Summary
Bacteroidetes are a phylum of Gram-negative bacteria abundant in mammalian-associated polymicrobial communities, where they impact digestion, immunity and resistance to infection. Despite extensive competition at high cell density that occurs in these settings, cell contact-dependent mechanisms of interbacterial antagonism, such as the type VI secretion system (T6SS), have not been defined in this group of organisms. Herein we report the bioinformatic and functional characterization of a T6SS-like pathway in diverse Bacteroidetes. Using prominent human gut commensal and soil-associated species, we demonstrate that these systems localize dynamically within the cell, export antibacterial proteins, and target competitor bacteria. The Bacteroidetes system is a distinct pathway with marked differences in gene content and high evolutionary divergence from the canonical T6S pathway. Our findings offer a potential molecular explanation for the abundance of Bacteroidetes in polymicrobial environments, the observed stability of Bacteroidetes in healthy humans, and the barrier presented by the microbiota against pathogens.
Introduction
The Bacteroidetes are a phylum of Gram-negative bacteria that can be isolated from diverse natural habitats (Thomas et al., 2011). Though they include agriculturally and medically relevant pathogens, as well as representatives that play important roles in critical environmental processes such as bioremediation, the phylum is relatively poorly studied. Bacteroidetes are perhaps most appreciated for their intimate association with humans and other mammals, as abundant residents of the gastrointestinal (GI) tract. In this ecosystem, bacteria form dense microbial communities that can exceed 1013 cells per milliliter (Lozupone et al., 2012; Qin et al., 2010; Smith et al., 2006). Bacteroidetes constitute 20–80% of the fecal microbiota of most adult humans. and are largely represented by two genera, Bacteroides and Prevotella (Human Microbiome Project Consortium, 2012). Members of this phylum generally act as mutualists by aiding in the digestion of complex carbohydrates, promoting gut development, modulating the immune system, and protecting against colonization by pathogens (Round and Mazmanian, 2009; Smith et al., 2006; Thomas et al., 2011). As metabolically pliable organisms, Bacteroidetes also help to support a diverse gut community through syntrophic interactions with other microbes (Fischbach and Sonnenburg, 2011; Rey et al., 2010).
Evidence suggests that the capacity of a bacterium to survive in a polymicrobial environment is related to the elaboration of interbacterial antagonistic factors. Studies performed primarily on Proteobacteria have shown that Gram-negative organisms can utilize soluble products as well as contact-dependent mechanisms to compete with other bacteria (Hayes et al., 2014; Riley and Wertz, 2002). Although Bacteroidetes occupy numerous polymicrobial niches, including the human gut, to our knowledge contact-dependent antagonistic pathways have not yet been characterized in this phylum.
The type VI secretion system (T6SS) is a pathway that grants Gram-negative bacteria the capacity to translocate substrates to a wide range of recipient cells (Coulthurst, 2013). Initially speculated to participate strictly in host cell interactions, it is now clear that the more common function of the system is to deliver proteins from the cytoplasm of a donor cell to the periplasm of a Gram-negative recipient (Schwarz et al., 2010a). Substrates transported in a T6S-dependent manner include antibacterial effectors with diverse activities such as phospholipases, peptidoglycan hydrolases, nucleases, and membrane pore-forming proteins (Benz and Meinhart, 2014; Russell et al., 2014). The pathway appears to lack a mechanism for discriminating self from non-self; thus, bacteria with active T6SSs possess immunity proteins that interact with, and inactivate, the effector molecules (Hood et al., 2010; Russell et al., 2011). These interactions are allele specific, and cognate effector–immunity (E–I) pairs are generally encoded adjacently within predicted operons (English et al., 2012; Russell et al., 2012).
The T6S pathway requires the functions of 13 core proteins; unique subsets of these appear to have evolutionary relatedness to type IV secretion system (T4SS) components or to bacteriophage (Boyer et al., 2009; Cascales and Cambillau, 2012). Proteins within the subsets are generally encoded adjacent to each other and interact extensively, suggesting that although each of the 13 core genes is essential, the complete system may be composed of modular, functionally distinct sub-complexes. The T4S-related components, TssL and TssM, are integral membrane proteins that form a trans-envelope complex with an outer-membrane lipoprotein, TssJ (Aschtgen et al., 2010; Felisberto-Rodrigues et al., 2011; Ma et al., 2009). The bacteriophage-like protein TssC, in conjunction with TssB, forms a dynamic filamentous assembly with gross structural similarity to the bacteriophage sheath complex (Basler et al., 2012; Bonemann et al., 2009). Two other bacteriophage-related proteins, VgrG and Hcp, interact with non-overlapping sets of effectors, forming the basis for genetically distinct pathways for T6S-dependent substrate export (Shneider et al., 2013; Silverman et al., 2013; Whitney et al., 2014). VgrG is a phage tail spike-like protein that interacts with effectors via conserved adaptor domains, whereas Hcp is ring-shaped, bears structural homology to the major phage tail tube protein gpV, and interacts with effectors within its pore. Supporting the relationship of Hcp to gpV, Hcp rings have been observed to form higher order head-to-tail stacked structures in vivo, analogous to those observed in bacteriophage (Brunet et al., 2014).
Here, we report the bioinformatic and functional characterization of a T6S-like pathway in the phylum Bacteroidetes. We demonstrate that this pathway has the capacity to mediate cell contact-dependent intra- and inter-phyla bacterial antagonism. Although the pathway lacks conserved elements essential to the well characterized Proteobacterial T6SS, and components that are shared with Proteobacteria display considerable sequence divergence, we provide evidence that they function in a mechanistically similar manner. Several genera that possess the Bacteroidetes T6S-like pathway, including Bacteroides, Prevotella, and Porphyromonas, are abundant members of human-associated polymicrobial communities, suggesting that the pathway may have an important role in defining the composition of the microbiome (Falagas and Siakavellas, 2000).
Results
Bioinformatic characterization of a T6S-like gene cluster in Bacteroidetes
A generally applicable diagnostic secretion signal for T6S effectors is not available; however, genes encoding these proteins can often be identified by sequence-based approaches. Our group and others have noted that in many cases T6S effector and vgrG genes occur proximally and co-directionally on bacterial chromosomes (Barret et al., 2011; Russell et al., 2013; Zhang et al., 2012). We recently exploited this observation to identify a large superfamily of T6S-exported phospholipases. Interestingly, a search for homologs of this class of effectors revealed their presence in Bacteroidetes – a bacterial phylum that does not possess a characterized T6S pathway (Russell et al., 2013). Moreover, like the Proteobacterial effectors, those found in the Bacteroidetes reside in close proximity to apparent vgrG genes and adjacent to open reading frames (ORFs) encoding predicted periplasmic immunity determinants (Figure S1). Our observations concerning phospholipase effector distribution are supported by an exhaustive study of polymorphic toxin domains conducted by Aravind and colleagues, which found genes encoding putative T6 effectors of various catalytic classes represented in Bacteroidetes (Zhang et al., 2012).
