Abstract
Oxamniquine resistance evolved in the human blood fluke (Schistosoma mansoni) in Brazil in the 1970s. We crossed parental parasites differing ~500-fold in drug response, determined drug sensitivity and marker segregation in clonally-derived F2s, and identified a single QTL (LOD=31) on chromosome 6. A sulfotransferase was identified as the causative gene using RNAi knockdown and biochemical complementation assays and we subsequently demonstrated independent origins of loss-of-function mutations in field-derived and laboratory-selected resistant parasites. These results demonstrate the utility of linkage mapping in a human helminth parasite, while crystallographic analyses of protein-drug interactions illuminate the mode of drug action and provide a framework for rational design of oxamniquine derivatives that kill both S. mansoni and S. haematobium, the two species responsible for >99% of schistosomiasis cases worldwide.
In the absence of effective vaccines for human helminth infections, repeated rounds of mass treatment with drug monotherapies are typically used for control in most developing countries (1, 2). These programs bring enormous health benefits, but impose strong selection on parasite populations and resistance is suspected for several helminth species, including Onchocerca volvulus, cause of river blindness, and Wuchereria bancrofti, cause of lymphatic filariasis (3, 4). However, resistance to oxamniquine (OXA) in Schistosoma mansoni, a trematode parasite that infects 67 million people in Africa and South America (1), provides the first and clearest example of naturally selected drug resistance in a human helminth parasite. OXA was the first line drug in Brazil until the late 1990s, and remained in use until 2010. Resistant parasites were isolated from Brazilian patients in the 1970s (5, 6) and also selected in the laboratory from sensitive parasite lines (7). Resistance has a recessive basis and results in ~500-fold reduction in drug sensitivity (8). Genetic complementation experiments demonstrate that the same gene determines resistance in both field and laboratory-selected parasites, although whether the same mutations are responsible is unknown (9). OXA is species-specific (10, 11), killing S. mansoni (67 million cases) but not other schistosome species (S. haematobium, 119 million cases) in Africa or S. japonicum (1 million cases) in Asia (1).
We exploited the S. mansoni genome sequence (12, 13) and genetic map (14) to identify genome region(s) that underlie OXA-resistance and to determine the basis for species-specific drug action. The complete life cycle of S. mansoni can be maintained in the laboratory, while clonal expansion of larval parasites within the snail host allows production of thousands of genetically identical single sex parasites, making this organism well suited to linkage mapping methods. We crossed an OXA-sensitive (LE) (15) and an OXA-resistant parasite (HR) (8) derived by laboratory selection. We used an intercross design: F1 individuals were crossed to generate 388 F2s. At each stage individual parasite genotypes were isolated by infecting snails with a single miracidium larva (Fig. 1A). We measured OXA-resistance by exposing adult parasites to 500 µg/mL OXA and monitoring parasite survival. The F1s and 136/182 (74.7%) F2s were OXA-sensitive while 36/182 (25.3%) F2s were OXA-resistant, consistent with recessive inheritance.
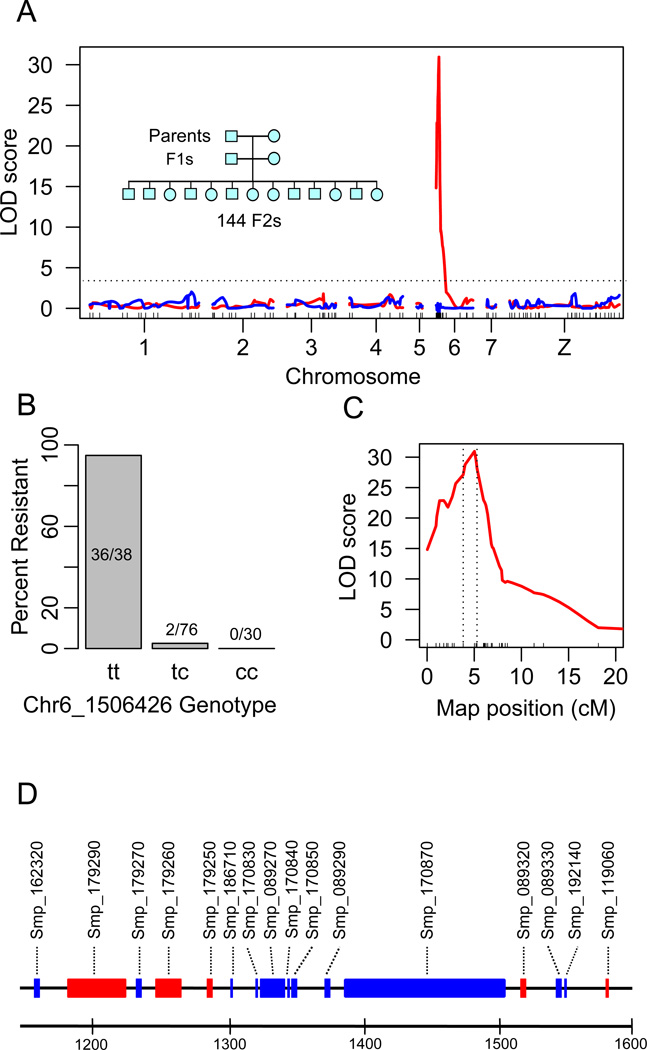
Fig. 1. Linkage mapping of OXA-resistance.
