Abstract
To study the controversial role of cytomegalovirus (CMV) in glioblastoma, we assessed the effects of murine CMV (MCMV) perinatal infection in a GFAP-cre; Nf1loxP/+; Trp53−/+ genetic mouse model of glioma (Mut3 mice). Early on after infection, MCMV antigen was predominantly localized in CD45+ lymphocytes in the brain with active viral replication and local areas of inflammation, but, by 7 weeks, there was a generalized loss of MCMV in brain, confirmed by bioluminescent imaging. MCMV-infected Mut3 mice exhibited a shorter survival time from their gliomas than control Mut3 mice perinatally infected with mock or with a different neurotropic virus. Animal survival was also significantly shortened when orthotopic gliomas were implanted in mice perinatally infected with MCMV versus controls. MCMV infection increased phosphorylated STAT3 (p-STAT3) levels in neural stem cells (NSC) harvested from Mut3 mice subventricular zone, and, in vivo, there was increased p-STAT3 in NSCs in MCMV-infected compared with control mice. Of relevance, human CMV (HCMV) also increased p-STAT3 and proliferation of patient-derived glioblastoma neurospheres, whereas a STAT3 inhibitor reversed this effect in vitro and in vivo. These findings thus associate CMV infection to a STAT3-dependent modulatory role in glioma formation/progression in the context of tumor suppressor mutations in mice and possibly in humans.
Introduction
Recent cancer genome studies have identified frequent genetic alterations in human glioblastomas (1, 2), the most common and aggressive glioma with a dismal prognosis (3). The causal role of several combinations of genetic mutations in glioblastoma formation have been shown in mice (4), although the mechanism by which these genetic alterations induce glioblastomas is not known. Several studies have shown the presence of CMV in glioblastoma (5–9), suggesting that the virus may participate in tumor pathogenesis. This remains controversial because other studies dispute the association between CMV and tumor (10–12). In addition, CMV has not been proven oncogenic, although CMV can deregulate cancer-related signaling (13–15). To date, no reports have investigated how intact CMV can contribute to glioblastoma in an animal model.
HCMV is a large DNA β-herpesvirus that infects 40% to 100% of adults with a majority of infections occurring during the first year of life (16). It infects a wide range of tissues including the brain (17). Primary infection in immunocompetent humans is typically asymptomatic and eventually subsides into latency or low-level persistent infection in many cells, including brain cells. Murine CMV (MCMV) and HCMV share significant functional similarities, making MCMV an accepted model for HCMV infection and pathogenesis (18, 19). Following acute primary infection in immunocompetent hosts, both HCMV and MCMV become latent and are reactivated by inflammation and immune suppression. Like HCMV, MCMV is neurotropic, and it has been used to study neuropathogenesis (20).
To investigate the role of CMV in glioblastoma, we infected physiologically relevant neurosphere culture and mouse models with CMV. Here, we report that CMV infection contributes to malignant gliomas, likely through STAT3 signaling.
Materials and Methods
Cell culture and virus preparation
We cultured NIH/3T3 mouse fibroblasts (American Type Culture Collection) in Dulbeccos' Modified Eagle Media (DMEM) with 10% bovine calf serum and human foreskin fibroblasts (HFF) in DMEM with 10% FBS. We maintained the cultures with penicillin (100 U/mL) and streptomycin (10 µg/mL). MCMV lacking the m157 gene (21), MCMV-luciferase (22), and HCMV TR clinical strain (23) were kindly provided by Ulrich Koszinowski (Ludwig-Maximilians-Universitat, Munich, Germany), Michael Mach (Institute for Clinical and Molecular Virology, University Hospital, Erlangen Germany), and Elizabeth Fortunato (University of Idaho, Moscow, ID), respectively. We cultured MCMV and HCMV in NIH/3T3 and HFF cells, respectively, and measured viral titer as previously described (21, 22, 24). HSV1 was cultured and tittered in vitro as previously described (25).
In vitro virus infection and analysis
Neurosphere cultures from patient-derived glioblastomas (146, 157, and 112310) were kindly provided by I. Nakano (Ohio State University Medical Center, Columbus, OH; ref. 26). We established and maintained neurosphere cultures from mouse subventricular zone (SVZ) as previously described (27, 28), except using EGF from Daewoong Pharmaceutical Co., Ltd and TrypLE (Invitrogen). For infection, we seeded cells in 6-well plates and treated with CMV or mock (purified extract from uninfected fibroblasts) the next day. We infected mouse SVZ neurospheres with MCMV [multiplicity of infection (MOI), 1.0] or patient-derived human glioblastoma neurospheres with HCMV (MOI, 1.0) for 2 days. We harvested the cells at specific time points with radioimmunoprecipitation assay (RIPA) buffer (150 mmol/L NaCl, 50 mmol/L Tris, 1% NP-40, 0.1% SDS, 0.5% SDS) for protein lysates. We conducted Western blotting as previously described (29). Primary antibodies used were against HCMV IE1, HCMV pp65 (Abcam), p-STAT3, STAT3 (Cell Signal Technology), α-tubulin (GE Healthcare), or β-actin (Sigma). To quantify proliferation, we used a colorimetric MTS assay (Promega) according to manufacturer's protocol. Absorbance was measured 4 hours after addition of substrate. In addition, we used Countess Cell Counter (Invitrogen) according to manufacturer's protocol, as an additional way to quantify proliferation. To assess the effect of STAT3 activation, we treated glioblastoma neurospheres with 0.5 µmol/L of Stattic (Tocris Bioscience) dissolved in dimethyl sulfoxide (DMSO; final 0.1%) 5 hours after plating as well as when new media were added.
Mice, virus infection, and bioluminescence imaging
All mouse experiments and care were approved by the Institutional Animal Care and Use Committee of The Ohio State University. We bred Mut3 male mice (27) with B6CBAF1/J females (The Jackson Laboratory) to generate Mut3 mice. We bred the mice in a CMV-free animal vivarium and transferred pregnant females to an isolated vivarium that tolerated CMV infection. We intraperitoneally injected postnatal day 2 (P2) mice with 103 plaque-forming units (pfu) of MCMV Smith strain in 100 µL of PBS or PBS only (mock). Additional mice received 103 pfu of HSV1 F strain (30) via intraperitoneal injection in 100 µL of PBS. To prevent virus spreading, we housed MCMV- and mock-infected mice on separate sides of a mouse cage rack, always handled mock-infected mice before the virus-infected mice, and changed cages on different days. Both groups of virus-infected mice were never handled on the same day. We genotyped the mice between P9 and P12 as described (27) and confirmed the genotypes after harvesting their tissues. For bioluminescence imaging (BLI), we injected a small cohort of P2 mice with 1.5 × 103 pfu of MCMV-luciferase. We injected MCMV-luciferase mice with luciferin (10 mg/kg) 20 minutes before BLI. After anesthetizing the mice with a mixture of isofluorane and oxygen, we placed them supine in a Xenogen IVIS imager Model FA 448 (Caliper Life Sciences) to assess luciferase activity under manufacturer's instruction. We conducted BLI on the same mice each week until luciferase activity disappeared.
