Abstract
Isovaleric acidemia (IVA) is an autosomal recessive inborn error of the leucine metabolism that is caused by a deficiency of isovaleryl-CoA dehydrogenase (IVD). Recent application of tandem mass spectrometry to newborn screening has allowed a significant expansion of the recognition of individuals with IVD deficiency. Although many patients have been reported worldwide, there are no genetically confirmed patients in Korea. This study characterizes IVD mutations in seven Korean IVA patients from six unrelated families. Bi-directional sequencing analysis identified two novel variations affecting consensus splice sites (c.144+1G>T in intron 1 and c.457-3_2CA>GG in intron 4) and three novel variations altering coding sequences (c.149G>T; Arg21Leu, c.832A>G; Ser249Gly, and c.1135T>G; Phe350-Val). Five patients from four families were found to be compound heterozygotes while two unrelated patients were homozygous for the c.457-3_2CA>GG variation. Reverse-transcription polymerase chain reaction confirmed that both intron variations cause aberrant splicing. Furthermore, analysis of cultured lymphocyte extracts of the seven patients showed no detectable enzyme activity and reduced levels of IVD protein (<10.0% of control) in all samples. These results confirm IVD mutations in Korean patients with IVA and reveal that the mutation spectrum is different from previously reported patients.
Keywords: Isovaleric acidemia, IVD, Isovaleryl-CoA dehydrogenase, Acyl-CoA dehydrogenase, Mutation, Korean
Introduction
Isovaleric acidemia (IVA; MIM 243500) is an autosomal recessively inherited organic acidemia that is caused by a deficiency of the enzyme, isovaleryl-CoA dehydrogenase (IVD; E.C.1.3.99.10), which catalyzes the third step in the catabolism of the amino acid leucine [1,2]. A deficiency in IVD results in the failure of isovaleryl-CoA to be oxidized to 3-methylcrotonyl-CoA. IVA is considered to be a severe, potentially life-threatening disorder that manifests with acute neonatal encephalopathy in approximately half of affected individuals, and recurrent episodes of vomiting, lethargy, coma and varying degrees of developmental delay in the other half of patients [3].
IVA due to an IVD deficiency has been reported at a frequency of 1:250,000 and 1:365,000 births in the United States and Taiwan, respectively, but is more common in Germany, where the prevalence is 1:62,500 [4,5]. However, recent application of tandem mass spectrometry (MS/MS) for the analysis of the acylcarnitine profile of blood spots obtained for newborn screening has allowed a significant expansion of the recognition of individuals with an IVD deficiency. Nevertheless, there is only one report of IVA in Korea, and the overall incidence of IVA is unknown [6].
IVD, a member of the family of acyl-CoA dehydrogenase, is encoded in the nuclear genome in precursor form. The precursor peptides are synthesized in the cytoplasm, transported into the mitochondria, and processed to homotetramers, with each monomer containing a noncovalently but tightly bound flavin adenine dinucleotide molecule [7]. The length of its precursor protein and mature sequence are 424 and 394 amino acid residues, respectively [8].
The human IVD gene is located on chromosome 15q14-15, and consists of 12 exons spanning 15 kb of the DNA. Thus far, over 25 mutations in the IVD gene have been characterized in patients with IVA, including missense/nonsense mutations, splicing mutation and frame shifts [4,9]. A significant proportion of mutant alleles lead to abnormal splicing of the IVD RNA and subsequent complete lack of the IVD protein [10]. Almost half of the mutant IVD alleles sequenced from infants diagnosed by newborn screening have been found to contain a common recurring missense mutation (c.932C>T; Ala282Val) and who have a novel mild and potentially asymptomatic phenotype of IVA [4]. However, most of the biological characteristics of IVA including clinical findings, biochemical features and genetic profiles have been in western populations; no molecular and functional investigations have been performed in Korean patients. This study reports the genetic basis of IVA in Korean patients and characterizes their clinical phenotype and biochemical profiles, and IVD protein expression and enzymatic activity in patient cells.
Materials and methods
Subjects
Seven IVA patients from six unrelated families were recruited from various regions of the Republic of Korea. Biochemical diagnosis was based on the detection of isovalerylglycine and other metabolites in urine as well as isovalerylcarnitine in plasma and blood spots. Patients PHI and PHB were brothers. The elder brother (patient PHB) was not diagnosed until his younger brother (patient PHI) was confirmed to have IVA from biochemical studies. Patients PHJ, CSJ, and YJH were identified by newborn screening with MS/MS. Patient OMK did not undergo MS/MS screening but experienced acute symptoms at 10 days of age including vomiting and a sweaty foot odor. She was confirmed to have IVA on repeat MS/MS screening of blood and organic acid analysis of urine. Her elder brother died of metabolic acidosis at age three without a definitive diagnosis. Patient BHJ showed recurrent vomiting and poor feeding without any specific diagnosis. At age 4, he was diagnosed as having IVA by organic acid analysis of urine. Patients PHJ, CSJ, and YJH were diagnosed with IVA through neonatal screening and remained without symptoms. Table 1 gives a summary of the clinical data of the seven patients.