(A) OXA-resistance QTL on chr. 6. The red line shows LOD scores plotted across the genome following fine mapping of the chr 6 QTL region. The blue line shows LOD scores when the marker (Chr6_1506426) showing the peak LOD is used as a covariate. The inset shows the three generation intercross design. (B) Frequencies of resistant parasites in different genotypic classes at marker Chr6_1506426. (C) Close up of Chr 6 QTL peak. The dotted vertical lines show the 1.8-LOD support interval (439kb, 16 genes) for the location of causative gene. (D) Gene content of the 1.8-LOD support interval. The scale is in kb, genes transcribed on the forward (blue) and reverse strand (red) are shown.
We genotyped parents, F1s and 144 F2s using 62 microsatellite markers (14) distributed at 20 cM (± 15 cM) intervals across the genome (Table S1). We identified a single quantitative trait locus (QTL) near the end of chromosome (chr.) 6 (Logarithm Of Odds (LOD)=11.5). The peak LOD was observed at the terminal marker genotyped, making gene location uncertain. The 1.8-LOD QTL support interval measured 5448 kb (0–5,448,149 bp) and contained 184 genes. To fine map this region, we sequenced the genomes of the two parents and two F1s to 11–29× coverage (Table S2), identifying 558,078 high confidence SNPs (1.5 every kb) that showed Mendelian segregation in the F1s. 6,909 were within the chr. 6 QTL, including 5,241 SNPs showing fixed differences between the OXA-resistant and sensitive parents. We genotyped the F2s using 48 SNPs, including 17 in the QTL region, as well as an additional 9 microsatellite markers surrounding the QTL peak (Table S1). Inclusion of these markers increased the peak LOD score to 31 (Fig. 1C), narrowed the 1.8-LOD QTL support interval to 439 kb (position 1,149,128–1,587,670bp) and reduced the number of genes to 16 (Fig. 1D). A secondary screen, in which the marker showing the highest LOD was used as a covariate, removed the chr. 6 QTL and failed to reveal further QTLs, consistent with monogenic trait inheritance (Fig. 1A).
The QTL contains several strong candidate loci. The parasite enzyme that activates OXA has the properties of a sulfotransferase and is found in a 30 kDa fraction of soluble worm extracts (16). Three of 16 genes within the QTL are annotated as “cell wall integrity and stress response” or “NAD-dependent epimerase/dehydratase” but show structural similarity to sulfotransferases using HHpred (17) and express a predicted protein with a molecular weight close to the expected size (25–35 kDa) (Table S3).
We determined the gene content of the QTL using the genome sequences from the parental parasites. The same genes were present in both parents within the QTL region, ruling out a gene or exon deletion as the cause of OXA-resistance. We used three approaches to prioritize candidate genes. First, we reasoned that the gene(s) involved would contain fixed non-synonymous differences (or indels) between the parents. Seven of 16 genes fulfilled this criterion, including one (Smp_089320) of three genes showing homology to sulfotransferases. Second, we measured transcript abundance in OXA-sensitive and resistant parental parasites using RNAseq. Six of 16 genes within the QTL, including two with homology to sulfotransferases (Smp_089330, and Smp_119060), showed no detectable expression (Table S3). Finally, we examined the size of predicted gene products. Four of 16 products were the size expected (25–35 kDa) for the OXA-activating factor (16). Only one gene (Smp_089320) fulfilled all three selection criteria (Table S3). We also conducted functional analyses on five additional genes within or adjacent to the QTL that satisfied at least two of our selection criteria.
