Abstract
A key role for phosphorylation of Smad2 by TGFß superfamily ligands in the axial patterning of early embryos is well established. The regulation and role of Smad2 signaling in post-neurula embryonic patterning, however, is less well understood. While a variety of TGFß superfamily ligands are implicated in various stages of anterior-posterior patterning, the ligand GDF11 has been shown to have a particular role in post-gastrula patterning in the mouse. Mouse GDF11 is specifically localized to the developing tail and is essential for normal posterior axial patterning. Mature GDF11 ligand is inhibited by its own prodomain, and extracellular proteolysis of this prodomain is thought to be necessary for GDF11 activity. The contribution of this proteolytic regulatory mechanism to Smad activation during embryogenesis in vivo, and to the development of posterior pattern, has not been characterized. We investigate here the role of Xenopus GDF11 in the activation of Smad2 during the development of tailbud stage embryos, and the role of this activation in larval development. We also demonstrate that the activity of BMP-1/Tolloid -like proteases is necessary for the normal GDF11-dependent activation of Smad2 phosphorylation during post-gastrula development. These data demonstrate that GDF11 has a central role in the activation of Smad2 phosphorylation in tailbud stage Xenopus embryos, and provide the first evidence that BMP-1/Tolloid-mediated prodomain cleavage is important for activation of GDF11 in vivo.
1. Introduction
1.1 Cleavage regulation of TGFß ligands
Signaling by TGFβ family ligands is essential for regulating multiple processes during vertebrate development, tissue homeostasis, and tissue repair (Chang et al., 2002). TGFβ superfamily ligands, which include activins, nodals, BMPs, GDFs, and canonical TGFβs, are produced and secreted into the endoplasmic reticulum as inactive proforms, which are then cleaved intracellularly or extracellularly into a mature, active region and a prodomain. For many TGFß superfamily members, the ligand is maintained in a latent form following maturation by ongoing non-covalent association with its own prodomain. Biological activation of the ligand then requires release from the associated prodomain, either by proteolytic cleavage of the prodomain or by interaction with specific extracellular regulators. While the biological role of latent ligand activation is now well understood for the canonical TGFß ligands, the significance of prodomain association for the regulation of ligand activity in vivo is less clear for other members of the superfamily.
1.2. Smad2 activation during Xenopus embryogenesis
During early vertebrate embryogenesis, Smad2 signaling, controlled by a subset of TGF-β ligands that includes the nodal, activin, and Vg-related ligands, is essential for germ layer specification and axial patterning. In the pre-gastrula frog embryo, the activation of Smad2 signaling can be tracked with phosphorylation state specific antibodies, and is distributed in a pattern consistent with its role in mesendodermal specification and patterning. While a variety of data suggest that Smad2-regulating ligands are important for post-gastrula patterning of the primary body axis, the nature of this regulation is still only partially understood.
1.3 GDF11 in development
In the mouse, the Smad2- activating ligand GDF11 has been shown to be essential for anterior-posterior patterning. GDF11 encodes a TGFβ family ligand that is most closely related to GDF8/myostatin, a known negative regulator of muscle mass (Gamer et al., 1999; McPherron et al., 1999; Nakashima et al., 1999). Mouse GDF11 is first expressed in the developing tailbud as early as 7.5 days post-coitum (Gamer et al., 1999; McPherron et al., 1999; Nakashima et al., 1999). GDF11 null mice display a characteristic anterior homeotic transformation of the thoracic and lumbar vertebrae, and a corresponding shift in Hox gene expression domains (McPherron et al., 1999). Mice lacking both copies of the ActRIIB gene (a type II receptor) and one copy of ActRIIA (a second type II receptor) also display anterior homeotic transformation of vertebral identity (Oh et al., 2002), suggesting that these receptors are necessary for anterior-posterior specification by GDF11 gene (Oh et al., 2002). Smad2 phosphorylation is locally activated in the tailbud of the day 10.5 mouse embryo, but the regulation and function of this phosphorylation is not known (de Sousa Lopes et al., 2003).