Table 1.
Clinical and biochemical characteristics of Korean IVA patients
| Patient | Sex/ age |
Age at diagnosis |
Symptoms onset |
Growth retardation |
Developmental delay |
Sweaty foot odor |
C5-Acyl- carnitinea |
Urine isovaleryl- glycineb |
Plasma isovaleryl- carnitinec |
Identified IVD gene mutations |
|---|---|---|---|---|---|---|---|---|---|---|
| PHBd | M/15 yr | 8 yr | 10 days | − | + | + | ND | 2962.8 | ND | c.144+1G>T homozygote |
| PHId | M/11 yr | 4 yr | 5 days | − | + | + | ND | 9010.0 | ND | c.144+1G>T homozygote |
| PHJ | M/5yr | 7 days | − | − | − | − | 33.8 | 1608.2 | 7.70 (<1.4) | Ser249Gly/Phe350Val |
| CSJ | M/15 m | 1 m | − | − | − | − | NI | 465.3 | 7.97 (<1.2) | Ser249Gly/Phe350Val |
| OMK | F/16 m | 22 days | 10 days | − | + | + | 4.75 | 469.9 | ND | c.457-3_2CA>GG homozygote |
| BHJ | M/7 yr | 3 yr | 5 days | − | + | + | ND | 5351.2 | 6.40 (<0.56) | c.457-3_2CA>GG homozygote |
| YJH | M/32 m | 15 days | 7 days | − | − | + | NI | 9212.9 | 7.78 (<0.33) | Arg21Leu/c.457-3_2CA>GG |
Abbreviations used: ND, not done; NI, not informed.
NBS blood spots, reference range: <1.2 µmol/L.
Reference range: 0.2–10.1 mmol/mol creatinine.
Reference range (nmol/mL) is in parentheses.
Patients PHB and PHI are brothers.
Biochemical analysis
Standardized filter papers soaked with 100 µL of whole blood from patients PHJ, CSJ, OMK, and YJH were analyzed with MS/MS as previously described [11]. Plasma and urine samples were collected from the patients and subjected to organic acid analysis by gas chromatography (GC)–MS.
Mutation analysis
After obtaining informed consent from the parents, blood samples were collected from the patients and their family members. Genomic DNA was isolated from peripheral blood leukocytes using the Wizard genomic DNA purification kit according to the manufacturer’s instruction (Promega, Madison, WI, USA). All 12 exons and their flanking intron sequences of the IVD gene were amplified by polymerase chain reaction (PCR) using appropriate primers designed by the authors (available upon request) and a thermal cycler (Model 9700, Applied Biosystems, Foster City, CA). Five microliters of amplification product were treated with 10U shrimp alkaline phosphatase and 2 U exonuclease I (USB Corp., Cleveland, OH) and direct sequencing was then performed using the Big- Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) on the ABI Prism 3100 genetic analyzer (Applied Biosystems). In addition, IVD sequences from 50 control individuals were analyzed in their entirety. Potential mutations were defined by exclusion from the Human Gene Mutation Databse (http://www.hgmd.cf.ac.uk) and the previously reported mutations on PubMed (http://www.ncbi.nlm.nih.gov/PubMed/).
Analysis of splicing prediction
The genomic sequences spanning an interval of 50 nucleotides up- and downstream of each substitution were evaluated by information analysis of splice donor and acceptor sites [9,12]. The individual information content of each natural and variant primary and secondary splice site was analyzed by measuring their Ri value expressed in bits of information using the Scan program (from the Laboratory of Computational and Experimental Biology at the National Institutes of Health) [13]. The effects of nucleotide substitutions on splicing information were predicted from Ri values.
Reverse transcription-PCR (RT-PCR)
To determine the effect of the identified mutations on mRNA splicing, each mutation containing segment comprising the appropriate exon was amplified by RT-PCR from mRNA isolated from EBV-transformed patient lymphocytes. Total RNA was extracted using Qiagen RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. One microgram of total RNA from each sample was reverse transcribed using an aliquot of the reverse transcription reaction (1 µL) that included 1× PCR buffer, 2.5 mM MgCl2, 0.2 mM of each dNTP, and 0.5 U AMV reverse transcriptase (Roche Diagnostics GmbH, Mannheim, Germany). Ten microliters of the reaction mixture were amplified by PCR using appropriate primers. To confirm the aberrant splicing by c.144+1G>T and by c.457-3_2CA>GG variations in introns 1 and 4, respectively, a cDNA fragment was amplified using either a forward primer in the 5′-untranslated region (5′-agagctggctcagtttcagc-3) and a reverse primer in exon 4 (5′-atctcctccatcaccagcac) or a forward primer in exon 2 (5′-cagcaatgagttcaagaacctg) and a reverse primer in exon 6 (5′-gggccattagtgatccagaa-3′). GAPDH mRNA was also amplified as a control with a primer set (GAPDH5, 5′-gtggatattgttgccatca-3′; GAPDH3, 5′-gactccacgacttactca-3′).
