaps and ambiguous amino acid residues were trimmed from the alignments as follows: VP7 (residues 321 to 324 were removed), VP4 (residue 775 was removed), VP6 (not trimmed), VP1 (residues 1074 to 1088 were removed), VP2 (residues 1 to 14, 37 to 43, and 879 to 890 were removed), VP3 (residues 1, 2, and 835 were removed), NSP1 (residues 1 to 43 were removed), NSP2 (not trimmed), NSP3 (residues 2 to 4 were removed), NSP4 (not trimmed), NSP5 (residue 17 was removed), and NSP6 (not trimmed). DC2920 was excluded due to ambiguous sites. Negative-control concatenated alignments were created by randomly shuffling the sequences of the second protein in all pairs (i.e., for all 66 alignments). The 132 concatenated alignments were exported from Geneious Pro v5.5.2 as individual NEXUS files and then analyzed for amino acid covariation using MISTIC (35). Mutual information scores were calculated for each amino acid position of the input alignment compared to all other amino acid positions in the alignment. The mutual information scores for the nonshuffled experimental alignments ranged from 120.61 (highest level of covariation) to −17.11 (lowest level of covariation). Only intermolecular connections with mutual information scores of >64.9 were quantitated (35). The mutual information scores of the intermolecular connections calculated for shuffled, negative-control alignments were <12.0. For VP4, the three-dimensional locations of intermolecularly covarying amino acid residues that correlated with codon dN/dS ratios of >1 were mapped onto the atomic structure (PDB no. 3IYU) using UCSF Chimera v1.8 (36, 37). The figures were prepared using Adobe Illustrator CS5 (Adobe Systems).
Nucleotide sequence accession numbers.
The Genbank accession numbers for strains sequenced in this study are KC580000 to KC580010, KC579559 to KC579569, KC579686 to KC579696, KC580292 to KC580302, KC579978 to KC579988, KC580363 to KC580373, KC580194 to KC580204, KC580092 to KC580102, KC579898 to KC579908, KC580022 to KC580032, KC579820 to KC579830, KC580440 to KC580450, KC580126 to KC580136, KC580260 to KC580270, KC579719 to KC579729, KC580374 to KC580384, KC579548 to KC579558, KC579526 to KC579536, KC579909 to KC579919, KC580183 to KC580193, KC579776 to KC579786, KC580562 to KC580572, KC580341 to KC580351, KC580618 to KC580628, KC579955 to KC579965, KC579592 to KC579602, KC579614 to KC579624, KC579581 to KC579591, KC579675 to KC579685, KC579798 to KC579808, KC580216 to KC580226, KC580418 to KC580428, KC579493 to KC579503, KC580521 to KC580531, KC579625 to KC579635, KC580429 to KC580439, KC580115 to KC580125, KC579730 to KC579740, KC580596 to KC580606, KC580249 to KC580259, KC580532 to KC580542, KC580498 to KC580508, KC580238 to KC580248, KC579765 to KC579775, KC580011 to KC580021, KC580081 to KC580091, KC580385 to KC580395, KC579570 to KC579580, KC580451 to KC580461, KC580227 to KC580237, KC579603 to KC579613, KC580396 to KC580406, KC579850 to KC579860, KC579741 to KC579751, KC580352 to KC580362, KC579470 to KC579480, KC579515 to KC579525, KC580629 to KC580639, KC580573 to KC580583, KC579809 to KC579819, KC579787 to KC579797, KC579697 to KC579707, KC579989 to KC579999, KC580205 to KC580215, KC579504 to KC579514, KC580607 to KC580617, KC580160 to KC580170, KC580303 to KC580313, KC580407 to KC580417, KC442909 to KC442919, KC579708 to KC579718, KC580048 to KC580061, KC580064 to KC580068, KC580271 to KC580284, KC580287 to KC580291, KC579861 to KC579874, KC580103 to KC580114, KC579940 to KC579954, KC580486 to KC580497, KC579886 to KC579897, KC579481 to KC579492, KC580474 to KC580485, KC580509 to KC580520, KC580171 to KC580182, KC580069 to KC580080, KC580584 to KC580595, KC579920 to KC579939, KC579636 to KC579656, KC580033 to KC580047, KC580314 to KC580329, KC580148 to KC580159, KC579657 to KC579674, KC580543 to KC580561, and KC579831 to KC579849.
RESULTS
Genome sequencing of DC RVs found in G1P[8]-positive fecal specimens.
During the years 1974 to 1991, fecal specimens were collected from infants and young children who were hospitalized with acute gastroenteritis at Children's Hospital National Medical Center in Washington, DC (18–21). Total RNA was extracted from RV-positive specimens and classified into preliminary G/P types using a microtiter plate hybridization-based PCR-ELISA (23) (Table 1). Of the 428 RNA samples that tested G1P[8] positive, 230 representatives were sent to JCVI for RT-PCR and next-generation sequencing (Table 2). Single nearly complete RV genome sequences were determined for 73 of the 230 RNA samples. These sequences showed little heterogeneity in nucleotide reads, suggesting that the fecal specimens each contained a single dominant strain (Table 2). However, 21 of the RNA samples yielded mixed sequences for one or more viral genes, indicating that the fecal specimens each contained at least two different RVs and that the child was probably coinfected (Table 2). Altogether, nucleotide sequences were deduced for the open reading frames (and in some cases for the 5′ and 3′ untranslated regions) for 1,124 RV genes (see Tables S1 and S2 in the supplemental material).
Each viral gene was genotyped based upon its nucleotide sequence using the RotaC Web server (http://rotac.regatools.be/) (references 27 and 11 and data not shown). Of the RVs in the 73 nonmixed specimens, 72 had the genotype level GC G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 and 1 (DC799) had the GC G3-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1. Of the RVs in the 21 mixed specimens, 19 exhibited the genotype level GC G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 while 2 (DC1127 and DC1230) showed the GC G4-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1. As such, all RVs sequenced in this study belong to genogroup 1; no genogroup 2 viruses or intergenogroup reassortants were found. When combined with the results of our previous studies, genome sequences now exist for 135 genogroup 1 strains from the Washington, DC, fecal-specimen collection (72 G1P[8], 52 G3P[8], and 11 G4P[8] RVs), and numerous additional individual gene sequences are available for genogroup 1 RVs from the 21 mixed specimens (25, 26).
Inter- and intragenotypic diversity of the DC RV VP7 genes.
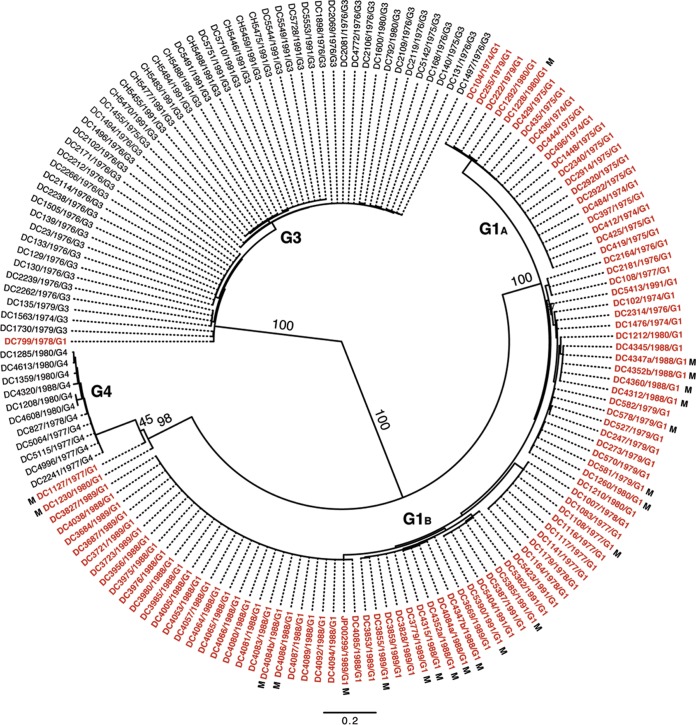
To determine the extent of inter- and intragenotypic diversity among the VP7 genes of the sequenced DC RVs, a maximum-likelihood phylogenetic tree was constructed (Fig. 1; see Table S3 in the supplemental material). The resultant radial dendrogram showed three major branches corresponding to VP7 genotypes G1, G3, and G4. Consistent with the results of RotaC, the VP7 gene of DC799 clustered in the phylogenetic tree with those of G3 strains, suggesting that this specimen was previously mistyped. Likewise, the VP7 genes of DC1230 and DC1127 clustered with those of G4 strains, thereby confirming the RotaC results. The DC RV G1 VP7 genes formed two distinct branches, which we designated G1A and G1B VP7 alleles. G1A VP7 alleles were detected only in DC RVs collected during the years 1974, 1975, 1979, and 1980, whereas G1B VP7 alleles were detected in DC RVs from nearly every collection year (i.e., 1974, 1976 to 1980, 1988 to 1989, and 1991). G1 DC RVs containing either of these allele types cocirculated in the Washington, DC, community during 1974, 1979, and 1980. The vast majority of mixed fecal specimens contained RVs with G1B VP7 alleles. For three of the mixed fecal specimens (DC4347, DC4352, and DC4084), we were able to resolve two separate G1B VP7 gene sequences (a and b).
FIG 1.
Inter- and intragenotypic diversity of the VP7 genes of the DC RVs. The maximum-likelihood phylogenetic tree was created using nucleotides 49 to 1020 of all 159 available DC RV VP7 gene sequences. The tree is midpoint rooted and shown in radial format. Bootstrap values are shown as percentages for key nodes, and horizontal branch lengths are drawn to scale (nucleotide substitutions per base). Four major groupings representing G1 (alleles G1A and G1B), G3, and G4 genotypes are labeled. Individual strain names are shown at the tips of the branches, along with their year of isolation and G type determined by PCR-ELISA. The VP7 gene sequences deduced in references 25 and 26 are shown in black, while those deduced in the current study are shown in red. VP7 gene sequences from mixed (M) fecal specimens are indicated.
