Abstract
T-20 (enfuvirtide) resistance is caused by the N43D primary resistance mutation at its presumed binding site at the N-terminal heptad repeat (N-HR) of gp41, accompanied by the S138A secondary mutation at the C-terminal HR of gp41 (C-HR). We have discovered that modifying T-20 to include S138A (T-20S138A) allows it to efficiently block wild-type and T20-resistant viruses, by a mechanism that involves improved binding of T-20S138A to the N-HR that contains the N43D primary mutation. To determine how HIV-1 in turn escapes T-20S138A we used a dose escalation method to select T-20S138A-resistant HIV-1 starting with either wild-type (HIV-1WT) or T-20-resistant (HIV-1N43D/S138A) virus. We found that when starting with WT background, I37N and L44M emerged in the N-HR of gp41, and N126K in the C-HR. However, when starting with HIV-1N43D/S138A, L33S and I69L emerged in N-HR, and E137K in C-HR. T-20S138A-resistant recombinant HIV-1 showed cross-resistance to other T-20 derivatives, but not to C34 derivatives, suggesting that T-20S138A suppressed HIV-1 replication by a similar mechanism to T-20. Furthermore, E137K enhanced viral replication kinetics and restored binding affinity with N-HR containing N43D, indicating that it acts as a secondary, compensatory mutation. We therefore introduced E137K into T-20S138A (T-20E137K/S138A) and revealed that T-20E137K/S138A moderately suppressed replication of T-20S138A-resistant HIV-1. T-20E137K/S138A retained activity to HIV-1 without L33S, which seems to be a key mutation for T-20 derivatives. Our data demonstrate that secondary mutations can be consistently used for the design of peptide inhibitors that block replication of HIV resistant to fusion inhibitors.
Keywords: resistance, HIV-1, gp41, T-20, mutation, fusion inhibitor
1. Introduction
Human immunodeficiency virus type 1 (HIV-1) fusion to host cell membrane is mediated by formation of a six-helix bundle of the transmembrane subunit gp41 (Chan et al., 1997). Peptides corresponding to amino acid sequences of the gp41 carboxyl-terminal heptad repeat (C-HR) inhibit the HIV-1 fusion by acting as decoys and interfering with the formation of the six-helix bundle (Chan et al., 1998, Malashkevich et al., 1998). Although modified peptides such as SC34EK (Nishikawa et al., 2009), T-2635 (Dwyer et al., 2008), and D-peptides (Welch et al., 2007), and small molecules (Debnath et al., 1999) have been developed, T-20 (enfuvirtide) is the only fusion inhibitor approved for HIV therapy. It is a 36 amino acid peptide derived from the sequence of C-HR of gp41. It is thought to bind at the N-HR domain of gp41and interfere with the C-HR-N-HR interactions required for membrane fusion and injection of virus into the host cell. T-20 has potent anti-HIV-1 activity and effectively suppresses replication of HIV-1 in vivo (Kilby et al., 1998, Lalezari et al., 2003, Lazzarin et al., 2003). However, HIV-1 rapidly develops resistance through mutations in the amino-terminal HR (N-HR) of gp41, especially in the region between L33 to L45, which is thought to be the binding site of T-20 (Aquaro et al., 2006, Cardoso et al., 2007, He et al., 2008). Among these residues, N43D in the N-HR is one of the representative mutations for resistance to T-20 (Bai et al., 2008, Cabrera et al., 2006, Oliveira et al., 2009, Izumi et al., 2009, Ueno et al., 2009). Interestingly, most variants show impaired replication fitness, and thus often go on to acquire secondary mutations, such as S138A (Xu et al., 2005), in the C-HR region of gp41 that corresponds to the sequence of T-20. We and others have recently demonstrated that S138A functions as secondary resistance mutation and enhances resistance to T-20 by restoring impaired replication kinetics of T-20-resistant variants that contain primary mutations in the N-HR region, most notably N43D (Izumi et al., 2009, Watabe et al., 2009).
To preempt this escape strategy, we have previously designed a peptide analog of T-20 with the S138A change incorporated in it (T-20S138A; Fig. 1A) and showed that this peptide significantly suppresses replication of T-20-resistant HIV-1 through enhancement of binding affinity to mutated N-HR, such as N-HRN43D (Izumi et al., 2009). Using circular dichroism (CD) and structural analyses, we also demonstrated that the S138A change provided increased stability to the six-helix bundle (Watabe et al., 2009). In subsequent studies, we validated our approach on another peptide-based fusion inhibitor, C34. In this case, we designed a variant of C34 carrying a secondary escape mutation, N126K, selected for the induction of C34 resistance (Nameki et al., 2005) and also present in HIV-1 isolates from T-20 experienced patients (Baldwin et al., 2004, Cabrera et al., 2006, Svicher et al., 2008). We showed that this C34 variant can effectively inhibit replication of C34-resistant HIV-1. These studies provided the proof of principle that it is possible to design improved peptide-based fusion inhibitors that are efficient against a major mechanism of drug resistance through introduction of resistance-associated mutation(s).
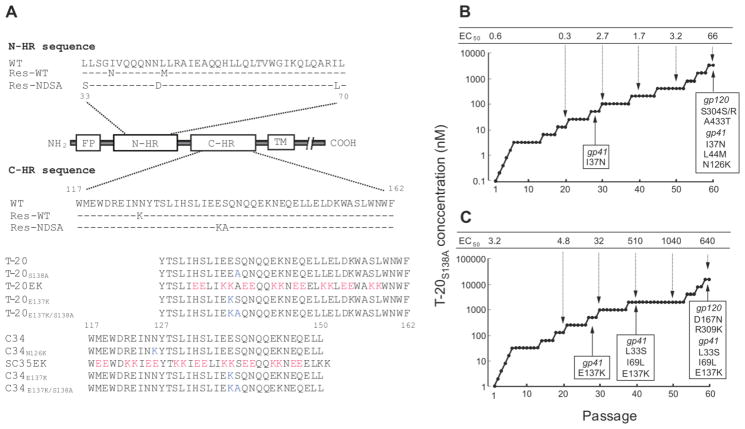
FIG 1. Domains of gp41 and induction of T-20S138A-resistant HIV-1.
(A) Domains of gp41, substitutions observed during in vitro passage with T-20S138A, and amino acid sequences of T-20- and C34-based peptides used in this study. The locations of the fusion peptide (FP), amino-terminal heptad region (N-HR), carboxyl-terminal heptad region (C-HR), transmembrane domain (TM), and C-HR-derived peptides are shown. The residue numbers of T-20 and C34 correspond to their positions in gp41. Substitutions of N- and C-HR in gp41 of wild-type (WT) and T-20S138A-resistant HIV-1 are shown. Res.-WT and Res.-NDSA indicate resistant HIV-1 that were initially selected from wild-type and HIV-1N43D/S138A, respectively. (B, C) Induction of T-20S138A-resistant HIV-1 by dose-escalating selection in MT-2 cells. Induction of resistant HIV-1 was carried out for a total of 60 passages of HIV-1WT (B) and HIV-1N43D/S138A (C), in 0.1 nM and 1 nM of T-20S138A, respectively. At the indicated passages, proviral DNA was sequenced, and the EC50 values of the HIV-1 variants were determined using the MAGI assay. To improve the replication kinetics, substitution of D36G was introduced into the NL4-3 background used in this study (wild-type virus) (Izumi et al., 2009, Mink et al., 2005).