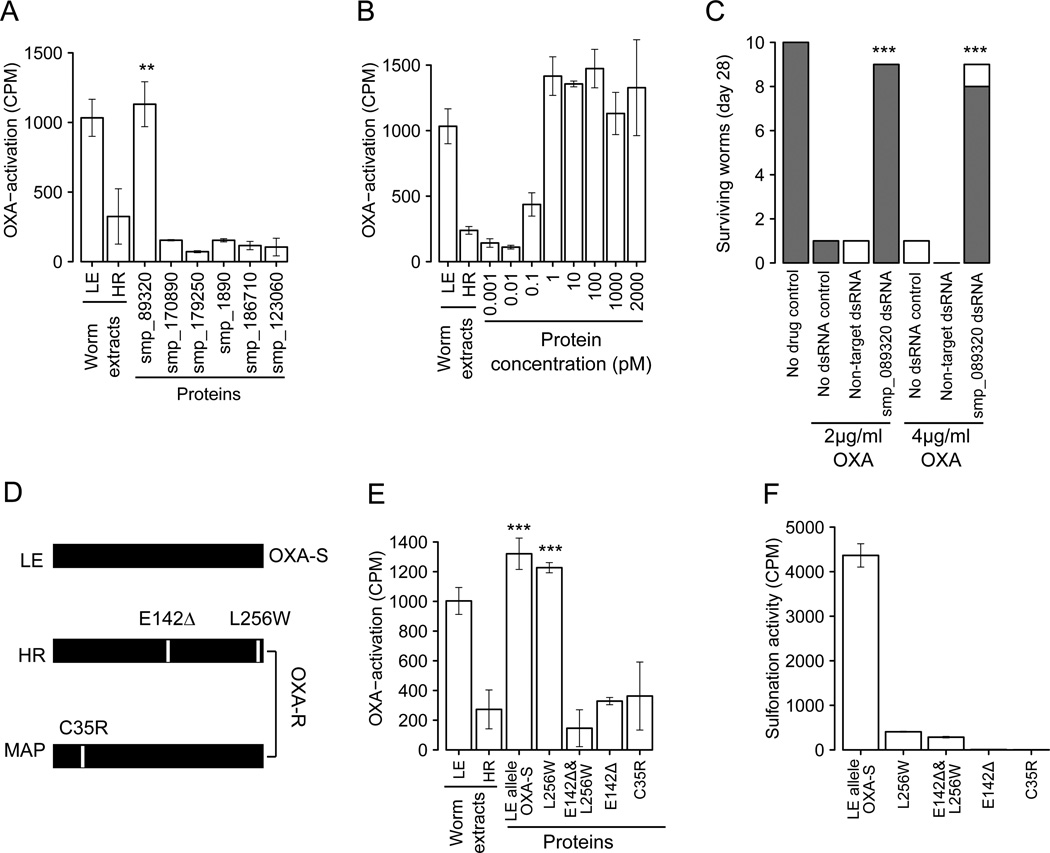
OXA is a pro-drug that is enzymatically converted into its active form in sensitive but not resistant parasites (18). We used a biochemical complementation assay to determine which of the candidate genes expresses a protein that activates OXA. We quantified OXA activation by measuring covalent binding of tritiated OXA to S. mansoni macromolecules in worm extracts (19). We produced recombinant proteins encoded by the six candidate genes, but only the recombinant Smp_089320 protein from the OXA-sensitive parent activated OXA in resistant worm extracts (Fig. 2A). Activation required minimal (1 pM) amounts of Smp_089320 protein (Fig. 2B). We confirmed the involvement of Smp_089320 in OXA-resistance in cultured parasites using RNAi. DsRNA targetting Smp_089320 reduced expression by 97% (±1.5%). OXA-sensitive (LE) parasites became resistant to 2 or 4 µg/mL OXA following knockdown of Smp_089320 (Fig. 2C, Movie S1). Hence, both biochemical complementation assays and RNAi implicate involvement of Smp_089320 in OXA-resistance.