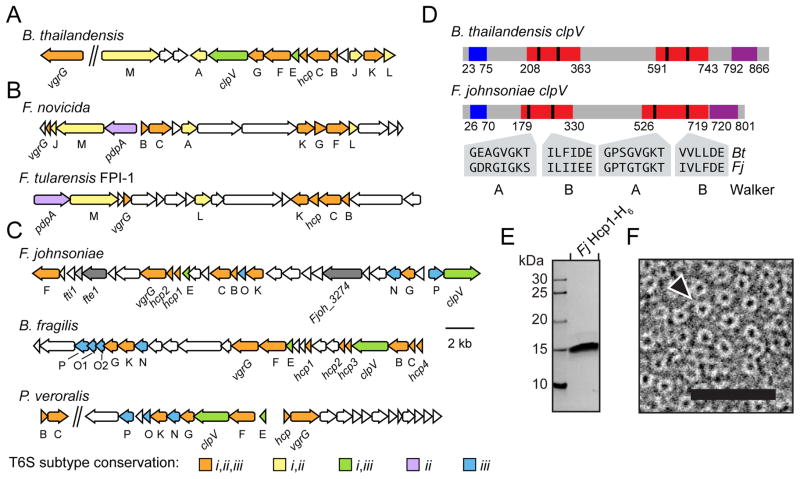
Given the abundance and widespread nature of antibacterial T6S effector genes in Bacteroidetes, we postulated that these proteins participate in interbacterial interactions via a yet uncharacterized T6S-like pathway. Proteobacterial T6S gene clusters often include effector loci; thus, to identify a T6S-like pathway in Bacteroidetes, we searched in the vicinity of putative effector genes for elements that could constitute a secretion system. This led to the identification of a cluster of twelve genes, including vgrG, with orthologs invariantly found in species with predicted effectors (Figure 1A–C and Table S1). Supporting the hypothesis that this gene cluster encodes a T6-like pathway, among the products of the twelve genes, we found a predicted ATPase with domain architecture similar to the Proteobacterial T6S core ATPase, ClpV (Figure 1D) (Schlieker et al., 2005).
Figure 1. T6S-like gene clusters are found within the Bacteroidetes.
(A–C) Gene content and conservation both within and between selected representative members of T6SSi (A), T6SSii (B), and T6SSiii (C). Genes with commonly used tss nomenclature are abbreviated to a single letter. The Francisella tularensis FPI is depicted owing to its status as the only T6SSii to be characterized; however, this system lacks clear homologs of tssA and tssJ, which are present in representative T6SSii systems such the F. novicida gene cluster shown. Locus tags of the regions shown: B. thailandensis E264 BTH_I2705 and BTH_I2954-2968 (A); F. novicida U112 FTN_0037-0054, F. tularensis SCHU FTT_1344-1361c (B); F. johnsoniae UW101 Fjoh_3254-328, B. fragilis NCTC 9343 BF9343_1918-1943, P. veroralis F0319 HMPREF0973_02422-02423 and HMPREF0973_02441-02466. Genes encoding F. johnsoniae T6SSiii substrates identified in this study (dark grey) and a validated immunity locus (light grey) are labeled. For sequence alignments of T6SSiii TssB,C,E,F,G,K proteins, including those depicted, see File S1.
(D) Comparison of domain organization of ClpV homologs from T6SSi (B. thailandensis, Bt) and T6SSiii (F. johnsoniae, Fj) pathways. Colors denote homologous domains: blue, Clp N; red, AAA+; purple, ClpB D2. Sequences highlight the conservation of motifs implicated in ATP binding and hydrolysis within the Walker A and B motifs.
(E) SDS-PAGE analysis of purified Fjoh_3262 bearing a C-terminal hexahistidine tag (Fj Hcp1–H6). Proteins were visualized by Coomassie Blue staining.
(F) F. johnsoniae Hcp1 is a ring-shaped molecule with dimensions similar to Proteobacterial Hcp proteins. Transmission electron micrograph of purified Fj Hcp1–H6 (E) negatively stained with uranyl formate. The arrowhead indicates a typical ring-like assembly. Scale bar, 40 nm.
Homology searches of the remaining conserved elements of the Bacteroidetes gene cluster failed to identify corresponding Proteobacterial T6S proteins. Given the evolutionarily distance between Bacteroidetes and Proteobacteria, we posited that conservation between the constituents of this putative secretion system and the Proteobacterial T6SS might be below the detection limit of non-iterative approaches. By applying iterative search algorithms such as jackHMMER and PSI-BLAST (Altschul et al., 1997; Johnson et al., 2010), we found that six additional genes in the Bacteroidetes cluster encode proteins bearing distant homology to core elements of the Proteobacterial T6SS, TssB, C, E, F, G, and K (Figure 1C and File S1).
In total, our sequence-based searches identified eight of the thirteen putative functional orthologs of the Proteobacterial T6SS encoded within the Bacteroidetes gene cluster (Table S1). Included in these eight components are each of the identified proteins of the bacteriophage-like module of the T6SS, with the exception of Hcp. In the Proteobacterial T6SS, Hcp proteins are required for effector recognition and export, thus the apparent absence of this conserved component was unexpected (Silverman et al., 2013). To ensure we had not overlooked a cryptic Hcp functional ortholog, we turned to structural prediction algorithms, which can identify highly divergent proteins, or convergent proteins, by their common folds. Indeed, using the Phyre remote homology modeling server, we found that one of the remaining four conserved genes in the Bacteroidetes cluster is predicted to encode a protein that adopts a structure with a high degree of similarity to Proteobacterial Hcp proteins (>90% confidence) (Kelley and Sternberg, 2009). To further probe this predicted relatedness, we heterologously expressed and purified a member of this putative Hcp-like protein family encoded within the T6S-like gene cluster of Flavobacterium johnsoniae, a soil-dwelling member of the Bacteroidetes phylum(Figure 1E) (McBride et al., 2009). Visualization of this protein by negative stain transmission electron microscopy demonstrated that it adopts the characteristic ring shape and approximate dimensions of Proteobacterial Hcp proteins (Figure 1F). Together, our data suggest that a conserved gene cluster within the Bacteroidetes phylum encodes a T6S-like pathway. Henceforth, we refer to this pathway as T6SS subtype 3 (T6SSiii), with the intent to distinguish it from the general Proteobacterial and Francisella pathogenicity island-like systems, herein constituting subtypes 1 and 2 (T6SSi,ii), respectively (Figure 1A–C) (Boyer et al., 2009; Broms et al., 2010).