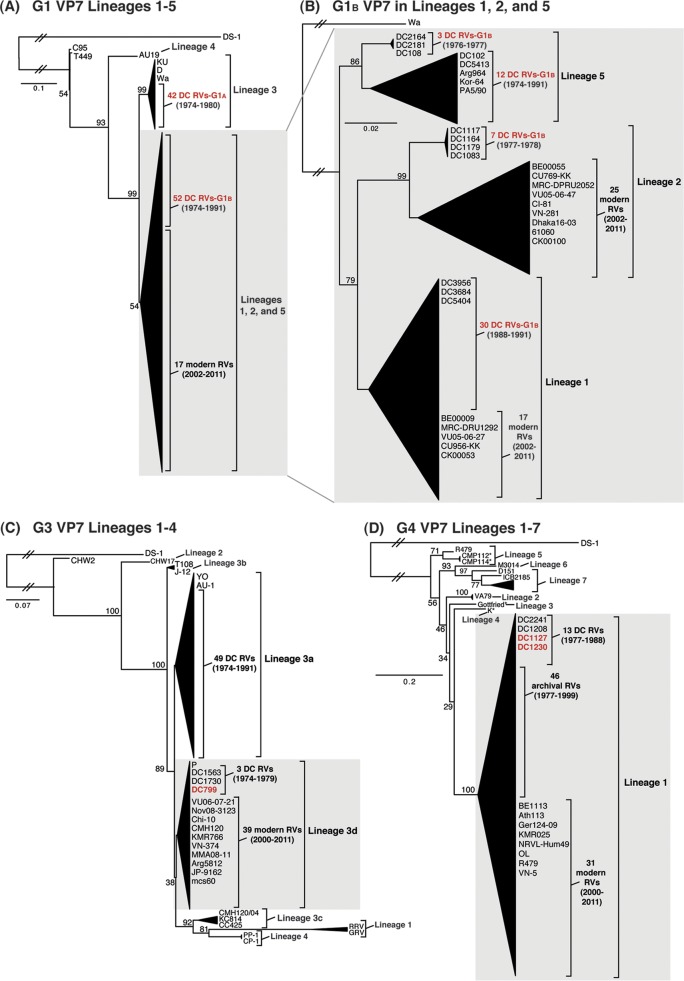
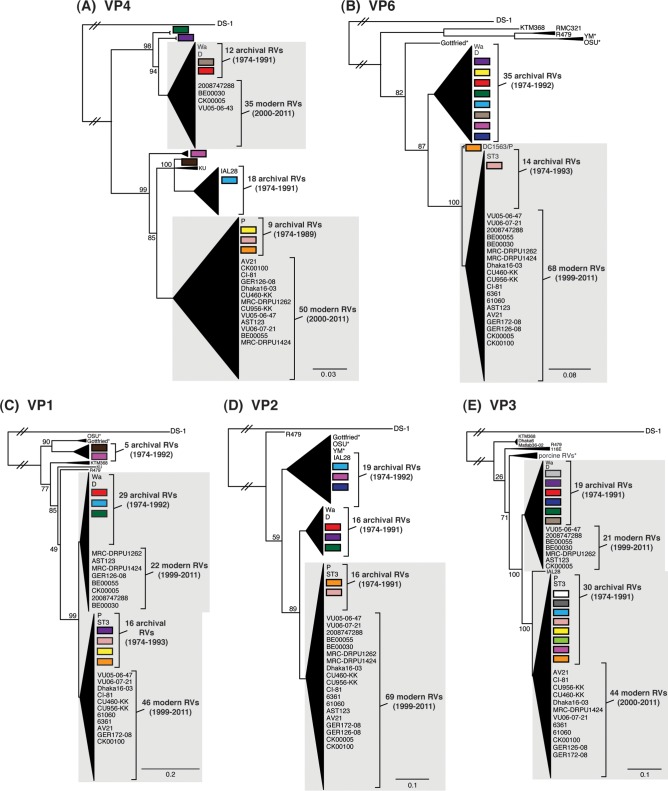
To elucidate the genetic relatedness of the archival DC RV VP7 genes to those of other strains, we constructed intragenotypic maximum-likelihood phylogenetic trees using sequences available in GenBank (Fig. 2; see Tables S4 to S6 in the supplemental material). The G1A VP7 alleles of the DC RVs clustered exclusively within lineage 3, along with the G1 VP7 genes of culture-adapted archival strains from the United States (Wa and D) and Japan (KU) (38–40). In contrast, the G1B VP7 alleles were found within phylogenetic lineages 1, 2, and 5, which were distinct from lineage 3 (Fig. 2A; see Table S4 in the supplemental material). Specifically, the G1B VP7 alleles of some of the DC RVs from 1974 to 1991 (e.g., DC2164, DC2181, DC108, DC102, and DC5413) fell into lineage 5 (Fig. 2B). These G1B VP7 alleles clustered with (or very near) the VP7 genes of wild-type archival G1P[8] strains that were isolated during the years 1982 to 1990 from Korea (Kor-64) and Italy (PA5/90), as well as with VP7 genes of culture-adapted strains from Argentina (Arg964 and A20) (41–43) (Fig. 2B; see Table S4 in the supplemental material). However, the G1B VP7 alleles of other DC RVs were found within lineages 1 and 2 and were closely related (>94.5% nucleotide identity) to the G1 VP7 genes of modern wild-type human RV strains isolated at various locations around the world, including the United States (VU05-06-47 and VU05-06-27), Belgium (BE00055 and BE00009), South Africa (MRC-DPRU2052 and MRC-DPRU1262), Thailand (CU769-KK and CU956-KK), South Korea (CI-81), Vietnam (VN-281), Bangladesh (Dhaka16-03), India (61060), and Australia (CK00100 and CK00053) (17, 44–48) (Fig. 2B; see Table S4 in the supplemental material).
FIG 2.
Comparison of the archival DC RV VP7 genes to those of other strains. Maximum-likelihood intragenotypic phylogenetic trees were constructed using the VP7 gene nucleotide sequences. All horizontal branch lengths are drawn to scale (nucleotide substitutions per base), bootstrap values are shown as percentages for key nodes, and lineages/sublineages are labeled. The relative locations of archival and modern RV genes in each tree are indicated, and select RV strains are listed by name. Strains sequenced in this study are shown in red. (A and B) G1 VP7 gene trees were created using nucleotides 73 to 970. (C) G3 VP7 gene tree created using nucleotides 1 to 988. (D) G4 VP7 gene tree created using nucleotides 61 to 807.
For 49 of the 51 previously sequenced G3P[8] DC RVs, the VP7 genes clustered within sublineage 3a and were similar to those of culture-adapted archival strains from Japan (YO and AU-1) and distinct from those of contemporary human strains (39, 46, 49, 50) (Fig. 2C; see Table S5 in the supplemental material). However, two of the previously sequenced G3P[8] DC RVs from 1974 and 1979 (DC1563 and DC1730, respectively) and one newly sequenced G3P[8] DC RV from 1979 (DC799) had VP7 genes that clustered along with modern G3 VP7 genes within lineage 3d of the phylogenetic tree (Fig. 2C; see Table S5 in the supplemental material). In fact, the VP7 genes of these three DC RVs (DC1563, DC1730, and DC799) had >97.3% nucleotide sequence identity to the G3 VP7 genes of wild-type human RVs isolated over the last 10 years from numerous locations, such as the United States (VU06-07-21), Russia (Nov08-3123), China (Chi-10), Thailand (CMH120), South Korea (KMR766), Vietnam (VN-374), Myanmar (MMA08-11), Argentina (Arg5812), Japan (JP-9162), and India (mcs60) (46, 51–56) (Fig. 2C; see Table S5 in the supplemental material).
The 11 previously sequenced G4P[8] DC RVs had VP7 genes that clustered within lineage 1, which is comprised of VP7 genes from nearly all archival and modern human G4 strains (25) (Fig. 2D; see Table S6 in the supplemental material). The two G4 VP7 genes sequenced from mixed fecal specimens in this study (DC1127 and DC1230) were found within lineage 1 (Fig. 2D). As such, the DC RV G4 VP7 genes are related (>88.7% nucleotide identity) to those of recently isolated wild-type human strains from locations including Belgium (BE1113), Greece (Ath113), Germany (GER124-09), South Korea (KMR025), Ireland (NRVL-Hum49), Nicaragua (OL), China (R479), and Vietnam (VN-5) (53, 57–61) (Fig. 2D; see Table S6 in the supplemental material). Together, these results suggest that some of the DC RV G1, G3, and G4 VP7 genes are likely ancestors of the genes of contemporary human strains and that other DC RV genes have since gone extinct in the virus population.
Intragenotypic diversity of the DC RV VP1-VP4, VP6, and NSP1-NSP5/6 genes.