It remains unknown to this date how HIV-1 develops further resistance to T-20S138A. Moreover, it is not known whether we can expand our strategy and modify T-20S138A to include the secondary mutation(s) that emerge during the selection of T-20S138A-resistant HIV, resulting in a strategy that is applicable to the design of peptides customized to address viral resistance mutations. Hence, in the current study we selected T-20S138A-resistant HIV-1 in vitro by a dose-escalating method. We revealed that the resistance mutations that emerged during selection experiments with wild-type or T-20-resistant HIV-1 are located in both the N-HR and the C-HR regions. Furthermore, the I37N and L33S mutations appeared to act as primary mutations for wild-type and T-20-resistant HIV-1, respectively. E137K, a C-HR mutation located in the T-20 sequence, improved replication kinetics and enhanced affinity to N-HR, indicating that E137K acts as a secondary mutation. Introducing the E137K change into the T-20S138A (T-20E137K/S138A) resulted into a peptide inhibitor effective against T-20S138A-resistant variants, suggesting that secondary or compensatory mutations can be widely applicable to the design of next generation peptide-based inhibitors that are active against HIV-1 resistant to earlier generation fusion-targeting drugs.
2. Materials and methods
2.1. Cells and viruses
MT-2 and 293T cells were grown in RPMI 1640 medium and Dulbecco’s modified Eagle medium-based culture medium, respectively. HeLa-CD4-LTR-β-gal cells were kindly provided by Dr. M. Emerman through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease (Bethesda, MD), and used for the drug susceptibility assay, as previously described(Nameki et al., 2005, Nishikawa et al., 2009). Recombinant infectious HIV-1 clones carrying various mutations were generated through site-directed mutagenesis of the pNL4-3 plasmid, as previously described (Nameki et al., 2005, Nishikawa et al., 2009). Each molecular clone was transfected into 293T cells with TransIT (Madison, WI). After 48 h, the supernatants were harvested and stored at −80 °C.
2.2. Antiviral agents
The peptides used in this study (Fig. 1A) were chemically synthesized using standard Fmoc-based solid-phase techniques, as previously described (Oishi et al., 2008, Otaka et al., 2002). An HIV-1 reverse transcriptase inhibitor, 2′,3′-dideoxycytidine (ddC) was purchased from Sigma-Aldrich Japan (Tokyo, Japan) and used as a control.
2.3. Determination of drug susceptibility
Peptide sensitivity of infectious clones was determined by the multinuclear activation of galactosidase indicator (MAGI) assay as previously described (Nameki et al., 2005, Nishikawa et al., 2009). Briefly, the target cells (HeLa-CD4-LTR-β-gal; 104 cells/well) were plated in flat 96-well microtiter culture plates. On the following day, the cells were inoculated with the HIV-1 clones (60 MAGI units/well, resulting into 60 blue cells after 48 h incubation) and cultured in the presence of various concentrations of drugs in fresh medium. Forty-eight hours after virus exposure, all the blue cells stained with X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) were counted in each well. The activity of test compounds was determined as the concentration that reduced HIV-1 infection by 50% (50% effective concentration [EC50]).
2.4. Induction of HIV-1 variants resistant to T-20S138A
MT-2 cells were exposed to HIV-1 and cultured in the presence of T-20S138A. Cultures were incubated at 37 °C until an extensive cytopathic effect (CPE) was observed. The culture supernatants were used for further passages in MT-2 cells in the presence of two-fold increasing concentrations of T-20S138A when massive CPEs were seen in the earlier periods. Each passage usually took 5–7 days. The timing is highly dependent on the type of specific mutations introduced, as previously reported (Nameki et al., 2005, Shimura et al., 2010). For example, a passage that follows introduction of novel mutation(s) should shorten the passage period to perhaps 4–5 days. However, there will be longer delays for passages where there are no novel mutations or when there is appearance of only secondary mutations. The dose-escalation process was repeated until resistant variants were obtained. This selection was carried out for a total of 60 passages (approximately 1 year). At the indicated passages (Fig. 1B and C), the sequence of the env region was determined by direct sequencing of the proviral DNA extracted from the infected MT-2 cells.
2.5. Viral replication kinetics assay
MT-2 cells (105 cells/1 mL) were infected with each virus preparation (500 MAGI units) for 16 h. Infected cells were then washed and cultured in a final volume of 3 mL. The culture supernatants were collected on day 2 through day 5 post-infection, and amounts of p24 antigen were determined.
2.6. CD spectroscopy
Each peptide was incubated at 37 °C for 30 min (the final concentrations of peptides were 10 μM in phosphate buffered saline [PBS]; pH 7.4). CD spectra were recorded on an AVIV model 202 spectropolarimeter (Aviv Instruments, Proterion Corporation, Piscataway, NJ) with a 1 mm path-length cuvette at 25 °C as the average of eight scans. The thermal stability was assessed by monitoring the change in the CD signal at 222 nm. The midpoint of the thermal unfolding transition (melting temperature [Tm]) of each complex was determined as previously described (Izumi et al., 2009).
3. Results
3.1. Selection of HIV-1 resistant to T-20S138A
An HIV-1NL4-3 strain containing a D36G substitution, which improves replication kinetics, was used as a wild-type virus (HIV-1WT) and for the construction of various mutants, as described (Izumi et al., 2009, Mink et al., 2005). HIV-1WT or T-20-resistant HIV-1N43D/S138A were used for selection of T-20S138A-resistant HIV-1. MT-2 cells were infected with HIV-1WT and HIV-1N43D/S138A, and incubated in the presence of T-20S138A at the initial concentrations of 0.1 nM and 1 nM, respectively. At the indicated passages, the sequence of the env region was determined by direct sequencing of the proviral DNA extracted from the infected MT-2 cells. During the selection, mutations in the gp41 were observed and are shown in Fig. 1B and C.
In the selection with HIV-1WT (Fig. 1B), at passage 28 (P-28), when T-20S138A concentration was 51.2 nM (P-28, 51.2 nM), isoleucine at position 37 in the gp41 was substituted to asparagine (I37N). At P-60 (3.3 μM), L44M and N126K in the gp41 further emerged. On the other hand, in the selection with T-20-resistant HIV-1N43D/S138A (Fig. 1C), at P-28 (512 nM) and at P-40 (2 μM), E137K in the gp41, and L33S and I69L in the gp41 emerged, respectively. The emergence of the I69L mutation in diverse HIV-1 strains has been previously reported (Eshleman et al., 2007). At P-60, the resistance of selected viruses from HIV-1WT and HIV-1N43D/S138A to T-20S138A, reached approximately 110- and 200-fold, respectively. These results indicate that even though T-20S138A was active against T-20 resistant variants, resistant HIV-1 emerged relatively rapidly compared with the next generation fusion inhibitors, such as SC34EK, which required 120 passages to acquire the resistance (Shimura et al., 2010).
3.2. Susceptibility of T-20S138A-resistant HIV-1 to T-20 and C34 derivatives
To validate our resistance data we used site-directed mutagenesis to prepare recombinant HIV-1 with the T-20S138A-resistance mutations and examined its susceptibility to T-20 and C34 derivatives with MAGI assay (Table 1). We also used as controls the modified α-helix T-20- and C34- peptide inhibitors, T-20EK (Oishi et al., 2008) and SC35EK (Nishikawa et al., 2009, Shimura et al., 2010), respectively, which are more efficient in vitro replication inhibitors of T-20-resistant HIV-1 than T-20 or C34. Finally, we also used as a control C34N126K, a modified version of C34 that includes the resistance-associated N126K substitution that effectively suppress replication of C34-resistant HIV-1 in vitro (Izumi et al., 2009).
TABLE 1.