In order to better understand the relationship of the three T6SS groups, we performed phylogenetic analyses on the shared elements TssC and TssF. Phylogenetic trees generated from conserved regions of these proteins exhibited similar topologies, suggesting that the genes encoding them have been co-inherited (Figures 2A and S2A and Files S1–3). In each tree, all Bacteroidetes components comprise a unique clade, distinct from Proteobacterial T6S homologs. While some proteins encoded by species in the phylum Acidobacteria are also found in this clade, analysis of the genomic context of these homologs indicates they reside in gene clusters that lack conserved Bacteroidetes components (Figure S2B). We therefore restrict our definition of T6SSiii to those systems that reside in Bacteroidetes (Figure 2B and Files S1 and S4). Interestingly, T6SSiii gene clusters lack homologs of the T6SSi proteins, TssA, L, M, and J. A gene encoding a putative TssM protein was recently suggested to reside in a B. cellulosilyiticus T6S gene cluster(Coyne et al., 2014). However, we note that homologs of this gene are neither generally found within or adjacent to T6SSiii clusters across the Bacteroidetes phylum, nor are they encoded in the genomes of all organisms that possess the T6SSiii pathway (data not shown). Our data suggest the entirety of the T6SSi trans-envelope sub-complex – including tssM – is absent from T6SSiii. In summary, our data suggest that a phylogenetically distinct T6S-like pathway – composed of an assemblage of proteins distinct from those required for the function of Proteobacterial T6SSs – is found within members of the phyla Bacteroidetes.
Figure 2. T6SSiii is phylogenetically distinct from T6SSi and T6SSii.
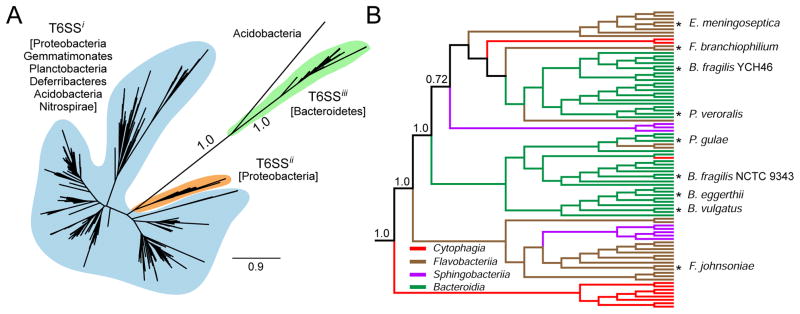
(A) Maximum likelihood (ML) phylogenetic tree generated from a partial alignment of 686 representative TssC sequences spanning the diversity present in T6SSi, T6SSii, and T6SSiii gene clusters. Phyla represented by each system are indicated. Branch support values derived from aBayes analysis for the T6SSiii clade are shown. Scale bar represents amino acid changes per site. A similar tree for TssF is provided in Figure S2a.
(B) ML tree created from a partial alignment of TssC sequences found within T6SSiii gene clusters. The tree is rooted with Acidobacterial TssC sequences. Lengths do not reflect evolutionary distance. Colors trace the Class from which each sequence is derived. Nodes representing TssC sequences of organisms discussed in the text and those of particular significance are marked with an asterisk. Branch support values were generated by aBayes analysis. Partial sequence alignments and branch supports corresponding to phylogenetic trees in panels (A) and (B) and Figure S2 are provided in Files S1–4.
A T6SSiii pathway exports antibacterial effectors
T6SSs are functionally versatile and can deliver effectors to other bacteria, to eukaryotic host cells, or to both of these cell types. We identified a number of predicted antibacterial effectors associated with T6SSiii gene clusters in Bacteroidetes, suggesting that this system might possess the capacity to mediate interbacterial antagonism (data not shown). To first establish whether the T6SSiii gene cluster encodes a secretory pathway responsible for the export of effector proteins, we conducted secretome measurements using F. johnsoniae. We chose this organism because it is genetically tractable, easily maintained under aerobic conditions in the laboratory (McBride et al., 2009; Rhodes et al., 2011), and it possesses a T6SSiii gene cluster that is highly representative of the system in other Bacteroidetes, including the human gut-associated commensal species B. fragilis, B. vulgatus, and B. eggerthii (Figure 2B).
To determine the contribution of the T6SSiii pathway to the secretome of F. johnsoniae, we compared the culture supernatant of a strain bearing an in-frame deletion of its predicted tssC homolog, Fjoh_3266, to the wild-type parental strain using mass spectrometry. This analysis identified six proteins that were undetected in F. johnsoniae ΔtssC, but met our criteria for inclusion in the wild-type secretome (Table 1 and Table S2). Strikingly, the two most abundant of these proteins were Fjoh_3262 and Fjoh_3260, the Hcp and VgrG homologs encoded within the T6SSiii gene cluster, respectively (Figure 1C). This finding parallels similar secretome studies of T6SSi pathways, which generally find Hcp- and VgrG-family proteins as the major components of the T6SS-dependent substrate pool (Fritsch et al., 2013; Hood et al., 2010; Russell et al., 2012).
Table 1.
Secreted proteins not detected the F. johnsoniae ΔtssC secretome
| Locus Taga | Name | Abundanceb | Unique peptides detected | Signal peptidec | T6SSiii clusterd | Molecular weight | Predicted/determined function |
|---|---|---|---|---|---|---|---|
| Fjoh_3262 | Hcp | 133 ± 3.5e | 10 | N | Y | 14.5 | Secreted T6S structural component |
| Fjoh_3260 | VgrG | 13.3 ± 1.5 | 5 | N | Y | 66.0 | Secreted T6S structural component |
| Fjoh_3206 | RemH | 11.7 ± 2.3 | 4 | Y | N | 16.9 | Gliding motility |
| Fjoh_0984 | RemF | 5 ± 1.0 | 4 | Y | N | 16.8 | Gliding motility |
| Fjoh_3274 | – | 4 ± 1.0 | 3 | N | Y | 102 | T6SSiii effector |
| Fjoh_3257 | Fte1 | 3 ± 1.7 | 2 | N | Y | 62.7 | T6SSiii effector |
Locus tags derived from F. johnsoniae UW101 (NCBI Accession NC_009441.1).