MCMV PCR, ELISA, and, plaque formation assay
After deeply anesthetizing mice by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (20 mg/kg) mixture, we obtained approximately 0.5 mL of blood sample via the facial vein. We then intracardially perfused the mice with ice-cold PBS and immediately dissected out the ear, salivary gland, cerebellum, cerebral cortex, subventricular zone, and hippocampus. We purified gDNA from the tissues using a DNeasy kit (Qiagen) and assessed MCMV glycopro-tein B (GB) gene by PCR using the primers in Supplementary Table S1. We also extracted serum from the blood samples and assessed CMV seroconversion by using a MCMV ELISA kit (XpressBio) under manufacturer's instructions. For plaque formation assay, after deeply anesthetizing mice by the ketamine and xylazine mixture, we decapitated mice and aseptically removed the brain. We sagittally divided the brain, placed in a preweighed vial containing DMEM, and sonicated at full power for 15 seconds to disrupt the tissue. We plated resulting lysate in 10-fold dilutions on confluent NIH/3T3 and conducted plaque formation assay as previously described (21). To isolate total RNA, we processed whole brain tumors from symptomatic mice using RNeasy Lipid Tissue Kit (Qiagen). DNA was removed using on-column DNase degradation. We synthesized cDNA from 1 µg of total RNA and carried out reverse transcriptase (RT)-PCR. Samples without RT enzyme were used as negative control to ensure that there was no DNA contamination.
Syngeneic orthotopic and ectopic mouse model of glioblastoma
We deeply anesthetized 8-week-old female B6CBAF1/J mice (Jackson Laboratory) by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (20 mg/kg) mixture and positioned them on stereotactic injection frames. After locally shaving hairs on the head, we drilled a small hole and injected 1.0 × 105 of neurosphere-forming glioblastoma cells into the striatum. The stereotactic coordination of intracranial injection was 0.5 mm anterior, 2.0 mm lateral to the bregma, and 2.75 mm intraparenchymally. We sacrificed the mice either 4 weeks after the injection or when they showed a neurological symptom of brain tumor as previously described (27). For the ectopic model, 8-week-old athymic mice (National Cancer Institute, Bethesda, MD) were temporarily anesthetized with isofluorane. Two hundred fifty thousand human brain tumor stem cells that were infected with HCMV (MOI, 1.0) or mock-infected for 1 week were injected into flanks of mice in an equal volume of Matrigel (BD Biosciences) for a total volume of 200 µL. After tumors were palpable, they were measured every 4 days with calipers. Volume of tumor was determined via this formula: (p/6) length × width2.
Histology
After anesthetizing mice by the ketamine and xylazine injection, we intracardially perfused the mice with ice-cold PBS followed by 4% paraformaldehyde (PFA), immediately dissected out the brain, and post-fixed in 4% PFA overnight. We then cut the brain into sagittal halves, processed for paraffin embedding, and cut into 6-µm sections. We stained every fifth section with hematoxylin and eosin. Board-certified neuropathologists who were blinded to infection status evaluated the slides for tumor and any other abnormalities. We conducted immunohistochemistry (IHC) as previously described (27–29, 31). To assess MCMV immunoreactivity, we used a polyclonal antibody against CMV that did not detect CMV-mediated Fc-like receptor (Supplementary Fig. S1). Antibodies used were against CMV (chicken, Virusys), MCMV IE1 (Croma 101 antibody; ref. 32; mouse, from Stipan Jonjic, University of Rijeka), GFP (chicken, Aves lab), Dcx (goat, Santa Cruz Biotechnology), CD45 (mouse, BD Pharmingen), or p-STAT3 (rabbit, Cell Signaling Technology). We used MOM detection kits (Vector Laboratories) when mouse-derived antibodies were used. When Croma101 antibody was used, we specially processed tissue sections as described (7), including pepsin treatment and blocking for avidin and biotin, and used a Fab fragment of secondary antibody against mouse IgG (Jackson Immunoresearch). To assess specificity of chicken anti-CMV antibody, we treated the sections with Fc receptor blocker (Innovex Biosciences), according to manufacturer's instruction. For p-STAT3 IHC, we additionally added SignalStain Enhancer (Cell Signaling Technology) for 40 minutes to detect primary antibody. We captured microscopic images by using Olympus BX51 microscope (Olympus America Inc.) equipped with DP72 camera, DP2-BSW S/W, and X-cite 120Q fluorescence lamp (Lumen Dynamics). For high-resolution analysis (1-µm optical slice, 8-µm Z-stack), we captured images with Zeiss LSM510 confocal microscope system (Carl Zeiss Inc.) equipped with 63× oil-immersion lens. We counted MCMV-, p-STAT3-, or proliferating cell nuclear antigen (PCNA)-positive cells in 3 or 4 0.44 × 0.33 nm2 fields by using Meta-Morph software (Universal Imaging Corporation) and displayed as mean ± SEM.
Statistical analyses
Mouse survival data were plotted (Kaplan–Meier survival curves) and analyzed using log-rank test in GraphPad Prism S/W (GraphPad Software, Inc.). For the in vitro assays, we used 2-sample Student t test or one-way ANOVA when more than 2 groups were compared. All error bars indicate SEM.
Results
Perinatal MCMV infection leads to viral infection in mouse brain
To test a potential role for CMV in glioblastoma pathogenesis, we used the Mut3 mouse model (GFAP-cre; Nf1loxP/+; Trp53−/+), which spontaneously develop high-grade astrocytomas (World Health Organization's grade III anaplastic astrocytoma and grade IV glioblastoma) with almost complete penetrance by adulthood (28). We infected Mut3 and wild-type (wt) mice with a nonlethal dose of MCMV-Δm157 strain or MCMV-luciferase on postnatal day 2 (P2). MCMV-infected mice were behaviorally and symptomatically indistinguishable from mock-infected mice before tumor development. We detected MCMV-specific glycoprotein B (GB) gene in multiple tissues, including the brain, from MCMV- but not mock-infected mice (Fig. 1A). At 3 weeks of age, scattered MCMV immunoreactivity was evident in multiple brain regions in MCMV-, but not mock-infected brains (Fig. 1B, top). MCMV-immunopositive brains exhibited inflammatory infiltrates (Fig. 1C), and a majority of MCMV-immunoreactivity was in CD45+ leukocytes (Fig. 1D). Brain extracts from MCMV-infected mice formed plaques in 3T3 cells (Fig. 1E), indicating MCMV replication. By 7 weeks of age, MCMV-immunopositive cells were mainly restricted to a few brain regions, including the hippo-campus and SVZ (Fig. 1B, bottom). BLI of mice infected with MCMV-luciferase showed a generalized loss of MCMV-mediated gene expression by P57 (Fig. 1F). These data were consistent with an initial level of active MCMV infection/inflammation in brain, followed by loss of MCMV gene expression at later time points.
Figure 1.