Enzyme assay and Western blot
IVD activities in cultured lymphocyte extracts of the seven patients were determined using the anaerobic electron-transferring flavoprotein fluorescence reduction assay and 50 µM isovaleryl-CoA as substrate as previously described [14,15]. Western blotting was performed with 50 µg of the total protein from cultured lymphocytes extracts, as described previously [16].
Molecular modeling of IVD mutations
To investigate the effect of the mutations identified on IVD structure, we used the crystal structure with the PDB code 1IVH [17]. A model of the IVD monomer mutants were generated with biopolymer module of Insight II package of modeling software from Molecular Simulations, Inc. (San Diego, CA). A side chain conformation of IVD mutant was determined using the Auto-rotamer command in Biopolymer module of Insight II package. The three-dimensional model structure of IVD was visualized with PYMOL (http://www.pymol.org).
Results
Biochemical characteristics of patients with IVA
Three patients (PHJ, CSJ, and YJH) were identified through newborn screening using MS/MS while the others who did not undergo newborn screening were diagnosed after presenting with typical clinical symptoms including a sweaty foot odor. Organic acid analysis of urine in all patients demonstrated an elevation of isovalerylgycine level with a peak concentration range of 465–9213 mmol/mol creatinine (reference range: 0.2–10.1). Biochemical characteristics of each patient are detailed in Table 1.
Enzyme assay and Western blot
Enzymatic assay with the sensitive and specific ETF fluorescent reduction assay showed IVD activity to be completely deficient in cultured lymphoblast from all patients. The IVD signal from lymphoblast extracts from patients PHJ and CSJ were approximately 10% of the intensity of the IVD signal in normal cell extracts. The lymphocyte extracts from the patients PHI, PHB, OMK, BHJ, and YJH contained no immunodetectable IVD protein.
Mutation analysis
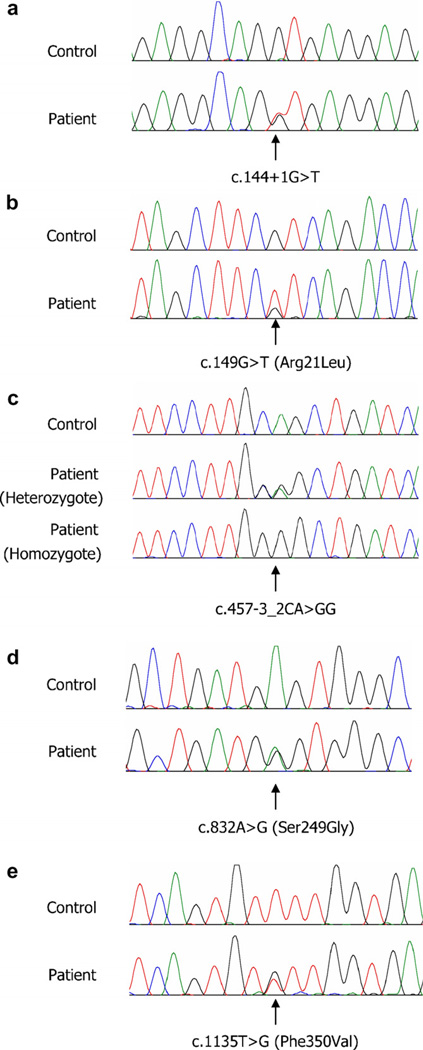
Bi-directional sequencing of the IVD exons and intron/exon junctions identified two novel variations affecting consensus splice sites (c.144+1G>T in intron 1 and c.457-3_2CA>GG in intron 4) along with three novel variations altering coding sequences (c.149G>T; Arg21Leu, c.832A>G; Ser249Gly, and c.1135T>G; Phe350Val) (Fig. 1). The nucleotide numbers were designated using GenBank entry NM_002225 and the substituted amino acid numbers were described according to the report of Matsubara et al. [18]. Patient PHI and his brother were compound heterozygous for c.144+1G>T and Ser249Gly variations. The Ser249Gly variation was also found in the patients PHJ and CSJ with the Phe350Val variation. Patients OMK and BHJ were homozygous for the c.457-3_2CA>GG variation. The c.457-3_2CA>GG variation was also found in the patient YJH who also had a missense variation in exon 2 (Arg21Leu). These five novel mutations were not detected in 100 control chromosomes.
Fig. 1.