Average spectral counts of three technical replicates.
Signal peptide predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP).
The T6SSiii gene cluster comprises two apparent divergently transcribed operons (Fjoh_3254-Fjoh_3281), with the terminal gene of each operon encoding a conserved T6SSIII element.
Standard deviation of three technical replicates.
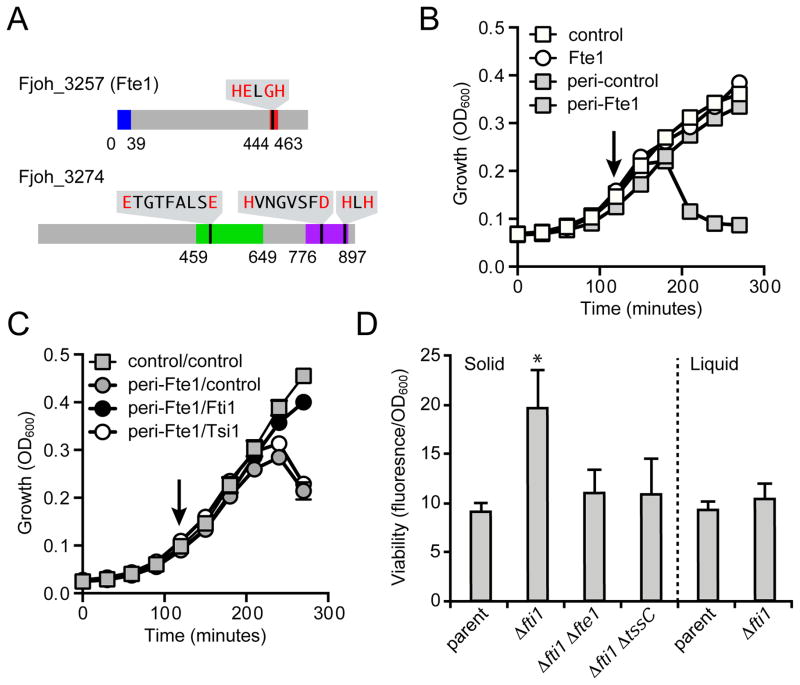
Of the remaining four proteins, two are encoded by hypothetical ORFs present within the T6SSiii gene cluster (Fjoh_3257 and Fjoh_3274), whereas the remaining two are implicated in gliding motility and possess predicted signal sequences (Figure 1C, Table 1) (Rhodes et al., 2011). While the latter may have yet unrecognized roles relevant to T6SSiii, for the purposes of identifying secreted effectors we focused on Fjoh_3257 and Fjoh_3274, which do not contain predicted signal peptides. Notably, these proteins both possess domains found in known or predicted T6SSi effectors. Fjoh_3257 contains an HEXGH motif found in zinc metalloproteases fused to T6SSi-exported VgrG proteins and Fjoh_3274 contains both a central glycoside hydrolase domain and a C-terminal zinc-dependent peptidoglycan endopeptidase motif (Figure 3A) (Pukatzki et al., 2007; Russell et al., 2012). Based on these data, we hypothesized that Fjoh_3257 and Fjoh_3274 are T6SSiii-exported effectors that exert toxicity in the periplasm of target cells.
Figure 3. F. johnsoniae T6SSiii exports an antibacterial protein that is encoded adjacent to a cognate immunity determinant.
(A) Domain organization of the putative substrates of F. johnsoniae T6SSiii. PAAR-like (blue), zinc-dependent metalloprotease (red), glycoside hydrolase (green), and zinc-dependent peptidoglycan amidase (purple) domains are indicated. Expanded amino acid sequences in each domain correspond to conserved motifs and invariant or critical catalytic residues (red).
(B–C) Growth of E. coli strains harboring the indicated expression vectors. Empty vectors (control) and vectors that introduce an N-terminal Sec signal peptide (peri) are indicated. Cells were induced to express predicted immunity proteins (C) at time 0 and Fte1 at the indicated time point (arrow). Type VI secretion immunity protein 1 (Tsi1) is used as a non-cognate immunity control. Error bars represent ± standard deviation (SD) (n = 3). Expression data for (B) are provided in Figure S3.
(D) Intercellular self-intoxication of the indicated F. johnsoniae strains as measured by propidium iodide staining. Liquid cultures were grown with vigorous shaking, which inhibits the formation of the prolonged cell–cell contacts required for T6-mediated interactions (Hood et al., 2010; Leroux et al., 2012). Error bars represent ± standard deviation (SD) (n = 3). Samples differing significantly from parent as measured by a two-tailed T-test are indicated by asterisks (p < 0.01).
To test the hypothesis that the T6SSiii pathway exports antibacterial effectors, we further investigated Fjoh_3257. When directed to the periplasm of Escherichia coli, Fjoh_3257 induced significant cell lysis, whereas the native protein – predicted to localize to the cytoplasm – did not exhibit apparent toxicity (Figures 3B and S3). T6SSi delivers effectors directly to the periplasm of recipient cells. If T6SSiii functions similarly, our data suggest Fjoh_3257, henceforth referred to as Flavobacterium type VI secretion effector 1 (Fte1), could promote intercellular toxicity and thus necessarily associate with a cognate immunity determinant. Moreover, we would expect this immunity protein to reside in the periplasmic compartment, as T6SSi effector inactivation invariably occurs via direct interaction with immunity (Benz et al., 2012; Dong et al., 2013a; Russell et al., 2011; Zhang et al., 2013). We identified a gene encoding a protein matching the predicted immunity criteria directly downstream of fte1, Fjoh_3256 (hereafter referred to as Flavobacterium type VI secretion immunity 1, or fti1). Co-expression of Fti1 specifically abrogated the lytic effects of Fte1, indicating that Fte1-Fti1 comprise an antibacterial effector–immunity pair of T6SSiii in F. johnsoniae (Figure 3C).
To test whether Fte1 exerts antibacterial activity in a T6SSiii-dependent manner, we measured the cellular integrity of F. johnsoniae strains lacking fti1. When propagated on a solid substratum, a condition conducive to prolonged cell contact, we observed increased membrane permeability in the Δfti1 strain (Figure 3D). This phenotype was abrogated by concomitant deletion of tssC, fte1, or by growth in liquid media. Together with our secretome studies, these data strongly suggest the capacity of T6SSiii to participate in interbacterial interactions through the export of antibacterial effectors.
