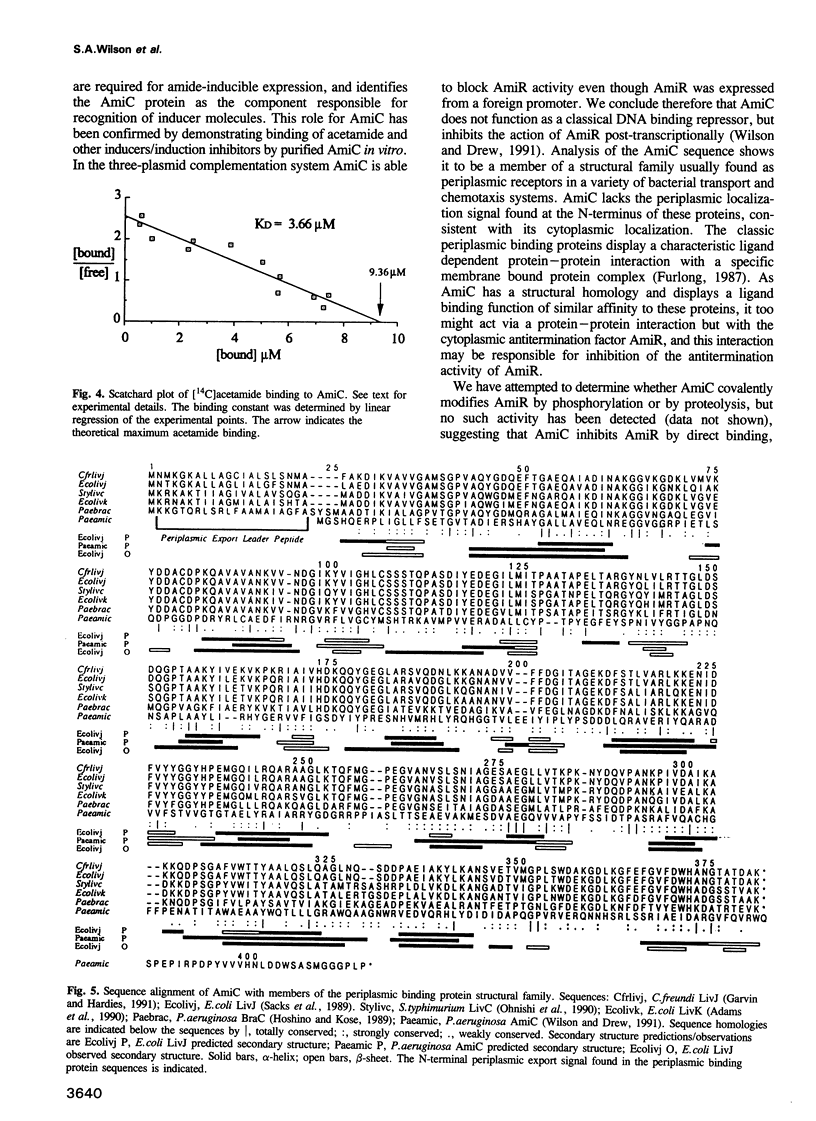
Abstract
Amide-inducible expression of the aliphatic amidase system of Pseudomonas aeruginosa can be reconstituted in Escherichia coli with only the amidase structural gene amiE, the negative regulator amiC and the positive regulator amiR, a transcription antitermination factor. Complementation experiments in E. coli suggest that negative control of amidase expression by AmiC is mediated by a protein-protein interaction with AmiR. Purified AmiC binds acetamide with a KD of 3.7 microM in equilibrium dialysis studies, and therefore AmiC appears to be the sensory partner of the AmiC/AmiR pair of regulatory proteins, responding to the presence of amides. Sequence analysis techniques suggest that AmiC is a member of the structural family of periplasmic binding proteins, but has a distinct and novel cytoplasmic role.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. D., Wagner L. M., Graddis T. J., Landick R., Antonucci T. K., Gibson A. L., Oxender D. L. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J Biol Chem. 1990 Jul 15;265(20):11436–11443. [PubMed] [Google Scholar]
- Amster-Choder O., Houman F., Wright A. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell. 1989 Sep 8;58(5):847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- BRAMMAR W. J., CLARKE P. H. INDUCTION AND REPRESSION OF PSEUDOMONAS AERUGINOSA AMIDASE. J Gen Microbiol. 1964 Dec;37:307–319. doi: 10.1099/00221287-37-3-307. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brammar W. J., Charles I. G., Matfield M., Liu C. P., Drew R. E., Clarke P. H. The nucleotide sequence of the amiE gene of Pseudomonas aeruginosa. FEBS Lett. 1987 May 11;215(2):291–294. doi: 10.1016/0014-5793(87)80164-3. [DOI] [PubMed] [Google Scholar]
- Clarke P. H., Drew R. E., Turberville C., Brammar W. J., Ambler R. P., Auffret A. D. Alignment of cloned amiE gene of Pseudomonas aeruginosa with the N-terminal sequence of amidase. Biosci Rep. 1981 Apr;1(4):299–307. doi: 10.1007/BF01114869. [DOI] [PubMed] [Google Scholar]
- Cousens D. J., Clarke P. H., Drew R. The amidase regulatory gene (amiR) of Pseudomonas aeruginosa. J Gen Microbiol. 1987 Aug;133(8):2041–2052. doi: 10.1099/00221287-133-8-2041. [DOI] [PubMed] [Google Scholar]
- Drew R. E., Clarke P. H., Brammar W. J. The construction in vitro of derivatives of bacteriophage lambda carrying the amidase genes of Pseudomonas aeruginosa. Mol Gen Genet. 1980 Jan;177(2):311–320. doi: 10.1007/BF00267444. [DOI] [PubMed] [Google Scholar]
- Drew R., Lowe N. Positive control of Pseudomonas aeruginosa amidase synthesis is mediated by a transcription anti-termination mechanism. J Gen Microbiol. 1989 Apr;135(4):817–823. doi: 10.1099/00221287-135-4-817. [DOI] [PubMed] [Google Scholar]