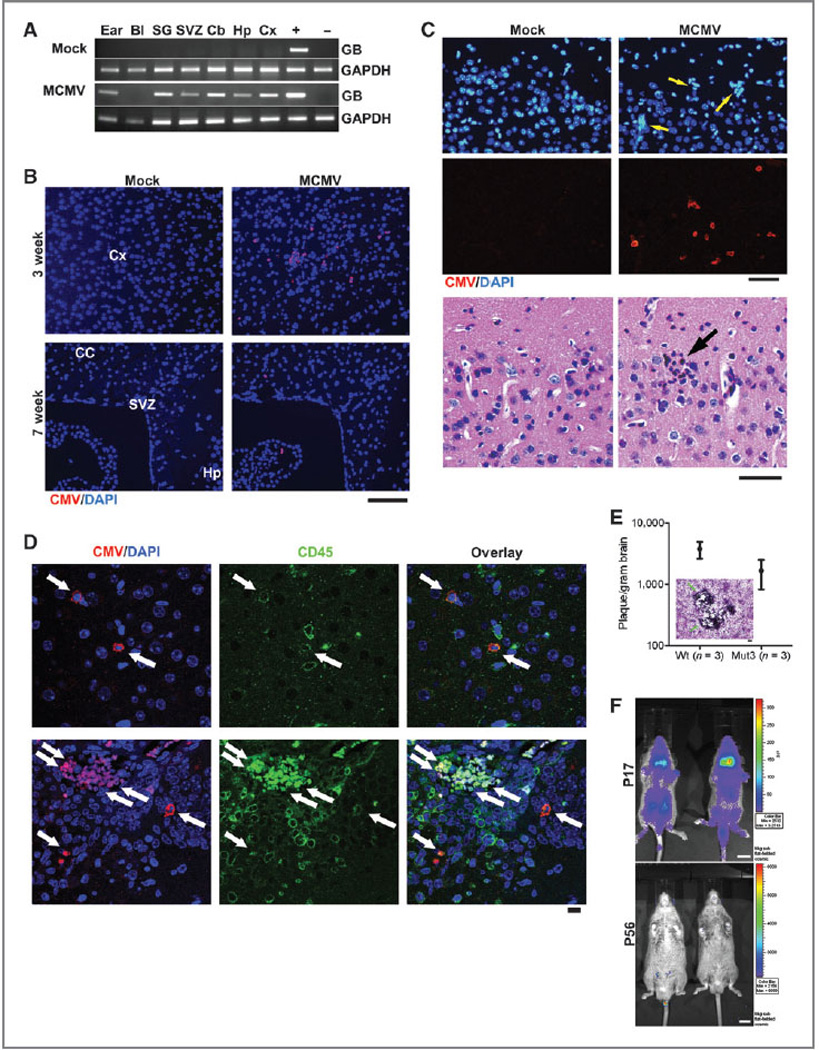
Perinatal MCMV infection resulted in productive virus replication and CMV immunoreactivity in CD45-positive cells in the brain. A, PCR analysis for MCMV glycoprotein B (GB) gene in genomic DNA obtained from PBS-perfused mock-versus MCMV-infected, 8-week-old mice. Representative data from n = 3 per group are shown. Bl, blood; SG, salivary gland; Cb, cerebellum; Hp, hippocampus; Cx, cortex. + and −, gDNA from MCMV-infected and -negative cells, respectively. B, IHC data for CMV in the brains of mock-versus MCMV-infected mice. Representative data from n = 3 per group are shown. Cx, cortex; CC, corpus callosum; Hp, hippocampus. Scale bar, 100 mm. C, top, IHC images show ectopic cellularity (blue, arrows) close to CMV-positive cells (red) in the cortex of 3-week-old wt mice infected with MCMV at P2, which were absent in mock-infected mice. Bottom, hematoxylin and eosin-stained brain sections adjacent to the sections in top images contained an inflammatory focus (arrow) in the MCMV mice, which was absent in mock-infected mice. Representative data from n = 3 per group are shown. In brain sections containing both the dentate gyrus and SVZ, 3.67 × 1.45 (mean ± SEM) of inflammatory foci were detected per section. Scale bars, 50 mm. D, confocal images of IHC for CMV and CD45 display immunoreactivity in the brains from 2-week-old wt mice. Cells double-positive for both CMV and CD45 are noted by arrows. Representative data from n = 7–8 images on n = 3 brains per group are shown. Scale bar, 10 mm. E, brain extract of 2-week-old mice infected with MCMV formed virus-induced plaques in 3T3 cells. There was no significant difference between wt and Mut3 (P = 0.16). Error bars indicate SEM. Inset, a representative image of plaque formation (arrows) in 3T3 cells. Scale bar, 100 mm. No plaque was formed by brain extract of mock-infected mice (n = 3 per group, not shown). F, bioluminescent live imaging on P17 and adult (P56) mice infected with MCMV-luciferase. Representative data from n = 3 per group are shown. Scale bar, 1 cm.
Perinatal MCMV infection specifically shortens the survival of Mut3 mice
We next determined the outcome of perinatal MCMV infection of Mut3 mice. MCMV-infected Mut3 mice died significantly earlier than mock-infected Mut3 mice both as a group and in the subset of mice with confirmed gliomas (Fig. 2A). Infection of mice with HSV1, another neurotropic herpesvirus, did not significantly alter survivorship. A majority of gliomas in MCMV-infected Mut3 mice were glioblastomas with necrosis and increased microvascular density (Fig. 2B and Supplementary Table S2). MCMV-infected wt mice did not develop brain tumors, nor did they live less than mock-infected wt mice (data not shown). IHC and RT-PCR showed that gliomas from MCMV-infected mice contained MCMV immunoreactivity (Fig. 2C) and GB gene expression (Fig. 2D), which were not detected in tumors from mock-infected Mut3 mice. These findings indicated that MCMV shortened survival in the context of Mut3 mutations.
Figure 2.
