Abstract
The controlled attachment of synthetic groups to proteins is important for a number of fields, including therapeutics, where antibody-drug conjugates are an emerging area of biologic medicines. We have previously reported a site-specific protein modification method using a transamination reaction that chemoselectively oxidizes the N-terminal amine of a polypeptide chain to a ketone or an aldehyde group. The newly introduced carbonyl can be used for conjugation to a synthetic group in one location through the formation of an oxime or a hydrazone linkage. To expand the scope of this reaction, we have used a combinatorial peptide library screening platform as a method to explore new transamination reagents while simultaneously identifying their optimal N-terminal sequences. N-methylpyridinium-4-carboxaldehyde benzenesulfonate salt (Rapoport's salt, RS) was identified as a highly effective transamination reagent when paired with glutamate-terminal peptides and proteins. This finding establishes RS as a transamination reagent that is particularly well suited for antibody modification. Using a known therapeutic antibody, herceptin, it was demonstrated that RS can be used to modify the heavy chains of the wild type antibody, or both the heavy and the light chains after N-terminal sequence mutation to add glutamate residues.
INTRODUCTION
The chemical modification of proteins is an important tool for a wide range of fields, including cell biology research,1,2,3 the construction of new biomaterials,4 and the development of novel therapeutics.5,6 The pharmaceutical industry has been particularly interested in antibody-drug conjugates (ADCs), with multiple products clinically approved and several more currently in advanced trials.7,8 Ideally, ADCs will need to be prepared using site-selective bioconjugation reactions that can control the stoichiometry and position of the attached drugs. However, antibodies are particularly difficult to modify in a controlled manner due to their large size, multiple chains, glycosylation, and structurally important disulfide bonds. Traditional methods such as lysine modification9 are indiscriminate given the abundance of these residues (up to 100 copies),8 leading to heterogeneous mixtures that complicate pharmacokinetic characterization. Even site-specific bioconjugation reactions such as periodate oxidation of N-terminal serine or threoine residues are often complicated for modification of antibodies as in this case the glycans will be oxidized.10 In some cases, selective modification can be achieved through the alkylation of cysteine residues arising from the partial reduction of the interchain disulfide bonds.11 Current alternative methods for site-specific antibody modification also include genetic mutation to alter the number of solvent-accessible cysteines,12,13 the introduction of unnatural amino acids,14 or recognition tags for enzymatic modification.15 While these methods can already be used successfully, the growing interest in ADCs as commercial treatments provides a need for a whole series of readily-scalable and functional group tolerant methods that can provide well-defined conjugates with control over attachment stoichiometry.
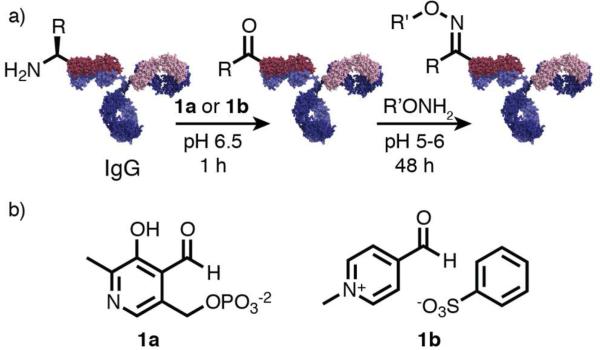
We have previously reported a site-specific transamination reaction that introduces a new ketone group at the N-terminus of proteins through incubation with pyridoxal 5’-phosphate (PLP, 1a).16,17 The carbonyl groups introduced by this reaction are not naturally occurring functionalities in proteins, and can therefore be used as unique points of attachment for synthetic groups through the formation of hydrazone or stable oxime bonds,18,19 Figure 1a. Although the side chain of the N-terminal residue does not participate directly in the transamination mechanism, the reaction yield was found to vary significantly depending on the amino acid in the N-terminal position.20 Given this situation, we previously developed a combinatorial peptide library screening platform to identify highly reactive sequences towards PLP-mediated transamination, leading to the identification of Ala-Lys N-terminal motifs.21 In the present work, this new bioconjugation development tool was used as a way to identify a new protein transamination reagent, N-methylpyridinium-4-carboxaldehyde benzenesulfonate salt (RS,22 1b), while simultaneously revealing glutamate-rich sequences as particularly reactive substrates for this reagent. This finding renders this approach particularly amenable to antibody substrates, since many human IgG1 isotypes, which are promising therapuetics, contain at least one glutamate-terminal chain.23,24,25
Figure 1.
Site-specific protein modification can be acheived using transamination reagents (1a or 1b) that oxidize the N-terminal amine to a ketone or an aldehyde group. The newly introduced carbonyl group is not natively found on proteins, and thus can be used for conjugation to a synthetic group (R'ONH2) through the formation of an oxime linkage. (a) Shown is a monoclonal antibody, which has two identical light chains and two identical heavy chains. This provides four N-termini as potential sites of attachment (only one shown). (b) Previous work used pyridoxal 5’-phosphate (PLP, 1a) as a transamination reagent, and this work identifies N-methylpyridinium-4-carboxalydehyde benzenesulfonate salt (Rapoport's salt, 1b) as a highly effective transamination reagent for acid-rich N-terminal sequences.