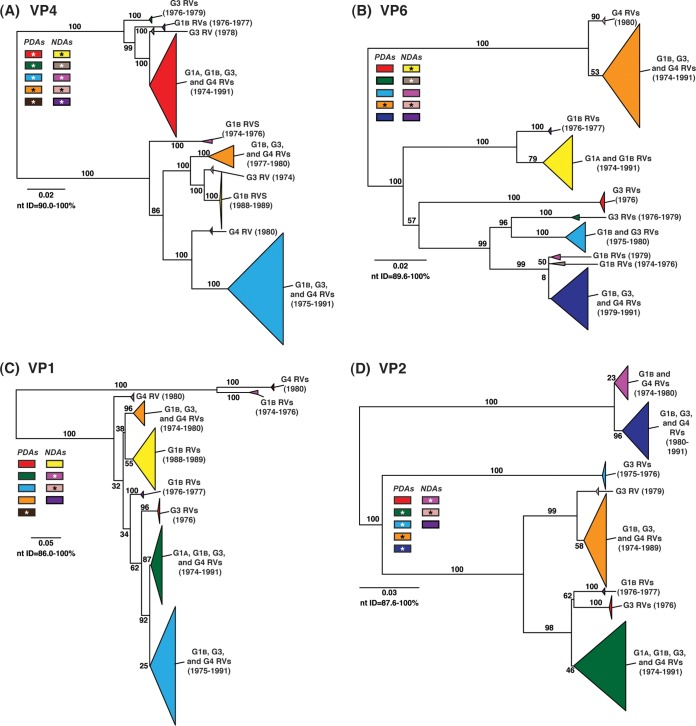
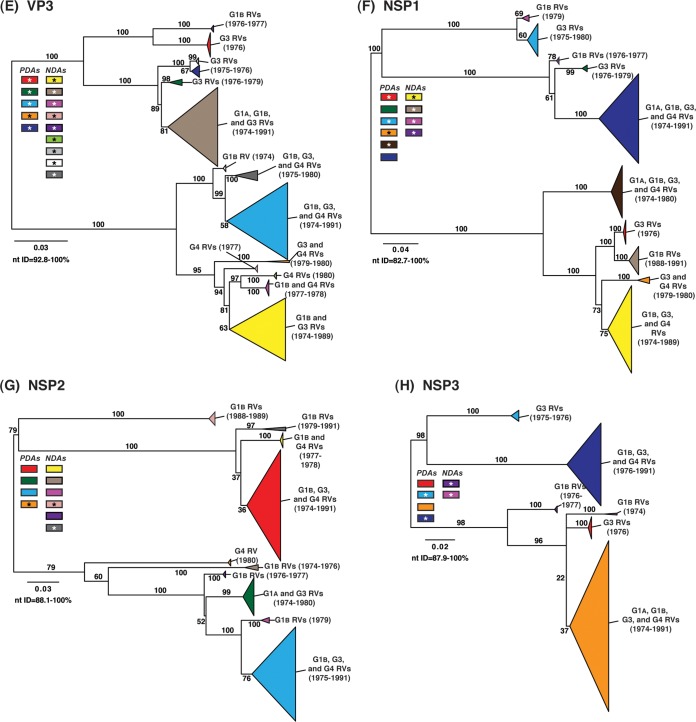
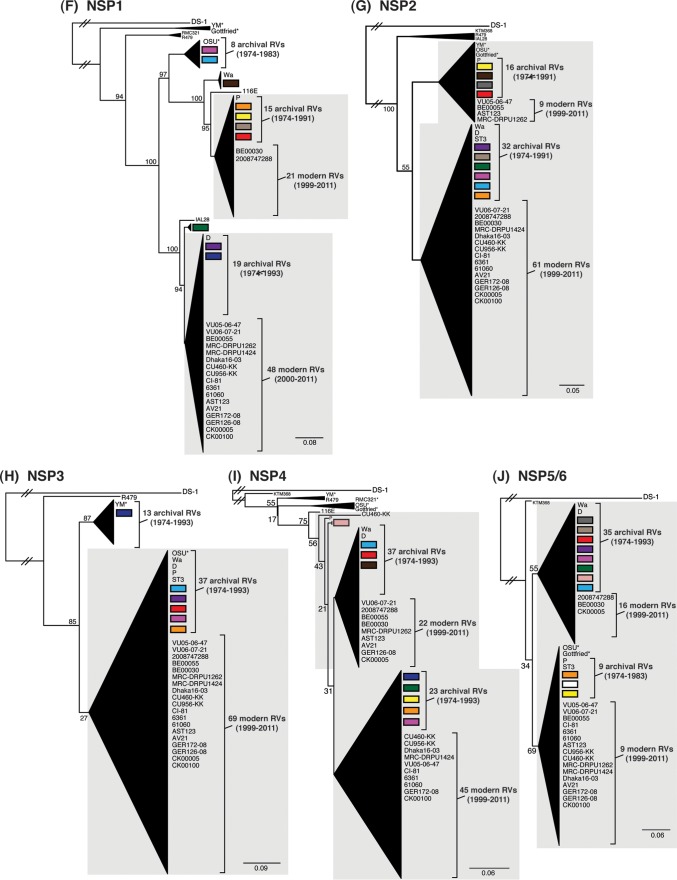
To determine the level of intragenotypic diversity among the VP1 to VP4, VP6, and NSP1 to NSP5/6 genes of the >135 sequenced DC RVs, maximum-likelihood phylogenetic trees were constructed (Fig. 3A to J; see Table S3 in the supplemental material). Based upon the clustering of sequences in the phylogenetic trees, 6 to 14 allele groupings were defined for each viral gene. These groupings showed interallelic nucleotide identity ranges of 83 to 98% and are color coded in the figures for ease of interpretation and discussion. About half of the newly sequenced DC RV genes clustered into previously defined allele (PDA) groupings (red, green, cyan, orange, navy, and brown), which were originally determined based upon the phylogenetic analysis of genes from the 51 G3P[8] and 11 G4P[8] DC RVs (25, 26) (Fig. 3A to J). For example, the red, orange, and cyan allele groupings in Fig. 3A are comprised of VP4 genes from previously sequenced G3P[8] and G4P[8] viruses, as well as genes of the G1P[8] DC RVs sequenced in the current study. However, other newly sequenced DC RV genes did not cluster in the phylogenetic trees with the PDAs, and instead, they formed clear monophyletic groupings, which we designated newly defined alleles (NDAs) and color yellow, beige, pink, pale pink, purple, light green, light gray, white, or dark gray. For instance, the yellow, beige, pink, and purple allele groupings shown in Fig. 3B were exclusively comprised of VP6 genes from the G1P[8] DC RVs sequenced here; the VP6 genes from the G3P[8] or G4P[8] DC RVs were not found within these groupings. A few genes from the previously sequenced G3P[8] and G4P[8] DC RVs were redefined as a result of the new analyses. Many of the PDA and NDA groupings were supported by strong bootstrap values (>70%) in the phylogenetic trees made with all available DC RV sequences (Fig. 3, asterisks). Still, those groupings that showed weak bootstrap values in the trees generated with all 135 to 167 gene sequences showed strong bootstrap values in subtrees that were outgrouped to the nearest ancestor, supporting their genetic divergence (data not shown).
FIG 3.
Intragenotypic diversity of the DC RV VP1 to VP4, VP6, and NSP1 to NSP5/6 genes. Maximum-likelihood intragenotypic phylogenetic trees were constructed for each gene and are midpoint rooted. The following nucleotides were used for each gene: VP4 gene, nucleotides 10 to 2337 (A); VP6 gene, nucleotides 30 to 1217 (B); VP1 gene, nucleotides 20 to 3242 (C); VP2 gene, nucleotides 139 to 2650 (D); VP3 gene, nucleotides 57 to 2556 (E); NSP1 gene, nucleotides 162 to 1491 (F); NSP2 gene, nucleotides 49 to 1000 (G); NSP3 gene, nucleotides 35 to 967 (H); NSP4 gene, nucleotides 42 to 569 (I); and NSP5/6 gene, nucleotides 22 to 615 (J). Horizontal branch lengths are drawn to scale (nucleotide substitutions per base), and bootstrap values are shown as percentages for key nodes. Monophyletic lineages representing allele groupings were collapsed (triangles) and colored coded. PDAs from McDonald et al. (25, 26) are colored red, green, cyan, orange, navy, or brown. NDAs are colored yellow, tan, dark pink, pale pink, purple, light green, light gray, white, or dark gray. Alleles defined for each gene are summarized as rectangles, with those having bootstrap support of >70% are indicated with asterisks. The G genotype and years of circulation for RVs with that particular allele are noted.
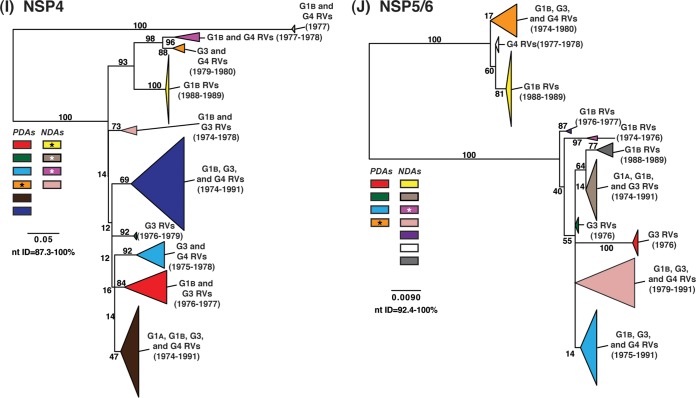
To elucidate the relatedness of the DC RV VP1 to VP4, VP6, and NSP1 to NSP5/6 genes to those of other archival and contemporary strains, we constructed maximum-likelihood phylogenetic trees using sequences available in GenBank (Fig. 4; see Table S7 in the supplemental material). The sequences chosen for this analysis included those of wild-type and culture-adapted human and animal RVs that have either genotype P[8] VP4 genes and/or genotype 1 internal protein genes (see Table S7 in the supplemental material). The results showed that some of the DC RV alleles were phylogenetically distinct from the genes of modern wild-type human strains, whereas other alleles clustered very close to them (Fig. 4; see Table S7 in the supplemental material). For example, most of the VP2 and VP6 alleles from the archival DC RVs were located in lineages that were separate from those containing genes of human RVs isolated more recently (Fig. 4B and D). However, VP2 and VP6 DC RV alleles, designated by the colors orange and pale pink, exhibited close genetic relationships (>95.5% nucleotide identity) with the genes of contemporary human strains isolated from locations such as the United States (VU05-06-47, VU06-07-21, and 2008747288), Belgium (BE00055 and BE00030), South Africa (MRC-DPRU1262), Cameroon (MRC-DPRU1424), Bangladesh (Dhaka16-03), Thailand (CU460-KK and CU956-KK), South Korea (CI-81), India (6361 and 61060), Italy (AST123 and AV21), Germany (GER172-08 and GER126-08), and Australia (CK00005 and CK00100) (17, 44–47, 62–64) (Fig. 4B and D; see Table S7 in the supplemental material). In addition, the DC RV alleles for many other genes were also very similar to the genes of the aforementioned contemporary human RVs: VP4 (beige, red, yellow, pale pink, and orange), VP1 (red, cyan, green, purple, pale pink, yellow, and orange), VP3 (all colors), NSP1 (orange, yellow, beige, red, purple, and navy), NSP2 (all colors), NSP3 (cyan, purple, red, pink, and orange), NSP4 (all colors), and NSP5/6 (all colors) (Fig. 4A, C, and E to J; see Table S7 in the supplemental material).
FIG 4.
Comparison of the VP1 to VP4, VP6, and NSP1 to NSP5/6 genes from the DC RVs to those of other strains. Maximum-likelihood intragenotypic phylogenetic trees were constructed for each gene and are outgroup rooted to the genes of strain DS-1. The following nucleotides were used for each gene: VP4 gene, nucleotides 10 to 2337 (A); VP6 gene, nucleotides 24 to 1217 (B); VP1 gene, nucleotides 19 to 3267 (C); VP2 gene, nucleotides 155 to 2701 (D); VP3 gene, nucleotides 50 to 2557 (E); NSP1 gene, nucleotides 32 to 1492 (F); NSP2 gene, nucleotides 47 to 1000 (G); NSP3 gene, nucleotides 35 to 967 (H); NSP4 gene, nucleotides 42 to 569 (I); and NSP5/6 gene, nucleotides 21 to 614 (J). Horizontal branch lengths are drawn to scale (nucleotide substitutions per base), bootstrap values are shown as percentages for key nodes, and monophyletic groupings were collapsed (triangles). The relative locations of the subgenotype alleles defined for the DC RVs in the trees are shown as colored rectangles. The relative locations of archival and modern RV genes in each tree are indicated, and select RV strains are listed. The asterisks indicate animal RV strains.