Antiviral activity of C-HR-derived peptides against gp41 recombinant viruses.
| EC50 (nM)
|
||||||
|---|---|---|---|---|---|---|
| T-20 | T-20S138A | T-20EK | C34 | C34N126K | SC35EK | |
| HIV-1WT* | 2.4 ± 0.6 | 0.6 ± 0.1 | 1.9 ± 0.5 | 2.1 ± 0.7 | 1.6 ± 0.5 | 2.4 ± 0.9 |
| HIV-1I37N | 47 ± 6.9 (20) | 4.3 ± 1.3 (7.2) | 21 ± 2.4 (11) | 3.3 ± 1.1 (1.6) | 1.9 ± 0.1 (1.2) | 1.0 ± 0.4 (0.4) |
| HIV-1L44M | 4.1 ± 1.2 (1.7) | 0.7 ± 0.2 (1.2) | 2.2 ± 0.6 (1.2) | 1.1 ± 0.3 (0.5) | 0.8 ± 0.2 (0.5) | 0.6 ± 0.2 (0.3) |
| HIV-1N126K | 4.4 ± 1.3 (1.8) | 1.2 ± 0.4 (2.0) | 2.8 ± 0.2 (1.5) | 6.3 ± 1.2 (3.0) | 1.5 ± 0.2 (0.9) | 3.3 ± 0.2 (1.4) |
| HIV-1I37N/N126K | 660 ± 180 (275) | 16 ± 4.8 (27) | 14 ± 5.1 (7.4) | 20 ± 4.5 (9.5) | 3.4 ± 0.4 (2.1) | 2.9 ± 0.3 (1.2) |
| HIV-1I37N/L44M/N126K | > 1000 (> 417) | 130 ± 40 (220) | 240 ± 95 (126) | 66 ± 23 (31) | 4.0 ± 0.8 (2.5) | 1.1 ± 0.1 (0.5) |
| HIV-1L33S | 23 ± 5.5 (9.6) | 3.1 ± 0.6 (5.2) | 13 ± 2.6 (6.8) | 3.2 ± 1.1 (1.5) | 2.1 ± 0.1 (1.3) | 3.0 ± 0.8 (1.2) |
| HIV-1N43D | 49 ± 10 (20) | 3.5 ± 0.9 (5.8) | 4.1 ± 1.2 (2.2) | 4.4 ± 0.4 (2.1) | 1.4 ± 0.1 (0.8) | 0.4 ± 0.2 (0.2) |
| HIV-1I69L | 2.1 ± 0.5 (0.9) | 0.5 ± 0.2 (0.8) | 2.2 ± 0.4 (1.2) | 2.7 ± 0.2 (1.3) | 2.2 ± 0.5 (1.4) | 2.7 ± 0.5 (1.1) |
| HIV-1E137K | 2.0 ± 0.3 (0.8) | 0.7 ± 0.1 (1.2) | 2.5 ± 0.4 (1.3) | 2.6 ± 0.2 (1.2) | 2.3 ± 0.7 (1.4) | 3.1 ± 0.8 (1.3) |
| HIV-1N43D/S138A | 84 ± 16 (35) | 3.2 ± 1.0 (5.3) | 3.4 ± 1.1 (1.8) | 2.7 ± 0.2 (1.3) | 1.6 ± 0.5 (1.0) | 0.3 ± 0.1 (0.1) |
| HIV-1L33S/N43D/S138A | > 1000 (> 417) | 550 ± 72 (174) | 330 ± 94 (14) | 30 ± 9.2 (2.6) | 4.2 ± 1.2 (0.4) | 0.9 ± 0.3 (0.4) |
| HIV-1N43D/E137K/S138A | 110 ± 31 (46) | 14 ± 4.7 (23) | 7.0 ± 2.4 (3.7) | 7.4 ± 1.9 (3.5) | 2.1 ± 0.7 (1.3) | 1.9 ± 0.6 (0.8) |
| HIV-1L33S/N43D/E137K/S138A | > 1000 (> 417) | > 1000 (> 1667) | > 1000 (> 526) | 31 ± 5.0 (15) | 6.7 ± 1.7 (4.2) | 1.2 ± 0.2 (0.5) |
| HIV-1L33S/N43D/I69L/E137K/S138A | > 1000 (> 417) | > 1000 (> 1667) | > 1000 (> 526) | 50 ± 12 (24) | 28±7.1 (17.5) | 1.0± 0.9 (0.4) |
Anti-HIV activity was determined using the MAGI assay. Fifty percent effective concentration (EC50) values and SD were obtained from the results of at least three independent experiments. Shown in parentheses are the fold-increases in resistance (increase in EC50 value) calculated by comparison to a wild-type virus (HIV-1WT). Increases of over 10-fold are indicated in bold.
To improve the replication kinetics, substitution of D36G, observed in majority of HIV-1 strains, was introduced into the NL4-3 background used in this study (wild-type virus; HIV-1WT) (Izumi et al., 2009, Mink et al., 2005).
Selected mutations I37N and L33S provided various levels of resistance to T-20 and its derivatives, T-20S138A and T-20EK, apparently acting as primary mutations to peptides with a T-20 backbone (Table 1). Other mutations, L44M, I69L, and E137K, which were observed in wild-type HIV-1 as polymorphisms (Kuiken et al., 2010, Loutfy et al., 2007), conferred little resistance to all peptide fusion inhibitors tested. However, introduction of L44M to HIV-1I37N/N126K (HIV-1I37N/L44M/N126K) remarkably enhanced resistance to T-20 derivatives. This was consistent with previous studies that also reported a resistance enhancement (1.8-fold) by L44M to T-20 (Loutfy et al., 2007). Collectively, these data suggest that L44M has as a role in HIV-1 resistance as a secondary mutation. All peptides sufficiently suppressed HIV-1I69L, suggesting that I69L may be a secondary mutation or a polymorphism. N126K conferred only marginal resistance (<3-fold) to all peptide fusion inhibitors, but in the background of I37N (HIV-1I37N/N126K) it enhanced resistance to T-20, T-20S138A, and C34. L33S, which was originally reported as a C34 resistance associated mutation (Armand-Ugon et al., 2003), significantly enhanced resistance in the background of N43D/S138A mutations (HIV-1L33S/N43D/S138A). Similar to the N126K mutation, E137K also enhanced resistance by N43D/S138A (HIV-1N43D/E137K/S138A) and L33S/N43D/S138A (HIV-1L33S/N43D/E137K/S138A) to T-20S138A, T-20, and T-20EK. These results indicate that L33S and I37N appear to be primary mutations for T-20 derivatives.
3.3. Effect of substitutions in the gp120 on peptide susceptibility
Polymorphisms in the gp120 that influence co-receptor usage may influence T-20 susceptibility (Labrosse et al., 2003, Reeves et al., 2002). Meanwhile, others reported that T-20 susceptibility was not influenced by co-receptor usage (Cilliers et al., 2004, Melby et al., 2006). Resistance induction experiments performed in this study revealed that most laboratory strains with in vitro resistance to fusion inhibitors acquired substitutions in both the gp120 and the gp41 (Armand-Ugon et al., 2003, Eggink et al., 2011, Fikkert et al., 2002, Izumi et al., 2010, Nameki et al., 2005, Shimura et al., 2010). However, most substitutions showed little impact on resistance, and only contributed to a small enhancement of replication capacity (Eggink et al., 2011, Izumi et al., 2010, Nameki et al., 2005, Shimura et al., 2010). In the present study, we examined peptide susceptibility of cloned viruses that contain all Env substitutions observed in the selection (both gp120 and gp41). Most substitutions in the gp120 attenuated resistance to fusion inhibitors (Table 3). Therefore, in vitro experiments showed that substitutions in the gp120 are not likely associated with resistance.
TABLE 3.