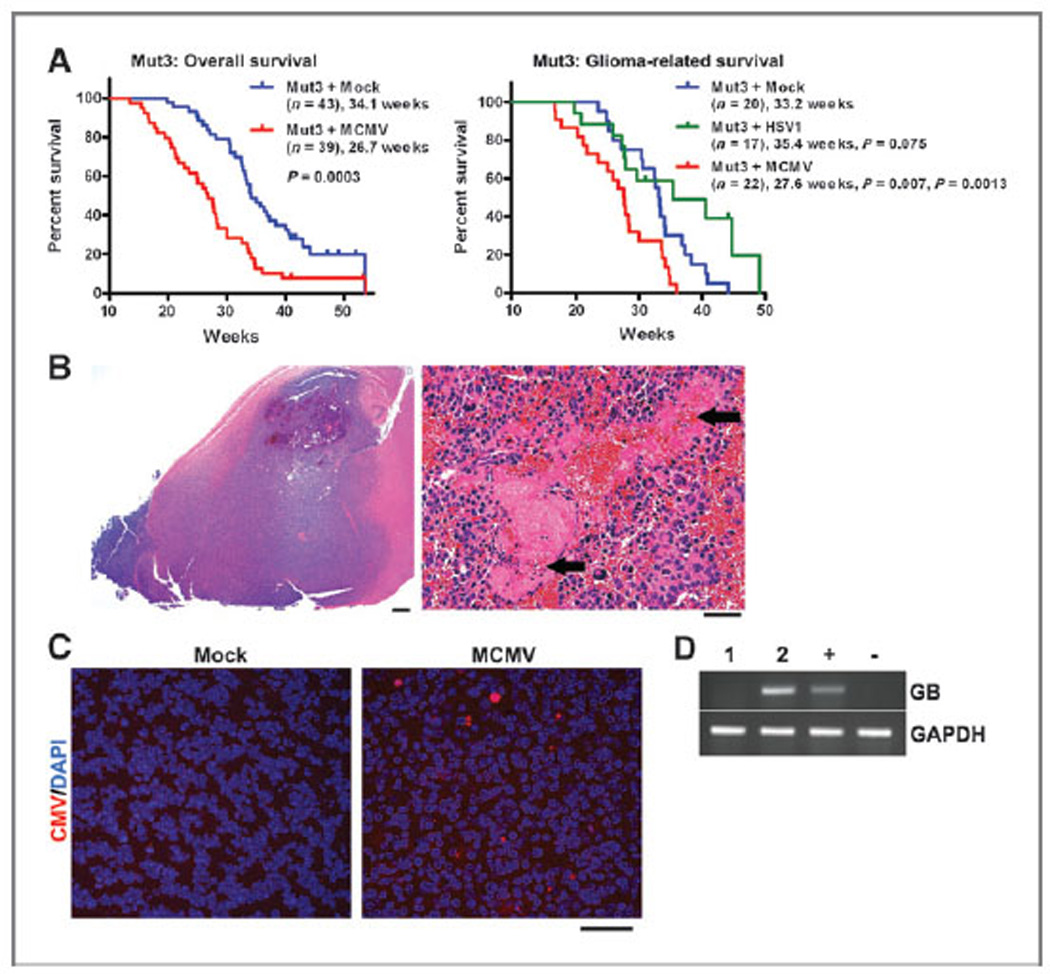
Perinatal MCMV infection shortens the survival of Mut3 mice. A, Kaplan–Meier curves display overall (left) and histologically confirmed glioma (right) survivals of Mut3 mice. Labels indicate infection groups, mouse numbers, mean survival time, and P values against mock and HSV1 (first and second P values, respectively). B, left, a representative hematoxylin and eosin-stained brain section from an MCMV-infected Mut3 mouse that harbored a grade IV astrocytoma (glioblastoma) spreading extensively throughout the forebrain. Scale bar, 500 mm. Right, a higher magnification image from the same tumor displaying extensive necrosis (arrows). Scale bar, 50 mm. C, IHC images for CMV in gliomas from mock-versus MCMV-infected Mut3 mice. Representative data from n = 4 per group are shown. About 41.3 × 16.2 of CMV-positive cells (mean ± SEM) were detected per nm2 of randomly chosen tumor field (n = 4) in gliomas from MCMV-infected Mut3 mice, but none from gliomas from mock-infected Mut3 mice. Scale bar, 50 mm. D, RT-PCR analysis for MCMV glycoprotein B (GB) in gliomas from mock- (1) versus MCMV-infected (2) Mut3 mice (n = 1). + and −, MCMV-infected and -negative cells, respectively.
The MCMV-infected brain microenvironment influences glioblastoma survival. The Mut3 mouse data showed MCMV presence in brain CD45 cells before glioma development. This suggested that the brain environment in MCMV-infected mice was influencing the timing of glioma formation/progression. To examine this, we used a syngeneic orthotopic model. Intracranial injection of Nf1:Trp53:Pten mouse glioblastoma cells (27) into the striatum of wt mice recapitulates glioblastoma histology (33). We infected wt (same strain as Mut3) mice with mock or MCMV at P2 and intracranially injected MCMV-free mouse glioblastoma cells (27) at 8 weeks of age. MCMV-infected wt recipients died significantly earlier than control (Fig. 3A). Mean survival was decreased by 1.8 weeks, an approximately 19% shortened survival similar to data from Mut3. Symptomatic mice in both groups died with glioblastomas (not shown). MCMV immunoreactivity co-localized with CD45-positive leukocytes in tumors from MCMV- but not mock-infected mice (Fig. 3B).
Figure 3.
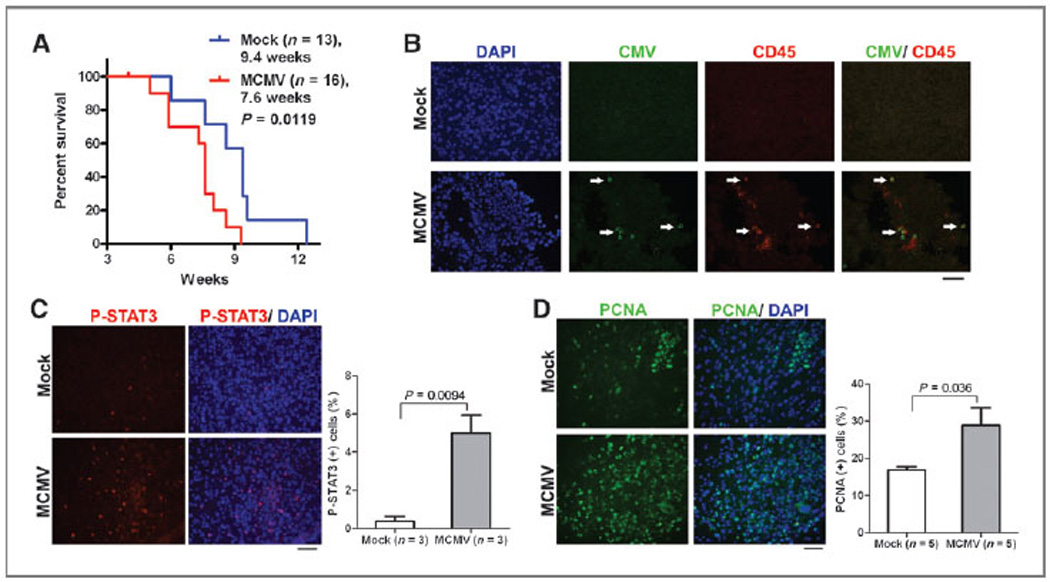
MCMV infection shortened survival of an orthotopic mouse model of glioma with STAT3 activation and increased PCNA. A, Kaplan–Meier curves display the survival of mock-versus MCMV-infected wt mice after orthotopic injection of mouse glioblastoma cells. Labels indicate infection groups, mouse numbers, mean survival time (weeks after tumor cell injection), and P values. B, IHC data for CMV and CD45 in gliomas derived from mock-versus MCMV-infected wt mice orthotopically injected with mouse glioblastoma cells. Arrows point to double-positive cells. Representative data from n = 3 per group are shown. Scale bar, 50 mm. C, left, IHC data for p-STAT3 in gliomas derived from mock-versus MCMV-infected wt mice 4 weeks after orthotopic injection with mouse glioblastoma cells. Representative data from n = 3 per group are shown. Scale bar, 50 mm. Right, blind cell counting displays ratio of p-STAT3–positive cells in the tumor regions. D, left, IHC data for PCNA in gliomas derived from mock-versus MCMV-infected wt mice 4 weeks after orthotopic injection with mouse glioblastoma cells. Representative data from n = 5 per group are shown. Scale bar, 50 mm. Right, blind cell counting displays ratio of PCNA-positive cells in the tumor regions.