Novel mutations in the IVD gene identified in Korean patients with isovaleric acidemia. (a) A G–T transversion at the consensus splicing donor site in the intron 1, (b) a G–T transversion at nucleotide 149 in exon 2 resulting in the substitution of Arg21 with Leu (Arg50Leu based on the precursor reference protein, NP_002216), (c) a CA–GG replacement at the consensus splicing acceptor site in intron 4, (d) an A–G transition at nucleotide 832 in exon 8 resulting in the substitution of Ser249 with Gly (Ser278Gly based on the precursor reference protein, NP_002216), (e) a T–G transversion at nucleotide 1135 in exon 11 resulting in the substitution of Phe350 with Val (Phe379Val based on the precursor reference protein, NP_002216).
Prediction of aberrant splicing
As summarized in Table 2, the c.144+1G>T variation weakened the natural splicing donor site of intron 1 (initial Ri 11.1, final Ri 3.3) and strengthened a 379 nucleotides downstream, cryptic donor site of exon 1 (6.5-bit). The c.457-3_2CA>GG variation inactivated the natural acceptor site for the splicing of intron 4 (initial Ri 7.9, final Ri −6.2) and used a pre-existing 4.9-bit cryptic site, 66 nucleotides upstream of exon 6.
Table 2.
Splicing prediction results of IVD gene mutations
| Nucleotide change | Affected splicing site | Primary splicing site | Secondary splicing site | % Binding (residual splicing) |
||
|---|---|---|---|---|---|---|
| Position relative to natural site |
Initial Ri → final Ri (bits) |
Position relative to natural site |
Ri, Natural – Ri, Mutant (bits) |
|||
| c.144+1G>T | Intron 1, donor | +1, G → T | 11.1 → 3.3 | 379 | 6.5 → 6.5 | 100 |
| c.149G>T (Arg21Leu) | Intron 1, acceptor | 149G → T | 8.5 → 8.5 | 9 | 7.3 → 10.4 | 887.3 |
| c.457-3_2CA>GG | Intron 4, acceptor | −2, A → G | 7.9 → −6.2 | 229 | 4.9 → 4.9 | 100 |
| c.832A>G (Ser249Gly) | Intron 8, donor | 832A → G | 9.2 → 9.2 | −46 | 0.9 → 3.9 | 830.7 |
| c.1135T>G (Phe350Val) | Intron 11, donor | 1135T → G | 4.2 → 3.3 | 100 | ||
The Arg21Leu variation had no influence on the Ri value of the adjacent natural acceptor site (8.5-bit), but strengthened a pre-existing cryptic acceptor site at position 153 (initial Ri 7.3, final Ri 10.4), nine nucleotides downstream from the natural splice site. The Ser249Gly variation did not alter the Ri value of the adjacent donor site (9.2-bit) but created a cryptic site, 46 nucleotides upstream from the natural splice donor site of exon 8. The Phe350-Val variation slightly weakened the pre-existing cryptic site (initial Ri 4.2, final Ri 3.3), 21 nucleotides downstream of exon 11.
RT-PCR analysis
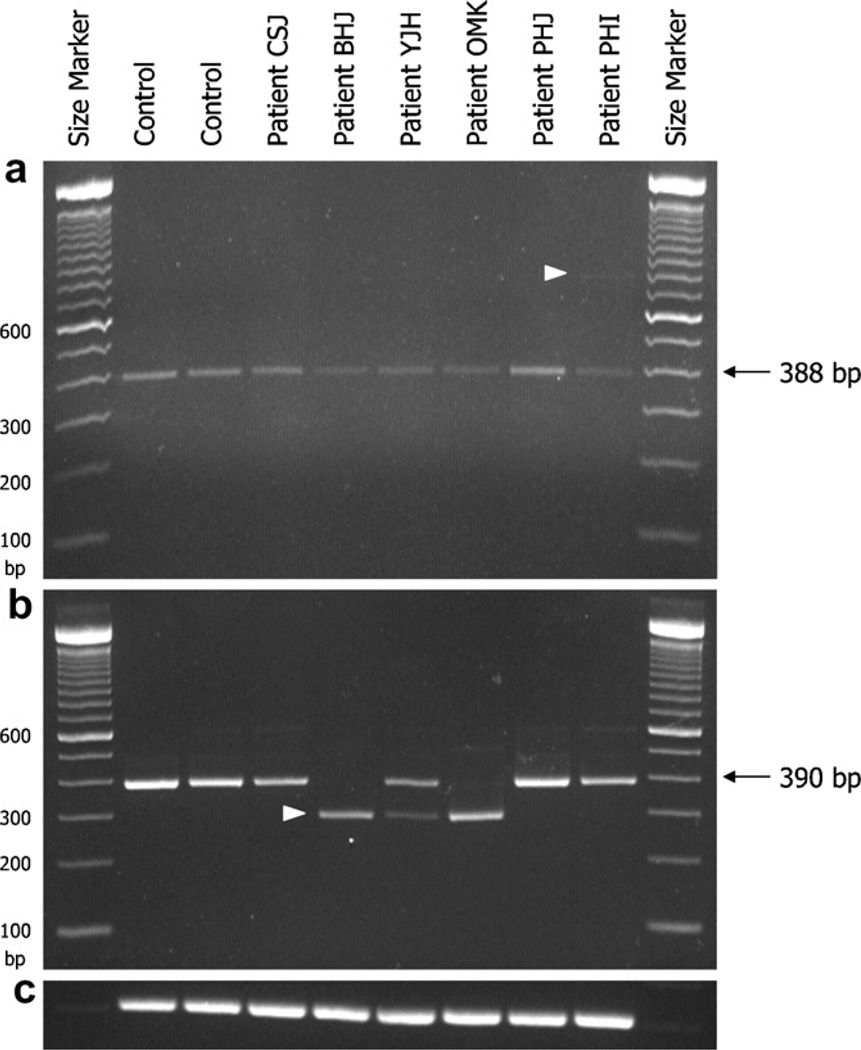
As predicted by splice site analysis, an extra band of larger size (767 bp) in addition to a normal sized band (388 bp) was visualized in patient PHI with the c.144+1G>T variation (Fig. 2a). Sequence analysis showed the 388 bp fragment to represent the normally spliced mRNA including exons 1, 2, 3, and a part of exon 4. The longer PCR product (767 bp) was generated by an addition of a part of intron 1 (379 bp) to normal mRNA. To determine the effect of the c.457-3_2CA>GG variation in intron 4, cDNA was amplified with a primer pair flanking exons 2 and 6. The 296 bp-sized band obtained was 94 bp smaller than normal transcript (390 bp) and lacked exon 5 (Fig. 2b).
Fig. 2.