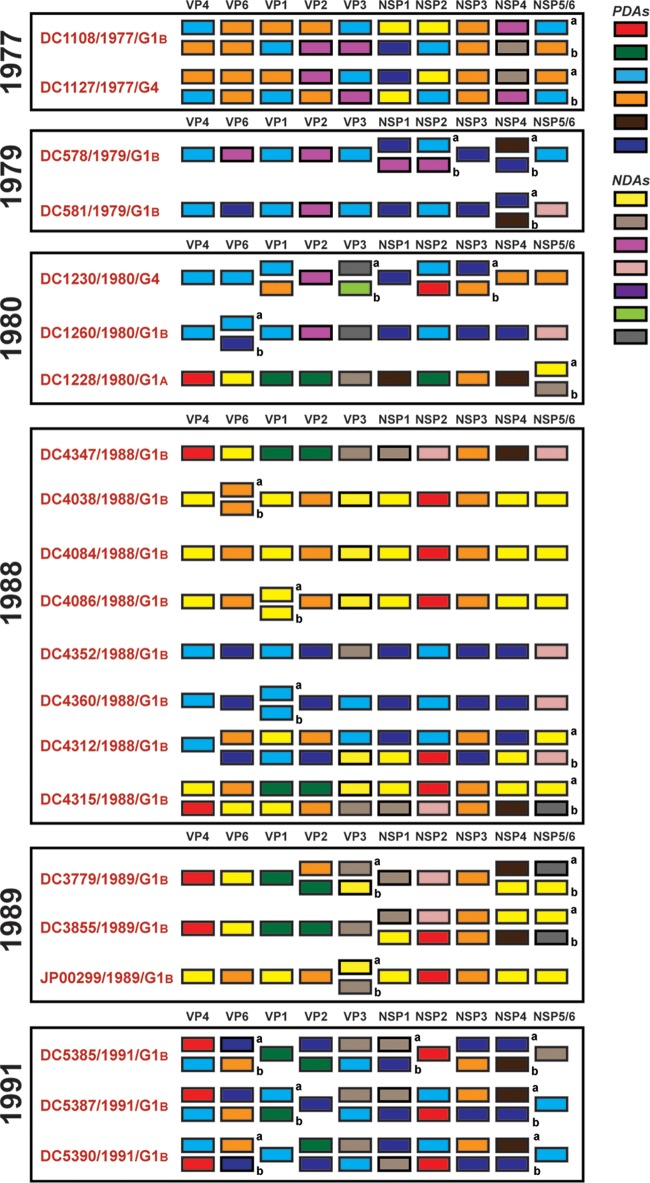
Persistent and transient allele level GCs of genogroup 1 DC RVs.
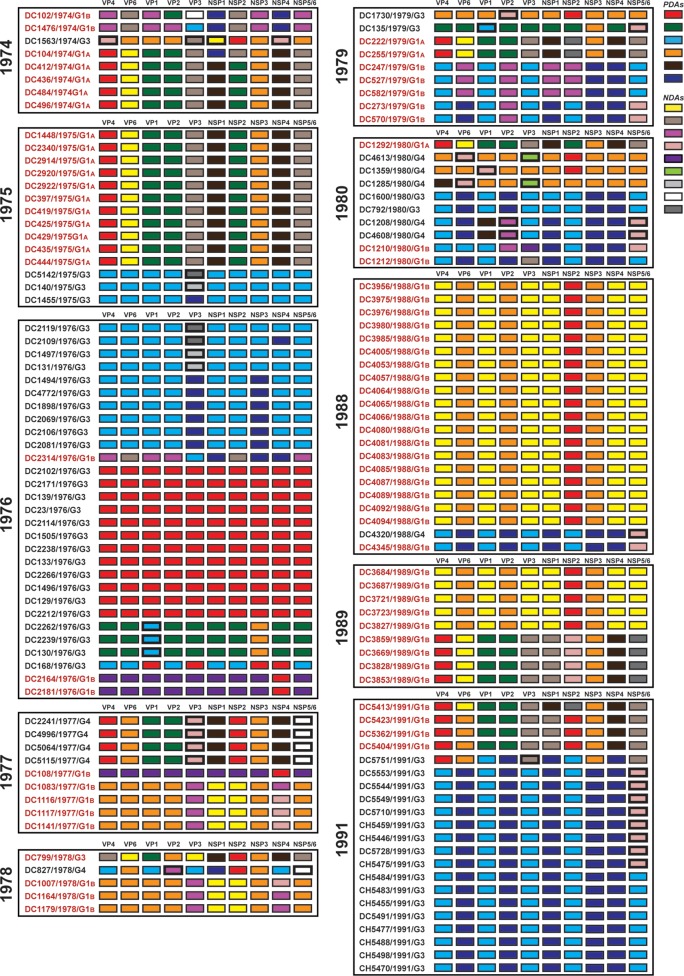
To further investigate the relationships among the archival DC RVs and to identify putative gene reassortment events, the color-coded alleles of each isolate were plotted horizontally, while the isolates were listed vertically by year of collection (Fig. 5 and 6). For the RVs in the mixed fecal specimens, it was not possible to determine which allele belonged to which coinfecting genogroup 1 strain (Fig. 6). However, for RVs in the nonmixed fecal specimens, this approach revealed their allele level GCs (Fig. 5). The results showed that genogroup 1 DC RVs isolated in any given season could have very different GCs when analyzed at the allele level. For instance, only 4 of the 7 G1P[8] viruses sequenced from 1974 (DC412, DC436, DC484, and DC496) showed identical allele level GCs. The isolate DC104 showed an allele level GC similar to those of DC412, DC436, DC484, and DC496, but it had NSP1 and NSP2 gene alleles (navy and purple, respectively) that were homologous to those of DC102 and DC1476. As such, DC104 is a putative intragenogroup reassortant strain that had a mostly DC412-like genetic background. Similar trends can be seen for the other collection years, where 2 to 9 different allele level GCs could be found and many putative gene reassortment events could be inferred.
FIG 5.
Allele level GCs of DC RVs from nonmixed specimens viewed by year of collection. The schematic illustrates the color coding of each gene for each sequenced DC RV strain based on the phylogenies shown in Fig. 1 and 3. The protein encoded by each viral gene (excluding VP7) is shown at the top. The strain name, its year of isolation, and its G genotype are shown on the left of the corresponding genome. PDAs from McDonald et al. (25, 26) are represented by rectangles colored red, green, cyan, orange, navy, or brown. NDAs are represented by rectangles colored yellow, tan, dark pink, pale pink, purple, light green, light gray, white, or dark gray. Allele designations of G3P[8] and G4P[8] DC RVs that were redefined based on the results of this study are outlined in thick lines. Strains circulating in the same year are boxed.
FIG 6.
Allele level GCs of DC RVs from mixed specimens viewed by year of collection. The schematic illustrates the color coding of each gene for each sequenced DC RV strain based on the phylogenies shown in Fig. 1 and 3. The protein encoded by each viral gene (excluding VP7) is shown at the top. The strain name, its year of isolation, and its G genotype are shown on the left of the corresponding genome. PDAs from McDonald et al. (25, 26) are represented by rectangles colored red, green, cyan, orange, navy, or brown. NDAs are represented by rectangles colored yellow, tan, dark pink, pale pink, purple, light green, gray, white, or dark gray. Strains circulating in the same year are boxed. Lowercase letters (a and b) indicate the two alleles found for homologous genes. DC4347 and DC4352 had two different VP7 gene sequences (Fig. 1).
Despite the observation that gene reassortment occurred among DC RVs, we found that viruses with some of the allele level GCs persisted in the community (Fig. 7). For example, the GC represented by the 1974 strain DC412 was indistinguishable at the allele level from that of DC1292, isolated 7 years later in 1980. Moreover, the 1979 strains DC222 and DC255 had the same allele level GC as DC5413, which was found 13 years later in 1991. Viruses with other allele level GCs seemed more transient, being found in only one collection year. However, the viruses with these transient GCs generally shared at least one allele designation with those having persistent GCs. For instance, strains from 1977 (e.g., DC2241), 1989 (e.g., DC3859), and 1991 (e.g., DC5423) had the same alleles for their VP4, VP1, VP2, NSP3, and NSP4 genes. This result suggests that certain sets of genes stayed together even in the context of reassortant strains. These data are consistent with the hypothesis that still unknown restrictions existed to limit gene reassortment among some of the genogroup 1 DC RVs.
FIG 7.
Allele level GCs of DC RVs from nonmixed specimens ordered by similarities. The schematic illustrates the color coding of each gene for each sequenced DC RV strain based on the phylogenies shown in Fig. 1 and 3. The protein encoded by each viral gene (excluding VP7) is shown at the top. The strain name, its year of isolation, and its G genotype are shown on the left of the corresponding genome. PDAs from McDonald et al. (25, 26) are represented by rectangles colored red, green, cyan, orange, navy, or brown. NDAs are represented by rectangles colored yellow, tan, dark pink, pale pink, purple, light green, light gray, white, or dark gray. Allele designations of G3P[8] and G4P[8] DC RVs that were redefined based on the results of this study are outlined in thick lines. Strains with similar allele level gene constellations are boxed. Allele level GCs are separated by lines. The numbers of years for which viruses with persistent allele level GCs were found are shown.
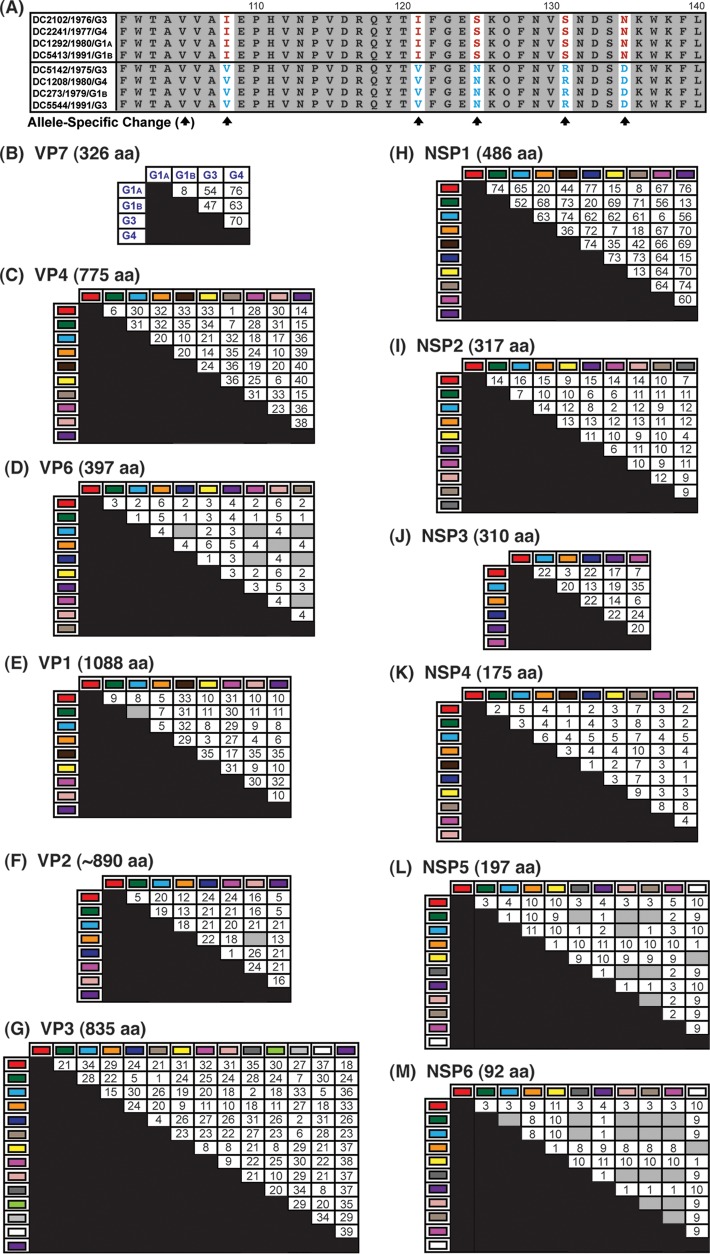
Allele-specific amino acid changes in DC RV proteins are involved in extensive covariance networks.