STAT3 is considered a critical mediator of glioblastoma and its activation results in proliferation and cell-cycle progression (34). To determine whether MCMV-infection altered STAT3 signaling in brains, we analyzed asymptomatic mice, 4 weeks after orthotopic injection of the tumor cells. At this stage, although gliomas were detected in the brain, the recipient mice did not exhibit behavioral abnormalities. We found a significant increase in p-STAT3 in tumors from MCMV-versus mock-infected asymptomatic mice (Fig. 3C). Because enhanced STAT3 activation increases proliferation, we examined PCNA at this time point. PCNA was significantly elevated in glioblastomas from MCMV-versus mock-infected wt mice (Fig. 3D). Increased glioblastoma proliferation as a function of p-STAT3 induced by MCMV suggests a potential mechanism for the observed acceleration of glioblastoma-induced death.
MCMV-mediated STAT3 activation in Mut3 mouse brains before glioma onset
We thus further tested whether perinatal MCMV led to STAT3 activation before glioblastoma detection in Mut3 mice. In vitro MCMV infection increased p-STAT3 levels in NSC cultures from 4-week-old, asymptomatic Mut3 mice (Fig. 4A). In vivo, p-STAT3 was observed in the subgranular zone (SGZ) and SVZ of asymptomatic MCMV-infected Mut3 brains but not of mock-infected Mut3 or MCMV-infected wt brains of the same age (Fig. 4B). The p-STAT3 immunoreactivity co-localized with a type A NSC marker Dcx (Fig. 4C), indicating that MCMV infection can potentiate STAT3 activation in a cell type reported to give rise to glioma (4, 27, 28). These data indicate that MCMV infection increased STAT3 activation in Mut3 mouse NSCs before glioma onset.
Figure 4.
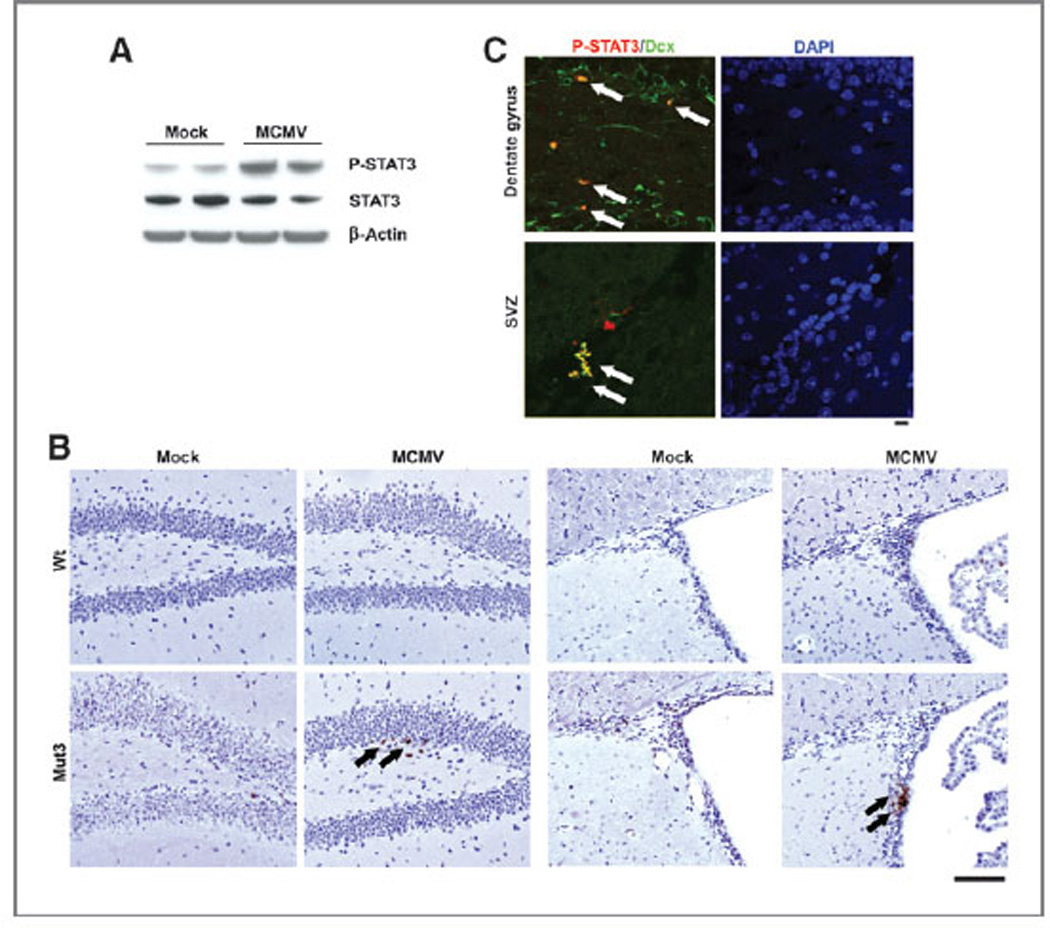
p-STAT3 in NSCs in MCMV-infected Mut3 brains. A, p-STAT3, STAT3, and β-actin Western blotting using NSC cultures derived from the SVZ of 4-week-old Mut3 mice. The cultures were infected with MOI of 1.0 of MCMV and incubated for 2 days. Representative data from n = 4 per group are shown. Densitometric scanning reveals 2.75-fold more p-STAT3/β-actin ratio in MCMV than in mock samples. B, p-STAT3 IHC staining (arrows) in the dentate gyrus SGZ (left) and SVZ (right) of 7-week-old mock-versus MCMV-infected wt or Mut3 mice. Representative data from n = 3 per group are shown. Scale bar, 100 mm. C, confocal images of IHC for p-STAT3 and Dcx in the brain of 7-week-old MCMV-infected Mut3 mice. Dually immunoreactive cells are labeled with arrows. Representative data from n = 3 per group are shown. Scale bar, 10 mm.
Human CMV infection specifically activates STAT3 in patient-derived glioblastoma neurospheres
The previous studies showed that MCMV increased STAT3 activation in Mut3 NSCs before glioma onset in association with increased proliferation of Mut3 glioblastomas. To determine human relevance, we used HCMV to infect human glioblastoma neurospheres. In vitro infection of patient-derived glioblastoma neurospheres with HCMV but not with a highly related virus herpes simplex virus 1 (HSV1) elevated p-STAT3 (Fig. 5A). There was upregulation of HCMV IE1 and pp65 in infected patient-derived glioblastoma neurospheres, indicative of active replication (Fig. 5B). There was a visible increase in the number and size of patient-derived glioblastoma neurospheres after HCMV infection (Fig. 5C). Quantitatively, HCMV infection significantly increased cell numbers in patient-derived glioblastoma neurospheres cultures (Fig. 5D). These findings thus showed that HCMV also increased patient-derived glioblastoma neurospheres growth and was associated with STAT3 activation.
Figure 5.