RT-PCR analysis for aberrant splicing. (a) RT-PCR with primers encompassing the 5′ untranslated region through exon 4 shows a normal 388 bp product in all patients and two controls. A faint band (767 bp, arrow) is observed only from mRNA from patient PHI with the c.144+1G>T mutation. It is of note that the band densities in patients PHI, OMK, BHJ, and YJH with one or two splicing mutations are weaker than those in patients with only missense mutations or in controls. (b) A 390-bp band is expected with primers encompassing exon 2–6. In patients OMK and BHJ homozygous for the c.457-3_2CA>GG mutation, only 296-bp band is observed. In patient YJH with Arg21Leu and c.457-3_2CA>GG mutations, a 296 bp and 390-bp bands are amplified but the 296-bp band is fainter than the 390 bp band. (c) GAPDH as a RNA control.
Discussion
In the present study, five novel mutations in the IVD gene were identified in six unrelated Korean patients with IVA. Two led to abnormal splicing while three were missense mutations predicted to affect enzymatic function and/or stability. Over 25 mutations in the IVD gene are currently listed in the Human Gene Mutation database (http://www.hgmd.cf.ac.uk/ac/), most of them have been identified in Caucasians. Recently, Lin et al. reported that the incidence of IVA in Taiwan is approximately 1/365,000, which is lower than in Germany or United States [5].
A c.932C>T (Ala282Val) mutation has recently been reported to be the most frequent mutation found in Caucasian IVA patients, and has been associated with a mild and potentially asymptomatic phenotype. This mutation was not found in the current study. Instead, c.457-3_2CA>GG was found to be a recurring mutation in Korean IVA patients (5 out of 12 mutant alleles). Interestingly, this mutation was found in three patients not known to be related from Jeju Island, the largest island in Korea, suggesting a possible founder effect in this population.
Patient PHI and his brother share a splice junction mutation in position +1 of intron 1 of the IVD gene and show abnormal IVD species in lymphoblast mRNA consistent with aberrant splicing. The efficacy of splicing is known to be influenced by the conserved elements at the 5′ splice site, the splicing lariat branch site, the polypyrimidine tract, and the 3′ splice site of pre-mRNA [19]. Mutations that disrupt a 5′ splice site have been shown to cause skipping of the exon that follows it, activation of a cryptic splice site that leads to 5′-alternative splicing, or intron retention [20]. Cells from patient PHI contain an mRNA species that retains intron 1 due to activation of a cryptic splice site 379 nucleotides downstream of this mutation. The resultant message is predicted to lead to production of an abnormal protein and a premature termination codon. Additional larger IVD species found in this patient’s mRNA suggest that additional aberrant splicing events occur as a result of the splice junction mutation.
The c.457-3_2CA>GG intron mutation appears to have inactivated the natural acceptor site for splicing of intron 4 and induced skipping of exon 5 during pre-mRNA splicing. Interestingly, an earlier study reported that IVD sequences could not be amplified from a cell line with an intron 4 acceptor site mutation (c.457-2A>G) suggesting that the IVD mRNA is unstable without exon 5 [9].