N-terminal transamination using PLP has previously been shown to modify monoclonal antibodies.26 However, the yields were not high and elevated temperatures were required, limiting the practical application of this approach. Using Rapoport's salt (RS) as a transamination reagent, the site-selective modification of the heavy chains of anti-HER2 human IgG1 (which have N-terminal glutamate residues) could be achieved with significantly greater efficiency.
The stoichiometry of drug attachment has been identified as a critical parameter for ADC efficacy, with two to four attachments reported as being optimal in at least some cases.8,27 Since antibodies have four N-termini (two identical heavy chains and two identical light chain units), we hypothesized that the sequence dependence of this reaction could give control over the total number of attachments. Antibodies such as anti-HER2 have one set of glutamate-terminal chains, for up to two attachment sites, but by mutating the antibody terminal sequences, the modification of both sets of chains also proved possible, enabling the attachment of up to four groups. The attachment of small molecules to the N-termini via oxime formation did not disrupt the antigen binding ability, as determined by flow cytometry. Overall, these findings establish RS as a facile and readily scalable method to obtain antibody conjugates in good yield and with control over the number of attached groups.
RESULTS AND DISCUSSION
Although we have found PLP to be an effective transamination reagent in a number of different contexts, we found its reactivity can suffer from batch-to-batch variability. In addition, the preference for positive charges near the N-terminal positions to obtain optimal activity may be difficult to achieve in some cases.21 We thus sought alternative aldehyde reagents that possessed similar reactivity, but might be more applicable to large scale protein modification reactions.
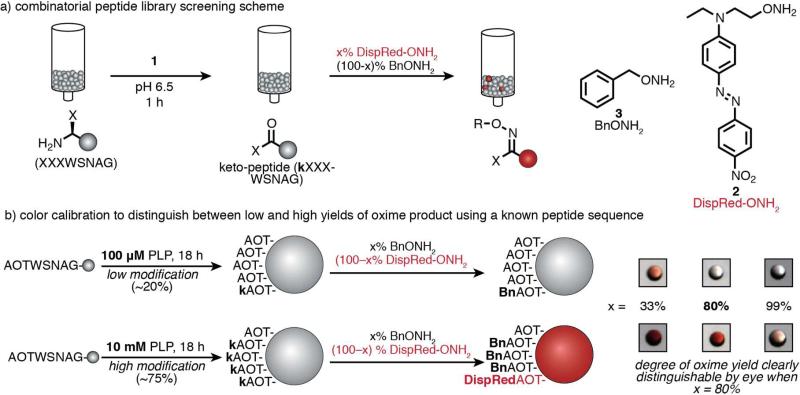
In our original report of site-specific N-terminal transamination on proteins, a number of common aldehydes were screened for transamination ability, and PLP was identified as a uniquely effective reagent.16 However, this screen was carried out using angiotensin as a single substrate. Given our later findings in terms of the dependence of PLP-mediated transamination on the N-terminal sequence, it seemed likely that this initial screen could have failed to identify other, more practical aldehyde reagents that could achieve similarly high levels of transamination, but with other sequence preferences. Such combinations would most readily be identified using combinatorial peptide library screening platforms that can evaluate the transamination ability of candidate reagents against all combinations of N-terminal sequences simultaneously (Figure 2a).
Figure 2.
(a) A one-bead-one-sequence combinatorial peptide library in which the three N-terminal residues were varied was used to screen potential transamination reagents (1) against all possible N-terminal sequences simultaneously. After transamination, the keto-group-containing peptides were identified through the covalent attachment of a visible dye (DispRed-ONH2, 2) through oxime formation. (b) To adjust the sensitivity of the colorimetric detection method, a colorless alkoxyamine (BnONH2, 3) was combined with DispRed-ONH2 in varying ratios. A peptide sequence (AOT-terminal, with similar reactivity to AKT-termini) and a transamination reagent with known reactivity (PLP, 1a) were used to generate beads with high (75%) and low (20%) levels of modification. The greatest visual distinction between beads bearing high and low modification levels was observed when using 80:20 ratio of 3 to 2.
Synthesis of the combinatorial peptide library and assay calibration for transamination screening
A one-bead-one-sequence combinatorial peptide library was first prepared using split-and-pool synthesis.28 The capping of a small portion of the growing peptide chains during the synthesis of the variable positions provided a truncation ladder for sequencing beads of interest by intact ion mass spectrometry.21 Because we sought to identify short motifs that could easily be incorporated into proteins, the diversity in the library was limited to the three N-terminal positions. The variable positions included each of the 20 natural amino acids, resulting in an 8,000 member library.