Antiviral activity of C-HR-derived peptides against gp160 recombinant viruses.
| compound | EC50 (nM) |
|---|---|
| ddC | 771±272 |
| T-20 derivatives | |
| T20 | >10000 (NA) |
| T20EK | 2729±1113 (NA) |
| T20S138A | 3126±453 (NA) |
| T20E137K | 2761±1477 (NA) |
| T20E137K/S138A | 203±54 (0.6) |
| C34 derivatives | |
| C34 | 171.0±106 (3.4) |
| C34N126K | 25.9±4.6 (NA) |
| SC34EK | 1.0±0.8 (1) |
| C34E137K | 7.0±4.4 (0.4) |
| C34E137K/S138A | 0.3±0.1 (0.3) |
Anti-HIV activity was determined using the MAGI assay. Fifty percent effective concentration (EC50) values and SD were obtained from the results of at least three independent experiments. Shown in parentheses are the fold-increases in resistance (increase in EC50 value) calculated by comparison to the resistant clone with mutations only in gp41 (HIV-1L33S/N43D/I69L/E137K/S138A). To improve the replication kinetics, substitution of D36G, observed in majority of HIV-1 strains, was introduced into the NL4-3 background used in this study(Izumi et al., 2009, Mink et al., 2005). NA: not available. ddC, is dideoxycytidine.
3.4. Influence of mutations in the gp41 on HIV-1 replication
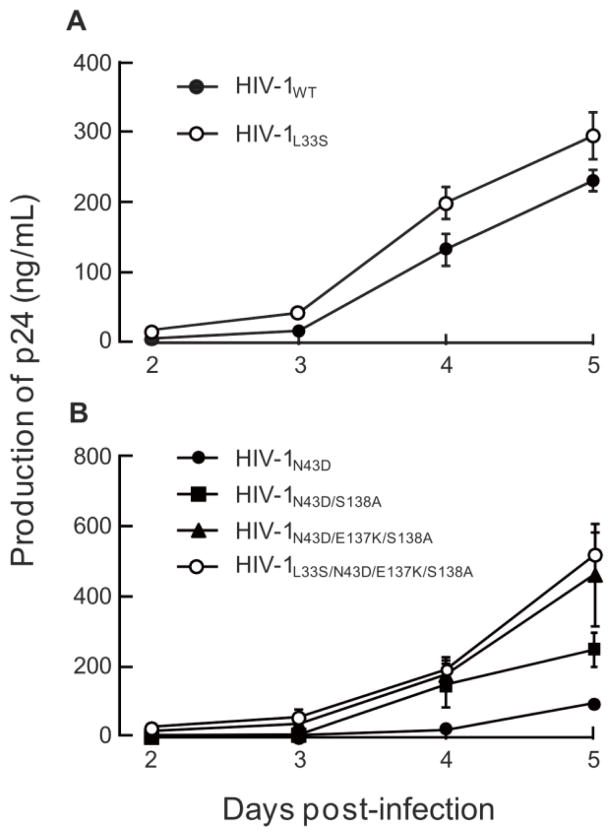
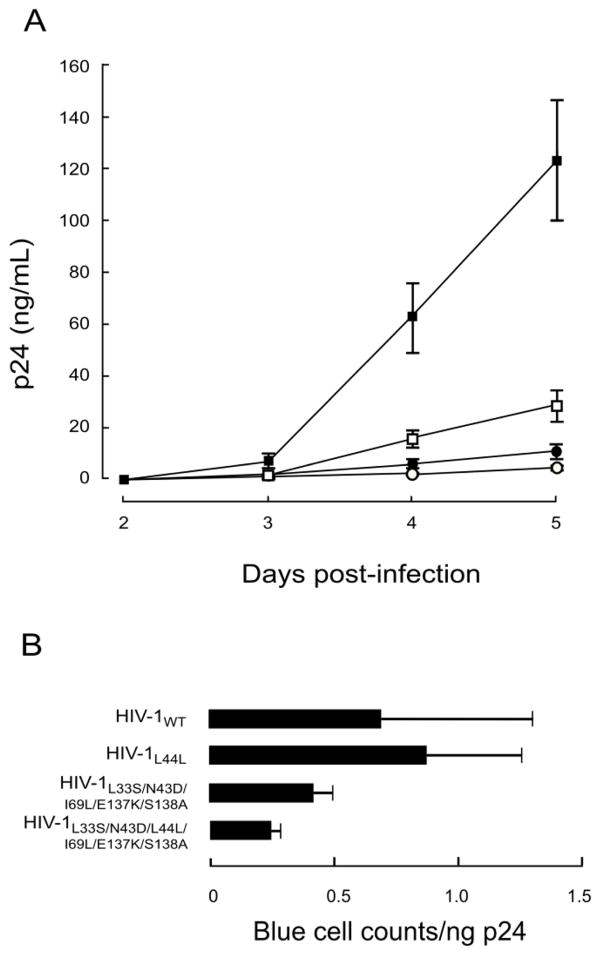
To address the effects of mutations on HIV-1 replication, we examined the replication kinetics of T-20S138A-resistant HIV-1N43D/S138A variants. Consistent with a previous report (Lohrengel et al., 2005), the L33S mutation did not significantly affect the replication kinetics and infectivity compared with those of HIV-1WT (Fig. 2A). The S138A mutation restored the replication kinetics of HIV-1N43D (Fig. 2B), as previously described (Izumi et al., 2009). E137K was also associated with N43D mutation in vivo (Svicher et al., 2008), and restored infectivity impaired by N43D (Tolstrup et al., 2007). Introduction of E137K into N43D/S138A enhanced the replication kinetics, and further addition of L33S to N43D/E137K/S138A resulted in equivalent replication kinetics compared with HIV-1N43D/E137K/S138A (Fig. 2B) as observed in HIV-1WT based mutants. During the passage of HIV-1N43D/S138A, a synonymous mutation at amino acid position L44, TTG to CTG, was observed. Interestingly, LTTG44LCTG enhanced viral replication kinetics through enhanced stability of the Rev-responsive element (RRE) secondary structure (Ueno et al., 2009). Therefore, we examined the viral replication kinetics of mutants with LTTG44LCTG, and compared HIV-1WT, with HIV-1L44L-CTG, and HIV-1L33S/N43D/L44L-CTG/E137K/S138A with HIV-1L33S/N43D/L44L-CTG/I69L/E137K/S138A. As expected, the presence of LTTG44LCTG enhanced replication in all viruses. Surprisingly, mutants with resistance mutations showed enhanced replication kinetics as determined by the p24 production assay of culture supernatants (Fig. 4A). Therefore, we further examined infectivity using the MAGI assay and determined that the infectivity of resistance variants containing LTTG44LCTG was reduced compared with HIV-1WT (Fig. 4B). These results indicate that the primary mutation, L33S, possesses less ability to attenuate HIV-1 replication, while I69L, S138A, and E137K enhance replication kinetics of T-20-resistant HIV-1 to a comparable level of HIV-1WT.
FIG 2. Replication kinetics of T-20S138A-resistant variants.
Replication kinetics of T-20S138A-resistant recombinant variants that introduced L33S mutation (A), or combinations of L33S, E137K, and S138A mutations in HIV-1N43D (B). To improve replication kinetics, the D36G polymorphism was introduced into the NL4-3 background used in this study (HIV-1WT). Supernatants from infected MT-2 cells were collected on days 2–7 and the amount of p24 produced was determined. Representative results are shown as mean values with standard deviations estimated from three independent experiments.
FIG 4. Effect of secondary mutations in the N-HR on (A) replication kinetics and (B) infectivity.
LTTG44LCTG was introduced into HIV-1WT and T-20S138A resistant HIV-1 (HIV-1L33S/N43D/L44L-CTG/I69L/E137K/S138A). Replication kinetics were determined by measuring p24 production in culture supernatants. HIV-1WT (open circles), HIV-1L44L (closed circles) HIV-1L33S/N43D/I69L/E137K/S138A (open squares), and HIV-1L33S/N43D/L44L-CTG/I69L/E137K/S138A (closed squares). LTTG44LCTG introduction statistically enhanced both replication of HIV-1WT and HIV-1L33S/N43D/L44L-CTG/I69L/E137K/S138A (student t-test, p < 0.01 on day 4 and 5). Relative infectivity (blue cell counts in MAGI cells divided by amount of p24) was calculated (B). Error bars indicate SD of three determinations. Decrease of infectivity between HIV-1L33S/N43D/I69L/E137K/S138A and HIV-1L33S/N43D/L44L-CTG/I69L/E137K/S138A) were statistically significant (student t-test, p < 0.05).