1.4 Regulation of GDF11 activity
GDF11 ligand produced in cultured cells is regulated by an inhibitory interaction with its own prodomain. Latent ligand can activated in vitro by cleavage of this prodomain by BMP-1/Tolloid family astacin metalloproteases (Ge et al., 2005; Wolfman et al., 2003). This activation is entirely dependent on a single conserved aspartate residue in the GDF11 prodomain at the BMP-1 cleavage site, and mutation of this site generates a protein that cannot be released from inhibition by BMP-1 (Ge et al., 2005; Wolfman et al., 2003). Thus, in addition to the standard components of the TGFβ signaling pathway, which are shared by many ligands and broadly expressed, GDF11 is further regulated by a ligand-specific proteolytic event. While the function of this prodomain inhibition/BMP-1 cleavage mechanism for GDF11 has been characterized in vitro and in cell culture, it has yet to be shown to be an important regulatory mechanism in vivo.
In this work, we have cloned and characterized the regulation and function of Xenopus GDF11 (xGDF11) in the post-neurula development of the larval tailbud. xGDF11, like its mouse orthologue, is expressed in the developing tailbud, as well as in the developing brain and branchial arches, during tailbud stages. Inhibition of xGDF11 expression using antisense morpholino oligonucleotides results in a dramatic reduction in Smad2 phosphorylation in tailbud embryos. Smad2 phosphorylation is locally activated in the tailbud, and this activation is completely dependent on GDF11. Inhibition of xGDF11-dependent Smad2 activation in the tailbud by several approaches results in a dramatic shortening of the tail. The Xenopus BMP-1/Tolloid family proteases, Xolloid and Xolloid-related, colocalize with xGDF11 in the tailbud (Dale et al., 2002; Goodman et al., 1998). Interfering with the function of these proteases, like GDF11 knockdown, inhibits Smad2 activation in the developing tail. These data provide the first in vivo evidence that the endogenous pattern of activity of a TGFß superfamily ligand is dependent on co-expressed BMP-1/Tolloid protease activity, and that BMP-1/Tolloid-mediated regulation of GDF11 is important for endogenous patterning of the embryo at tailbud stages.
2. Results
2.1 Endogenous patterns of p-Smad2 signaling during tailbud stages
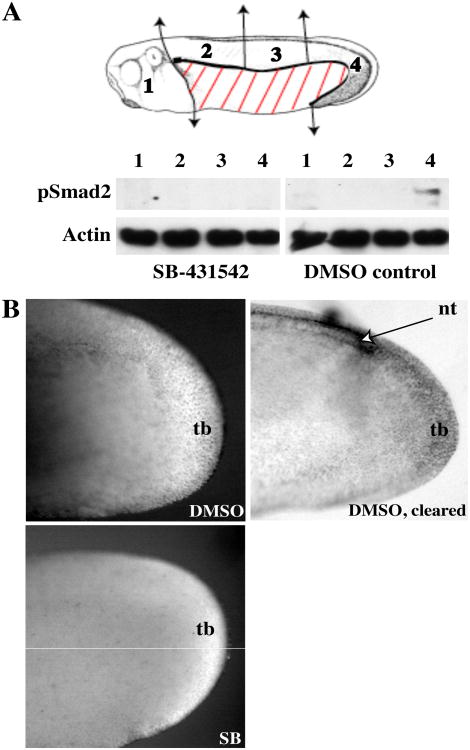
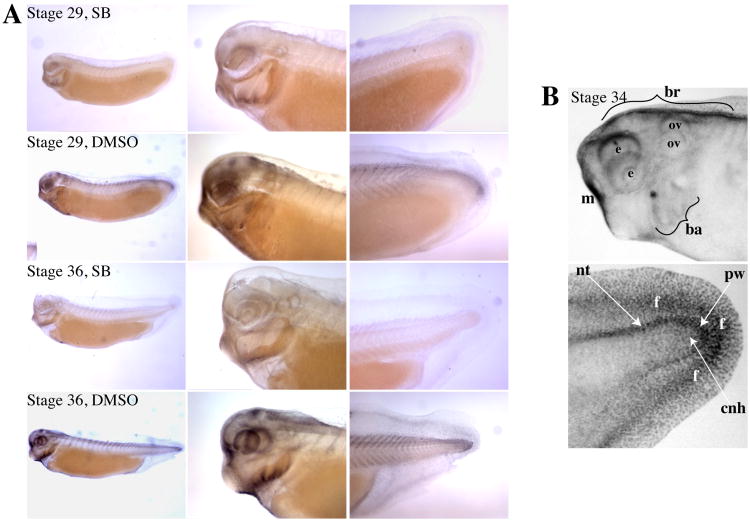
While the pattern of endogenous p-Smad2 activation during germ layer formation has been extensively described (Lee et al., 2001; Schohl and Fagotto, 2002), the pattern and regulation of Smad2 activation after gastrulation has not been studied in detail. Anti-pSmad2 Western blot analysis of tissues dissected from tailbud stage embryos along the anterior-posterior axis revealed a marked posterior enrichment of p-Smad2 by stage 26 (Fig. 1A). Treatment of embryos with the ALK4/5/7 inhibitor SB-431542 completely inhibited the Western blot