To determine whether the polypeptides encoded by the alleles had specific changes, amino acid sequence alignments were created for each of the 12 viral proteins (VP1 to VP4, VP6, VP7, and NSP1 to NSP6). Alignments of homologous proteins were compared to each other in pairwise allele combinations (i.e., red VP4 versus cyan VP4) (Fig. 8A). In this manner, we were able to quantitate amino acids that showed both complete intra-allelic conservation and complete interallelic variation. The results showed that the DC RV alleles generally encoded different proteins with numerous allele-specific amino acid changes (Fig. 8B to M). Only a few alleles for 3 of the DC RV genes were found to encode identical polypeptides: VP6 (cyan versus navy versus pink versus beige), VP2 (orange versus pale pink), and NSP5 (green versus gray versus pale pink versus beige; cyan versus pale pink; yellow versus white) and NSP6 (green versus gray versus pale pink versus beige versus pink versus cyan; orange versus white) (Fig. 8D, F, L, and M, gray boxes). On average, VP6, NSP4, NSP5, and NSP6 exhibited the fewest allele-specific amino acid changes, while NSP1, VP3, and VP4 showed the highest number of changes (Fig. 8B, C, F, G, K, and L). When the length of the protein is taken into account (i.e., the number of allele-specific changes divided by the total number of amino acids), VP6, VP1, and VP2 could be considered the most conserved proteins and NSP1, NSP3, and NSP6 the most variable.
FIG 8.
Allele-specific amino acid changes in DC RV proteins. (A) Schematic showing red versus cyan VP4 (residues 101 to 140) as an example of the strategy used to identify allele-specific changes (arrows). (B to M) Positions with both complete intra-allelic amino acid conservation and complete interallelic amino acid variations were quantitated and are shown in the matrices. The total length of each protein in amino acids (aa) is shown above the corresponding matrix. For VP7, the G1A and G1B alleles were also compared to genotype G3 and G4 proteins. The gray boxes indicate that no allele-specific changes were identified.
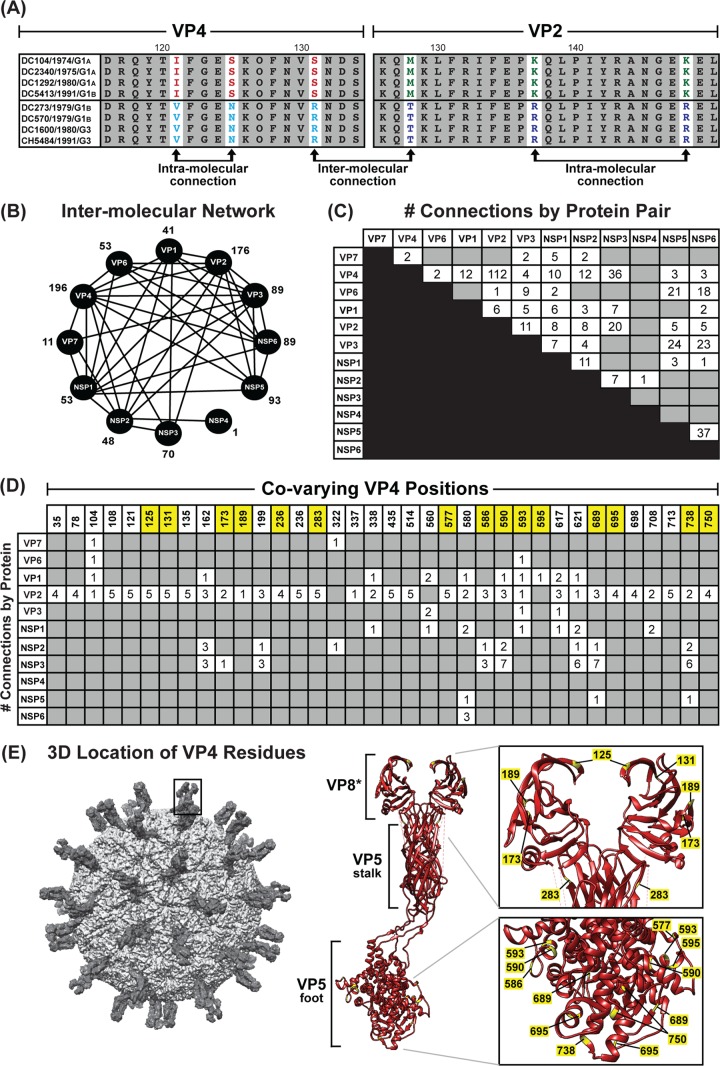
We next wondered whether the allele-specific amino acid changes detected in one viral protein correlated with changes in a different viral protein, as such intermolecular covariation is indicative of protein coadaptation (6, 7). To test this idea, a mutual information-based approach was used to quantitate the levels of interdependence between all possible positions of concatenated amino acid sequence alignments (Fig. 9A). The results of this analysis revealed an extensive genome-wide covariance network among the 12 proteins of the DC RVs (Fig. 9B). Each viral protein showed intermolecular connections with at least one other viral protein; usually, the amino acids of several proteins covaried together. The highest number of intermolecularly covarying amino acids was found for VP4 (Fig. 9B and C and Table 3). More specifically, 36 positions of VP4 covaried with amino acid positions of VP1, VP2, VP3, VP6, VP7, NSP1, NSP2, NSP3, NSP5, and/or NSP6 (Fig. 9B and C). Of the 195 total VP4 intermolecular connections, 112 were with VP2 (Fig. 9C and D).
FIG 9.
Amino acid covariation among DC RV proteins. (A) Schematic showing an example of correlated amino acid changes within (intramolecular connections) and between (intermolecular connection) two different viral proteins (VP4 residues 116 to 131 versus VP2 residues 126 to 150). (B) Intermolecular covariation network. Each viral protein is represented by a black circle. Lines connecting the circles indicate that the two proteins showed intermolecularly covarying amino acid positions with mutual information scores of >64.9. The total number of intermolecular connections made by each protein is shown next to the circle. (C) Matrix showing the number of intermolecular connections for each protein pair with mutual information scores of >64.9. The gray boxes indicate that no connections were found. (D) Matrix showing the covarying amino acid positions of VP4 and the number of connections each position makes with another viral protein. VP4 positions highlighted in yellow have codons with dN/dS ratios of >1. (E) The RV virion is shown on the far left, with a VP4 spike outlined. The atomic structure of VP4 is shown in ribbon representation (PDB number 3IYU) in the middle, with VP8* and VP5 stalk/foot subdomains labeled. Residues in yellow are those from panel C. A magnified view of the VP8* and VP5 foot subdomains is shown on the right, with the positions of amino acids labeled.
To investigate whether the identified sites of amino acid covariation could be a reflection of selection acting on the gene (i.e., the RNA molecule) at the nucleotide level, we analyzed the ratio of nonsynonymous-to-synonymous substitution rates (dN/dS) for each codon of the DC RV genomes (Table 3). We found that many, but not all, of the covarying amino acid positions were encoded by codons with dN/dS ratios of >1, suggesting that noncoding mutations are tolerated in the codon and that selection likely acts on the protein rather than the RNA. For VP4, 15 of the 36 covarying amino acid positions were associated with codons that showed dN/dS ratios of >1 (Fig. 9D and Table 3). The three-dimensional locations of these 15 VP4 amino acid positions were mapped onto the atomic structure of the trypsin-activated protein, which is comprised of a dimeric VP8* head, a dimeric VP5 stalk, and a trimeric VP5 foot (37). None of the covarying residues with codon dN/dS ratios of >1 are in the VP5 stalk region. However, residues 125, 131, 173, and 189 are located in the globular VP8* domain; residue 283 is located at the very tip of the VP5 stalk; and residues 577, 586, 590, 593, 595, 689, 695, 738, and 750 lie within the VP5 foot (Fig. 9E). Residue 236 is removed from VP4 following trypsin activation and is not able to be mapped. The significance of the covariation between these sites on VP4 and the amino acids of other viral proteins is currently unknown. Nevertheless, the results presented here suggest that the DC RV proteins may be coadapted, but not necessarily in a manner reflective of their physical interactions. The results further suggest that protein coadaptation may have influenced the maintenance of allele level GCs for the archival DC RVs.
TABLE 3.