T6SSiii mediates interspecies bacterial antagonism
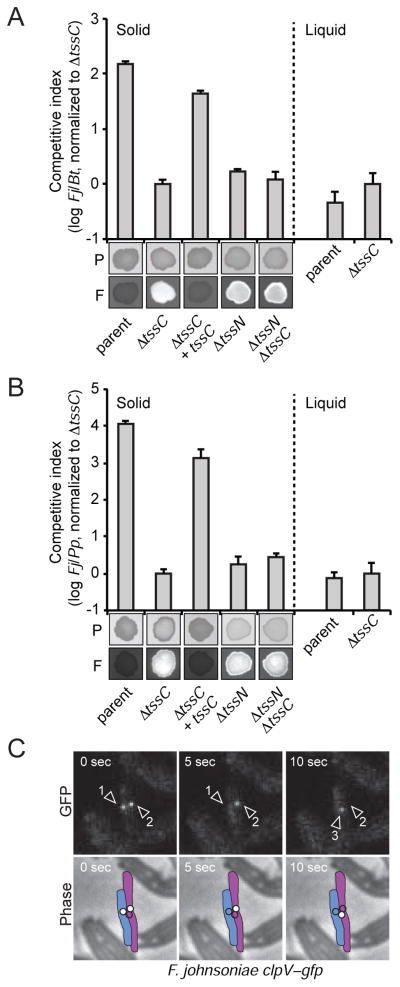
Interbacterial T6SSi has been observed to be a crucial determinant of fitness during polymicrobial growth. Under contact-promoting conditions, its inactivation generally leads to significant defects in the capacity to outcompete other organisms in co-culture. To determine whether T6SSiii also functions in interspecies antagonism we grew wild-type F. johnsoniae and derivative strains with either Burkholderia thailandensis or Pseudomonas putida under T6-conducive conditions. The inactivation of T6SSiii by a deletion of tssC in F. johnsoniae greatly impacted the outcome of these growth competitions, allowing for significant expansion of the competitor population (Figure 4A and 4B). This phenotype could be complemented by the introduction of an extra-chromosomal copy of tssC, demonstrating that the observed change in fitness was not due to mutant polarity. Moreover, wild-type and ΔtssC displayed equal fitness in liquid growth medium, consistent with the known requirement for intimate cell–cell contact in T6S-dependent interactions. We further examined interspecies co-cultures containing F. johnsoniae lacking the T6SSiii-restricted component, tssN (Fjoh_3277, Table S1). F. johnsoniae ΔtssN antagonized B. thailandensis and P. putida to an equivalent degree as ΔtssC or a strain bearing deletions in both tssC and tssN, consistent with our hypothesis that these genes encode essential elements of the same pathway (Figure 4A and 4B). Overall, our data strongly suggest that the T6SSiii pathway mediates interbacterial antagonism in a manner analogous to T6SSi, yet using a distinct complement of proteins.
Figure 4. F. johnsoniae utilizes a dynamic T6SSiii apparatus to target competitor organisms.
(A–B) Growth experiments measuring fitness of the indicated F. johnsoniae (Fj) strains in co-culture for 20 hours with fluorescently-labeled competitors B. thailandensis (A, Bt) or P. putida (B, Pp). Competitive index is defined as the change in F. johnsoniae/competitor colony forming units between initiation and harvest of the co-culture. Experiments were performed under contact-promoting (solid) and contact-inhibiting (liquid) conditions. Qualitative analysis of competition outcome by photographic (P) and fluorescence (F) imaging is shown for corresponding samples grown under contact-promoting conditions after 48 hours of co-culture. Error bars represent ± SD (n = 3).
(C) Micrograph series depicting a 10s time course of wild-type F. johnsoniae clpV–gfp. Phase and GFP channels are presented separately. Three dynamic foci were observed over the duration of the experiment (arrowheads), and the presence or absence of each focus in each frame is schematized in the phase micrographs with white or unfilled circles, respectively. Full-length movies that include the region represented are available as Supplemental Files (Movies S1 and S2). See also Figure S4.
The T6SSiii apparatus exhibits dynamic behavior
Antibacterial effectors released by the T6SSi pathway are operative on Gram-negative recipients only if they are delivered across the outer membrane by the translocation machinery. Owing to this feature of the system, the apparatus must behave dynamically in order to sample localizations that orient the system toward competitor cells. Green fluorescent protein (GFP) fusions to the C-terminus of ClpV proteins have served as a convenient means to visualize this behavior of T6SSi systems. Since an apparent ClpV ortholog is identifiable in T6SSiii gene clusters, we sought to monitor the subcellular localization and dynamic behavior of this protein as a way to further interrogate the mechanistic similarity between T6SSi and T6SSiii pathways. We began by generating a strain of F. johnsoniae bearing a functional clpV–gfp fusion at the native clpV locus (Figure S4). Visualization of this strain using time-lapse fluorescence microscopy revealed punctate foci appearing, disappearing, and frequently reappearing at different subcellular locations, on a rapid time scale (Figure 4C and Movies S1 and S2). Inactivation of the system in F. johnsoniae through the deletion of tssC abrogated these foci, similar to what has been observed in the T6SSi pathway. It is worth noting that T6SSiii ClpV exhibits punctate localization and dynamic behavior in the absence of an apparent TssM homolog, whereas in T6SSi systems, TssM proteins are required for ClpV dynamics. Taken together with our bioinformatic, secretomic, and phenotypic data, these findings strongly suggest that the T6SSiii pathway functions in a manner mechanistically similar to T6SSi despite a highly divergent and unique assemblage of core components.
Bacteroides fragilis targets B. thetaiotaomicron via T6SSiii
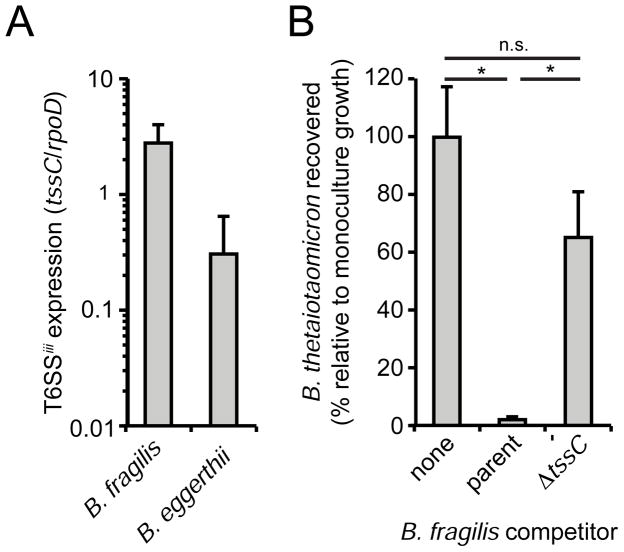
Motivated by our characterization of the T6SSiii pathway in F. johnsoniae, we sought to explore the relevance of our findings to human-associated Bacteroidetes. Our analyses indicate T6SSiii gene clusters are present in many members of the genus Bacteroides, including numerous prominent human gut residents (Figure 2B and File S1). To probe the potential for the T6SSiii pathway to influence the behavior of these organisms in a physiological setting, we colonized germfree mice with a community containing B. fragilis, B. eggerthii, and the Proteobacterium E. coli, and measured T6SSiii expression. Quantitative reverse transcriptase (RT)-PCR of the cecal contents from these mice revealed the tssC gene in both organisms is expressed, at levels approaching (B. eggerthii) or exceeding (B. fragilis) the housekeeping transcript rpoD (Figure 5A).