3.5. Circular dichroism
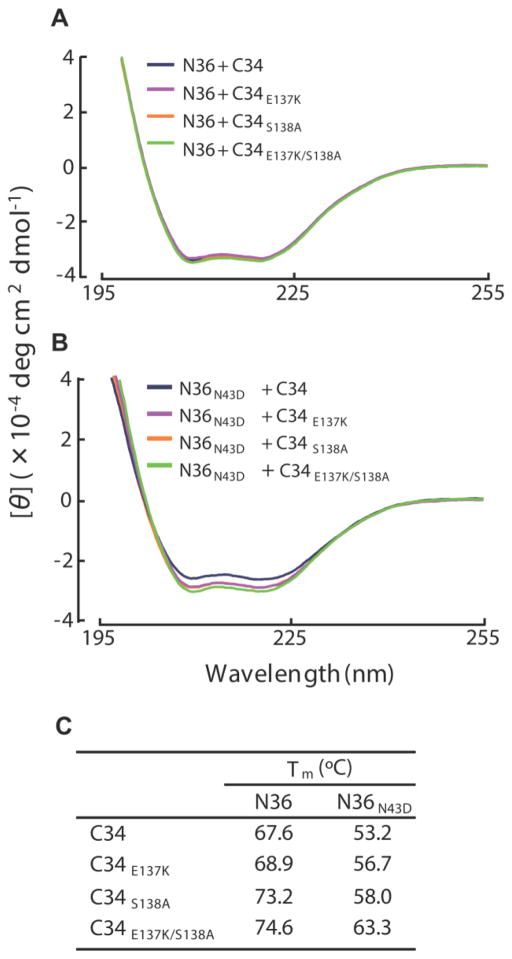
To clarify the effect of E137K substitutions on peptide binding, we examined the binding affinities of E137K-containing C-HR peptides to N-HR using CD analysis. CD spectra reveal the presence of stable α-helical structures of six-helix bundles that are required for biological activity and are thought to mechanistically and thermodynamically correlate with HIV-1 fusion (Bianchi et al., 2005). Since in vitro T-20 does not interact with the N36 peptide (amino acid positions 35–70 of the N-HR), we used instead peptide C34 with E137K and/or S138A substitutions (Fig. 1A). We found that mixtures of C34E137K, C34S138A, or C34E137K/S138A with N36 or N36N43D showed sufficient and comparable α-helicity at 25°C (Fig. 3A and B). We also determined the thermal stability of the helical complexes formed by the N36 and C34 peptides, which is also an indication of the binding affinity of these peptides. Hence, we measured and compared the melting temperatures (Tm) of various complexes, which indicates the 50% disruption of the six-helix bundle (Fig. 3C). Complexes of N36 and C34 containing the S138A and E137K/S138A substitutions (N36/C34S138A and N36/C34E137K/S138A, respectively), showed higher thermal stability than N36/C34. Similarly, S138A and E137K/S138A restored the binding affinity of C34 to N36N43D. These results indicate that E137K acts as a compensatory mutation for the T-20S138A-resistance primary mutation, causing enhancement of replication kinetics.
FIG 3. CD spectra (A, B) and thermal stability (C) of N36/C34 complexes.
Peptide sequences used in this study are shown in FIG 1A and have also been previously described (Izumi et al., 2009). CD spectra of C34E137K, C34S138A, and C34E137K/S138A complexes with N36 (A) and N36N43D (B) are shown. Equimolar amounts (10 μM) of the N- and C-HR peptides were incubated at 37 °C for 30 min in PBS. The CD spectra of each mixture were then collected at 25 °C using a Jasco (Model J-710) spectropolarimeter. (C) Thermal stabilities, defined as the midpoint of the thermal unfolding transition (Tm) values, of the potential six-helix bundles of N- and C-HR peptides, were determined.
3.6. Antiviral activity of E137K-modified peptides
Recently, we demonstrated that introduction of the S138A secondary mutation to T-20 (T-20S138A) enhanced binding to mutated N-HR and suppresses resistance of T-20-resistance variants (Izumi et al., 2009). Similarly, as shown in Figure 3, E137K enhanced binding affinity with N-HR, suggesting that introduction of E137K to T-20 may enhance the antiviral activity of T-20. We synthesized T-20 and T-20S138A variants containing the E137K change (T-20E137K and T-20E137K/S138A) (Fig. 1A) and examined their anti-HIV activity against T-20S138A-resistant HIV-1 (Table 2). All peptides exhibited potent antiviral activity against HIV-1WT. HIV-1L33S/N43D/S138A and HIV-1I37N/L44M/N126K showed high resistance to T-20E137K, indicating that the resistance mechanism of T-20E137K is similar to that of T-20S138A. On the other hand, T-20E137K/S138A (Table 2) maintained some antiviral activity against HIV-1L33S/N43D/S138A, HIV-1L33S/N43D/E137K/S138A, and HIV-1I37N/L44M/N126K compared with other T-20 derivatives including electrostatically constrained T-20EK (Tables 1and Fig. 1). C34E137K and C34E137K/S138A significantly suppressed all HIV-1 variants tested except for HIV-1I37N/L44M/N126K by C34E137K. These results indicate that peptides with resistant mutations may sustain their activity against particular resistant variants.
TABLE 2.
Antiviral activity of E137K-induced C-HR peptides against T-20S138A-resistant variants.
| EC50 (nM)
|
||||
|---|---|---|---|---|
| T-20E137K | T-20E137K/S138A | C34E137K | C34E137K/S138A | |
| HIV-1WT* | 0.8 ± 0.2 | 0.5 ± 0.1 | 1.0 ± 0.3 | 0.7 ± 0.2 |
| HIV-1L33S | 13 ± 3.2 (16) | 2.2 ± 0.4 (4.5) | 0.7 ± 0.2 (0.7) | 0.5 ± 0.1 (0.7) |
| HIV-1N43D/S138A | 4.2 ± 0.7 (5.3) | 0.7 ± 0.2 (1.4) | 0.3± 0.1 (0.3) | 0.4 ± 0.1 (0.6) |
| HIV-1L33S/N43D/S138A | 700 ± 150 (880) | 45 ± 9.9 (90) | 2.3 ± 0.4 (2.3) | 0.5 ± 0.2 (0.7) |
| HIV-1N43D/E137K/S138A | 12 ± 3.6 (15) | 2.4 ± 0.8 (4.8) | 0.2 ± 0.1 (0.2) | 0.4 ± 0.1 (0.6) |
| HIV-1L33S/N43D/E137K/S138A | 480 ± 47 (600) | 36 ± 3.1 (72) | 3.8 ± 1.3 (3.8) | 1.0 ± 0.4 (1.4) |
| HIV-1L33S/N43D/I69L/E137K/S138A | 1808 ± 852 (2260) | 157±83 (314) | 4±2 (4) | 1.0±0.4 (1.4) |
| HIV-1I37N/L44M/N126K | 200 ± 24 (250) | 30 ± 8.7 (60) | 17 ± 3.8 (17) | 2.2 ± 0.3 (3.1) |
Anti-HIV activity was determined using the MAGI assay. Fifty percent effective concentration (EC50) values and SD were obtained from the results of at least three independent experiments. Shown in parentheses are the fold-increases in resistance (increase in EC50 value) calculated by comparison to a wild-type virus (HIV-1WT). Increases of over 10-fold are indicated in bold.