To identify the library sequences that had been transaminated to form a keto-peptide, a colorful, Disperse Red-based alkoxyamine reagent (2, DispRed-ONH2)21 was used in a subsequent oxime-formation reaction. The resulting beads possessing the desired oxime product thus took on a bright red color. In our previous work, however, it was found that the color on the beads saturated at ~30% oxime yield, making it difficult to identify the highest yielding reaction combinations.21 For the next generation of this screening technique, we recalibrated the detection scheme such that a visually red bead would correspond only to a high oxime yield (Figure 2b). To do this, reaction conditions were established that would result in beads with a low and high level of modification using a known transamination reagent (PLP) and peptide sequence. The peptide used was AOTWSNAG (where O stands for ornithine), which was found to have similar reactivity to a previously reported AKT sequence.21 With this sequence, incubation with 100 μM PLP for 18 h resulted in 20% oxime yield (representing a low modification case), as verified by LC-MS quantification of the corresponding benzyloxime. Incubation with 10 mM PLP for 18 h resulted in 75% yield, representing a high conversion case. Then DispRed-ONH2 was combined with different ratios of a colorless alkoxyamine (benzylalkoxyamine, BnONH2) to find a ratio that resulted in the greatest visual contrast between low and high oxime yields. With a low stringency ratio (a low proportion of BnONH2, 33% ), beads with both high and low levels of modification resulted in a red color, and with a high stringency detection ratio (99% BnONH2) neither case had a red color. An intermediate ratio was found (80% BnONH2), wherein only the highly transaminated beads were evident. Thus, this ratio of alkoxyamines was used for subsequent library screening to identify transamination conditions that resulted in maximal yields.
Identification of a new transamination reagent for glutamate-terminal sequences
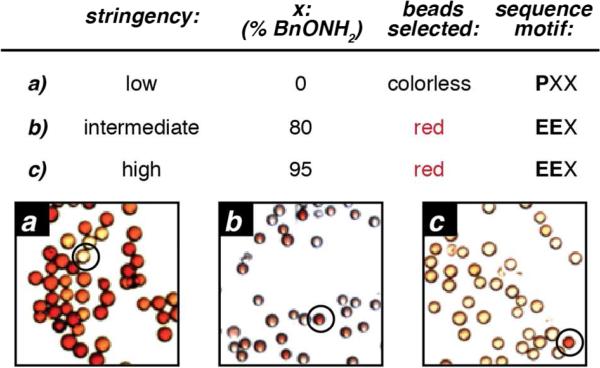
Portions of the library were incubated with 10 mM concentrations of a series of aldehyde reagents at pH 6.5. Substrates screened included salicylaldehyde, 2-hydroxy-5-nitrobenzaldehyde, benzaldehyde, glyoxylic acid, and N-methylpyridinium-4-carboxaldehyde (Rapoport's salt, 1b). In the case of Rapoport's salt (RS), it was found that a large proportion of the beads turned red using the low stringency assay conditions, indicating that most sequences had at least some degree of reactivity (Figure 3a). This reagent is particularly promising for transamination reactions because it is inexpensive (particularly if synthesized in-house)22 and the reagent can be purified before use via recrystallization from acetonitrile. Relative to PLP, this reagent is also more amenable to structural modifications, which could be explored as ways to increase reactivity in future studies. RS has been reported previously as a transamination reagent for small molecules in organic solution29,30 suggesting that it could serve as a protein transamination reagent as well. The other aldehydes screened had lower activity and some suffered from limited aqueous solubility. In some cases initial leads were followed, but found to lack reactivity upon verification.
Figure 3.
Library screening to identify reactive sequences towards Rapoport's salt-mediated transamination. The library was treated with 10 mM RS for 1 h at pH 6.5, followed by oxime formation with the specified ratios of BnONH2 (x%) and DispRed-ONH2 (100–x%). The active sequences were identified by selecting and sequencing the red beads in the library. When a low stringency screen was applied (a) many beads had a red color, indicating that many sequences form some degree of oxime product. In this case, the beads that remained colorless were selected, and proline was identified as an N-terminal residue that prevented reactivity. To identify sequences that led to high levels of oxime product, more stringent screens were used that could distinguish low from high levels of oxime yield (b, c). Sequencing the red beads from these screens identified glutamate-terminal sequences as a common reactive motif.
Since most sequences showed some reactivity towards RS using the low stringency assay conditions (Figure 3a), the few remaining colorless beads were selected to see which sequences showed no transamination reactivity. Proline in the N-terminal position was identified as the common motif among the colorless beads. This result was not unexpected given that as a secondary amine, proline is not able to undergo the standard transamination mechanism. Previous studies with PLP did show appreciable reactivity using the transamination/oximation procedure,20 although the resulting product has not yet been fully characterized and was not observed with RS.
To identify the optimal N-terminal sequences for RS-mediated transamination, higher stringency detections of oxime yield were applied to the library (Figure 3b). After reaction with 10 mM RS for 1 h in pH 6.5 phosphate buffer followed by oxime formation with a 80:20 ratio of 3 to 2, a small number of red beads were observed. These beads were collected and sequenced, and the common pattern identified was multiple glutamate residues at the N-terminus (the full list of sequences appears in Supporting Information, Figure S2). The screen was next repeated using a higher ratio of the colorless alkoxyamine BnONH2 (95%) to provide an even more stringent filter for high oxime yield (Figure 3c). Again the same reactive motif was identified by sequencing these red beads: glutamate in the N-terminal and second positions, with less consensus at the third position.
Another question about reactivity that could be answered using the library was whether residues in the second or third positions could significantly reduce the high reactivity of glutamate-terminal sequences. Knowledge of whether certain neighboring residues should be avoided is particularly important for incorporation of the reactive motif onto protein substrates. To address this question, a glutamate-terminal subset of the library, in which all peptides had E as the N-terminal residue and the second and third positions were varied (EXX), was screened. Sequencing the colorless beads after transamination and oxime formation on this subset of the library revealed a consensus motif that had proline in the second position (Supporting Information, Figure S2). Thus the library screening identified glutamate-terminal sequences as highly reactive, and sequences with proline in the N-terminal or second position as less reactive towards RS-mediated transamination. Although the third position had a lesser effect on reactivity, we selected EES as the highly reactive sequence to test on peptide and protein substrates. PES and EPS were used as representative non-reactive sequences to explore how much the change of a single amino acid could affect the reactivity.