Intermolecularly covarying amino acid positions for DC RV proteins and codons with nonsynonymous-to-synonymous substitution rate (dN/dS) ratios of >1
| Protein | Status | Amino acid/codon position(s)a |
|---|---|---|
| VP7 | Covarying | 29, 316 |
| dN/dS > 1 | 11, 16, 19, 22, 26, 37, 38, 41, 45, 47, 48, 66, 74, 75, 87, 90, 96, 97, 101, 105, 116, 119, 120, 123, 125, 126, 146, 147, 149, 159, 178, 179, 193, 211, 212, 213, 217, 220, 221, 227, 233, 237, 238, 239, 242, 267, 268, 278, 303, 309, 316 | |
| VP4 | Covarying | 35, 78, 104, 108, 121, 125, 131, 135, 162, 173, 189, 199, 236, 254, 283, 322, 337, 338, 435, 514, 560, 577, 580, 586, 590, 593, 595, 617, 621, 689, 695, 698, 708, 713, 738, 750 |
| dN/dS > 1 | 125, 131, 144, 145, 146, 173, 189, 236, 281, 283, 303, 311, 365, 577, 586, 590, 593, 594, 595, 689, 695, 738, 739, 750 | |
| VP6 | Covarying | 55, 80, 130, 199, 252 |
| dN/dS > 1 | 54, 55, 129 | |
| VP1 | Covarying | 95, 298, 430, 555, 648, 813, 973 |
| dN/dS > 1 | 120, 156, 171, 210, 298, 285, 440, 444, 554, 555, 779, 821, 823, 859, 865, 886, 888, 893, 999, 1049, 1050 | |
| VP2 | Covarying | 27, 28, 35, 39, 62, 66, 68, 127, 136, 159, 239, 406, 416, 570, 729 |
| dN/dS > 1 | 46, 66, 68, 107, 119, 127, 128, 136, 143, 113, 228, 239, 532, 559, 570, 653, 775 | |
| VP3 | Covarying | 89, 103, 203, 214, 217, 310, 324, 363, 490, 504, 639, 762, 812, 826 |
| dN/dS > 1 | 115, 139, 200, 277, 301, 328, 346, 356, 363, 373, 452, 454, 521, 568, 681, 683, 707, 733, 735, 748, 762, 812, 814 | |
| NSP1 | Covarying | 103, 139, 146, 160, 200, 214, 216, 225, 303, 326, 385, 391, 399, 401, 422, 434, 473, 477, 485 |
| dN/dS > 1 | 90, 103, 111, 112, 119, 121, 139, 146, 163, 165, 166, 180, 188, 189, 192, 200, 230, 233, 251, 254, 265, 271, 272, 289, 301, 307, 314, 326, 338, 357, 359, 386, 389, 402, 408, 434, 436, 474, 476, 477 | |
| NSP2 | Covarying | 48, 64, 75, 91, 105, 136, 150, 175, 187, 189, 197, 200, 201, 248, 293, 307 |
| dN/dS > 1 | 24, 36, 48, 64, 75, 82, 93, 108, 136, 197, 200, 202, 248, 279, 293 | |
| NSP3 | Covarying | 9, 65, 79, 141, 155, 191, 229, 233, 235, 252, 255, 268, 278, 300, 301, 308 |
| dN/dS > 1 | 9, 65, 76, 79, 104, 229, 233, 240, 255, 257, 278, 300, 308 | |
| NSP4 | Covarying | 72 |
| dN/dS > 1 | 70, 72, 135, 161, 169 | |
| NSP5 | Covarying | 37, 41, 43, 45, 108, 121, 126, 131, 142, 188 |
| dN/dS > 1 | 45, 131, 132 | |
| NSP6 | Covarying | 18, 21, 25, 26, 54, 67, 73, 75, 76, 82, 88 |
| dN/dS > 1 | 21 |
The numbers are based on those of strain Wa. Positions involved in the covariance network and that have a codon dN/dS ratio of >1 are highlighted in boldface.
DISCUSSION
RVs are important causes of pediatric gastroenteritis worldwide, and infections are particularly devastating for children living in geographical regions where there is limited access to medical care (65). Although two live attenuated RV vaccines are now in widespread use, their efficacy is limited in some developing countries, raising fears about whether they will protect the most vulnerable children from disease (66). Moreover, reports of vaccine-associated RV gastroenteritis and the observed capacity of vaccine strains to reassort genes with circulating pathogenic strains have elicited safety concerns (57, 67–74). As such, genetically stable RV vaccines that show broader efficacy and increased safety profiles may be needed. Comparative genomic studies, such as that described here, are expected to provide key information about RV evolution, thereby informing the design of future vaccines. The goal of the current study was to better understand human RV evolution by analyzing the genome sequences of strains found in archival fecal specimens that were collected over an 18-year time span at a single hospital in Washington, DC. Although we did not have access to clinical or demographic information for the associated patients, we predict that the RVs in the fecal specimens represent wild-type pathogenic strains, as they caused gastroenteritis severe enough that the children sought medical treatment (18–21). We have previously reported on the sequencing and analysis of 51 G3P[8] and 11 G4P[8] RVs from the same archival fecal collection (25, 26). Here, we sequenced >73 additional strains from this collection, including 72 human RVs with G1P[8] specificity. All of the G1, G3, and G4 RVs sequenced to date from this collection were found to have identical genotype level GCs of P[8]-I1-C1-R1-A1-N1-T1-E1-H1, consistent with genogroup 1 strains. However, by constructing intragenotypic phylogenetic trees for each gene, we found that some of these archival genogroup 1 viruses were very different at the allele level (Fig. 3 and 5). Novel gene alleles (i.e., NDAs) were found in the G1P[8] DC RV population; these alleles were absent in the G3P[8] and G4P[8] viruses (Fig. 3). This result suggests that the diversity of the DC RVs during the years 1974 to 1991 was far greater than what we had previously predicted. Moreover, by comparing these new gene alleles to the genes of other strains from around the world, we also discovered that many of them are strikingly similar (∼98% nucleotide sequence identity) to the genes of contemporary human strains (Fig. 2 and 4). These observations suggest that the alleles identified in the current study represent important, never-before-seen, ancestral precursors to modern-day strains. Ongoing studies in our laboratories are using Bayesian inference methodologies and these new gene sequences to calculate rates of nucleotide substitution, thereby providing genetic-drift estimates for this significant pediatric pathogen (75).
The results of this study corroborate and extend our previous findings regarding the existence of preferred allele level GCs in the DC RV population (25, 26). Specifically, we found that that the individual gene alleles of a given RV did not necessarily correlate with the year in which the specimen was isolated. Some of the DC RVs found in the same year (i.e., G1 viruses from 1974) had gene alleles with <83% nucleotide identity, while those found >12 years apart (i.e., G1 viruses from 1974 and 1991) showed alleles with >97% nucleotide identity (Fig. 3). Even more, we found that some of the DC RVs exhibited identical allele level GCs, despite being isolated in disparate years. Thus, these data provide additional evidence that (i) human RVs with distinct and stable GCs can cocirculate in a given community and (ii) gene reassortment among cocirculating strains is less frequent than what would be predicted based upon chance alone. However, the results of this study extend those previous findings by delving more deeply into the mechanistic basis for the observed GCs in the analyzed DC RV population. Because our previous sequencing studies relied on Sanger sequencing, we were not able to resolve the viruses in the mixed G3/G4P[8]-positive specimens (25, 26). Here, we employed next-generation techniques, which allowed us to determine the genome sequences of DC RVs in the G1P[8]-positive fecal specimens in the absence of primer-derived biases. Using this approach, we discovered that 21 of the 93 specimens for which genome sequences were deduced (∼23%) contained two or more genogroup 1 strains (Table 2). The allele designations of the individual genes for RVs in the mixed specimens were representative of those found in the nonmixed specimens (Fig. 5 and 6). This result suggests that some of the children were coinfected and that the genogroup 1 DC RVs had ample opportunities for genetic exchange.
While future laboratory-based mechanistic experiments will be required to untangle the factors contributing to GC maintenance, some clues about viral fitness and selection pressures may be gleaned from analyzing the nucleotide and amino acid sequences. In this study, we created pairwise amino acid sequence alignments for the DC RVs and found that the vast majority of identified allele groupings were supported by specific, albeit conservative, changes in the encoded viral proteins (Fig. 8). Even more, by concatenating the alignments of two or more viral proteins, we noticed that allele-specific changes in one viral protein often correlated with allele-specific changes in a different viral protein (Fig. 9). Such intermolecular amino acid covariation has been described for numerous viruses and is indicative of functional/physical protein coadaptation (7, 76–78). For the DC RV proteins, we quantitated the levels of interdependence between all possible positions of concatenated amino acid sequence alignments using a mutual information-based approach (35). The results showed an extensive covariance network involving amino acid positions in all 12 RV proteins (Fig. 9 and Table 3). Importantly, no connections were found in negative-control alignments, which were created by mismatching (i.e., shuffling) the protein sequences from different viral strains.
The covariation analysis revealed that the number of intermolecular connections was highest between the viral attachment protein, VP4, and the core shell protein, VP2, for the DC RVs. This result was unexpected, because these two structural proteins are not thought to directly bind to each other. It is possible that these proteins do in fact interact at some unknown point in the viral replication cycle. Another possibility is that these viral proteins do not interact but that they can independently, yet synergistically, affect viral fitness (6). For instance, the negative fitness consequences of a loss-of-function mutation in VP4 (e.g., one that decreases viral entry rates) might be counteracted by a simultaneous gain-of-function mutation in VP2 (e.g., one that increases viral assembly rates). Still another possibility is that the correlated intermolecular amino acid changes in VP4 and VP2 are simply a reflection of covarying changes in the RNA molecules themselves (79). For instance, RNA sequence or structural elements involving the codons of the VP4 and VP2 genes could interact during the assortment/packaging process (80). To investigate this idea, we calculated the coding and noncoding substitution rates for each codon of the DC RV genomes (Table 3). We found that many, but not all, of the amino acid positions involved in the identified covariance network showed higher levels of nonsynonymous changes than of synonymous changes in their codons (i.e., dN/dS ratios of >1). For these particular residues, it is expected that selection pressures are most likely acting on the viral proteins (32–34). Nonetheless, the importance of RNA sequence/structures in GC maintenance, even for these sites, cannot be excluded based on the existing data.
Altogether, the results of this study enhance our understanding of human RV genetic diversity and raise important questions about the dynamic interplay between mutation accumulation and gene reassortment during the evolution of this pediatric pathogen. We propose that covarying amino acid residues of human RVs represent molecular fitness determinants that influence GC maintenance and temper gene reassortment. In the future, these sites could be manipulated to create new live attenuated vaccine candidates that have decreased capacity to reassort with naturally circulating strains. Moreover, covarying amino acid positions in viral proteins might shed new light on potential protein-protein interaction interfaces, which could be specifically targeted by rationally designed antiviral drugs (6). As such, this comparative genomic work represents a first step in the continuum of research that could lead to new treatments for human RVs and possibly for other segmented RNA viral pathogens, as well.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge additional McDonald laboratory members (Crystal Boudreaux, Courie Cohen, Kris Ivey, Allison McKell, Joe Pechacek, and Priya Srihari) for helpful discussions and critical readings of the manuscript. We also thank Rebecca Halpin, Nadia Fedorova, Timothy Stockwell, and Susmita Shrivastava for help with genome sequencing, assembly, and annotation.
S.Z., T.A.T., P.W.M., and S.M.M. were supported by laboratory start-up funds from the Virginia Tech Carilion Research Institute. T.A.T. was also supported by funds from the Virginia Tech Carilion School of Medicine. J.T.P. and A.F.D. were supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. This project was also supported in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract no. HHSN272200900007C).
Footnotes
Published ahead of print 18 June 2014
This paper is dedicated to the memory of Albert Z. Kapikian, scientist, mentor, and friend.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01562-14.