Figure 5. The T6SSiii pathway is expressed and active in the genus Bacteroides.
(A) Quantitative RT-PCR analysis of the in vivo expression of tssC for the indicated bacteria. Expression was measured in cecal samples from germfree mice colonized for one week with B. fragilis, B. eggerthii, and E. coli and normalized using species-specific primers for the housekeeping sigma factor, rpoD.
(B) Growth competitions measuring viability of B. thetaiotaomicron after 24 h in the presence of the indicated B. fragilis strains on solid media. B. thetaiotaomicron growth was normalized to values obtained in the absence of B. fragilis. Viability was determined by colony forming units. Error bars represent ± SD (n = 3). Significance as indicated by asterisks was measured by a two-tailed T-test (p<0.002), n.s.; no statistical difference.
Expression of tssC in the mammalian gut environment led us to hypothesize that the T6SSiii pathway could be employed by Bacteroides to target other Gram-negative human gut microbes, including other species of Bacteroides. To test this hypothesis, assessed the ability of wild-type B. fragilis to inhibit growth of the prominent human gut commensal B. thetaiotaomicron, which lacks a T6SS. Growth competition experiments revealed that B. fragilis reduces B. thetaiotaomicron growth by approximately two orders of magnitude (Figure 5B). Strikingly, this activity is almost entirely T6SSiii-dependent, as an in-frame, unmarked deletion of tssC (BF9343_1941) in B. fragilis renders this species largely unable to reduce B. thetaiotaomicron growth. These data demonstrate the capacity of T6SSiii to act between prominent human gut-associated members of the genus Bacteroides.
Discussion
With the bioinformatic and functional characterization of T6SSiii, it is now evident that the Bacteroidetes possess a means for contact-dependent interbacterial antagonism. This is in-line with the observation that Bacteroidetes frequently occupy contact-promoting, polymicrobial niches (Thomas et al., 2011). Indeed, many of the organisms we identify the T6SSiii pathway within, including Porphyromonas, Prevotella and Bacteroides spp, are highly adapted host-associated obligate anaerobes that predominate – as pathogens or commensals – within the most densely populated polymicrobial sites in the human body (Falagas and Siakavellas, 2000; Smith et al., 2006). Thus, within sites such as the GI tract, oral cavity, and the vagina, where bacteria with T6SSiii are abundant, the pathway may play a broad role in defining community composition.
By analogy with T6SSi and T6SSii, it is reasonable to speculate that T6SSiii has the capacity to mediate host cell interactions in addition to its now established role in interbacterial antagonism. Certain T6SSi and T6SSii pathways appear to specialize in either bacterial or host cell targeting, whereas others can act on both cell types (Hood et al., 2010; MacIntyre et al., 2010; Pukatzki et al., 2007; Schwarz et al., 2010b). Target range appears to be dictated, at least in part, by the specific complement and corresponding activities of the effectors transported by a system. For example, recent reports suggest that by virtue of structural conservation among the phospholipid constituents of cellular membranes, T6S effectors belonging to the Tle phospholipase superfamily can confer both anti-bacterial and anti-eukaryotic activity (Dong et al., 2013b; Jiang et al., 2014; Russell et al., 2013). While members of the Tle superfamily are among the many apparent effectors of T6SSiii, the preponderance of predicted effectors that target peptidoglycan, a molecule found exclusively in bacteria, indicates that interbacterial antagonism is likely the basal function of the T6SSiii pathway.
Despite lacking several T6SSi core components, including TssJ, L, and M, our observations suggest that T6SSiii functions in a fundamentally analogous manner. Both systems exhibit dynamic behavior, target effectors to the periplasm of recipients, and abundantly export VgrG and Hcp-family proteins. There are several conceivable explanations for these observations. One possibility is that the unique T6SSiii components, TssN, TssO, and TssP functionally substitute for the missing components. This model is supported by the prediction that these components, like TssL and M, are integral membrane proteins (Ma et al., 2009) (Aschtgen et al., 2012). However, TssJ is a predicted lipoprotein that requires localization to the outer membrane for function, and so far a T6SSiii-conserved predicted outer membrane-localized protein has not been identified. It is worth noting that TssJ, L, and M interact stably to form a trans-envelope complex (Cascales and Cambillau, 2012). While it has been postulated that this complex facilitates the passage of bacteriophage-like proteins and effectors out of the recipient cell, there are little experimental data to support this notion. It is therefore not yet possible to rule out a model whereby the components shared between T6SSi-iii – namely those belonging to the bacteriophage-like sub-complex – represent the minimal structural assemblage of the T6SS. Distinguishing essential structural components from proteins with critical regulatory roles, for example, can be challenging (Hsu et al., 2009; Silverman et al., 2011). Understanding the functional significance of the varied complement of core elements associated with T6SSi-iii will ultimately require both detailed biochemical approaches aimed at defining more precisely the roles of the individual proteins and ultra-structural characterization of the system.
While T6SSi and T6SSiii are divergent, predicted effector proteins in T6SSiii-encoding organisms are often closely related to homologs in organisms possessing T6SSi. For example, homologs of Fte1 are readily identified in Acinetobacter spp as well as strains of E. coli. These homologs, like Fte1, are encoded adjacent to predicted periplasmic immunity proteins as well as VgrG, suggesting that they likely possess common modes of toxicity and export. The relative ease with which homologs of effectors that transit the T6SSi and T6SSiii pathways can be identified is further indicative of the similarity between these two systems and suggests they might share a common pool of potential effectors exchanged through horizontal gene transfer.