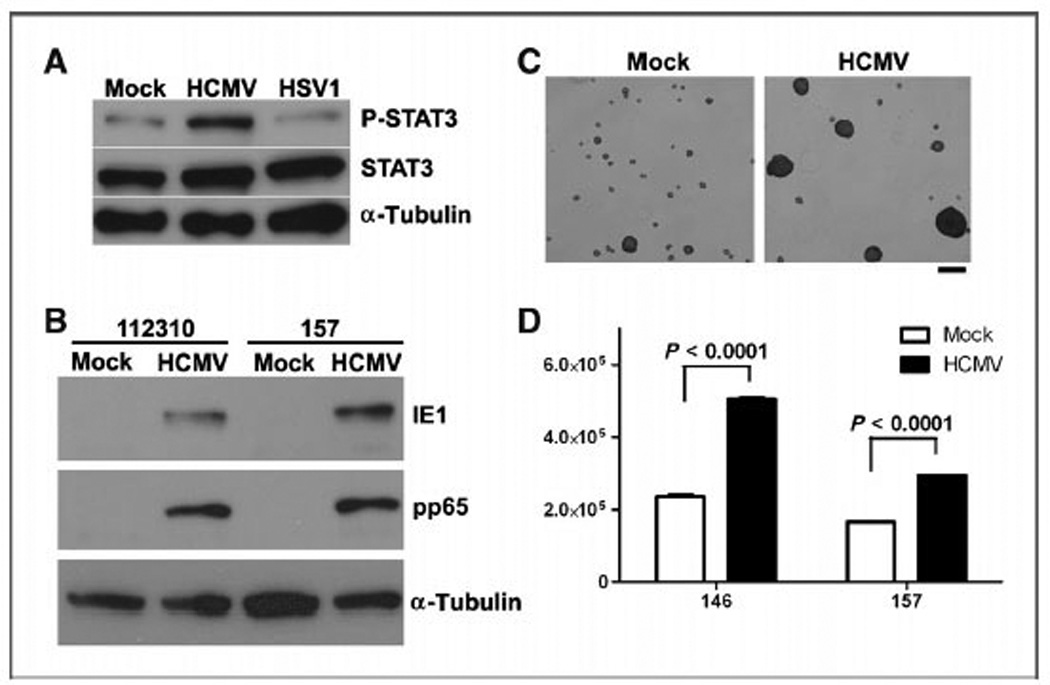
HCMV increased p-STAT3 and the proliferation of human glioblastoma neurosphere cultures. A, p-STAT3, STAT3, and α-tubulin Western blotting using a patient-derived human glioblastoma neurosphere culture, 2 days after in vitro infection with MOI of 1.0 of HCMV or HSV1. Representative data from n = 3 per group are shown. Densitometric scanning reveals 2.43-fold more p-STAT3/α-tubulin ratio in HCMV than in mock samples. B, HCMV-specific IE1 and pp65 Western blotting using 2 patient-derived human glioblastoma neurosphere cultures (112310 and 157), 2 days after in vitro infection with MOI of 1.0 of HCMV. Representative data from n = 3 per group are shown. C, representative images of a patient-derived human glioblastoma neurosphere culture 7 days after in vitro infection with MOI of 1.0 of HCMV are shown. Scale bar, 100 mm. D, two patient-derived human glioblastoma neurosphere cultures (146 and 157) were infected with HCMV and cell numbers were counted (n = 3 per group).
STAT3 inhibition blocks HCMV stimulation of patient-derived glioblastoma neurosphere growth
Finally, we asked whether inhibition of STAT3 activation decreased the observed stimulation of HCMV on patient-derived glioblastoma neurosphere growth in vitro and in vivo. Using 2 different patient-derived glioblastoma neurosphere lines, STAT3 inhibition significantly decreased the proliferation of HCMV-infected cells (Fig. 6A). Ectopic mock-infected glioblastoma implants grew less than HCMV-infected ones, but STAT3 inhibition led to similar growth, indicating that STAT3 inhibition abolished the HCMV-mediated proliferative advantage (Fig. 6B). STAT3 inhibition reduced the number of p-STAT3+ tumor cells (Fig. 6C). These data provided evidence that STAT3 activation was a likely mechanism for the observed effect of CMV on glioblastoma growth.
Figure 6.
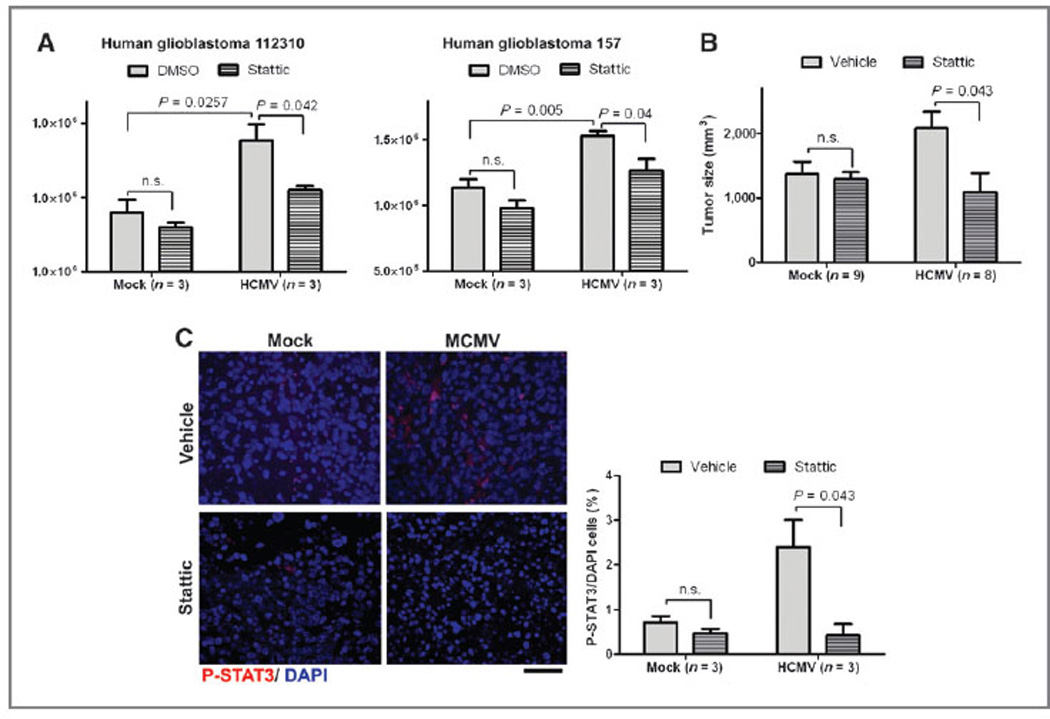
STAT3 inhibition abolished the HCMV-mediated proliferative advantage of GBM neurospheres. A, two patient-derived human glioblastoma neurosphere cultures (112310 and 157) were infected with HCMV (MOI, 1.0) and incubated with or without STAT3 inhibitor Stattic (0.5 mmol/L). Cell numbers were counted 7 days after incubation. B, either mock-infected or HCMV-infected (MOI, 1.0) patient-derived glioblastoma neurospheres were injected into flanks of athymic mice. Mice were i.p. injected with either Stattic (50 mg/kg) or vehicle every day. Tumors were measured every 4 days with calipers (P < 0.05, ANOVA). C, left, IHC data for p-STAT3 in gliomas from mouse flanks 26 days after injection with mock-versus HCMV-infected human glioblastoma cells after treatment with Stattic (50 mg/kg, 1 i.p. dose/d) or vehicle. Representative data from n = 3 per group are shown. Scale bar, 50 mm. Right, blind cell counting displays ratio of p-STAT3/40,6-diamidino-2-phenylindole (DAPI)-positive cells in the tumor regions.