Fig. 2. Identification of gene and mutations underlying OXA-resistance.
(A) Complementation assays to identify proteins that restore OXA-binding in resistant worm homogenates. Extracts of LE (OXA-sensitive) and HR (OXA-resistant) worms were positive and negative controls. Error bars show 1 SD in triplicate assays. Significant increases in activation relative to the HR control are shown (** p<0.01). (B) Complementation using serial dilution of Smp-089320 protein. (C) RNAi knockdown of Smp-089320. Surviving worms were categorized as vigorous (shaded) or unwell (white) (Movie S1). Significant increase in survival relative to treated controls are marked (*** p<0.001) (D) Smp-089320 alleles from LE (OXA-sensitive) and two OXA-resistant parasites (HR, laboratory-selected; MAP, field-derived). (E) Identification of OXA-resistance mutations. Proteins bearing different mutations were used in OXA-complementation assays. Significant increases in activation relative to the HR control are shown (*** p<0.001). (F) Sulfonation activity of Smp-089320 alleles.
Smp_089320 from the resistant parent (HR) differs at two positions from the sensitive parent (LE): a L256W substitution and a deletion of 142E (E142Δ) (Fig 2D). We conducted complementation assays using recombinant proteins containing either mutation to determine the residue responsible for OXA-resistance. Smp_089320-L256W activates OXA. However, Smp_089320-E142E∆ failed to activate OXA (Fig.2E), identifying this deletion as the cause of resistance in the cross. The OXA-resistant parasite (HR) was laboratory-selected (7) so to evaluate whether OXA-resistance observed in the field results from the same mutation, we tested an OXA-resistant parasite isolate (MAP) acquired from a Brazilian patient in 1978 (20). Smp_089320 from MAP carried a C35R substitution (Fig 2D) and the protein was unable to activate OXA (Fig. 2E). Hence, mutations causing loss-of-function in Smp_089320 can be independently derived in field- and laboratory-derived OXA-resistant parasites.
The sulfotransferase in sensitive worms extracts activates OXA by transferring sulfate groups from the universal sulfate donor 3’-phosphoadenosine-5’-phosphosulfate (PAPS) to the drug (16). To validate that Smp_089320 is a sulfotransferase, we performed a sulfonation assay (21) using quercetin (16) as substrate. Proteins encoded by Smp_089320 (LE allele), but not Smp_089320_E142∆ or Smp_089320_C35R, were able to transfer a sulfate group from [35S]-PAPS to quercetin (Fig. 2F) providing insights into the resistance mechanism and providing additional evidence for involvement of these mutations in OXA-resistance. Proteins encoded by Smp_089320_L256W show low sulfonation activity (<10% of LE parent), suggesting that this radical substitution may play a compensatory role restoring functionality with the normal substrate for this enzyme.