To improve the replication kinetics, substitution of D36G, observed in majority of HIV-1 strains, was introduced into the NL4-3 background used in this study (wild-type virus; HIV-1WT) (Izumi et al., 2009, Mink et al., 2005).
4. Discussion
The current study describes the introduction of resistance changes into the original and modified (T-20S138A) versions of the T-20 peptide-fusion inhibitor. We analyzed the new T-20 derivatives using both wild-type and T-20-resistant strains. We also identified through dose escalation experiments, T-20S138A-resistants. We found that T-20S138A-resistant HIV-1 showed cross-resistance only to the T-20 derivatives, but not to C34 derivatives. Through the CD anlysis, the N126K and E137K mutations in the C-HR may act as compensatory mutations for impaired interaction by a primary mutation, I37N and N43D in the N-HR, respectively. Since E137K is located within the T-20 sequence, we synthesized and characterized the activity of novel T-20-based peptides containing E137K (T-20E137K). Here we demonstrate that the introduction of a secondary resistance mutation (E137K) in the backbone of a peptide fusion inhibitor is a useful change that results into more potent fusion inhibitors, even for HIV-1 strains that are resistant to peptide fusion inhibitors.
Selection of T-20S138A-resistance starting with wild-type HIV-1 resulted in the emergence of I37N and L44M substitutions, which were located in the N-HR region that is thought to interact with T-20. Other substitutions at position 37 (I37T or I37K) also conferred resistance to T-20 and C34 (Nameki et al., 2005), suggesting that I37 in N-HR is critical for the attachment of C-HR-derived peptide fusion inhibitors. The L44M mutation has only been observed in subtype B HIV-1-infected patients treated with T-20 (Carmona et al., 2005), and conferred weak resistance to T-20 (Loutfy et al., 2007). In this study, L44M did not confer resistance to all peptide inhibitors; however, L44M in combination with other mutations (I37N/N126K) remarkably enhanced resistance to T-20S138A, suggesting that L44M serves as a secondary mutation to enhance resistance to T-20S138A. N126K also enhances resistance to some fusion inhibitors (Baldwin et al., 2004, Nameki et al., 2005, Eggink et al., 2008) by helping recover losses in intra-gp41 interactions that were caused by primary mutations, such as N43D.
When we selected T-20S138A-resistant HIV-1 (HIV-1N43D/S138A) we obtained a somehow different set of mutations that included L33S, which is located at the presumed T-20 binding site at N-HR, as well as I37N, N43D, and L44M. L33S was previously reported in HIV-1 variants resistant to T-20 (Fikkert et al., 2002), C34 (Armand-Ugon et al., 2003), and a membrane-anchored C-HR-derived peptide, M87 (Lohrengel et al., 2005). Although our work clearly demonstrates that L33S is involved in resistance to T-20 derivatives, it was not possible to discern whether L33S affected binding affinity to C-HR in the CD analyses because L33 was not in the sequence of the N36 N-HR peptide that we had to use in this study. As shown in Fig. 2, L33S did not significantly affect replication kinetics compared with HIV-1WT, suggesting that L33S might sustain binding affinity with C-HR to form a stable six-helix bundle. The L33S mutation is located in the loop of stem IIc of the RRE (Ueno et al., 2009). Hence, nucleotide changes for L33S do not require compensatory mutations to maintain secondary structure of the RRE. Therefore, it is likely that L33S has little effect on replication kinetics. In this study, L33S conferred little resistance to C34 in this study, while it was previously reported to confer up to 10-fold resistance (Armand-Ugon et al., 2003), suggesting that some other viral background might affect the resistance, since Armand et al. (Armand-Ugon et al., 2003) examined bulk virus samples obtained from the selection.
A prevalent polymorphism, E137K, which was associated with N43D in vivo (Svicher et al., 2008), has been proven to restore infectivity that has been impaired by N43D (Tolstrup et al., 2007). E137K did not affect susceptibility to all peptide fusion inhibitors by itself, but in combination with primary mutations, it remarkably enhanced resistance to T-20S138A. Moreover, introduction of the E137K change into N43D/S138A enhanced the viral replication kinetics as shown in Fig. 2. A possible hydrogen bond between K137 and D43 may partially restore the reported loss in six-helix bundle stability conferred by the N43D mutation (Bai et al., 2008), suggesting that E137K can compensate for losses in the interactions between N-HRN43D and C-HR. This hypothesis is consistent with our CD results presented in Fig. 3.
Because E137K restored binding affinity with N-HR similar to the S138A mutation, we expected that introduction of E137K into T-20 would effectively suppresses replication of T-20-resistant HIV-1. We examined the antiviral activity of E137K- and E137K/S138A-containing T-20 and C34 to T-20S138A-resistant HIV-1. We found that T-20E137K had similar antiviral activity with other T-20 derivatives such as T-20S138A and T-20E137K/S138A. Hence, we believe that the combination of few substitution secondary mutations can enhance the antiviral activity of peptide fusion inhibitors. Therefore, it is possible to design peptides that include the secondary mutations in the C-HR and use them by themselves and/or in combinations to block fusion inhibitor resistant viruses. Importantly, we have succesfully applied this strategy to suppress HIV-1 resistance to next generation fusion inhibitor SC34EK (Shimura et al., 2010).
In this study, we identified two distinct pathways to escape pressure of T-20S138A. Emergence of drug resistance mutants under drug pressure involves a stochastic selection. Nonetheless, the makeup of the final population depends on both the ability of specific populations to evade the drug, as well as their fitness that determines their representation in the escape population. There are several examples in the literature where HIV becomes resistant to the same drug by different mechanisms. For example, in the case of the most commonly used drugs that target HIV reverse transcriptase (RT), the virus can develop multidrug resistance by either the Q151M complex pathway (Kavlick et al., 1998, Shirasaka et al., 1995) or by accumulation of thymidine associated mutations (TAMs) (Hachiya et al., 2008, Kosalaraksa et al., 1999). We recently report some of background polymorphisms can also influence resistance pathways, such 172R/K in the RT region (Hachiya et al., 2012). In the case of the T-20S138A inhibitor, the N43D/S138A may also act as such polymorphisms despite the presence of primary mutations (Izumi et al., 2009) and preferentially affect the emergence of specific mutations.
5. Conclusion
As previously discussed (Izumi et al., 2009), although other developed peptide-based fusion inhibitors need many amino acid additions and/or substitutions for the enhancement of their antiviral activity (Chinnadurai et al., 2007, Eggink et al., 2008, Dwyer et al., 2007, Otaka et al., 2002), application of secondary mutations similar to T-20S138A and T-20E137K/S138A is straightforward. It is based on information from viral evolution studies under drug pressure that help design improved inhibitors.
Highlights.
T-20S138A, a resistance mutation introduced peptide, blocks T20-resistant HIV-1.
To determine how HIV-1 escapes, we induced T-20S138A-resistant HIV-1 in vitro.
We found that a unique mutation E137K in resistant HIV-1.
E137K introduced T-20S138A suppressed replication of T-20S138A-resistant HIV-1.
Mutations can be consistently useful for the design of peptide inhibitors.
Acknowledgments
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a grant for the Promotion of AIDS Research from the Ministry of Health, Labour and Welfare. Additional support was by National Institute of Health (NIH) research grants AI094715, AI076119, AI079801, and AI100890 (SGS). We are grateful to Biomedical Research Core (Tohoku University School of Medicine) for technical support.
Footnotes
The authors declare non-financial competing interests.