Discussion
There has been considerable controversy related to the role of CMV in glioma formation/progression. Several report HCMV in nearly 100% of glioblastoma (6–8). However, whether CMV is causal, inconsequential, or oncomodulatory remains unknown. In 2 glioma mouse models, we show that MCMV infection decreases mice survival by about 20%, a phenomenon not observed with another neurotropic virus. Analysis of tumors revealed CMV protein. Wt mice infected with MCMV did not develop gliomas and thus MCMV is not causal for tumors on its own. Instead, MCMV cooperates with tumor suppressor loss to accelerate glioma formation/proliferation. Therefore, these data indicate for the first time that a nononcogenic virus exhibits oncomodulatory effects in the context of a specific set of genetic mutations, at least in mice. This is the first animal model that uses the combination of CMV and tumor suppressor mutations to show their combined effect on glioma formation/progression.
We observed activation of STAT3 in vitro in human glioblastoma cells and neurosphere by HCMV. Pharmacologic inhibition of STAT3 with Stattic abrogated this enhanced proliferation. Analysis in an orthotopic model of glioma in MCMV-infected mice not only showed STAT3 activation in the tumor but also increased proliferation as evidenced by a significant increase in PCNA-positive cells. In vivo analysis of pretumorigenic MCMV-infected Mut3 mice reveal STAT3 activation that is not observed in age-matched mock-infected Mut3 mice or MCMV-infected wt mice. Interestingly, STAT3 activation is seen in the NSC population, a cell type that has been shown to give rise to gliomas (28). These data show that STAT3 activation may be a central mechanism through which CMV accelerates glioma formation/progression. Others have shown that STAT3 activation is critical for several glioblastoma phenotypes including cell proliferation (35). HCMV-encoded chemokine receptor US28 activated STAT3 in U373MG glioblastoma cells, 3T3 and HEK293T cell lines, and increased proliferation of 3T3 cells (36). Transgenic expression of the HCMV US28 gene in intestinal epithelial cells promoted neoplasia in mice (15). In vitro, CMV infection of glioma "stem" cells and monocytes has been shown to induce a variety of signaling pathways including STAT3 (37). In this study, for the first time, we provide evidence that CMV does upregulate STAT3 in vivo and that inhibition of STAT3 reverses the effects of CMV on glioblastoma growth in vivo. Currently, there are several STAT3 inhibitors being investigated in clinical trials (http://clinicaltrials.gov/). CMV-enhanced STAT3 activation provides a rational target as a therapeutic option for CMV-positive gliomas.
Our data suggest a mechanism whereby perinatal MCMV infection contributes to enhanced glioma formation, possibly through non–cell-autonomous effects on NSCs. This could be mediated through the observed increase in inflammatory cells, leading to increased STAT3 activation, a driver of increased aggressiveness of biologic behavior in gliomas. STAT3 activation in NSCs with tumor suppressor deficiency may provide an explanation for why an endemic CMV infection in humans results in only relatively rare cases of gliomas. Glioma formation in MCMV-free Mut3 mice and the absence of brain tumor in MCMV-infected wt mice clearly indicate that CMV alone is not causal for gliomagenesis. However, the endemic CMV infection in humans, detection of HCMV in most human patients with glioblastoma, and our data proving the role of CMV in glioblastoma cultures and in vivo models suggest that HCMV infection contributes to glioma onset/proliferation.
We have shown the first in vivo evidence that CMV is able to significantly modulate brain cancer in mouse models of glioma. CMV activates STAT3, which leads to an increase in proliferation of glioma and NSCs and ultimately a reduced survival of Mut3 and of an orthotopic mouse models. The described model of MCMV oncomodulation of enhanced gliomagenesis is significant to studying aspects relevant to gliomas such as immunomodulation and glioma formation as well as the use of anti-CMV therapies as a treatment for this tumor. It has been proposed that CMV reactivation by immunosuppression or inflammation may contribute to malignant gliomas (6, 8). Our mouse models of MCMV-mediated glioma will be useful tools to assess the relation-ship among CMV reactivation, immunomodulation, and malignant gliomas. In addition to glioblastoma, HCMV epitopes has been detected in several other cancer types, such as colon cancer (38), breast cancer (39), and medulloblastoma (40). Our models of MCMV infection may be more widely applicable to the study of other cancers in mouse models in which CMV is implicated. Not only will these models help elucidate the role of CMV in tumor biology but also may be useful to test anti-CMV therapies in a variety of cancers.
Supplementary Material
Acknowledgments
The authors thank S.O. Yoon, S.J. Baker, J. Trgovcich, J. Waldman, H. Kawasaki, and members of Dardinger Neurooncology Center and Solid Tumor Program for helpful discussion and suggestion; M. Mach, U. Koszinowski, and Elizabeth Fortunato for generously providing us viruses; I. Nakano for kindly providing us human glioblastoma neurospheres; Daewoong Pharmaceutical Co., Ltd for generously providing us EGF; Stipan Jonjic for kindly providing us Croma 101 antibody; and M, Reddehase, P. Zimmerman, T.H. Kim, J. Chen, B.W. Luikart, H. Duale, A. Chonhenchob, F. Jaynes, A. Fletchner, and M. Rawahneh for technical information and assistance; and Dr. Luis F. Parada (UT Southwestern Medical School) for providing Mut3 mice and discussions.