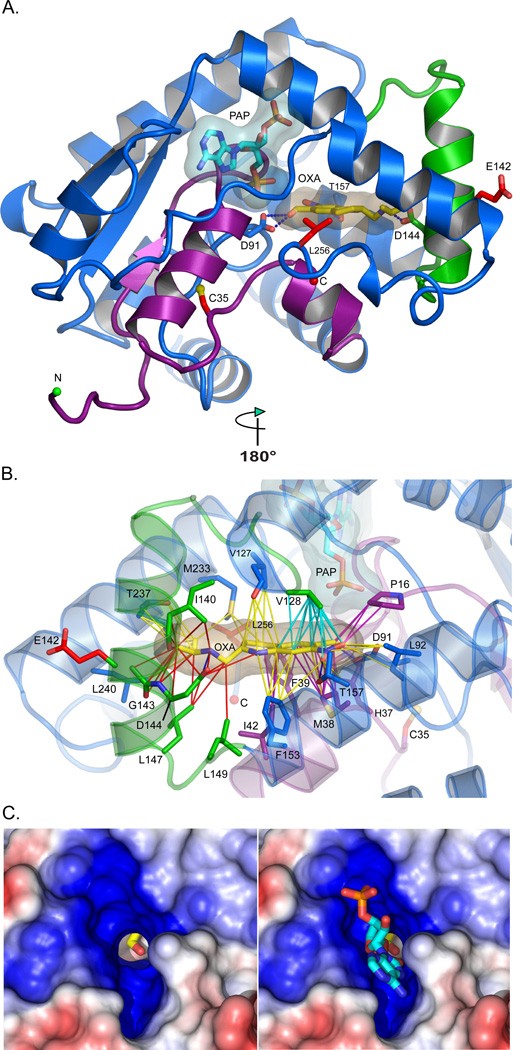
To understand the molecular bases for OXA action and resistance, the crystal structure of Smp_089320 from sensitive parasites was determined with PAP and OXA bound (Table S4, Fig S2). Smp_089320 is 40 residues shorter than human cyotsolic sulftransferases, differs in overall topology, and possesses an additional α-helix (green, Fig. 3A) (22). OXA binds in the central cavity of the L-shaped, predominantly α-helical enzyme (Fig. 3A, 3B). The hydroxyl group (the sulfonation target) is precisely positioned adjacent to a shaft running from the surface of the molecule, permitting the PAPS 5′-phosphosulfate access to the sequestered substrate (Fig. 3c). The relative positions of the accepting OXA and donating PAPS groups are consistent with the formation of a sulfonated OXA hydroxyl group (Fig. S1). The molecular bases for OXA-resistance are mutation-induced perturbations of enzyme structure that abrogate OXA binding and/or sulfonation. The C35R mutation is in a densely packed region between two critical helices involved in PAPS and OXA-binding (Fig. S3). Replacement of cysteine with a bulky arginine is predicted to disrupt ≤26 interactions, and to displace D91, which is critical for OXA binding and sulfonation (Fig. 3A, Fig. S3, Table S5). E142Δ occurs in a helix in intimate contact with the OXA tail and is predicted to disrupt 20–40 enzyme-OXA interactions (Fig. 3B, Table S5).
Fig. 3. Structure of Smp_089320 sulfotransferase.
(A) Smp_089320 sulfotransferase structure refined at 1.75 Å resolution. The depleted co-factor PAP and the pro-drug OXA are cyan and yellow sticks with semi-transparent envelopes as van der Waals surfaces. Residues 1–45 (purple) are major structural determinants of PAP and OXA binding. C35, E142, and L256 mutation sites are red sticks. D91, D144, and T157 (labeled) form hydrogen bonds (blue dashes) with OXA. (B) Contacts likely disrupted by the C35R and E142Δ mutations are purple and red lines, respectively. Cyan contacts could be compromised (Table S5). (C) Positive (blue) and negative (red) electrostatic potentials contoured at +/− 5kT around the PAP binding site. The OXA hydroxyl group to be sulfonated (Fig. S1) is visible in the PAPS 5′-phosphosulfate access shaft without (left) and with (right) PAP. The shaft permits the PAPS 5'-phosphosulfate to project into the cavity where the OXA hydroxyl group resides (Fig. S1).
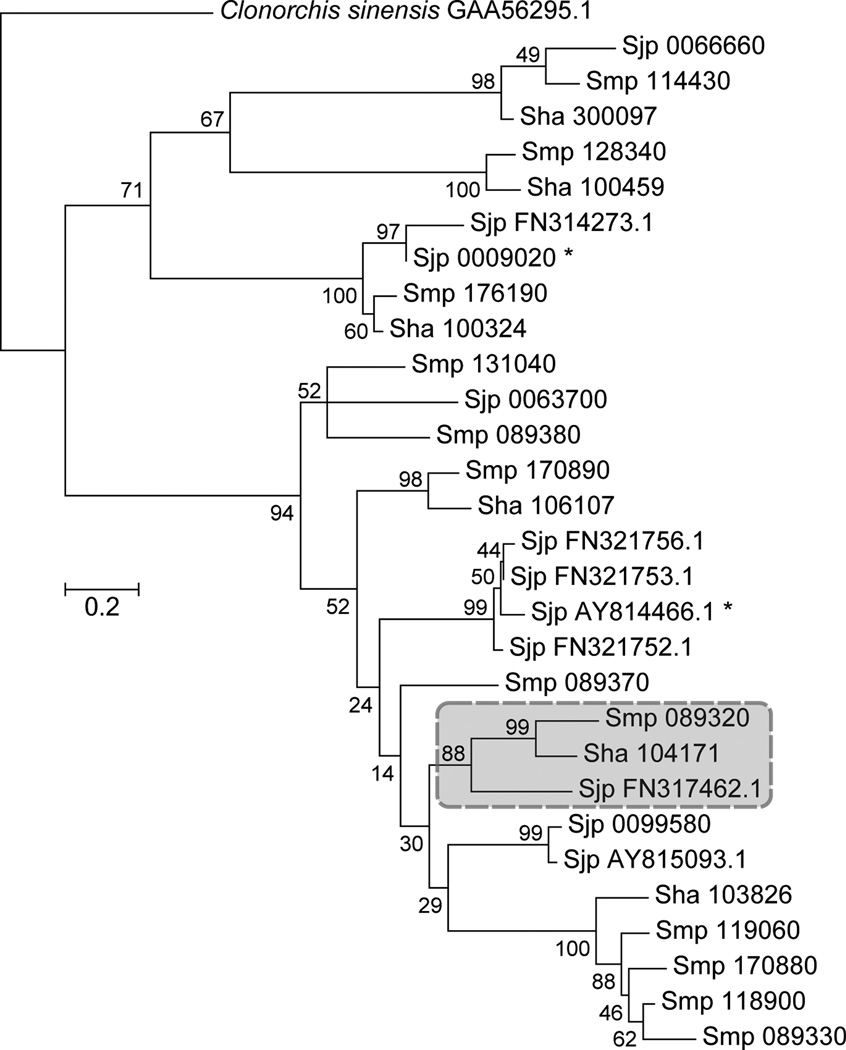