signal, confirming that this signal reflects endogenous pSmad2. Immunohistochemical anti-pSmad2 staining of stage 26 embryos revealed SB-431542 sensitive staining in the tailbud; clearing of the embryos revealed that, in addition to the tailbud itself, p-Smad2 was also present at the edges of the posterior neural tube, extending further anteriorly on the dorsal side (Fig.1B). We also examined the pattern of pSmad2 activation at later tailbud stages, when the IHC signaling can be seen better. At stages 29 and 36, nuclear pSmad2 was enriched in the tailbud proper, and also along the dorsal aspect of the posterior somites (Fig. 2A). By stage 36, a distinct line of staining was also visible in the posterior fin surrounding the tailbud. Cleared tails revealed that, at stage 34, p-Smad2 formed a gradient starting at the far posterior and extending anteriorly similar to stage 29; accordingly, tailbud staining was strongest in the posterior wall of the tailbud and the posterior neural tube, with weaker staining in the chordoneural hinge area, which is further anterior (Fig. 2B). (For a detailed description of these tailbud landmarks, see (Beck and Slack, 1998)) Anteriorly, p-Smad2 was seen in various structures, including the dorsal side of the brain, the dorsal eye, the prospective mouth, and the branchial arches (Fig. 2A).By stage 36, additional staining could be seen around the otic vesicle. These data suggest that pSmad2 activation participates in the development of a number of embryonic structures during tailbud stages, and shows a local activation in the tailbud itself that is sustained throughout tailbud stages.
Figure 1. p-Smad2 expression in stage 26 embryos.
(A) Embryos treated with 100 μM SB-431542 or 0.2% DMSO from stage 13 were dissected into regions 1-4 (head, anterior dorsal trunk, medial dorsal trunk, and posterior dorsal trunk/tailbud respectively) as indicated on the schematic and Western blotted for p-Smad2. Actin serves as a loading control. (B) Stage 26 embryos treated with SB-431542 or DMSO as in (A) were immunostained with anti-p-Smad2. Tailbuds of stained embryos were photographed without clearing (left column) or with clearing (right column, DMSO only; cleared SB-431542 tailbuds are not shown, as they were almost impossible to photograph due to lack of staining and contrast). tb: tailbud, nt: posterior neural tube.
Figure 2. p-Smad2 immunostaining at later tailbud stages.
(A) Tailbud stage embryos were treated with 100 μM SB-431542 or 0.2% DMSO for 22 hours prior to harvesting at stage 29 or 36. Embryos were stained with anti-p-Smad2 and cleared. (B) Stage 34 embryos stained for p-Smad2 were cleared to reveal specific structures in the head (top) and tail (bottom). Note that some bilateral structures can be seen twice in cleared heads. br:brain, e:eye, ov:otic vesicle, ba:branchial arches, m:mouth, tb:tailbud, f:fin, nt:posterior neural tube, pw:posterior wall of tailbud, cnh:chordoneural hinge.