A c.149G>C mutation in the IVD gene has previously been shown to lead to missplicing of exon 2 as well as alter the codon for Arg21 [9]. The c.149G>T mutation strengthened a pre-existing cryptic acceptor site at position 153 (final Ri 10.4) compared with c.149G>C mutation (Ri 9.8) on splicing prediction analysis. In contrast the c.149G>T (Arg21Leu) mutation found in one of the current patients did not lead to any apparent abnormally sized IVD message. This discrepancy might be due to a difference in the substituted base or to the difference of cell types used for RNA analysis.
Cell lines from all of the patients in this study were devoid of IVD activity and exhibited greatly reduced to no stable IVD protein expression. Not surprisingly, extracts of cells from patients with splicing mutations leading to aberrant splicing of IVD RNA showed the weakest IVD protein signal on Western blots. The levels of abnormal metabolites identified in patients, however, did not reliably differentiate between patients carrying splicing mutation and those with coding gene mutations. Thus, neither mutant genotype nor urine isovalerylglycine concentration provided any insight into the clinical course of patients, a conclusion reached in other patient populations as well [10,21].
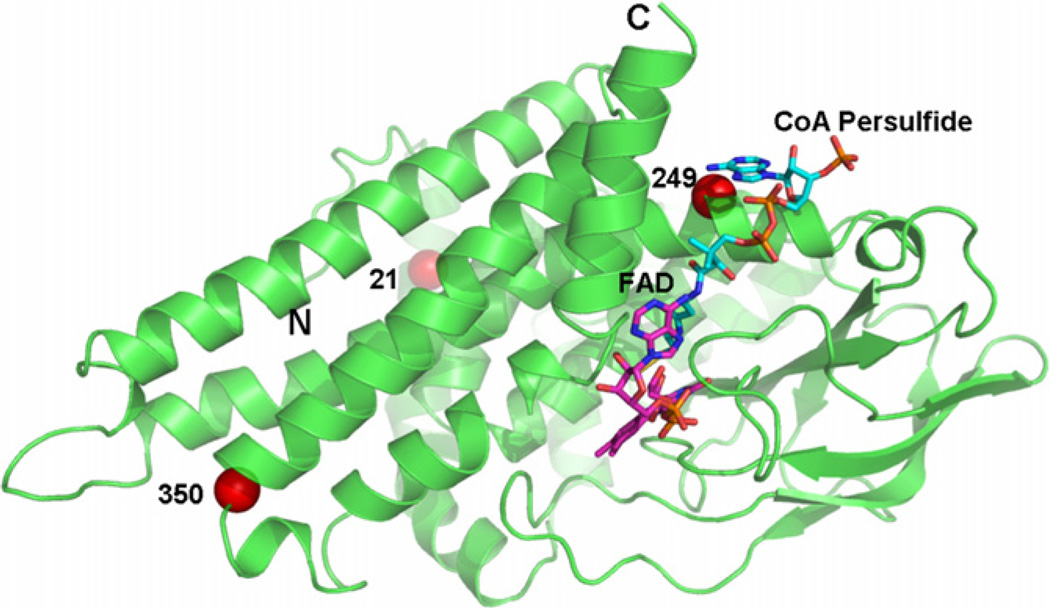
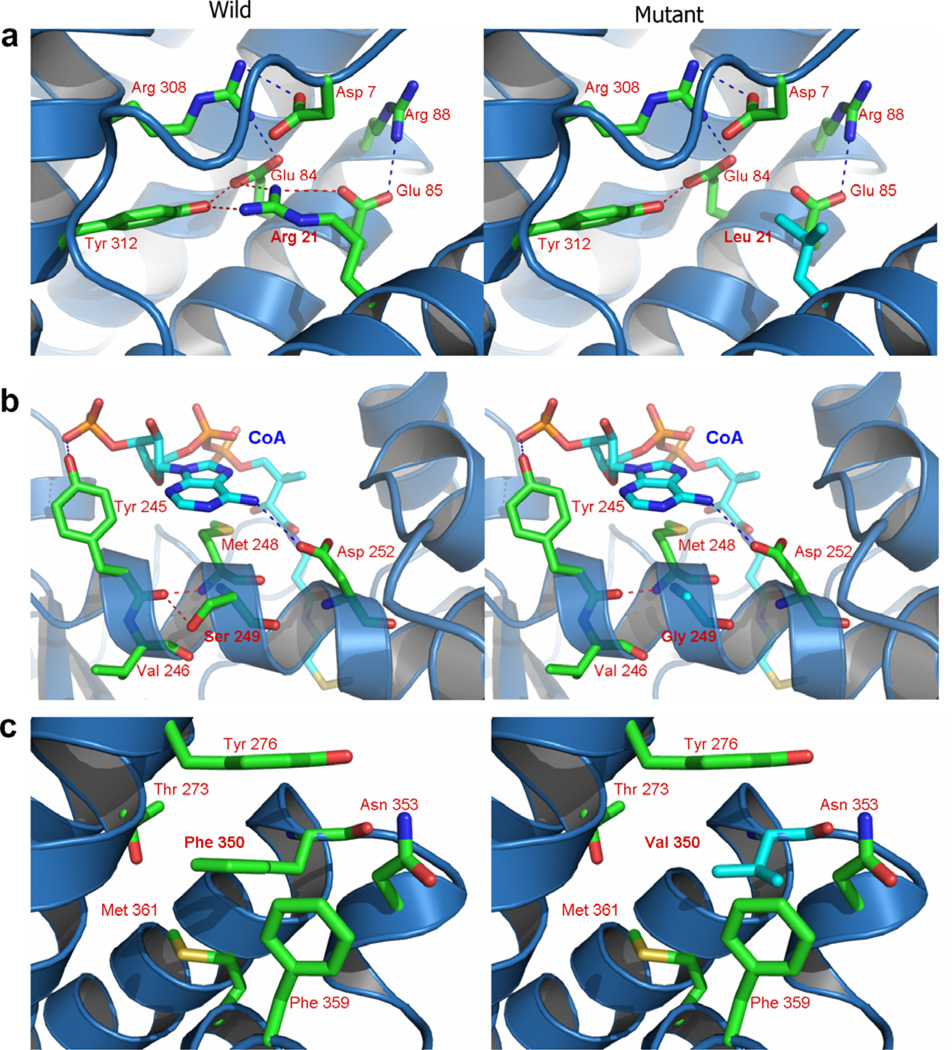
The locations of the three missense variants identified in this study on the three dimensional structure of an IVD monomer are depicted in Fig. 3. All three variations are located in α-helical regions. Arg21 belongs to the N-terminal α-helical domain (helix A). Both Ser249 and Phe350 belong to the C-terminal α-helical domain. However, Ser249 is located on α-helix G region while Phe350 is on α-helix J region. Residues 21 and 249 are normally buried deep within the protein while residue 350 is nearer the protein surface. By changing Arg21 into leucine, a