OXA kills S. mansoni but not the other human schistosome species S. haematobium and japonicum (10, 11). To investigate species-specific drug action, we conducted a phylogenetic analysis. We found 11 genes with homology to Smp_089320 in S. mansoni, six in S. haematobium and 11 in S. japonicum. Genes in S. haematobium (Sha_104171) and S. japonicum (Sjp_FN317462.1) share >50% homology with Smp_089320 and form a monophyletic group (Fig.4, Fig. S4–5) with relationships consistent with schistosome phylogeny (23). We sequenced PCR-products amplified from S. haematobium and S. japonicum cDNA showing that these genes are expressed in adult parasites. Comparison of S. mansoni and S. haematobium is of particular interest as these two species account for 186/187 million (99.5%) human schistosome infections (1). S. haematobium and S. mansoni proteins are 70% identical. Of the 16 residues that contact OXA, three differ in S. haematobium (Fig S5B, Fig. S6 Table S5). Two of these (T157 Sm → S166 Sh and 149L Sm → I157 Sh) are conservative substitutions not predicted to impact OXA binding. However, the third substitution (F39 Sm → Y54 Sh) results in a change in polarity and size that is predicted to negatively impact OXA binding (Fig. S6).
Fig. 4. Identification of homologous genes in S. haematobium and S. japonicum.
Maximum likelihood tree of aligned sequences (File S4) showing homology with Smp_089320. The tree is rooted with Clonorchis sinensis, branch lengths are numbers of substitutions/site, while % Bootstrap values (1000 replicates) are shown. The cluster containing the homologues of Smp_089320 in S haematobium (Sha) and S. japonicum (Sjp) is boxed, and is recovered using alternative phylogenetic methods (Fig S5A).
Linkage mapping has been outstandingly successful in protozoan parasites (24–26). Identification of the OXA-resistance locus, which we name SmSULT-OR (S. mansoni SULfoTransferase-OXA-Resistance), extends the utility of this approach to the most important of the human helminth parasites, paving the way for genetic analyses of biomedically important heritable traits including praziquantel resistance (27, 28), host specificity (29) and virulence (30). New drugs are urgently required for treatment of schistosomiasis because current treatments use praziquantel monotherapy to which partial resistance has been documented (27). Comparative analysis of SmSULT-OR and its S. haematobium homolog provides a framework for structure-based redesign of broad-spectrum OXA derivatives active against both S. mansoni and S. haematobium. Such a drug could be partnered with praziquantel to retard the onset of resistance. More generally, this work demonstrates how genome sequence data can be leveraged for functional genomic analyses of a biomedically important trait in a neglected human helminth parasite.