2.2 Cloning and characterization of a Xenopus GDF11 homolog
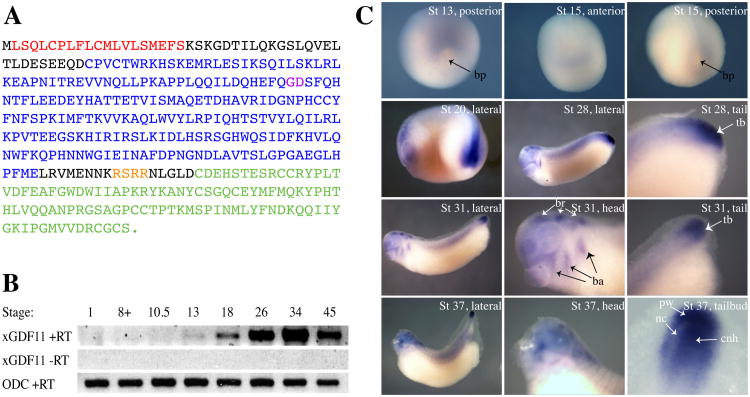
In early embryos, the nodal related ligands, Vg1, derriere, and activins, have been demonstrated to act through Smad2 activation to regulate mesoendodermal establishment and left-right patterning. Expression of most of these ligands drops sharply after gastrulation, however, and has largely disappeared by tailbud stages. Since we observed p-Smad2 in tailbud stage embryos in many places (tailbud, anterior neural tissue, branchial arches) where GDF11 is known to be expressed in the mouse during this period of embyrogenesis (McPherron et al., 1999; Nakashima et al., 1999), we reasoned that GDF11 would be a good candidate for the ligand initiating some or all of the p-Smad2 signal at these stages. We therefore used a combination of virtual cloning with available EST sequences and 5′RACE to clone full-length Xenopus laevis GDF11 (xGDF11). xGDF11 encodes a 384-aa protein that is 83% identical to human and mouse GDF11 (96% identity within the mature domain) and 62% identical to human GDF-8/myostatin. As expected, xGDF11 contains a putative signal peptide, TGF-b propeptide domain, tetrabasic cleavage site (RSRR) and TGF-b mature domain with nine cysteine residues, as well as a putative BMP-1/Tolloid cleavage site (G106-D107) (Fig. 3A). Alignment of xGDF11 to the recently completed X. tropicalis genome reveals a 98% identity with X. trop. GDF11, with 100% identity in the mature ligand region.
Figure 3. Expression of Xenopus GDF11.
(A) Sequence of Xenopus GDF11 protein. The following conserved regions are present: putative signal sequence (red), TGFß propeptide domain (blue), TGFß mature domain (green). Cleavage sites for BMP-1/Tolloid protease (GD, purple) and furin protease (RSRR, orange) are also indicated. (B) Temporal expression profile of xGDF11. RT-PCR was performed on whole embryo samples harvested at indicated stages. xGDF11 RT-PCR was performed on both +RT and -RT samples. Ornithine decarboxylase (ODC) was used as a loading control. (C) In situ hybridization for xGDF11 from stages 13-37. In stages 20-37, anterior is to the left, dorsal to the top. bp: blastopore, tb: tailbud, br:brain, ba: branchial arches, nc: neurenteric canal, pw: posterior wall of tailbud, cnh: chordoneural hinge.
RT-PCR analysis in whole embryo samples revealed that xGDF11 expression begins at stage 13 (the end of gastrulation) and increases during neurula and tailbud stages, reaching a peak at late tailbud stage (stage 34), after which expression declines slightly by stage 45 (feeding tadpole stages) (Fig. 3B). Whole-mount in situ hybridization revealed that expression of xGDF11 correlates with areas of p-Smad2 activation. At stage 13, xGDF11 is weakly expressed in prospective tail-forming region (Fig. 3C) (Gont et al., 1993). At stage 15 (neurula), xGDF11 is still expressed in the tail-forming region and is also beginning to be seen at the far anterior of the embryo (Fig. 3C). By Stage 20 (late neurula), xGDF11 expression has grown much stronger, especially in the tailbud region (Fig. 3C). At stage 28 (early tailbud), strong staining is seen broadly in the tailbud as well as in various head structures (Fig. 3C). By stage 31 (mid-tailbud), the tailbud expression domain has become more restricted to the far posterior, and the head staining has become more pronounced, with strong signal clearly visible in the brain, branchial arches, and the prospective mouth-forming region, with weaker expression in the eye. At stage 37 (late tailbud/early tadpole), this same staining pattern persists; furthermore, tailbud expression at this stage can be seen on both sides of the neurenteric canal, in the posterior wall of the tailbud as well as the chordoneural hinge (Fig. 3C). A sense control probe showed no staining (not shown).