Verification of RS-mediated transamination using peptides
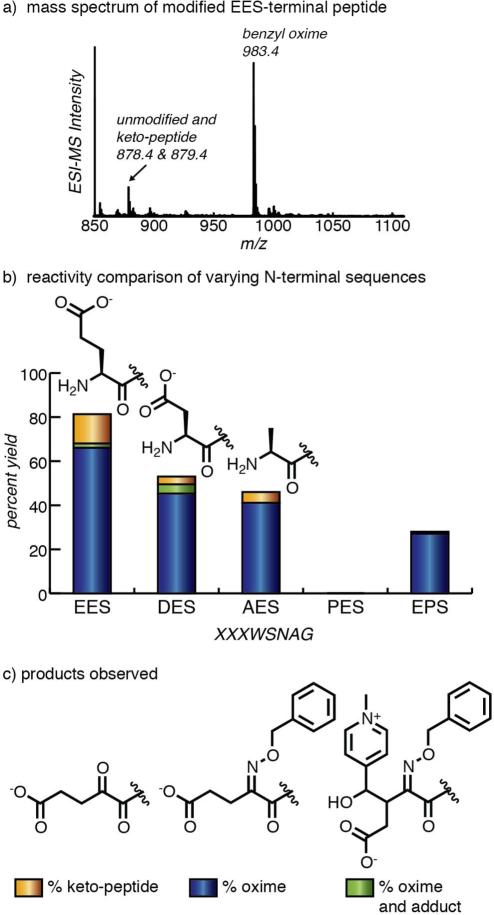
To verify the reactive and non-reactive sequence motifs identified by library screening, peptides of the form XXXWSNAG were synthesized. After synthesis, the peptides were treated with RS as described above, followed by benzylalkoxyamine for quantification of the oxime yield by LC-MS. The optimal sequence peptide, EESWSNAG, led to high modification yield, as seen in Figure 4a. A screen of the reaction conditions, varying the concentration of RS and the reaction time, was next performed. These screens (data shown in Figure S3) identified exposure to 100 mM RS for 1 h at pH 6.5 as the conditions resulting in high transamination yield with few byproducts. This reaction protocol was subsequently used to evaluate all of the peptide substrates. As seen in Figure 4b, the EE-terminal peptide resulted in over 80% transamination. Not all of the transaminated, keto-peptide species (shown in yellow) was converted to oxime (blue) under the oxime formation conditions used. A small amount of covalent addition of RS to the N-terminus was also observed (green), which is presumed to be an aldol-type addition. This byproduct was observed in higher yields with longer reaction times, but could be minimized through the use of shorter reaction times (45 min to 1 h, Figure S3).
Figure 4.
Verification of library screening results using peptides. Sequences were resynthesized in the form XXXWSNAG. (a) An ESI mass spectrum of EESWSNAG showed high conversion to the oxime product under the standard reaction conditions. (b) Various N-terminal sequences were subjected to the same reaction conditions and the product yields were quantified by LC-MS. The reported data represent the average of three replicate experiments (Figure S4). (c) Three products were observed: transaminated keto-peptide that did not form an oxime, the desired oxime product, and oxime with the addition of RS to the N-terminus (proposed structure drawn). Aspartate-terminal peptides were observed by mass spectrometry to decarboxylate during transamination. A proline-terminal sequence was confirmed to have no reactivity towards RS-mediated transamination. With proline in the second position, the reactivity of the E-terminal sequence was substantially decreased.
To test whether the EE-terminal motif was optimal only for RS or was highly reactive towards transamination in general, the modification of this sequence with PLP-mediated transamination was examined as well (Figure S3c). Under the same reaction conditions the yield was much higher using RS, indicating the library screening protocol had identified an optimal reagent/sequence pair. The observed reactivity of the EE-terminal peptides with RS was comparable to the AKT-terminal sequence we had previously identified as optimal for PLP.21 The N-terminal site-specificity of RS-mediated transamination was confirmed using tandem mass spectrometry for both the transaminated keto-peptide and the benzyl oxime (Figure S5, S6, and S7).
Due to their structural similarity, aspartate-terminal sequences were also examined. E- and D- terminal peptides were found to have significantly different yields towards RS-mediated transamination, as shown in Figure 4b. The mass spectra indicated that decarboxylation of the aspartate-terminal peptide occured during the transamination reaction, as has been observed previously using PLP.20 Comparison with an alanine-terminal peptide showed that the aspartate-terminal sequence had a more similar reactivity to alanine (which it resembles after decarboxylation) than glutamate. Therefore we concluded that optimal sequences for RS-mediated transamination are glutamate-terminal sequences, and that D and E are not interchangable for the N-terminal position for this reaction.
The nonreactive sequences identified by library screening were also verified on resynthesized peptides. The proline-terminal peptide (PES) resulted in neither transamination nor oxime formation. The glutamate-terminal sequence with proline in the second position (EPS) did result in some oxime product, but at a yield that was significantly less than that of the multiglutamate-terminal peptide. This result underscores the impact of a single amino acid substitution in an internal sequence position.