- Farin F., Clarke P. H. Positive regulation of amidase synthesis in Pseudomonas aeruginosa. J Bacteriol. 1978 Aug;135(2):379–392. doi: 10.1128/jb.135.2.379-392.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Hoshino T., Kose K. Cloning and nucleotide sequence of braC, the structural gene for the leucine-, isoleucine-, and valine-binding protein of Pseudomonas aeruginosa PAO. J Bacteriol. 1989 Nov;171(11):6300–6306. doi: 10.1128/jb.171.11.6300-6306.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houman F., Diaz-Torres M. R., Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990 Sep 21;62(6):1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. A new approach to protein fold recognition. Nature. 1992 Jul 2;358(6381):86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- KELLY M., CLARKE P. H. An inducible amidase produced by a strain of Pseudomonas aeruginosa. J Gen Microbiol. 1962 Feb;27:305–316. doi: 10.1099/00221287-27-2-305. [DOI] [PubMed] [Google Scholar]
- Lowe N., Rice P. M., Drew R. E. Nucleotide sequence of the aliphatic amidase regulator gene (amiR) of Pseudomonas aeruginosa. FEBS Lett. 1989 Mar 27;246(1-2):39–43. doi: 10.1016/0014-5793(89)80249-2. [DOI] [PubMed] [Google Scholar]
- Mowbray S. L., Petsko G. A. The x-ray structure of the periplasmic galactose binding protein from Salmonella typhimurium at 3.0-A resolution. J Biol Chem. 1983 Jul 10;258(13):7991–7997. doi: 10.2210/pdb1gbp/pdb. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Nakazima A., Matsubara K., Kiritani K. Cloning and nucleotide sequences of livB and livC, the structural genes encoding binding proteins of the high-affinity branched-chain amino acid transport in Salmonella typhimurium. J Biochem. 1990 Feb;107(2):202–208. doi: 10.1093/oxfordjournals.jbchem.a123026. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack J. S., Saper M. A., Quiocho F. A. Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. J Mol Biol. 1989 Mar 5;206(1):171–191. doi: 10.1016/0022-2836(89)90531-7. [DOI] [PubMed] [Google Scholar]
- Schnetz K., Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988 Oct;7(10):3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S. A., Chayen N. E., Hemmings A. M., Drew R. E., Pearl L. H. Crystallization of and preliminary X-ray data for the negative regulator (AmiC) of the amidase operon of Pseudomonas aeruginosa. J Mol Biol. 1991 Dec 20;222(4):869–871. doi: 10.1016/0022-2836(91)90579-u. [DOI] [PubMed] [Google Scholar]
- Wilson S., Drew R. Cloning and DNA sequence of amiC, a new gene regulating expression of the Pseudomonas aeruginosa aliphatic amidase, and purification of the amiC product. J Bacteriol. 1991 Aug;173(16):4914–4921. doi: 10.1128/jb.173.16.4914-4921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukin R. S. Evidence for a conformational change in the Escherichia coli maltose receptor by excited-state fluorescence lifetime data. Biochemistry. 1979 May 29;18(11):2139–2145. doi: 10.1021/bi00578a001. [DOI] [PubMed] [Google Scholar]