Supplementary Material
Acknowledgements
Funded by NIH (5R21-AI096277, 5R21-AI072704, R01-AI097576) and WHO (HQNTD1206356) in facilities constructed with support from Research Facilities Improvement Program Grant (C06 RR013556) from NCRR. Work at WTSI was supported by the Wellcome Trust (grant 098051). PJH was funded by the Robert A. Welch Foundation (AQ-1399). B. glabrata snails were supplied by Fred Lewis (Biomedical Research Institute, Rockville, MD) under NIH-NIAID Contract No. HHSN272201000005I. We thank Winka Le Clec’h and Cristiana Valle (capillary sequencing), Nancy Holroyd (coordinated genome sequencing), Quentin Bickle (RNAi support), and George Anderson (movie compilation). The X-ray Crystallography Core Laboratory is supported by the UTHSCSA Office of the Vice President for Research. The authors declare no competing financial interests.
Data deposition: Illumina sequence reads (http://www.ebi.ac.uk/ena/ accession # ERP000160); Genbank Accessions (KF733459-61), RNAseq data (http://www.ncbi.nlm.nih.gov/geo/, GSE51847), Structural data (http://www.wwpdb.org/, Accession 4MUA, 4MUB).
Footnotes
Supplementary Materials:
Materials and Methods
Figures S1-S6
Tables S1-S5
Movie S1
References (31– 57)
References
- 1.Hotez PJ, et al. In: Disease Control Priorities in Developing Countries. ed. 2nd. Jamison DT, editor. Washington (DC): The International Bank for Reconstruction and Development/The World Bank Group; 2006. [PubMed] [Google Scholar]
- 2.Hotez PJ, Fenwick A, Savioli L, Molyneux DH. Rescuing the bottom billion through control of neglected tropical diseases. Lancet. 2009;373:1570–1575. doi: 10.1016/S0140-6736(09)60233-6. [DOI] [PubMed] [Google Scholar]
- 3.Osei-Atweneboana MY, et al. Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus. PLoS Negl Trop. Dis. 2011;5:e998. doi: 10.1371/journal.pntd.0000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwab AE, Boakye DA, Kyelem D, Prichard RK. Detection of benzimidazole resistance-associated mutations in the filarial nematode Wuchereria bancrofti and evidence for selection by albendazole and ivermectin combination treatment. Am. J. Trop. Med. Hyg. 2005;73:234–238. [PubMed] [Google Scholar]
- 5.Katz N, Dias EP, Araujo N, Souza CP. Estudo de uma cepa de Schistosoma mansoni resistente a agentes esquistossomicidas. Rev Soc Bras Med Trop. 1973;7:381–387. [Google Scholar]
- 6.Gentile R, Oliveira G. Brazilian studies on the genetics of Schistosoma mansoni. Acta Trop. 2008;108:175–178. doi: 10.1016/j.actatropica.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers SH, Bueding E. Hycanthone resistance: development in Schistosoma mansoni. Science. 1971;172:1057–1058. doi: 10.1126/science.172.3987.1057. [DOI] [PubMed] [Google Scholar]
- 8.Cioli D, Pica-Mattoccia L, Moroni R. Schistosoma mansoni: hycanthone/oxamniquine resistance is controlled by a single autosomal recessive gene. Exp. Parasitol. 1992;75:425–432. doi: 10.1016/0014-4894(92)90255-9. [DOI] [PubMed] [Google Scholar]
- 9.Pica-Mattoccia L, Dias LC, Moroni R, Cioli D. Schistosoma mansoni: genetic complementation analysis shows that two independent hycanthone/oxamniquine-resistant strains are mutated in the same gene. Exp. Parasitol. 1993;77:445–449. doi: 10.1006/expr.1993.1104. [DOI] [PubMed] [Google Scholar]
- 10.Pica-Mattoccia L, Novi A, Cioli D. Enzymatic basis for the lack of oxamniquine activity in Schistosoma haematobium infections. Parasitol. Res. 1997;83:687–689. doi: 10.1007/s004360050320. [DOI] [PubMed] [Google Scholar]
- 11.Foster R, Cheetham BL. Studies with the schistosomicide oxamniquine (UK-4271). I. Activity in rodents and in vitro. Trans. R. Soc. Trop. Med. Hyg. 1973;67:674–684. doi: 10.1016/0035-9203(73)90038-2. [DOI] [PubMed] [Google Scholar]