Grant Support
This study was funded by NIH P30NS045758-06, R01NS064607 (B. Kaur), a Viral Oncology Program Grant, the Dardinger Neurooncology Fund, the Jeffrey Thomas Hayden Foundation (E.A. Chiocca), OSU MBCG Program grant (E.A. Chiocca and C.-H. Kwon), and OSU CCC Start-up Fund (C.-H. Kwon)
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: R.L. Price, C. Cook, E.A. Chiocca, C.-H. Kwon Development of methodology: R.L. Price, C.-H. Kwon
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): R.L. Price, K. Bingmer, M.O. Nowicki, T. Hollon, E. Murnan, C. Alvarez-Breckenridge, B. Kaur, C.-H. Kwon
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): R.L. Price, J. Song, T.H. Kim, X. Mo, T. Hollon, S. Fernandez, A. Rivera, M. Oglesbee, C. Cook, E.A. Chiocca, C.-H. Kwon
Writing, review, and/or revision of the manuscript: R.L. Price, J. Song, T.H. Kim, X. Mo, T. Hollon, C. Alvarez-Breckenridge, M. Oglesbee, C. Cook, E.A. Chiocca, C.-H. Kwon
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): R.L. Price, J.-Y. Yi, E. Murnan, E.A. Chiocca, C.-H. Kwon
Study supervision: E.A. Chiocca, C.-H. Kwon
References
- 1.TCGA-Research-Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleihues P, Cavenee WK, editors. Pathology and genetics of tumours of the nervous system. Lyon, France: IARC Press; 2000. [Google Scholar]
- 4.Kwon C-H. Pten-deficient mouse models for high-grade astrocytomas. In: Meir EV, editor. CNS cancer: models, markers, prognostic factors, targets, and therapeutic approaches. New York, NY: Humana Press; 2009. pp. 77–92. [Google Scholar]
- 5.Miller G. A viral link to glioblastoma? Science. 2009;323:30–31. doi: 10.1126/science.323.5910.30. [DOI] [PubMed] [Google Scholar]
- 6.Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- 7.Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116:79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell DA, Xie W, Schmittling R, Learn C, Friedman A, McLendon RE, et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro Oncol. 2008;10:10–18. doi: 10.1215/15228517-2007-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prins RM, Cloughesy TF, Liau LM. Cytomegalovirus immunity after vaccination with autologous glioblastoma lysate. N Engl J Med. 2008;359:539–541. doi: 10.1056/NEJMc0804818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau SK, Chen YY, Chen WG, Diamond DJ, Mamelak AN, Zaia JA, et al. Lack of association of cytomegalovirus with human brain tumors. Mod Pathol. 2005;18:838–843. doi: 10.1038/modpathol.3800352. [DOI] [PubMed] [Google Scholar]
- 11.Poltermann S, Schlehofer B, Steindorf K, Schnitzler P, Geletneky K, Schlehofer JR. Lack of association of herpesviruses with brain tumors. J Neurovirol. 2006;12:90–99. doi: 10.1080/13550280600654573. [DOI] [PubMed] [Google Scholar]
- 12.Sabatier J, Uro-Coste E, Pommepuy I, Labrousse F, Allart S, Tremoulet M, et al. Detection of human cytomegalovirus genome and gene pro-ducts in central nervous system tumours. Br J Cancer. 2005;92:747–750. doi: 10.1038/sj.bjc.6602339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cinatl J, Scholz M, Kotchetkov R, Vogel JU, Doerr HW. Molecular mechanisms of the modulatory effects of HCMV infection in tumor cell biology. Trends Mol Med. 2004;10:19–23. doi: 10.1016/j.molmed.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bongers G, Maussang D, Muniz LR, Noriega VM, Fraile-Ramos A, Barker N, et al. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J Clin Invest. 2010;120:3969–3978. doi: 10.1172/JCI42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CA. Congenital cytomegalovirus infection. Curr Probl Pediatr Adolesc Health Care. 2003;33:70–93. doi: 10.1067/mps.2003.3. [DOI] [PubMed] [Google Scholar]
- 17.Sinzger C, Jahn G. Human cytomegalovirus cell tropism and pathogenesis. Intervirology. 1996;39:302–319. doi: 10.1159/000150502. [DOI] [PubMed] [Google Scholar]
- 18.Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddehase MJ, Simon CO, Seckert CK, Lemmermann N, Grzimek NK. Murine model of cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol. 2008;325:315–331. doi: 10.1007/978-3-540-77349-8_18. [DOI] [PubMed] [Google Scholar]
- 20.Tsutsui Y, Kashiwai A, Kawamura N, Aiba-Masago S, Kosugi I. Pro-longed infection of mouse brain neurons with murine cytomegalovirus after pre- and perinatal infection. Arch Virol. 1995;140:1725–1736. doi: 10.1007/BF01384337. [DOI] [PubMed] [Google Scholar]
- 21.Bubic I, Wagner M, Krmpotic A, Saulig T, Kim S, Yokoyama WM, et al. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol. 2004;78:7536–7544. doi: 10.1128/JVI.78.14.7536-7544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klenovsek K, Weisel F, Schneider A, Appelt U, Jonjic S, Messerle M, et al. Protection from CMV infection in immunodeficient hosts by adoptive transfer of memory B cells. Blood. 2007;110:3472–3479. doi: 10.1182/blood-2007-06-095414. [DOI] [PubMed] [Google Scholar]
- 23.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Britt WJ. Human cytomegalovirus: propagation, quantification, and storage. Curr Protoc Microbiol. 2010;Chapter 14(Unit 14E.3) doi: 10.1002/9780471729259.mc14e03s18. [DOI] [PubMed] [Google Scholar]
- 25.Terada K, Wakimoto H, Tyminski E, Chiocca EA, Saeki Y. Development of a rapid method to generate multiple oncolytic HSV vectors and their in vivo evaluation using syngeneic mouse tumor models. Gene Ther. 2006;13:705–714. doi: 10.1038/sj.gt.3302717. [DOI] [PubMed] [Google Scholar]
- 26.Nakano I, Joshi K, Visnyei K, Hu B, Watanabe M, Lam D, et al. Siomycin A targets brain tumor stem cells partially through a MELK-mediated pathway. Neuro Oncol. 2011;13:622–634. doi: 10.1093/neuonc/nor023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, et al. Pten haploinsufficiency accelerates formation of high grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser MM, Zhu X, Kwon CH, Uhlmann EJ, Gutmann DH, Baker SJ. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 2004;64:7773–7779. doi: 10.1158/0008-5472.CAN-04-2487. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Hur T, Rosen-Wolff A, Lamade W, Darai G, Becker Y. HSV-1 DNA sequence determining intraperitoneal pathogenicity in mice is required for transcription of viral immediate-early genes in macrophages. Virology. 1988;163:397–404. doi: 10.1016/0042-6822(88)90280-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Kwon CH, Lin L, Li Y, Parada LF. Inducible site-specific recombination in neural stem/progenitor cells. Genesis. 2009;47:122–131. doi: 10.1002/dvg.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trgovcich J, Stimac D, Polic B, Krmpotic A, Pernjak-Pugel E, Tomac J, et al. Immune responses and cytokine induction in the development of severe hepatitis during acute infections with murine cytomegalovirus. Arch Virol. 2000;145:2601–2618. doi: 10.1007/s007050070010. [DOI] [PubMed] [Google Scholar]
- 33.Kim TH, Song J, Alcantara Llaguno SR, Murnan E, Liyanarachchi S, Palanichamy K, et al. Suppression of peroxiredoxin 4 in glioblastoma cells increases apoptosis and reduces tumor growth. PLoS One. 2012;7:e42818. doi: 10.1371/journal.pone.0042818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brantley EC, Benveniste EN. Signal transducer and activator of tran-scription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slinger E, Maussang D, Schreiber A, Siderius M, Rahbar A, Fraile-Ramos A, et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal. 2010;3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 37.Dziurzynski K, Wei J, Qiao W, Hatiboglu MA, Kong LY, Wu A, et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Cancer Res. 2011;17:4642–4649. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harkins L, Volk AL, Samanta M, Mikolaenko I, Britt WJ, Bland KI, et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–1563. doi: 10.1016/S0140-6736(02)11524-8. [DOI] [PubMed] [Google Scholar]
- 39.Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, Wang W, et al. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baryawno N, Rahbar A, Wolmer-Solberg N, Taher C, Odeberg J, Darabi A, et al. Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J Clin Invest. 2011;121:4043–4055. doi: 10.1172/JCI57147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