Chain-specific modification of Anti-HER2 Human IgG1
Given that the heavy chain of human anti-HER2 IgG1 (Herceptin)31 has an N-terminal glutamate residue, the wild-type antibody provided a suitable substrate to test RS-mediated transamination on a protein. The wild-type anti-HER2 antibody (denoted as wild-type, WT), which was commercially obtained from Eureka Therapuetics (Emeryville, CA), has a EVQ-terminal heavy chain and a DIQ-terminal light chain.31 We also expressed an N-terminal mutant of the antibody in HEK cells to introduce additional reactivity using the EES optimal sequence on both chains. For the transamination reaction, the antibodies were incubated with a freshly prepared 100 mM solution of RS for 60 min in pH 6.5 phosphate buffer at 37 °C. After transamination and removal of the excess small molecule, the antibodies were incubated with BnONH2 for 48 h at RT in pH 5.5 phosphate buffer. The modification of each chain was then analyzed and quantified using mass spectrometry.
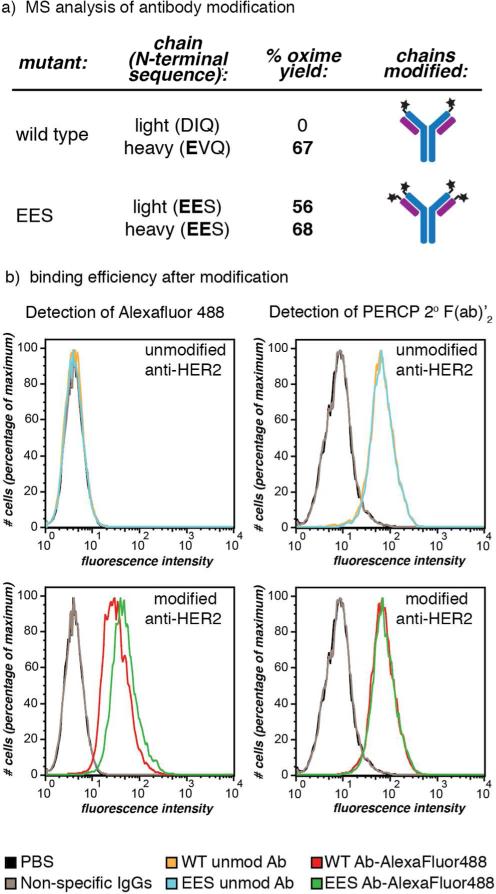
The heavy chain wild-type sequence provided 67% conversion to the oxime product, in which 15% also included the RS adduct (Figure S9). In contrast, no modification was observed for the light chain (Figure 5a). This is likely because the steric environment of the folded protein reduces the accessibility of the already less-reactive substrate (DIQ terminal sequence). The complete lack of modification also clearly demonstrated that RS does not react with lysine side chain amines or other residues. The net result of this experiment is that RS-mediated transamination of the wild-type sequence allowed the selective modification of only the heavy chain. We have not yet carried out studies to determine whether the presence of the small percentage of adduct affects the biological properties of the antibody. This reactivity pattern was also observed on another widely used human IgG1, anti-hTNFα (Invivogen, San Diego, CA), which also has a glutamate-terminal heavy chain and an aspartate-terminal light chain (Figure S13).32
Figure 5.
IgG modification using RS-mediated transamination. (a) Wild-type anti-HER2 human IgG1 antibodies, as well as mutants with EES N-terminal sequences, were treated with RS, followed by oxime formation with BnONH2. The heavy and light chains were separated for mass spectrometry quantification of the benzyl oxime product. The wild-type antibody showed modification of the heavy chain and no modification of the light chain, while the mutant with EES on both sets of termini exhibited modification of both chains. (b) The antigen binding ability of wild-type (WT) and EES-terminal anti-HER2 antibodies post-modification (transaminated by RS, followed by oxime formation with AlexaFluor488-ONH2) was confirmed using flow cytometry. MCF7 clone 18 cells (a HER2 positive cell line) were subjected to modified (bottom histograms) and unmodified (top) wild-type and EES-terminal antibodies. By direct detection of the AlexaFluor488, the binding of modifed antibodies was seen (bottom left), while the unmodified showed no fluorescent shift (top left). In order to compare the binding of the modified antibodies to unmodified, the unmodified species were detected using a secondary detection method: anti-human IgG PerCP-conjugated 2° F(ab’)2 (right histograms). Both wild-type and EES-terminal antibodies modified with the fluorophore retained similar binding affinity and specificity to the unmodified ones. Non-specific human IgG1 and PBS were used as negative control agents. Jurkat cells (a HER2 negative cell line) did not bind to either unmodified or modified anti-HER2 antibodies (Figure S15).
To see if both antibody chains could be modified through sequence alteration, we prepared a herceptin analog in which all N-termini were extended by three residues to add the EES motif. These antibodies were modified on both the light and the heavy chains (56% and 68%, respectively). During the expression of this mutant, we observed some improper cleavage of the IL2 signal sequences, which led to the production of a small proportion of light and heavy chains that lacked the N-terminal EE groups. The resulting serine-terminal analogs were not modified to the same extent as the EES sequence on the heavy chain, and we observed no modification for the serine-terminal light chain (Figure S12). Overall, these experiments demonstrate that this new RS method can achieve a previously inaccessible degree of labeling control for one or both sets of antibody chain termini.