- 12.Protasio AV, et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop. Dis. 2012;6:e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berriman M, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Criscione CD, Valentim CL, Hirai H, LoVerde PT, Anderson TJ. Genomic linkage map of the human blood fluke Schistosoma mansoni. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-6-r71. R71-2009-10-6-r71. Epub 2009 Jun 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis FA, Stirewalt MA, Souza CP, Gazzinelli G. Large-scale laboratory maintenance of Schistosoma mansoni, with observations on three schistosome/snail host combinations. J. Parasitol. 1986;72:813–829. [PubMed] [Google Scholar]
- 16.Pica-Mattoccia L, et al. The schistosome enzyme that activates oxamniquine has the characteristics of a sulfotransferase. Mem. Inst. Oswaldo Cruz. 2006;101(Suppl 1):307–312. doi: 10.1590/s0074-02762006000900048. [DOI] [PubMed] [Google Scholar]
- 17.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cioli D, Pica-Mattoccia L, Rosenberg S, Archer S. Evidence for the mode of antischistosomal action of hycanthone. Life Sci. 1985;37:161–167. doi: 10.1016/0024-3205(85)90419-9. [DOI] [PubMed] [Google Scholar]
- 19.Pica-Mattoccia L, Archer S, Cioli D. Hycanthone resistance in schistosomes correlates with the lack of an enzymatic activity which produces the covalent binding of hycanthone to parasite macromolecules. Mol. Biochem. Parasitol. 1992;55:167–175. doi: 10.1016/0166-6851(92)90137-9. [DOI] [PubMed] [Google Scholar]
- 20.Drescher KM, et al. Response of drug resistant isolates of Schistosoma mansoni to antischistosomal agents. Mem. Inst. Oswaldo Cruz. 1993;88:89–95. doi: 10.1590/s0074-02761993000100014. [DOI] [PubMed] [Google Scholar]
- 21.Varin L, Barron D, Ibrahim RK. Enzymatic assay for flavonoid sulfotransferase. Anal. Biochem. 1987;161:176–180. doi: 10.1016/0003-2697(87)90669-5. [DOI] [PubMed] [Google Scholar]
- 22.Negishi M, et al. Structure and function of sulfotransferases. Arch. Biochem. Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- 23.Lawton SP, Hirai H, Ironside JE, Johnston DA, Rollinson D. Genomes and geography: genomic insights into the evolution and phylogeography of the genus Schistosoma. Parasit. Vectors. 2011;4 doi: 10.1186/1756-3305-4-131. 131-3305-4-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sibley LD. Development of forward genetics in Toxoplasma gondii. Int. J. Parasitol. 2009;39:915–924. doi: 10.1016/j.ijpara.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su X, Hayton K, Wellems TE. Genetic linkage and association analyses for trait mapping in Plasmodium falciparum. Nat. Rev. Genet. 2007;8:497–506. doi: 10.1038/nrg2126. [DOI] [PubMed] [Google Scholar]
- 26.Ranford-Cartwright LC, Mwangi JM. Analysis of malaria parasite phenotypes using experimental genetic crosses of Plasmodium falciparum. Int. J. Parasitol. 2012;42:529–534. doi: 10.1016/j.ijpara.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg RM. New approaches for understanding mechanisms of drug resistance in schistosomes. Parasitology. 2013:1–13. doi: 10.1017/S0031182013000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doenhoff MJ, Cioli D, Utzinger J. Praziquantel: mechanisms of action, resistance and new derivatives for schistosomiasis. Curr. Opin. Infect. Dis. 2008;21:659–667. doi: 10.1097/QCO.0b013e328318978f. [DOI] [PubMed] [Google Scholar]
- 29.Kalbe M, Haberl B, Hertel J, Haas W. Heredity of specific host-finding behaviour in Schistosoma mansoni miracidia. Parasitology. 2004;128:635–643. doi: 10.1017/s0031182004005037. [DOI] [PubMed] [Google Scholar]
- 30.Gower CM, Webster JP. Fitness of indirectly transmitted pathogens: restraint and constraint. Evolution. 2004;58:1178–1184. doi: 10.1111/j.0014-3820.2004.tb01698.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.