2.3 Effect of xGDF11 knockdown on signaling in the embryo
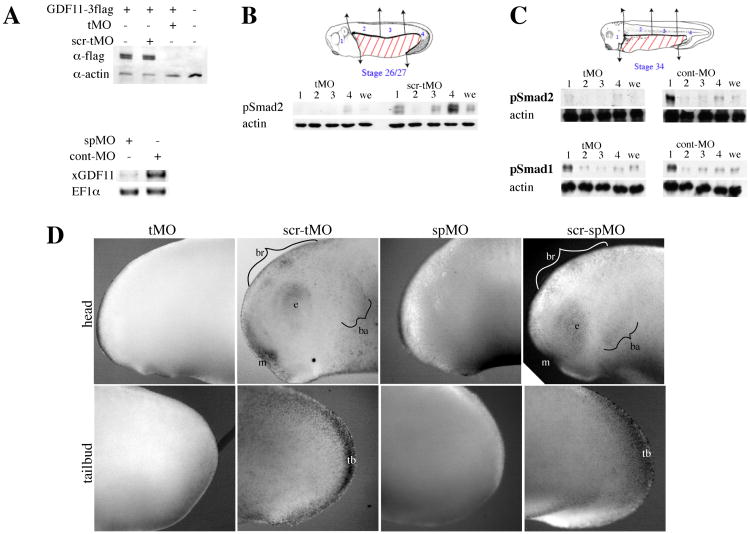
Since xGDF11 is expressed in the areas of the embryo where p-Smad2 was activated (Figs. 1, 2, 3), we asked how xGDF11 knockdown would affect p-Smad2 activation. To reduce endogenous xGDF11 expression, we generated two independent morpholino oligos. A morpholino targeting the xGDF11 translation start site (xGDF11 tMO) could efficiently prevent protein expression from injected, internally-tagged xGDF11 mRNA, while a scrambled control morpholino (xGDF11 scr-tMO) could not (Fig. 4A). A morpholino targeted to the exon1-intron1 splice site was also generated (xGDF11 spMO), which was capable of severely reducing the levels of mature endogenous xGDF11 mRNA in a stage 25 embryo, while a control morpholino (cont-MO) had no effect .
Figure 4. GDF11 knockdown results in loss of p-Smad2 at tailbud stages.
(A) Confirmation of xGDF11 morpholino oligos. Top panel: embryos were injected with 50 pg xGDF11-3flag mRNA along with 50 ng of tMO or scr-tMO and harvested at stage 10+ (uninjected controls were stage 11) for Western blotting against the flag epitope. Actin was used as a loading control. Bottom panel: embryos injected with 20 ng spMO or cont-MO were harvested at stage 25 for RT-PCR using primers spanning the splice site targeted by spMO. Elongation factor 1a (EF1a) was used as a loading control. (B) Effect of xGDF11 knockdown on p-Smad2 at stage 26/27. Embryos injected with 50 ng xGDF11 tMO or scr-tMO were dissected into regions 1-4 (head, anterior dorsal trunk, medial dorsal trunk, and posterior dorsal trunk/tailbud respectively) as indicated in the schematic and Western blotted for p-Smad2. we: whole embryo. Actin was used throughout as a loading control. (C) Effect of xGDF11 knockdown on p-Smad2 and p-Smad1 at stage 34. Embryos injected with 40 ng xGDF11 tMO or cont-MO were dissected into regions 1-4 as indicated in the schematic and Western blotted for p-Smad2 (top) and p-Smad1 (bottom). (D) xGDF11 knockdown eliminates endogenous p-Smad2 staining in multiple tissues at stage 25. Embryos were injected with 60 ng xGDF11 tMO, 60 ng xGDF11 scr-tMO, 20 ng xGDF11 spMO, or 20 ng xGDF11 scr-spMO and immunostained at stage 25 with anti-p-Smad2. br: brain, e:eye, ba:branchial arches, m:mouth, tb:tailbud.
To test the effects of xGDF11 knockdown on p-Smad2 levels, control or anti-xGDF11 MO injected embryos were dissected at stage 26/27 (early tailbud) embryos into head, anterior dorsal trunk, medial dorsal trunk, and posterior dorsal trunk+tailbud sections for Western blotting (Fig. 4B). (The ventral trunk region was discarded because it does not express p-Smad2 at this stage and contains large quantities of yolk, which distorts Western blots (data not shown)). In scr-tMO control, high levels of p-Smad2 are seen in the head and posterior regions of the embryo, with low levels in the medial trunk and virtually no signal in the anterior trunk (Fig. 4B). In contrast, xGDF11 tMO strongly reduce p-Smad2 in all of the regions of the embryo (Fig. 4B). Levels of total Smad2 were not affected (data not shown). xGDF11 spMO and scr-spMO control showed similar results (data not shown). xGDF11 tMO and xGDF11 spMO had no effect on endogenous pSmad2 activation at stage 10 (data not shown), consistent with the absence of endogenous xGDF11 and likely role of other ligands in pSmad2 activation at this stage (Birsoy et al., 2006; Onuma et al., 2002; Ramis et al., 2007; Whitman, 2001). We performed a similar dissection and Western blot at late tailbud stage (Stage 34) (Fig. 4C), and likewise saw a severe reduction in p-Smad2 in all regions of the embryo with xGDF11 tMO injection. At the same stage, MO injection had no effect on the level or distribution of p-Smad1 (Fig. 4C), consistent with the published data indicating that GDF11 signals through Smad2/3 and not Smad1/5/8 (Oh et al., 2002).