Since the N-termini of the antibody heavy and light chains flank the antigen binding domains (but are not part of the hypervariable loops, Figure 1a),33 it was important to confirm that the modification of these locations did not disrupt antigen binding. To evaluate this, we used AlexaFluor 488-ONH2 to label transaminated antibodies, and we used flow cytometry (Figure 5b) to compare the unmodified antibodies (top histograms) to their N-terminally modified counterparts (bottom histograms). In each experiment, the antibodies were incubated with HER2 overexpressing cells (MCF7 clone 18).34 The AlexaFlour 488 dyes were detected directly, and an anti-human IgG fluorescently labeled with peridinin chlorophyll protein (PerCP) was used as a secondary detection method. As shown in Figure 5b (right set of histograms), no significant disruption of binding was observed for any of the AlexaFluor 488 oxime conjugates. The left histograms of Figure 5b indicate that the fluorophore-labeled antibodies were specifically involved in the binding, as only species modified with the AlexaFluor dye showed fluorescent shifts in this channel. Jurkat cells, which are a HER2-negative cell line, were used as a negative control. Neither modified nor unmodified antibodies were observed to bind these cells. To demonstrate that it is possible to attach larger groups using RS-mediated transamination, we also used a 5,000 Da MW poly(ethylene glycol) alkoxyamine reagent35 to attach a polymer to the antibody (Figure S13).
Discussion
The peptide and protein studies presented herein demonstrate the potential of RS to be a highly practical protein modification method. There are other effective and long standing N-terminal modification reactions, such as the oxidation of N-terminal serine residues with periodate. However, this method is incompatible with a number of protein classes. For example, the glycans on antibodies will result in periodate-based cleavage as a side reaction,10 while the RS-mediated reaction does not suffer this competing pathway. Relative to other transamination reagents we have used, RS is inexpensive and easy to recrystallize to a high degree of purity. RS-mediated transamination enables the attachment of the synthetic group to the protein via an oxime linkage. We have found these N-terminal oximes to be stable over weeks, and even months, particularly when the protein is stored at 4 °C.19 We also have tested the stability of oximes formed with N-terminal pyruvamide groups in previous studies.36
As with many bioconjugation reactions, the purification of the modified product can be challenging. For applications that require it, the properties of the attached synthetic group will likely dictate the most successful strategy. As examples, hydrophobic ion chromatography (HIC) could be used to isolate antibody-drug conjugates,37 while ion exchange could be used to isolate bioconjugates with differing charge.38 Nonetheless, current efforts in our labs are complementing these studies by developing new techniques that can achieve bioconjugate purification in a more general fashion.
CONCLUSIONS
Through these studies, we have demonstrated that combinatorial peptide libraries can be used as a tool to accelerate the discovery of protein modification reactions. Given the importance of pairing a transamination reagent with its optimal N-terminal sequence, this method can be used to evaluate the transamination capability of candidate reagents and identify their optimal N-terminal sequences simultaneously. Currently we are synthesizing rationally designed pyridinium derivatives that are anticipated to have enhanced reactivity. Using the information obtained from library screening in the present work, we have demonstrated site-specific modification of monoclonal antibodies with good yields, and that it is possible to control whether the heavy chain, or the heavy and light chains are modified. RS-mediated transamination of antibodies (and other glutamate-terminal proteins) is facile and should be readily scalable, thus potentially providing a suitable method for the practical production of desired antibody conjugates.
EXPERIMENTAL SECTION
Solid-Phase Peptide and Combinatorial Library Synthesis
The combinatorial peptide library and individual peptides were synthesized following previously reported protocols, which are described in the SI text.21
General Procedure for Library Screening
Portions of resin-bound library (approx. 25 mg of resin at a time) were treated with 1 mL of freshly prepared 10 mM RS solution in 50 mM phosphate buffer, pH 6.5 with 0.02% NaN3 and 10% DMF at rt. After 1 h of reaction time, the resin samples were washed with three portions of deionized water, followed by three portions of DMF to remove residual aldehyde. The library was then incubated with 1 mL of a mixture of Disperse Red alkoxyamine and O-benzylhydroxylamine hydrochloride (BnONH2) in the specified ratio in a 1:1 H2O:DMF solution for 3 h at rt. The excess alkoxyamine was removed by rinsing with three portions of dichloromethane (DCM), followed by three portions of DMF. The beads were then rinsed with ethanol and transferred to a Petri dish for visual inspection. The beads were examined using a Leica S6D Microscope and L2 Light Source (Leica, Germany) equipped with a Moticam 2300 3.0 MP camera using Motic Image Plus 2.0 ML software for capturing images. Individual red beads were manually removed using a Pipet-Lite LTS L-20 pipet (Rainin, Oakland, CA) and transferred to PCR tubes for sequencing. The residual ethanol in the tubes was removed by pipetting.
General Procedure for Library Sequencing
The procedure for sequencing individual beads from the library is described in the SI text.