References
- Aquaro S, D’Arrigo R, Svicher V, Perri GD, Caputo SL, Visco-Comandini U, Santoro M, Bertoli A, Mazzotta F, Bonora S, Tozzi V, Bellagamba R, Zaccarelli M, Narciso P, Antinori A, Perno CF. Specific mutations in HIV-1 gp41 are associated with immunological success in HIV-1-infected patients receiving enfuvirtide treatment. J Antimicrob Chemother. 2006;58:714–722. doi: 10.1093/jac/dkl306. [DOI] [PubMed] [Google Scholar]
- Armand-Ugon M, Gutierrez A, Clotet B, Este JA. HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antiviral Res. 2003;59:137–142. doi: 10.1016/s0166-3542(03)00071-8. [DOI] [PubMed] [Google Scholar]
- Bai X, Wilson KL, Seedorff JE, Ahrens D, Green J, Davison DK, Jin L, Stanfield-Oakley SA, Mosier SM, Melby TE, Cammack N, Wang Z, Greenberg ML, Dwyer JJ. Impact of the enfuvirtide resistance mutation N43D and the associated baseline polymorphism E137K on peptide sensitivity and six-helix bundle structure. Biochemistry. 2008;47:6662–6670. doi: 10.1021/bi702509d. [DOI] [PubMed] [Google Scholar]
- Baldwin CE, Sanders RW, Deng Y, Jurriaans S, Lange JM, Lu M, Berkhout B. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J Virol. 2004;78:12428–12437. doi: 10.1128/JVI.78.22.12428-12437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Finotto M, Ingallinella P, Hrin R, Carella AV, Hou XS, Schleif WA, Miller MD, Geleziunas R, Pessi A. Covalent stabilization of coiled coils of the HIV gp41 N region yields extremely potent and broad inhibitors of viral infection. Proc Natl Acad Sci U S A. 2005;102:12903–12908. doi: 10.1073/pnas.0502449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera C, Marfil S, Garcia E, Martinez-Picado J, Bonjoch A, Bofill M, Moreno S, Ribera E, Domingo P, Clotet B, Ruiz L. Genetic evolution of gp41 reveals a highly exclusive relationship between codons 36, 38 and 43 in gp41 under long-term enfuvirtide-containing salvage regimen. AIDS. 2006;20:2075–2080. doi: 10.1097/QAD.0b013e3280102377. [DOI] [PubMed] [Google Scholar]
- Cardoso RM, Brunel FM, Ferguson S, Zwick M, Burton DR, Dawson PE, Wilson IA. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J Mol Biol. 2007;365:1533–1544. doi: 10.1016/j.jmb.2006.10.088. [DOI] [PubMed] [Google Scholar]
- Carmona R, Perez-Alvarez L, Munoz M, Casado G, Delgado E, Sierra M, Thomson M, Vega Y, Vazquez de Parga E, Contreras G, Medrano L, Najera R. Natural resistance-associated mutations to Enfuvirtide (T20) and polymorphisms in the gp41 region of different HIV-1 genetic forms from T20 naive patients. J Clin Virol. 2005;32:248–253. doi: 10.1016/j.jcv.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Chan DC, Chutkowski CT, Kim PS. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci U S A. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Chinnadurai R, Rajan D, Munch J, Kirchhoff F. Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J Virol. 2007;81:6563–6572. doi: 10.1128/JVI.02546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilliers T, Patience T, Pillay C, Papathanasopoulos M, Morris L. Sensitivity of HIV type 1 subtype C isolates to the entry inhibitor T-20. AIDS Res Hum Retroviruses. 2004;20:477–482. doi: 10.1089/088922204323087714. [DOI] [PubMed] [Google Scholar]
- Debnath AK, Radigan L, Jiang S. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J Med Chem. 1999;42:3203–3209. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- Dwyer JJ, Wilson KL, Davison DK, Freel SA, Seedorff JE, Wring SA, Tvermoes NA, Matthews TJ, Greenberg ML, Delmedico MK. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc Natl Acad Sci U S A. 2007;104:12772–12777. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer JJ, Wilson KL, Martin K, Seedorff JE, Hasan A, Medinas RJ, Davison DK, Feese MD, Richter HT, Kim H, Matthews TJ, Delmedico MK. Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency. Protein Sci. 2008;17:633–643. doi: 10.1110/ps.073307608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D, Baldwin CE, Deng Y, Langedijk JP, Lu M, Sanders RW, Berkhout B. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J Virol. 2008;82:6678–6688. doi: 10.1128/JVI.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D, Bontjer I, Langedijk JP, Berkhout B, Sanders RW. Resistance of human immunodeficiency virus type 1 to a third-generation fusion inhibitor requires multiple mutations in gp41 and is accompanied by a dramatic loss of gp41 function. J Virol. 2011;85:10785–10797. doi: 10.1128/JVI.05331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshleman SH, Hudelson SE, Bruce R, Lee T, Owens MR, Hackett J, Swanson P, Devare SG, Marlowe N. Analysis of HIV type 1 gp41 sequences in diverse HIV type 1 strains. AIDS Res Hum Retroviruses. 2007;23:1593–1598. doi: 10.1089/aid.2007.0130. [DOI] [PubMed] [Google Scholar]
- Fikkert V, Cherepanov P, Van Laethem K, Hantson A, Van Remoortel B, Pannecouque C, De Clercq E, Debyser Z, Vandamme AM, Witvrouw M. env chimeric virus technology for evaluating human immunodeficiency virus susceptibility to entry inhibitors. Antimicrob Agents Chemother. 2002;46:3954–3962. doi: 10.1128/AAC.46.12.3954-3962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya A, Kodama EN, Sarafianos SG, Schuckmann MM, Sakagami Y, Matsuoka M, Takiguchi M, Gatanaga H, Oka S. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J Virol. 2008;82:3261–3270. doi: 10.1128/JVI.01154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya A, Marchand B, Kirby KA, Michailidis E, Tu X, Palczewski K, Ong YT, Li Z, Griffin DT, Schuckmann MM, Tanuma J, Oka S, Singh K, Kodama EN, Sarafianos SG. HIV-1 reverse transcriptase (RT) polymorphism 172K suppresses the effect of clinically relevant drug resistance mutations to both nucleoside and non-nucleoside RT inhibitors. J Biol Chem. 2012;287:29988–29999. doi: 10.1074/jbc.M112.351551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xiao Y, Song H, Liang Q, Ju D, Chen X, Lu H, Jing W, Jiang S, Zhang L. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J Biol Chem. 2008;283:11126–11134. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- Izumi K, Kodama E, Shimura K, Sakagami Y, Watanabe K, Ito S, Watabe T, Terakawa Y, Nishikawa H, Sarafianos SG, Kitaura K, Oishi S, Fujii N, Matsuoka M. Design of Peptide-based Inhibitors for Human Immunodeficiency Virus Type 1 Strains Resistant to T-20. J Biol Chem. 2009;284:4914–4920. doi: 10.1074/jbc.M807169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K, Nakamura S, Nakano H, Shimura K, Sakagami Y, Oishi S, Uchiyama S, Ohkubo T, Kobayashi Y, Fujii N, Matsuoka M, Kodama EN. Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino-terminal heptad repeat. Antiviral Res. 2010;87:179–186. doi: 10.1016/j.antiviral.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Kavlick MF, Wyvill K, Yarchoan R, Mitsuya H. Emergence of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 variants, viral sequence variation, and disease progression in patients receiving antiretroviral chemotherapy. J Infect Dis. 1998;177:1506–1513. doi: 10.1086/515324. [DOI] [PubMed] [Google Scholar]
- Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- Kosalaraksa P, Kavlick MF, Maroun V, Le R, Mitsuya H. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an In vitro competitive HIV-1 replication assay. J Virol. 1999;73:5356–5363. doi: 10.1128/jvi.73.7.5356-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins JI, Rambaut A, Wolinsky S, Korber B. HIV Sequence Compendium 2010. Los Alamos, NM: Los Alamos National Laboratory, Theoretical Biology and Biophysics; 2010. [Google Scholar]
- Labrosse B, Labernardiere JL, Dam E, Trouplin V, Skrabal K, Clavel F, Mammano F. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J Virol. 2003;77:1610–1613. doi: 10.1128/JVI.77.2.1610-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalezari JP, Henry K, O’Hearn M, Montaner JS, Piliero PJ, Trottier B, Walmsley S, Cohen C, Kuritzkes DR, Eron JJ, Jr, Chung J, DeMasi R, Donatacci L, Drobnes C, Delehanty J, Salgo M. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N Engl J Med. 2003;348:2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- Lazzarin A, Clotet B, Cooper D, Reynes J, Arasteh K, Nelson M, Katlama C, Stellbrink HJ, Delfraissy JF, Lange J, Huson L, DeMasi R, Wat C, Delehanty J, Drobnes C, Salgo M. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N Engl J Med. 2003;348:2186–2195. doi: 10.1056/NEJMoa035211. [DOI] [PubMed] [Google Scholar]
- Lohrengel S, Hermann F, Hagmann I, Oberwinkler H, Scrivano L, Hoffmann C, von Laer D, Dittmar MT. Determinants of human immunodeficiency virus type 1 resistance to membrane-anchored gp41-derived peptides. J Virol. 2005;79:10237–10246. doi: 10.1128/JVI.79.16.10237-10246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutfy MR, Raboud JM, Montaner JS, Antoniou T, Wynhoven B, Smaill F, Rouleau D, Gill J, Schlech W, Brumme ZL, Mo T, Gough K, Rachlis A, Harrigan PR, Walmsley SL. Assay of HIV gp41 amino acid sequence to identify baseline variation and mutation development in patients with virologic failure on enfuvirtide. Antiviral Res. 2007;75:58–63. doi: 10.1016/j.antiviral.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Malashkevich VN, Chan DC, Chutkowski CT, Kim PS. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc Natl Acad Sci U S A. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby T, Sista P, DeMasi R, Kirkland T, Roberts N, Salgo M, Heilek-Snyder G, Cammack N, Matthews TJ, Greenberg ML. Characterization of envelope glycoprotein gp41 genotype and phenotypic susceptibility to enfuvirtide at baseline and on treatment in the phase III clinical trials TORO-1 and TORO-2. AIDS Res Hum Retroviruses. 2006;22:375–385. doi: 10.1089/aid.2006.22.375. [DOI] [PubMed] [Google Scholar]
- Mink M, Mosier SM, Janumpalli S, Davison D, Jin L, Melby T, Sista P, Erickson J, Lambert D, Stanfield-Oakley SA, Salgo M, Cammack N, Matthews T, Greenberg ML. Impact of human immunodeficiency virus type 1 gp41 amino acid substitutions selected during enfuvirtide treatment on gp41 binding and antiviral potency of enfuvirtide in vitro. J Virol. 2005;79:12447–12454. doi: 10.1128/JVI.79.19.12447-12454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki D, Kodama E, Ikeuchi M, Mabuchi N, Otaka A, Tamamura H, Ohno M, Fujii N, Matsuoka M. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J Virol. 2005;79:764–770. doi: 10.1128/JVI.79.2.764-770.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Nakamura S, Kodama E, Ito S, Kajiwara K, Izumi K, Sakagami Y, Oishi S, Ohkubo T, Kobayashi Y, Otaka A, Fujii N, Matsuoka M. Electrostatically constrained alpha-helical peptide inhibits replication of HIV-1 resistant to enfuvirtide. Int J Biochem Cell Biol. 2009;41:891–899. doi: 10.1016/j.biocel.2008.08.039. [DOI] [PubMed] [Google Scholar]
- Oishi S, Ito S, Nishikawa H, Watanabe K, Tanaka M, Ohno H, Izumi K, Sakagami Y, Kodama E, Matsuoka M, Fujii N. Design of a Novel HIV-1 Fusion Inhibitor That Displays a Minimal Interface for Binding Affinity. J Med Chem. 2008;51:388–391. doi: 10.1021/jm701109d. [DOI] [PubMed] [Google Scholar]
- Oliveira AC, Martins AN, Pires AF, Arruda MB, Tanuri A, Pereira HS, Brindeiro RM. Enfuvirtide (T-20) resistance-related mutations in HIV type 1 subtypes B, C, and F isolates from Brazilian patients failing HAART. AIDS Res Hum Retroviruses. 2009;25:193–198. doi: 10.1089/aid.2008.0160. [DOI] [PubMed] [Google Scholar]
- Otaka A, Nakamura M, Nameki D, Kodama E, Uchiyama S, Nakamura S, Nakano H, Tamamura H, Kobayashi Y, Matsuoka M, Fujii N. Remodeling of gp41-C34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells. Angew Chem Int Ed Engl. 2002;41:2937–2940. doi: 10.1002/1521-3773(20020816)41:16<2937::AID-ANIE2937>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, Sharron M, Pohlmann S, Sfakianos JN, Derdeyn CA, Blumenthal R, Hunter E, Doms RW. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci U S A. 2002;99:16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K, Nameki D, Kajiwara K, Watanabe K, Sakagami Y, Oishi S, Fujii N, Matsuoka M, Sarafianos SG, Kodama EN. Resistance profiles of novel electrostatically constrained HIV-1 fusion inhibitors. J Biol Chem. 2010;285:39471–39480. doi: 10.1074/jbc.M110.145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaka T, Kavlick MF, Ueno T, Gao WY, Kojima E, Alcaide ML, Chokekijchai S, Roy BM, Arnold E, Yarchoan R, et al. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci U S A. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svicher V, Aquaro S, D’Arrigo R, Artese A, Dimonte S, Alcaro S, Santoro MM, Di Perri G, Caputo SL, Bellagamba R, Zaccarelli M, Visco-Comandini U, Antinori A, Narciso P, Ceccherini-Silberstein F, Perno CF. Specific enfuvirtide-associated mutational pathways in HIV-1 Gp41 are significantly correlated with an increase in CD4(+) cell count, despite virological failure. J Infect Dis. 2008;197:1408–1418. doi: 10.1086/587693. [DOI] [PubMed] [Google Scholar]
- Tolstrup M, Selzer-Plon J, Laursen AL, Bertelsen L, Gerstoft J, Duch M, Pedersen FS, Ostergaard L. Full fusion competence rescue of the enfuvirtide resistant HIV-1 gp41 genotype (43D) by a prevalent polymorphism (137K) AIDS. 2007;21:519–521. doi: 10.1097/QAD.0b013e3280187558. [DOI] [PubMed] [Google Scholar]
- Ueno M, Kodama EN, Shimura K, Sakurai Y, Kajiwara K, Sakagami Y, Oishi S, Fujii N, Matsuoka M. Synonymous mutations in stem-loop III of Rev responsive elements enhance HIV-1 replication impaired by primary mutations for resistance to enfuvirtide. Antiviral Res. 2009;82:67–72. doi: 10.1016/j.antiviral.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Watabe T, Terakawa Y, Watanabe K, Ohno H, Nakano H, Nakatsu T, Kato H, Izumi K, Kodama E, Matsuoka M, Kitaura K, Oishi S, Fujii N. X-ray crystallographic study of an HIV-1 fusion inhibitor with the gp41 S138A substitution. J Mol Biol. 2009;392:657–665. doi: 10.1016/j.jmb.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. Potent D-peptide inhibitors of HIV-1 entry. Proc Natl Acad Sci U S A. 2007;104:16828–16833. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Pozniak A, Wildfire A, Stanfield-Oakley SA, Mosier SM, Ratcliffe D, Workman J, Joall A, Myers R, Smit E, Cane PA, Greenberg ML, Pillay D. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob Agents Chemother. 2005;49:1113–1119. doi: 10.1128/AAC.49.3.1113-1119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]