salt bridge between Arg21 and Glu85 is predicted to be weakened and the hydrogen bond between Arg21 and Tyr312 disrupted (Fig. 4a). The Ser249Gly variation may interfere with a hydrogen bond between this residue and Val246 (Fig. 4b). The hydrophobic interaction among Tyr276, Met361, and Phe359 is expected to be weakened as a result of the Phe350Val substitution, and π–π interactions between Phe350 and Tyr276 may be diminished (Fig. 4c).
Fig. 3.
Ribbon diagram of a monomer of human IVD with bound CoA persulfide depicting the locations of the amino acid replacements identified in Korean IVA patients. Substituted amino acid residues are denoted with red-colored balls. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Predicted structural changes in human IVD induced by patient mutations. (a) Changing of Arg21 to leucine is predicted to induce a conformation change in α-helix A, weakening the salt bridging between Arg21 and disrupting the hydrogen bond between Arg21 and Tyr312. (b) The Ser249Gly, is predicted to eliminate a hydrogen bond normally present between Ser249 with Val246 increasing flexibility of the α-helix associated with the IVD catalytic site. (c) In the Phe350Val, hydrophobic interactions among Tyr276, Met361 and Phe359 are predicted to be weakened and π–π interaction between Phe350 and Tyr276 decreased.
In conclusion, Korean patients with IVA have been shown to have mutations in the IVD gene but the mutation spectrum is different from previously reported ethnic groups. Therefore, mutation analysis of the IVD gene may be helpful for rapid confirmation of a biochemical diagnosis of IVA, prenatal diagnosis, and carrier detection.
Acknowledgments
This work was supported by the Samsung Biomedical Research Institute Grant, #SBRI C-A6-403-2 (Dr. Ki). Dr. Vockley was supported in part by NIH Grant RO1DK45482 and the Pennsylvania Department of Health, Tobacco Formula Funding.