General Procedure for RS-Mediated Transamination of Resin-Bound Peptide Substrates
Portions of resin-bound peptides (appx. 10 mg of resin) were treated with 1 mL of 100 mM freshly prepared RS solution (or the specified concentration) in 50 mM phosphate buffer, pH 6.5 with 0.02% NaN3 and 10% DMF at rt. After 1 h of reaction time, the resin was washed with three portions of deionized water, followed by three portions of DMF to remove residual aldehyde. The peptides were then incubated with 1 mL of a 250 mM BnONH2 solution in water for 3 h at rt. The resin was then rinsed with three portions of deionized water, followed by three portions of DMF. The peptides were then cleaved from the resin via incubation with 300 μL of a 100 mM sodium hydroxide solution for 30 min. The resulting peptide solution was added to 700 μL of 50 mM phosphate buffer (pH 6.5). The resulting solution was diluted 20-fold into dd-H2O for mass spectrometry analysis.
Antibody Expression
The construction of the heavy chain and light chain antibody expression plasmids, and the expression of these proteins in human embryonic kidney cells is described in the SI text.
General Procedure for RS-Mediated Transamination of Protein Substrates
Protein and RS stock solutions were prepared at twice the desired final concentration and mixed in equal volumes in a 1.5 mL Eppendorf tube. The final volume of each reaction was 100 μL. The 2x protein stock solutions were prepared at 0.5 - 1 mg/mL in 25 mM phosphate buffer at pH 6.5. The 2x RS stock solution (200 mM) was freshly prepared before each reaction in 25 mM phosphate buffer (with 0.02% NaN3), pH 6.5 from RS (recrystallized from acetonitrile). The reaction mixture was briefly agitated to ensure mixing and then incubated without further agitation at 37 °C for 1 h. Following the reaction, the excess aldehyde was removed using NAP Sephadex size exclusion columns (GE Healthcare, USA). The resulting keto-protein solution was then concentrated and buffer exchanged using 0.5 mL spin concentrators with a MWCO of 10 kDa (Millipore, Billerica, MA). The buffer exchange first involved the dilution of each sample to 500 μL with 25 mM phosphate buffer (pH 6.5). Each sample was then concentrated to 100 μL, and the process was repeated 3 times. The resulting keto-protein was then treated with the alkoxyamine stock solution of choice in a 1.5 mL Eppendorf tube and incubated at rt for 48 h. The alkoxyamine stock solutions, and their final concentrations used were: 125 mM BnONH2 (in water with the pH adjusted to 5.5), 50 mM PEG(2kDa)-ONH2 35 solution (in water), 5 mM PEG(5kDa)-ONH2 solution (in water), and 2 mM AlexaFluor488-ONH2 solution (in water and DMSO). For PEG and AlexaFluor488 modification, 100 mM of aniline was also used as a catalyst. After oxime formation, the NAP column and buffer exchange steps were again repeated to remove the excess alkoxyamine to stop the reaction. The reduction and capping of antibody chains for mass spectrometry analysis of modification of the heavy and light chains is described in the SI text.
Flow Cytometry of Antibody Mutants
The flow cytometry procedure for the determination of the HER2 binding ability of the unmodified and modified anti-HER2 IgG1 mutants is described in the SI text.
Supplementary Material
ACKNOWLEDGEMENTS
The reaction development and library screening aspects of this work were funded by the Energy Biosciences Institute at UC Berkeley. The antibody modification studies were funded by the DOD Breast Cancer Research Program [Grant BC016995] L.S.W. and K.S.P. were supported by a predoctoral fellowship from the N.S.F and from the Berkeley Chemical Biology Graduate Program (NRSA Training Grant 1 T32 GMO66698) C.N. was supported by a Howard Hughes Medical Institute International Student Research Fellowship. LC-MS instrumentation was acquired with National Institutes of Health Grant 1S10RR022393-01.