REFERENCES
- 1.Estes MK, Kapikian AZ. 2007. Rotaviruses and their replication, p 1917–1974 In Fields BN, Knipe DM, Howley PM, Griffen DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 2.Gouvea V, Brantly M. 1995. Is rotavirus a population of reassortants? Trends Microbiol. 3:159–162. 10.1016/S0966-842X(00)88908-8 [DOI] [PubMed] [Google Scholar]
- 3.Lauring AS, Frydman J, Andino R. 2013. The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 11:327–336. 10.1038/nrmicro3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao L. 1997. Evolution of sex and the molecular clock in RNA viruses. Gene 205:301–308. 10.1016/S0378-1119(97)00405-8 [DOI] [PubMed] [Google Scholar]
- 5.Domingo E, Escarmis C, Sevilla N, Moya A, Elena SF, Quer J, Novella IS, Holland JJ. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859–864 [DOI] [PubMed] [Google Scholar]
- 6.Codoner FM, Fares MA. 2008. Why should we care about molecular coevolution? Evol. Bioinform. Online 4:29–38 [PMC free article] [PubMed] [Google Scholar]
- 7.Donlin MJ, Szeto B, Gohara DW, Aurora R, Tavis JE. 2012. Genome-wide networks of amino acid covariances are common among viruses. J. Virol. 86:3050–3063. 10.1128/JVI.06857-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patton JT. 2012. Rotavirus diversity and evolution in the post-vaccine world. Discov. Med. 13:85–97 [PMC free article] [PubMed] [Google Scholar]
- 9.Matthijnssens J, Van Ranst M. 2012. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr. Opin. Virol. 2:426–433. 10.1016/j.coviro.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 10.Nakagomi O, Nakagomi T, Akatani K, Ikegami N. 1989. Identification of rotavirus genogroups by RNA-RNA hybridization. Mol. Cell Probes 3:251–261. 10.1016/0890-8508(89)90006-6 [DOI] [PubMed] [Google Scholar]
- 11.Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 82:3204–3219. 10.1128/JVI.02257-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esona MD, Banyai K, Foytich K, Freeman M, Mijatovic-Rustempasic S, Hull J, Kerin T, Steele AD, Armah GE, Geyer A, Page N, Agbaya VA, Forbi JC, Aminu M, Gautam R, Seheri LM, Nyangao J, Glass R, Bowen MD, Gentsch JR. 2011. Genomic characterization of human rotavirus G10 strains from the African Rotavirus Network: relationship to animal rotaviruses. Infect. Genet. Evol. 11:237–241. 10.1016/j.meegid.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 13.Esona MD, Geyer A, Page N, Trabelsi A, Fodha I, Aminu M, Agbaya VA, Tsion B, Kerin TK, Armah GE, Steele AD, Glass RI, Gentsch JR. 2009. Genomic characterization of human rotavirus G8 strains from the African rotavirus network: relationship to animal rotaviruses. J. Med. Virol. 81:937–951. 10.1002/jmv.21468 [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S, Adachi N, Gatheru Z, Nyangao J, Yamamoto D, Ishino M, Urushibara N, Kobayashi N. 2011. Whole-genome analysis reveals the complex evolutionary dynamics of Kenyan G2P[4] human rotavirus strains. J. Gen. Virol. 92:2201–2208. 10.1099/vir.0.033001-0 [DOI] [PubMed] [Google Scholar]
- 15.Jere KC, Mlera L, O'Neill HG, Potgieter AC, Page NA, Seheri ML, van Dijk AA. 2011. Whole genome analyses of African G2, G8, G9, and G12 rotavirus strains using sequence-independent amplification and 454(R) pyrosequencing. J. Med. Virol. 83:2018–2042. 10.1002/jmv.22207 [DOI] [PubMed] [Google Scholar]
- 16.Medici MC, Abelli LA, Martella V, Martinelli M, Lorusso E, Buonavoglia C, Dettori G, Chezzi C. 2007. Characterization of inter-genogroup reassortant rotavirus strains detected in hospitalized children in Italy. J. Med. Virol. 79:1406–1412. 10.1002/jmv.20878 [DOI] [PubMed] [Google Scholar]
- 17.Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, Iturriza-Gomara M, Iftekharuddin N, Azim T, Van Ranst M. 2007. Evolutionary history and global spread of the emerging G12 human rotaviruses. J. Virol. 81:2382–2390. 10.1128/JVI.01622-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandt CD, Kim HW, Rodriguez WJ, Arrobio JO, Jeffries BC, Stallings EP, Lewis C, Miles AJ, Chanock RM, Kapikian AZ, Parrott RH. 1983. Pediatric viral gastroenteritis during eight years of study. J. Clin. Microbiol. 18:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandt CD, Kim HW, Rodriguez WJ, Thomas L, Yolken RH, Arrobio JO, Kapikian AZ, Parrott RH, Chanock RM. 1981. Comparison of direct electron microscopy, immune electron microscopy, and rotavirus enzyme-linked immunosorbent assay for detection of gastroenteritis viruses in children. J. Clin. Microbiol. 13:976–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandt CD, Kim HW, Yolken RH, Kapikian AZ, Arrobio JO, Rodriguez WJ, Wyatt RG, Chanock RM, Parrott RH. 1979. Comparative epidemiology of two rotavirus serotypes and other viral agents associated with pediatric gastroenteritis. Am. J. Epidemiol. 110:243–254 [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez WJ, Kim HW, Brandt CD, Bise B, Kapikian AZ, Chanock RM, Curlin G, Parrott RH. 1980. Rotavirus gastroenteritis in the Washington, DC, area: incidence of cases resulting in admission to the hospital. Am. J. Dis. Child. 134:777–779. 10.1001/archpedi.1980.02130200047015 [DOI] [PubMed] [Google Scholar]
- 22.Davidson GP, Bishop RF, Townley RR, Holmes IH. 1975. Importance of a new virus in acute sporadic enteritis in children. Lancet i:242–246 [DOI] [PubMed] [Google Scholar]
- 23.Santos N, Honma S, Timenetsky MDC, Linhares AC, Ushijima H, Armah GE, Gentsch JR, Hoshino Y. 2008. Development of a microtiter plate hybridization-based PCR-enzyme-linked immunosorbent assay for identification of clinically relevant human group A rotavirus G and P genotypes. J. Clin. Microbiol. 46:462–469. 10.1128/JCM.01361-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. 2008. Viral genome sequencing by random priming methods. BMC Genomics 9:5. 10.1186/1471-2164-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald SM, Davis K, McAllen JK, Spiro DJ, Patton JT. 2011. Intra-genotypic diversity of archival G4P[8] human rotaviruses from Washington, DC. Infect. Genet. Evol. 11:1586–1594. 10.1016/j.meegid.2011.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald SM, Matthijnssens J, McAllen JK, Hine E, Overton L, Wang S, Lemey P, Zeller M, Van Ranst M, Spiro DJ, Patton JT. 2009. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog. 5:e1000634. 10.1371/journal.ppat.1000634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maes P, Matthijnssens J, Rahman M, Van Ranst M. 2009. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 9:238. 10.1186/1471-2180-9-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53:793–808. 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Nei M. 2000. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol. Biol. Evol. 17:1251–1258. 10.1093/oxfordjournals.molbev.a026408 [DOI] [PubMed] [Google Scholar]
- 32.Korber BT, Farber RM, Wolpert DH, Lapedes AS. 1993. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc. Natl. Acad. Sci. U. S. A. 90:7176–7180. 10.1073/pnas.90.15.7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 34.Ota T, Nei M. 1994. Variance and covariances of the numbers of synonymous and nonsynonymous substitutions per site. Mol. Biol. Evol. 11:613–619 [DOI] [PubMed] [Google Scholar]
- 35.Simonetti FL, Teppa E, Chernomoretz A, Nielsen M, Marino Buslje C. 2013. MISTIC: Mutual Information Server to Infer Coevolution. Nucleic Acids Res. 41:W8–W14. 10.1093/nar/gkt427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 37.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. 2011. Atomic model of an infectious rotavirus particle. EMBO J. 30:408–416. 10.1038/emboj.2010.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heiman EM, McDonald SM, Barro M, Taraporewala ZF, Bar-Magen T, Patton JT. 2008. Group A human rotavirus genomics: evidence that gene constellations are influenced by viral protein interactions. J. Virol. 82:11106–11116. 10.1128/JVI.01402-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urasawa T, Urasawa S, Taniguchi K. 1981. Sequential passages of human rotavirus in MA-104 cells. Microbiol. Immunol. 25:1025–1035. 10.1111/j.1348-0421.1981.tb00109.x [DOI] [PubMed] [Google Scholar]
- 40.Wyatt RG, James HD, Jr, Pittman AL, Hoshino Y, Greenberg HB, Kalica AR, Flores J, Kapikian AZ. 1983. Direct isolation in cell culture of human rotaviruses and their characterization into four serotypes. J. Clin. Microbiol. 18:310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arista S, Giammanco GM, De Grazia S, Ramirez S, Lo Biundo C, Colomba C, Cascio A, Martella V. 2006. Heterogeneity and temporal dynamics of evolution of G1 human rotaviruses in a settled population. J. Virol. 80:10724–10733. 10.1128/JVI.00340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barril P, Martinez L, Giordano M, Masachessi G, Isa M, Pavan J, Glikmann G, Nates S. 2013. Genetic and antigenic evolution profiles of G1 rotaviruses in Cordoba, Argentina, during a 27-year period (1980–2006). J. Med. Virol. 85:363–369. 10.1002/jmv.23462 [DOI] [PubMed] [Google Scholar]
- 43.Jin Q, Ward RL, Knowlton DR, Gabbay YB, Linhares AC, Rappaport R, Woods PA, Glass RI, Gentsch JR. 1996. Divergence of VP7 genes of G1 rotaviruses isolated from infants vaccinated with reassortant rhesus rotaviruses. Arch. Virol. 141:2057–2076. 10.1007/BF01718215 [DOI] [PubMed] [Google Scholar]
- 44.Arora R, Chhabra P, Chitambar SD. 2009. Genetic diversity of genotype G1 rotaviruses co-circulating in western India. Virus Res. 146:36–40. 10.1016/j.virusres.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 45.Cho MK, Jheong WH, Lee SG, Park CJ, Jung KH, Paik SY. 2013. Full genomic analysis of a human rotavirus G1P[8] strain isolated in South Korea. J. Med. Virol. 85:157–170. 10.1002/jmv.23366 [DOI] [PubMed] [Google Scholar]