The gene encoding one of the putative substrates identified in our study, Fjoh_3274, is found adjacent to a locus that encodes a small protein possessing a DUF4280 domain (Fjoh_3275). In Fjoh_3274 homologs found in other species, these open reading frames are often fused, suggesting their function is linked. Structure prediction algorithms indicate a strong likelihood that DUF4280 adopts a fold closely related to the PAAR domain, which forms a pyramidal structure that is thought to recruit effector proteins to the apparatus via interaction with VgrG (Shneider et al., 2013). Interestingly, proteins bearing DUF4280 are found in Gram-positive bacteria, a division of bacteria not known to possess a T6S-like pathway. Moreover, the genes encoding these proteins are often found adjacent to VgrG-like proteins. Our finding herein that the T6S pathway extends to the Gram-negative phylum Bacteroidetes raises the possibility that other organisms, even Gram-positive bacteria, may also possess related systems that have yet to be identified. By analogy, the antibacterial nature of the C-terminal polymorphic toxin domains of YD-repeat proteins was initially discovered in Gram-negative bacteria; however, more recent studies have found homologs of these toxin domains participate in interbacterial antagonism in Gram-positive organisms (Koskiniemi et al., 2013).
Colonization resistance is a property of the gut microbiota whereby it acts as a coherent, resilient entity that exhibits resistance to invading microbes (Stecher and Hardt, 2011). The importance of this property is exemplified by the enhanced susceptibility to pathogens observed following either depletion or dysbiosis of the gut microbial community during antibiotic treatment (Lawley and Walker, 2013). Notably, recent studies have also shown that individuals carry the same commensal strains in their gut microbiomes for years or decades, and that members of the Bacteroidetes exhibit the greatest stability (Faith et al., 2013). A complete molecular explanation for these observations will likely include metabolic exclusion (Turnbaugh et al., 2009), colonization of critical niches (Lee et al., 2013), and the production of diffusible antimicrobials such as bacteroicins (Pujol et al., 2011). We posit that antagonistic contact-dependent interactions mediated by the T6SSiii pathway are another important contributor to colonization resistance and commensal stability. Interestingly, Turnbaugh and colleagues recently identified a transcript corresponding to a core T6SSiii element (vgrG) derived from Bacteroidales in a metatranscriptome of fresh human fecal samples (Maurice et al., 2013). Moreover, another study showed that the physical environment of the gut is conducive to T6S-mediated interbacterial interactions (Fu et al., 2013). While experiments involving T6SSiii mutants within gut colonization models will be needed in order to directly establish its role in this environment, taken together with our demonstration of the antibacterial nature of the system, these findings are consistent with the hypothesis that contact-dependent interbacterial interactions occur among commensals in the human gut.
Experimental Procedures
Bacterial strains, plasmids, and growth conditions
F. johnsoniae, B. thailandensis, P. putida, B. fragilis, B. eggerthii, and B. thetaiotaomicron used in this study were derived from the sequenced strains UW101, E264, KT2440, NCTC 9343, ATCC 27754, and VPI-5482 respectively. E. coli strains used in this study included DH5α for plasmid maintenance and tri-parental conjugation of plasmids into F. johnsoniae and B. fragilis, Rosetta 2(DE3) (EMD Millipore) for toxicity experiments, BL21(DE3) pLysS for the expression and purification of Fjoh_3262, and Nissle 1917 for mouse colonization experiments. Growth conditions for all strains and plasmid and strain construction details are described in Supplemental Experimental Procedures.
Informatic analyses
ClpV- and VgrG-like proteins were identified by automated annotation from NCBI blast servers. Hcp-like proteins were initially found within F. johnsoniae by PHYRE 2.0 (Kelley and Sternberg, 2009), and were thereafter identified by homology using blastp analysis. Other T6SS homologs were identified in T6SSiii gene clusters by the application of the iterative search algorithm jackHmmer on the RefSeq protein database using seed proteins obtained from F. johnsoniae (Finn et al., 2011). Alignments, domain prediction, phylogenetic trees, and subcellular localization were determined as described in the Supplemental Experimental Procedures.
Secretome preparation and MS analysis
The F. johnsoniae secretome was obtained using previously described methods with modifications indicated in the Supplemental Experimental Procedures (Hood et al., 2010). The UniProt F. johnsoniae UW101 database was used as a reference for peptide identification using MaxQuant v1.4.1.2 (Cox and Mann, 2008). Relative abundance of proteins was assessed using spectral counting (Liu et al., 2004). Proteins were filtered such that all had at least two unique peptides detected and possessed an average of three spectral counts in wild-type replicates.
Cellular toxicity assays
E. coli toxicity assays were performed as described previously with minor modifications (Russell et al., 2011). For the analysis of T6SSiii-dependent Fte1 toxicity in F. johnsoniae, strains were grown on a nitrocellulose surface as monocultures for 20 h before analysis by propidium iodide staining. Full details are provided in Supplemental Experimental Procedures.
Bacterial competitions
Bacterial co-cultures were prepared as described in Supplemental Experimental Procedures and either spotted on nitrocellulose placed on solid media or sub-inoculated into liquid media. After 20 h (F. johnsoniae) or 24 h (Bacteroides) of competition, cells were harvested and plated on selective media for quantification of each organism. Fluorescence images and photographs were acquired for the F. johnsoniae experiments after 48 h. Full details are provided in Supplemental Experimental Procedures.
Fluorescence microscopy
Microscopy was performed as described previously (Leroux et al., 2012). F. johnsoniae cells were prepared for microscopy after growth in conditions similar to bacterial competition experiments and were visualized on 1.5 % w/v agarose phosphate-buffered saline pads. Automated image acquisition was performed at 5s intervals for 6 minutes. Full details are provided in Supplemental Experimental Procedures.
In vivo expression of T6SSiii
All animal experiments were performed using protocols approved by the Yale University Institutional Animal Care and Use Committee. Germ-free Swiss Webster mice were maintained in flexible plastic gnotobiotic isolators with a 12-hour light/dark cycle. Mice (n = 5/group) were individually caged and were provided with standard autoclaved mouse chow (5K67 LabDiet, Purina) ad libitum. On day 0, mice were gavaged orally with 2x108 CFU of each strain (B. fragilis, B. eggerthii, and E. coli). Animals were sacrificed on day 7 and samples were collected along the length of the gut. All samples were snap-frozen in liquid nitrogen and stored at −80°C.