We also performed p-Smad2 immunostaining on stage 25 embryos to ascertain which p-Smad2-positive tissues were xGDF11-dependent. We find that all of the p-Smad2 seen in stage 25 embryos was eliminated when xGDF11 was knocked down (Fig. 4D). Embryos injected with control morpholinos (scr-tMO or scr-spMO) showed specific nuclear p-Smad2 staining in brain, eye, prospective mouth, branchial arches, and tailbud, all of which did not stain in embryos injected with either xGDF11 tMO or xGDF11 spMO. Taken together, these data indicate that xGDF11 is required for the majority of p-Smad2 signaling in tailbud stage embryos, in both tailbud and multiple anterior structures.
2.4 Phenotypic effects of xGDF11 knockdown in Xenopus embryos
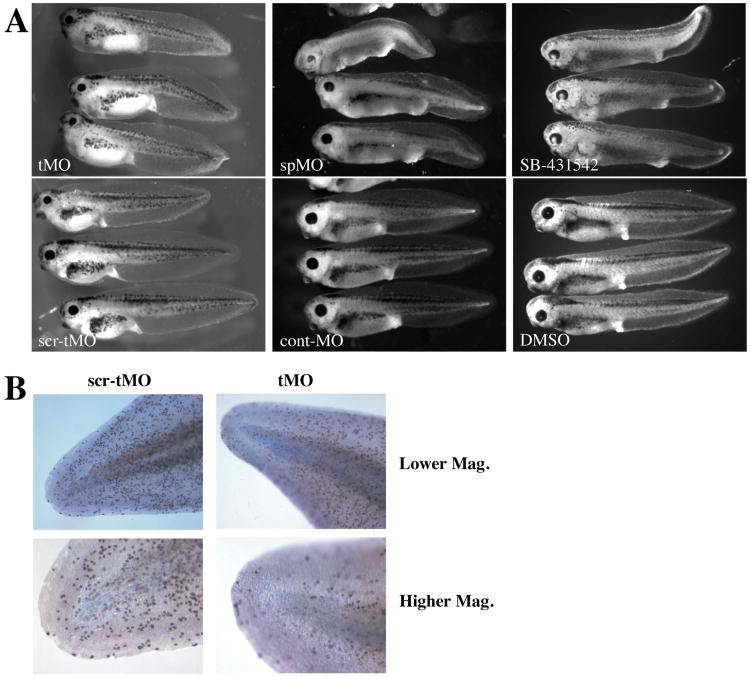
We next examined how knockdown of xGDF11 affected the phenotype of developing embryos. xGDF11 tMO and xGDF11 spMO each caused a clear shortening of the tadpole tail by stage 40-41 (Fig. 5A). Additionally, a reduction in the size of the head and head structures such as eyes was often observed. Embryos injected with various control morpholinos at equivalent doses always appeared wild-type. Tail defects could be observed shortly after the onset of tail elongation from the tailbud (data not shown). Expression of early tailbud markers (xbra, xcad3, xnot2) at stage 26 was normal in xGDF11 MO treated embryos (data not shown), indicating the knockdown of xGDF11 does not perturb the normal specification of the tailbud region. During tailbud outgrowth, a consistent reduction in cell proliferation was evident in the tailbud region of xGDF11 MO treated embryos relative to control tailbuds, when assessed by either phosphohistone-H3 staining (Fig. 5B) or by BrdU incorporation (data not shown). Embryos treated with SB-431542 at stage 13 (the onset of xGDF11 expression) displayed very similar axial elongation phenotypes as well as perturbed head structures, suggesting that loss of Smad2 signaling could be the cause of the morpholino phenotypes (Fig. 5A). In order to quantitate differences in tail length and trunk length between morpholino-injected and control embryos, we measured tail length (starting at the proctodaeum) and head+trunk length of stage 39-41 embryos. Although the severity of the morpholino phenotype varied by experiment (perhaps because morpholino effectiveness could be affected by differences in genetic background between batches), we always observed a statistically significant (p<0.005) decrease in tail length with xGDF11 knockdown (Table 1). Mean tail length of xGDF11 MO embryos was anywhere from 33-85% of controls . In contrast, the average length of the rest of the embryo (trunk+tail) was slightly greater for xGDF11 MO embryos, ranging from 105-120% (p<0.005 in 3 out of 5 experiments). Total length tended to be slightly lower than normal (72-96%, p<0.005 for 3 out of 5 batches, p<0.05 for the other 2), indicating that the increase in trunk length cannot completely compensate for the decrease in tail length. Consistent with this observation, there was a marked decrease in proliferative rate in tailbuds lacking GDF11 (data not shown).