- 46.McDonald SM, McKell AO, Rippinger CM, McAllen JK, Akopov A, Kirkness EF, Payne DC, Edwards KM, Chappell JD, Patton JT. 2012. Diversity and relationships of cocirculating modern human rotaviruses revealed using large-scale comparative genomics. J. Virol. 86:9148–9162. 10.1128/JVI.01105-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theamboonlers A, Maiklang O, Thongmee T, Chieochansin T, Vuthitanachot V, Poovorawan Y. 2014. Complete genotype constellation of human rotavirus group A circulating in Thailand, 2008–2011. Infect. Genet. Evol. 21:295–302. 10.1016/j.meegid.2013.11.020 [DOI] [PubMed] [Google Scholar]
- 48.Trinh QD, Nguyen TA, Phan TG, Khamrin P, Yan H, Hoang PL, Maneekarn N, Li Y, Yagyu F, Okitsu S, Ushijima H. 2007. Sequence analysis of the VP7 gene of human rotavirus G1 isolated in Japan, China, Thailand, and Vietnam in the context of changing distribution of rotavirus G-types. J. Med. Virol. 79:1009–1016. 10.1002/jmv.20920 [DOI] [PubMed] [Google Scholar]
- 49.Matthijnssens J, Joelsson DB, Warakomski DJ, Zhou T, Mathis PK, van Maanen MH, Ranheim TS, Ciarlet M. 2010. Molecular and biological characterization of the 5 human-bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine, RotaTeq. Virology 403:111–127. 10.1016/j.virol.2010.04.004 [DOI] [PubMed] [Google Scholar]
- 50.Nakagomi O, Nakagomi T, Hoshino Y, Flores J, Kapikian AZ. 1987. Genetic analysis of a human rotavirus that belongs to subgroup I but has an RNA pattern typical of subgroup II human rotaviruses. J. Clin. Microbiol. 25:1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan-it W, Thongprachum A, Dey SK, Phan TG, Khamrin P, Okitsu S, Nishimura S, Kobayashi M, Kikuta H, Baba T, Yamamoto A, Sugita K, Hashira S, Tajima T, Ishida S, Mizuguchi M, Ushijima H. 2011. Detection and genetic characterization of rotavirus infections in non-hospitalized children with acute gastroenteritis in Japan, 2007–2009. Infect. Genet. Evol. 11:415–422. 10.1016/j.meegid.2010.11.018 [DOI] [PubMed] [Google Scholar]
- 52.Khamrin P, Maneekarn N, Peerakome S, Tonusin S, Phan TG, Okitsu S, Ushijima H. 2007. Molecular characterization of rare G3P[9] rotavirus strains isolated from children hospitalized with acute gastroenteritis. J. Med. Virol. 79:843–851. 10.1002/jmv.20840 [DOI] [PubMed] [Google Scholar]
- 53.Min BS, Noh YJ, Shin JH, Baek SY, Kim JO, Min KI, Ryu SR, Kim BG, Kim DK, Lee SH, Min HK, Ahn BY, Park SN. 2004. Surveillance study (2000 to 2001) of G- and P-type human rotaviruses circulating in South Korea. J. Clin. Microbiol. 42:4297–4299. 10.1128/JCM.42.9.4297-4299.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukherjee A, Mullick S, Kobayashi N, Chawla-Sarkar M. 2012. The first identification of rare human group A rotavirus strain G3P[10] with severe infantile diarrhea in eastern India. Infect. Genet. Evol. 12:1933–1937. 10.1016/j.meegid.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 55.Stupka JA, Carvalho P, Amarilla AA, Massana M, Parra GI. 2009. National rotavirus surveillance in Argentina: high incidence of G9P[8] strains and detection of G4P[6] strains with porcine characteristics. Infect. Genet. Evol. 9:1225–1231. 10.1016/j.meegid.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 56.Wang YH, Kobayashi N, Zhou X, Nagashima S, Zhu ZR, Peng JS, Liu MQ, Hu Q, Zhou DJ, Watanabe S, Ishino M. 2009. Phylogenetic analysis of rotaviruses with predominant G3 and emerging G9 genotypes from adults and children in Wuhan, China. J. Med. Virol. 81:382–389. 10.1002/jmv.21387 [DOI] [PubMed] [Google Scholar]
- 57.Bucardo F, Rippinger CM, Svensson L, Patton JT. 2012. Vaccine-derived NSP2 segment in rotaviruses from vaccinated children with gastroenteritis in Nicaragua. Infect. Genet. Evol. 12:1282–1294. 10.1016/j.meegid.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pietsch C, Schuster V, Liebert UG. 2011. A hospital based study on inter- and intragenotypic diversity of human rotavirus A VP4 and VP7 gene segments, Germany. J. Clin. Virol. 50:136–141. 10.1016/j.jcv.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 59.Trinh QD, Pham NT, Nguyen TA, Phan TG, Yan H, Hoang Le P, Khamrin P, Maneekarn N, Li Y, Okitsu S, Mizuguchi M, Ushijima H. 2010. Sequence analysis of the VP7 gene of human rotaviruses G2 and G4 isolated in Japan, China, Thailand, and Vietnam during 2001–2003. J. Med. Virol. 82:878–885. 10.1002/jmv.21630 [DOI] [PubMed] [Google Scholar]
- 60.Wang YH, Kobayashi N, Zhou DJ, Yang ZQ, Zhou X, Peng JS, Zhu ZR, Zhao DF, Liu MQ, Gong J. 2007. Molecular epidemiologic analysis of group A rotaviruses in adults and children with diarrhea in Wuhan city, China, 2000–2006. Arch. Virol. 152:669–685. 10.1007/s00705-006-0904-y [DOI] [PubMed] [Google Scholar]
- 61.Zeller M, Patton JT, Heylen E, De Coster S, Ciarlet M, Van Ranst M, Matthijnssens J. 2012. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J. Clin. Microbiol. 50:966–976. 10.1128/JCM.05590-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ianiro G, Heylen E, Delogu R, Zeller M, Matthijnssens J, Ruggeri FM, Van Ranst M, Fiore L. 2013. Genetic diversity of G9P[8] rotavirus strains circulating in Italy in 2007 and 2010 as determined by whole genome sequencing. Infect. Genet. Evol. 16:426–432. 10.1016/j.meegid.2013.03.031 [DOI] [PubMed] [Google Scholar]
- 63.Nyaga MM, Jere KC, Peenze I, Mlera L, van Dijk AA, Seheri ML, Mphahlele MJ. 2013. Sequence analysis of the whole genomes of five African human G9 rotavirus strains. Infect. Genet. Evol. 16:62–77. 10.1016/j.meegid.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 64.Pietsch C, Liebert UG. 2009. Human infection with G12 rotaviruses, Germany. Emerg. Infect. Dis. 15:1512–1515. 10.3201/eid1509.090497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. 2012. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 12:136–141. 10.1016/S1473-3099(11)70253-5 [DOI] [PubMed] [Google Scholar]
- 66.Steele AD, Patel M, Parashar UD, Victor JC, Aguado T, Neuzil KM. 2009. Rotavirus vaccines for infants in developing countries in Africa and Asia: considerations from a World Health Organization-sponsored consultation. J. Infect. Dis. 200(Suppl 1):S63–S69. 10.1086/605042 [DOI] [PubMed] [Google Scholar]
- 67.Boom JA, Sahni LC, Payne DC, Gautam R, Lyde F, Mijatovic-Rustempasic S, Bowen MD, Tate JE, Rench MA, Gentsch JR, Parashar UD, Baker CJ. 2012. Symptomatic infection and detection of vaccine and vaccine-reassortant rotavirus strains in 5 children: a case series. J. Infect. Dis. 206:1275–1279. 10.1093/infdis/jis490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bowen MD, Payne DC. 2012. Rotavirus vaccine-derived shedding and viral reassortants. Expert Rev. Vaccines 11:1311–1314. 10.1586/erv.12.114 [DOI] [PubMed] [Google Scholar]
- 69.Donato CM, Ch'ng LS, Boniface KF, Crawford NW, Buttery JP, Lyon M, Bishop RF, Kirkwood CD. 2012. Identification of strains of RotaTeq rotavirus vaccine in infants with gastroenteritis following routine vaccination. J. Infect. Dis. 206:377–383. 10.1093/infdis/jis361 [DOI] [PubMed] [Google Scholar]
- 70.Hemming M, Vesikari T. 2014. Detection of rotateq vaccine-derived, double-reassortant rotavirus in a 7-year-old child with acute gastroenteritis. Pediatr. Infect. Dis. J. 33:655–656. 10.1097/INF.0000000000000221 [DOI] [PubMed] [Google Scholar]
- 71.Hemming M, Vesikari T. 2012. Vaccine-derived human-bovine double reassortant rotavirus in infants with acute gastroenteritis. Pediatr. Infect. Dis. J. 31:992–994. 10.1097/INF.0b013e31825d611e [DOI] [PubMed] [Google Scholar]
- 72.Patel NC, Hertel PM, Hanson IC, Krance RA, Crawford SE, Estes M, Paul ME. 2012. Chronic rotavirus infection in an infant with severe combined immunodeficiency: successful treatment by hematopoietic stem cell transplantation. Clin. Immunol. 142:399–401. 10.1016/j.clim.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Payne DC, Edwards KM, Bowen MD, Keckley E, Peters J, Esona MD, Teel EN, Kent D, Parashar UD, Gentsch JR. 2010. Sibling transmission of vaccine-derived rotavirus (RotaTeq) associated with rotavirus gastroenteritis. Pediatrics 125:e438–e341. 10.1542/peds.2009-1901 [DOI] [PubMed] [Google Scholar]
- 74.Werther RL, Crawford NW, Boniface K, Kirkwood CD, Smart JM. 2009. Rotavirus vaccine induced diarrhea in a child with severe combined immune deficiency. J. Allergy Clin. Immunol. 124:600. 10.1016/j.jaci.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 75.Guindon S. 2010. Bayesian estimation of divergence times from large sequence alignments. Mol. Biol. Evol. 27:1768–1781. 10.1093/molbev/msq060 [DOI] [PubMed] [Google Scholar]
- 76.Codoner FM, Fares MA, Elena SF. 2006. Adaptive covariation between the coat and movement proteins of prunus necrotic ringspot virus. J. Virol. 80:5833–5840. 10.1128/JVI.00122-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rolland M, Carlson JM, Manocheewa S, Swain JV, Lanxon-Cookson E, Deng W, Rousseau CM, Raugi DN, Learn GH, Maust BS, Coovadia H, Ndung'u T, Goulder PJ, Walker BD, Brander C, Heckerman DE, Mullins JI. 2010. Amino-acid co-variation in HIV-1 Gag subtype C: HLA-mediated selection pressure and compensatory dynamics. PLoS One 5:e12463. 10.1371/journal.pone.0012463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stobart CC, Lee AS, Lu X, Denison MR. 2012. Temperature-sensitive mutants and revertants in the coronavirus nonstructural protein 5 protease (3CLpro) define residues involved in long-distance communication and regulation of protease activity. J. Virol. 86:4801–4810. 10.1128/JVI.06754-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park C, Chen X, Yang JR, Zhang J. 2013. Differential requirements for mRNA folding partially explain why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U. S. A. 110:E678–E686. 10.1073/pnas.1218066110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald SM, Patton JT. 2011. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 19:136–144. 10.1016/j.tim.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.