References
- 1.Tanaka K, Budd MA, Efron ML, Isselbacher KJ. Isovaleric acidemia: a new genetic defect of leucine metabolism. Proc. Natl. Acad. Sci. USA. 1966;56:236–242. doi: 10.1073/pnas.56.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budd MA, Tanaka K, Holmes LB, Efron ML, Crawford JD, Isselbacher KJ. Isovaleric acidemia. Clinical features of a new genetic defect of leucine metabolism. N. Engl. J. Med. 1967;277:321–327. doi: 10.1056/NEJM196708172770701. [DOI] [PubMed] [Google Scholar]
- 3.Sweetman LWJ. Branched chain organic acidurias. In: Scriver CRBA, Valle D, Sly WS, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Diseases. Vol. 2. New York: McGraw-Hill; 2001. pp. 2125–2163. [Google Scholar]
- 4.Ensenauer R, Vockley J, Willard JM, Huey JC, Sass JO, Edland SD, Burton BK, Berry SA, Santer R, Grunert S, Koch HG, Marquardt I, Rinaldo P, Hahn S, Matern D. A common mutation is associated with a mild, potentially asymptomatic phenotype in patients with isovaleric acidemia diagnosed by newborn screening. Am. J. Hum. Genet. 2004;75:1136–1142. doi: 10.1086/426318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin WD, Wang CH, Lee CC, Lai CC, Tsai Y, Tsai FJ. Genetic mutation profile of isovaleric acidemia patients in Taiwan. Mol. Genet. Metab. 2007;90:134–139. doi: 10.1016/j.ymgme.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Cheon KS, Lee DH. Isovaleric acidemia in siblings diagnosed by organic acid analysis. J. Korean Pediatr. Soc. 2000;43:828–831. [Google Scholar]
- 7.Ikeda Y, Keese SM, Fenton WA, Tanaka K. Biosynthesis of four rat liver mitochondrial acyl-CoA dehydrogenases: in vitro synthesis, import into mitochondria, and processing of their precursors in a cell-free system and in cultured cells. Arch. Biochem. Biophys. 1987;252:662–674. doi: 10.1016/0003-9861(87)90072-5. [DOI] [PubMed] [Google Scholar]
- 8.Matsubara Y, Indo Y, Naito E, Ozasa H, Glassberg R, Vockley J, Ikeda Y, Kraus J, Tanaka K. Molecular cloning and nucleotide sequence of cDNAs encoding the precursors of rat long chain acylcoenzyme A, short chain acyl-coenzyme A, and isovaleryl-coenzyme A dehydrogenases. Sequence homology of four enzymes of the acyl-CoA dehydrogenase family. J. Biol. Chem. 1989;264:16321–16331. [PubMed] [Google Scholar]
- 9.Vockley J, Rogan PK, Anderson BD, Willard J, Seelan RS, Smith DI, Liu W. Exon skipping in IVD RNA processing in isovaleric acidemia caused by point mutations in the coding region of the IVD gene. Am. J. Hum. Genet. 2000;66:356–367. doi: 10.1086/302751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vockley J, Parimoo B, Tanaka K. Molecular characterization of four different classes of mutations in the isovaleryl-CoA dehydrogenase gene responsible for isovaleric acidemia. Am. J. Hum. Genet. 1991;49:147–157. [PMC free article] [PubMed] [Google Scholar]
- 11.Millington DS, Kodo N, Norwood DL, Roe CR. Tandem mass spectrometry: a new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J. Inherit. Metab. Dis. 1990;13:321–324. doi: 10.1007/BF01799385. [DOI] [PubMed] [Google Scholar]
- 12.Rogan PK, Faux BM, Schneider TD. Information analysis of human splice site mutations. Hum. Mutat. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 13.Schneider TD. Information content of individual genetic sequences. J. Theor. Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 14.Frerman FE, Goodman SI. Fluorometric assay of acyl-CoA dehydrogenases in normal and mutant human fibroblasts. Biochem. Med. 1985;33:38–44. doi: 10.1016/0006-2944(85)90124-3. [DOI] [PubMed] [Google Scholar]
- 15.Mohsen AW, Vockley J. Identification of the active site catalytic residue in human isovaleryl-CoA dehydrogenase. Biochemistry. 1995;34:10146–10152. doi: 10.1021/bi00032a007. [DOI] [PubMed] [Google Scholar]
- 16.Mohsen AW, Anderson BD, Volchenboum SL, Battaile KP, Tiffany K, Roberts D, Kim JJ, Vockley J. Characterization of molecular defects in isovaleryl-CoA dehydrogenase in patients with isovaleric acidemia. Biochemistry. 1998;37:10325–10335. doi: 10.1021/bi973096r. [DOI] [PubMed] [Google Scholar]
- 17.Tiffany KA, Roberts DL, Wang M, Paschke R, Mohsen AW, Vockley J, Kim JJ. Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity. Biochemistry. 1997;36:8455–8464. doi: 10.1021/bi970422u. [DOI] [PubMed] [Google Scholar]
- 18.Matsubara Y, Ito M, Glassberg R, Satyabhama S, Ikeda Y, Tanaka K. Nucleotide sequence of messenger RNA encoding human isovaleryl-coenzyme A dehydrogenase and its expression in isovaleric acidemia fibroblasts. J. Clin. Invest. 1990;85:1058–1064. doi: 10.1172/JCI114536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 20.Huang HD, Horng JT, Lin FM, Chang YC, Huang CC. SpliceInfo: an information repository for mRNA alternative splicing in human genome. Nucleic Acids Res. 2005;33:D80–D85. doi: 10.1093/nar/gki129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda Y, Keese SM, Tanaka K. Molecular heterogeneity of variant isovaleryl-CoA dehydrogenase from cultured isovaleric acidemia fibroblasts. Proc. Natl. Acad. Sci. USA. 1985;82:7081–7085. doi: 10.1073/pnas.82.20.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]