RNA extraction from mouse cecal samples, cDNA synthesis, and quantitative RT-PCR were performed using standard methods. Expression for each T6SS gene was normalized to rpoD expression levels in the same organisms using species-specific primers. Genomic DNA samples used to generate standards for quantitative RT-PCR were obtained by published methods (Degnan et al., 2014). Full details are provided in Supplemental Experimental Procedures.
Supplementary Material
Bacterial T6SS divides into three phylogenetically distinct subtypes (T6SSi-iii)
T6SSiii is restricted to Bacteroidetes and is composed of unique components
T6SSiii targets toxic effectors to competing Proteobacteria and other Bacteroidetes
Bacteroides fragilis T6SSiii targets B. thetaiotaomicron and is expressed in vivo
Acknowledgments
We thank M. McBride and L. Comstock for sharing reagents and protocols necessary for generating the F. johnsoniae and B. fragilis mutants used in our study, M. LeRoux and R. Kirkpatrick for assistance with fluorescence microscopy, and members of the Mougous and Goodman laboratories for helpful discussions. This work was supported by grants from the National Institutes of Health (NIH) (AI080609 and AI105268 to J.D.M.; DK089121, GM103574 and GM105456 to A.L.G). Research in the Gonen laboratory is supported by the Howard Hughes Medical Institute. A.B.R. was supported by the Josephine de Karman Fellowship Trust and the University of Washington Department of Microbiology Helen Whiteley Award. J.C.W. was supported by a postdoctoral research fellowship by the Canadian Institutes of Health Research. A.J.B. was supported by a grant from the National Science Foundation (DGE-1256082). S.C. was supported by a Howard Hughes Medical Institute Life Sciences Research Foundation Fellowship. J.D.M. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschtgen MS, Gavioli M, Dessen A, Lloubes R, Cascales E. The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol Microbiol. 2010;75:886–899. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- Aschtgen MS, Zoued A, Lloubes R, Journet L, Cascales E. The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 Type VI secretion system, is inserted by YidC. Microbiologyopen. 2012;1:71–82. doi: 10.1002/mbo3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret M, Egan F, Fargier E, Morrissey JP, O’Gara F. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology (Reading, England) 2011;157:1726–1739. doi: 10.1099/mic.0.048645-0. [DOI] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz J, Meinhart A. Antibacterial effector/immunity systems: it’s just the tip of the iceberg. Current opinion in microbiology. 2014;17:1–10. doi: 10.1016/j.mib.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Benz J, Sendlmeier C, Barends TR, Meinhart A. Structural insights into the effector-immunity system Tse1/Tsi1 from Pseudomonas aeruginosa. PLoS One. 2012;7:e40453. doi: 10.1371/journal.pone.0040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. The EMBO journal. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms JE, Sjostedt A, Lavander M. The Role of the Francisella Tularensis Pathogenicity Island in Type VI Secretion, Intracellular Survival, and Modulation of Host Cell Signaling. Frontiers in microbiology. 2010;1:136. doi: 10.3389/fmicb.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet YR, Henin J, Celia H, Cascales E. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO reports. 2014;15:315–321. doi: 10.1002/embr.201337936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci. 2012;367:1102–1111. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthurst SJ. The Type VI secretion system - a widespread and versatile cell targeting system. Research in microbiology. 2013;164:640–654. doi: 10.1016/j.resmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. Evidence of Extensive DNA Transfer between Bacteroidales Species within the Human Gut. MBio. 2014:5. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B(1)(2) analogs and compete in the gut. Cell host & microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Zhang H, Gao ZQ, Wang WJ, She Z, Liu GF, Shen YQ, Su XD, Dong YH. Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3. The Biochemical journal. 2013a doi: 10.1042/BJ20130193. [DOI] [PubMed] [Google Scholar]
- Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proceedings of the National Academy of Sciences of the United States of America. 2013b;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Molecular microbiology. 2012;86:921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Siakavellas E. Bacteroides, Prevotella, and Porphyromonas species: a review of antibiotic resistance and therapeutic options. Int J Antimicrob Agents. 2000;15:1–9. doi: 10.1016/s0924-8579(99)00164-8. [DOI] [PubMed] [Google Scholar]
- Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 2011;7:e1002386. doi: 10.1371/journal.ppat.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic acids research. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell host & microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch MJ, Trunk K, Diniz JA, Guo M, Trost M, Coulthurst SJ. Proteomic Identification of Novel Secreted Antibacterial Toxins of the Serratia marcescens Type VI Secretion System. Molecular & cellular proteomics : MCP. 2013;12:2735–2749. doi: 10.1074/mcp.M113.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell host & microbe. 2013;14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb Perspect Med. 2014:4. doi: 10.1101/cshperspect.a010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Schwarz S, Mougous JD. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:1111–1125. doi: 10.1111/j.1365-2958.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium T. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. A Pseudomonas aeruginosa Type VI Secretion Phospholipase D Effector Targets Both Prokaryotic and Eukaryotic Cells. Cell host & microbe. 2014;15:600–610. doi: 10.1016/j.chom.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical chemistry. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LS, Lin JS, Lai EM. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol. 2009;191:4316–4329. doi: 10.1128/JB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Applied and environmental microbiology. 2009;75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol A, Crost EH, Simon G, Barbe V, Vallenet D, Gomez A, Fons M. Characterization and distribution of the gene cluster encoding RumC, an anti-Clostridium perfringens bacteriocin produced in the gut. FEMS Microbiol Ecol. 2011;78:405–415. doi: 10.1111/j.1574-6941.2011.01176.x. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. Dissecting the in vivo metabolic potential of two human gut acetogens. The Journal of biological chemistry. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes RG, Pucker HG, McBride MJ. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant gliding motility genes remF, remG, remH, and remI. Journal of bacteriology. 2011;193:2418–2428. doi: 10.1128/JB.00117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annual review of microbiology. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nature reviews Microbiology. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell host & microbe. 2012;11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem. 2005;386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010a;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010b:6. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. Haemolysin Coregulated Protein Is an Exported Receptor and Chaperone of Type VI Secretion Substrates. Molecular cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Rocha ERJPB. The Medically Important Bacteroides spp. Health and Disease. 2006;7 [Google Scholar]
- Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Current opinion in microbiology. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut bacteroidetes: the food connection. Frontiers in microbiology. 2011;2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, et al. Genetically distinct pathways guide effector export through the type VI secretion system. Molecular microbiology. 2014;92:529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao ZQ, Wang WJ, Liu GF, Xu JH, Su XD, Dong YH. Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. The Journal of biological chemistry. 2013;288:5928–5939. doi: 10.1074/jbc.M112.434357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.