Footnotes
ASSOCIATED CONTENT
Supporting information, including further experimental details and supporting figures is available in the SI Text. This information is available free of charge via the internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.O'Hare HM, Johnsson K, Gautier A. Curr. Opin. Struct. Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 3.Cravatt BF, Wright AT, Kozarich JW. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 4.Witus LS, Francis MB. Acc. Chem. Res. 2011;44:774–783. doi: 10.1021/ar2001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zalipsky S. Bioconjugate Chem. 1995;6:150–165. doi: 10.1021/bc00032a002. [DOI] [PubMed] [Google Scholar]
- 6.Baker DP, Lin EY, Lin K, Pellegrini M, Petter RC, Chen LL, Arduini RM, Brickelmaier M, Wen D, Hess DM, Chen L, Grant D, Whitty A, Gill A, Lindner DJ, Pepinsky RB. Bioconjugate Chem. 2005;17:179–188. doi: 10.1021/bc050237q. [DOI] [PubMed] [Google Scholar]
- 7.Wu AM, Senter PD. Nat. Biotech. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 8.Alley SC, Okeley NM, Senter PD. Curr. Opin. Chem.Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 9.Hermanson GT. Bioconjugate Techniques. 1st ed. Academic Press; 1996. [Google Scholar]
- 10.Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC. Nat. Meth. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doronina SO, Toki BE, Torgov MY, Mendelsohn BA, Cerveny CG, Chace DF, DeBlanc RL, Gearing RP, Bovee TD, Siegall CB, Francisco JA, Wahl AF, Meyer DL, Senter PD. Nat. Biotech. 2003;21:778–784. doi: 10.1038/nbt832. [DOI] [PubMed] [Google Scholar]
- 12.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, Lu Y, Meng YG, Ng C, Yang J, Lee CC, Duenas E, Gorrell J, Katta V, Kim A, McDorman K, Flagella K, Venook R, Ross S, Spencer SD, Lee Wong W, Lowman HB, Vandlen R, Sliwkowski MX, Scheller RH, Polakis P, Mallet W. Nat. Biotech. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 13.McDonagh CF, Turcott E, Westendorf L, Webster JB, Alley SC, Kim K, Andreyka J, Stone I, Hamblett KJ, Francisco JA, Carter P. Prot. Engineer. Design Selec. 2006;19:299–307. doi: 10.1093/protein/gzl013. [DOI] [PubMed] [Google Scholar]
- 14.Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, Lu Y, Tran H, Seller AJ, Biroc SL, Szydlik A, Pinkstaff JK, Tian F, Sinha SC, Felding-Habermann B, Smider VV, Schultz PG. Proc. Natl. Acad. Sci. 2012;109:16101–16106. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P, Shui W, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Proc. Natl. Acad. Sci. 2009;106:3000–2005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. Angew. Chem. Int. Ed. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 17.Witus LS, Francis MB. Curr. Prot. Chem. Biol. 2010;3:1–10. doi: 10.1002/9780470559277.ch100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jencks WP. J. Am. Chem. Soc. 1959;81:475–481. [Google Scholar]
- 19.Kalia J, Raines RT. Angew. Chem. Int. Ed. 2008;47:7523–7526. doi: 10.1002/anie.200802651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheck RA, Dedeo MT, Iavarone AT, Francis MB. J. Am. Chem. Soc. 2008;130:11762–11770. doi: 10.1021/ja802495w. [DOI] [PubMed] [Google Scholar]
- 21.Witus LS, Moore T, Thuronyi BW, Esser-Kahn AP, Scheck RA, Iavarone AT, Francis MB. J. Am. Chem. Soc. 2010;132:16812–16817. doi: 10.1021/ja105429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckley TF, Rapoport H. J. Am. Chem. Soc. 1982;104:4446–4450. [Google Scholar]
- 23.Liu H, Gaza Bulseco G, Faldu D, Chumsae C, Sun J. J. Pharm. Sciences. 2008;97:2426–2447. doi: 10.1002/jps.21180. [DOI] [PubMed] [Google Scholar]
- 24.Chelius D, Jing K, Lueras A, Rehder DS, Dillon TM, Vizel A, Rajan RS, Li T, Treuheit MJ, Bondarenko PV. Anal. Chem. 2006;78:2370–2376. doi: 10.1021/ac051827k. [DOI] [PubMed] [Google Scholar]
- 25.Yu L, Vizel A, Huff MB, Young M, Remmele RL, Jr., He B. J. Pharm. Biomed. Anal. 2006;42:455–463. doi: 10.1016/j.jpba.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Scheck RA, Francis MB. ACS Chem. Biol. 2007;2:247–251. doi: 10.1021/cb6003959. [DOI] [PubMed] [Google Scholar]
- 27.Hamblett KJ, Senter PD, Chace DF, Sun MMC, Lenox J, Cerveny CG, Kissler KM, Bernhardt SX, Kopcha AK, Zabinski RF, Meyer DL, Francisco JA. Clinical Cancer Res. 2004;10:7063–7070. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 28.Lam KS, Lebl M, Krchňák V. Chem. Rev. 1997;97:411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 29.Giustiniano M, Pirali T, Massarotti A, Biletta B, Novellino E, Campiglia P, Sorba G, Tron G. Synthesis. 2010;2010:4107–4118. [Google Scholar]
- 30.Shi Q, Verdier-Pinard P, Brossi A, Hamel E, McPhail AT, Lee K-H. J. Med. Chem. 1997;40:961–966. doi: 10.1021/jm960663k. [DOI] [PubMed] [Google Scholar]
- 31.Cho H-S, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Leahy DJ. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 32.Marusic J, Podlipnik C, Jevsevar S, Kuzman D, Vesnaver G, Lah J. J. Biol. Chem. 2012;287:8613–8620. doi: 10.1074/jbc.M111.318451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris LJ, Skaletsky E, McPherson A. J. Molec. Biol. 1998;275:861–872. doi: 10.1006/jmbi.1997.1508. [DOI] [PubMed] [Google Scholar]
- 34.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe J-P, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo W-L, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlick TL, Ding Z, Kovacs EW, Francis MB. J. Am. Chem. Soc. 2005;127:3718–3723. doi: 10.1021/ja046239n. [DOI] [PubMed] [Google Scholar]
- 36.Esser-Kahn AP, Francis MB. Angew. Chem. Int. Ed. 2008;47:3751–3754. doi: 10.1002/anie.200705564. [DOI] [PubMed] [Google Scholar]
- 37.Wakankar A, Chen Y, Gokarn Y, Jacobson FS. mAbs. 2011;3:161–172. doi: 10.4161/mabs.3.2.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netirojjanakul C, Witus LS, Behrens CR, Weng C, Iavarone AT, Francis MB. Chem. Sci. 2013;4:266–272. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.