Figure 5. Effect of xGDF11 knockdown on embryonic phenotype.
(A) Phenotype of stage 41 embryos injected with 60 ng tMO versus scr-tMO (left), and of stage 40 embryos injected with 20 ng spMO or cont-MO (center), and stage 40 embryos treated with 100 mM SB-431542 or 0.1% DMSO at stage 13. B) Cell proliferation was measured by phospho-histone H3 staining of tails from at Stage 40 embryos treated with 60 ng xGDF11 tMO (right) or 60 ng xGDF11 scr-tMO left. High magnification is of each tail tip is shown at bottom. Data shown are typical of three separate experiments, a similar reduction in proliferation with xGDF11 knockdown was seen when measured using BrdU incorporation (not shown).
Table 1. Effect of xGDF11 knockdown on A-P body axis length.
| Total | Tail | Head+trunk | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| batch | xGDF11 MO | control MO | ng | stage | MO:control1 | p2 | MO:control1 | p2 | MO:control | p2 |
| 1 | tMO | scr-tMO | 50 | 40 | 95.56% | 7.87E-03 | 85.47% | 2.87E-06 | 108.82% | 9.81E-04 |
| 2 | tMO | cont-MO | 40 | 40 | 72.48% | 8.80E-07 | 33.04% | 4.97E-08 | 120.23% | 1.73E-01 |
| 3 | tMO | scr-tMO | 50 | 41 | 88.38% | 1.36E-05 | 76.67% | 1.45E-07 | 112.22% | 1.39E-04 |
| 4 | tMO | cont-MO | 40 | 39 | 94.36% | 1.04E-02 | 72.22% | 2.50E-09 | 110.26% | 1.23E-03 |
| 5 | spMO | cont-MO | 20 | 39-40 | 75.87% | 3.97E-06 | 45.13% | 1.44E-08 | 105.10% | 1.29E-01 |
xGDF11MO:control ratios are expressed as ratio of means within each batch.
P values are calculated using an unpaired, two-tailed Student's T-test
2.5 Regulation of xGDF11 in the embryo by BMP-1/Tolloid family proteases
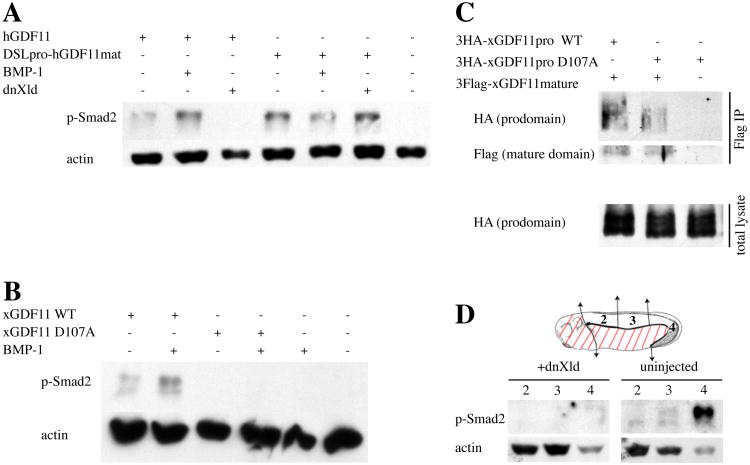
BMP-1/Tolloid-mediated cleavage of the GDF11 prodomain has been shown to regulate activation of GDF11 in cell culture (Ge et al., 2005), but a role for this regulation in vivo has not been directly tested. Since xGDF11 expression in the tailbud is strikingly co-localized with that of the Xenopus tolloid homologues, xolloid and xlr (Dale et al., 2002; Goodman et al., 1998), we examined the role of xGDF11 prodomain cleavage by BMP1/Xolloid proteases in the Xenopus embryo. To first establish that the role of GDF11 prodomain cleavage reported in cell culture can be reproduced in Xenopus embryos, we tested GDF11 activation of p-Smad2 in Xenopus animal cap explants. Expression of human GDF11 (hGDF11) in animal caps resulted in moderate p-Smad2 activation, and this signal was potentiated by co-injection of BMP-1 (Fig. 6A). Since animal caps express endogenous BMP-1/Tolloid activity, we blocked this endogenous activity using a dominant-negative form of the Xenopus Tolloid homolog Xolloid (dnXld), which has been shown to block the activity of all family members in Xenopus (Wardle et al., 1999). Co-injection of dnXld and hGDF11 resulted in the complete abrogation of p-Smad2 signaling (Fig. 6A). This effect of dnXld was dependent on the GDF11 prodomain, as p-Smad2 induced by a fusion protein containing the chick dorsalin prodomain and the hGDF11 mature domain (cDslpro-hGDF11mat) was not affected by either BMP-1 or dnXld (Fig. 6A).
Figure 6. Regulation of GDF11 signaling by BMP-1/Tolloid-dependent prodomain cleavage.
(A) Animal cap explants from embryos injected with mRNAs for hGDF11 (100 pg), DSLpro-hGDF11mat (100 pg), BMP-1 (500 pg), and dnXld (1 ng) were processed at stage 10.5 for Western blotting against p-Smad2 and actin. (B) Animal cap explants from embryos injected with mRNAs for xGDF11 WT (100 pg), xGDF11 D107A (100 pg), and BMP-1 (1 ng) were processed at stage 11 for Western blotting against p-Smad2 and actin. (C) Embryos were injected with HA-tagged xGDF11 prodo main (500 pg, either WT or D107A mutant) and Flag-tagged xGDF11 mature domain (500 pg), harvested at stage 9, immunoprecipitated with anti-Flag antibody, and detected by Western blotting against HA or Flag. (D) Regulation of endogenous p-Smad2 signal by blocking BMP-1/Tolloid function. Schematic of stage 26 embryo shows regions 2 (anterior dorsal trunk), 3 (medial dorsal trunk), and 4 (posterior dorsal trunk+tailbud) that were analyzed. Regions indicated with red lines were discarded. Embryos injected ventrally with 4 ng of dnXld or uninjected controls were dissected into regions 2, 3, and 4 at stage 26 and the pieces were analyzed for p-Smad2 by Western blotting. Actin was used as a loading control. Signal in the head was not analyzed in this experiment as the injection did not target the prospective head region since dnXld has early effects on head patterning independent of p-Smad2 and GDF11.
The activity of xGDF11, like that of hGDF11, was potentiated by co-expression of BMP-1 (Fig. 6B). Previous studies have identified an aspartate residue in the GDF11 prodomain that is essential for BMP-1 cleavage and activation of GDF11 (Ge et al., 2005; Wolfman et al., 2003). Mutation of the comparable aspartate (Asp 107) in xGDF11 to alanine (xGDF11 D107A) completely prevented activation of xGDF11 by either endogenous BMP1/xolloid activities or exogenous BMP-1 (Fig. 6B). Inhibition of GDF11 ligand signaling by the prodomain is thought to occur through direct non-covalent interaction of these two domains (Ge et al., 2005). Co-immunoprecipitation assays using HA-tagged xGDF11 prodomain and Flag-tagged xGDF11 mature domain confirmed that these domains did indeed interact in the embryo (Fig. 6C). Taken together, these biochemical data indicate that the that cleavage of the xGDF11 prodomain at D107 by BMP1/xolloid proteases relieves inhibitory interections between the ligand domain and the prodomain to allow ligand activation of p-Smad2.
To test whether BMP1/tolloid family proteases are important in the endogenous activation of p-Smad2 by xGDF11 in the tailbud, we targeted dnXolloid to the developing tailbud. dnXld mRNA was injected into the prospective posterior region of the the embryo, and at stage 26 anterior dorsal trunk, medial dorsal trunk, and posterior dorsal trunk+tailbud tissue was isolated for Western blotting (Fig. 6D). As in the case of xGDF11 knockdown, dnXld expression dramatically inhibited p-Smad2 activation, most notably preventing the strong pSmad2 activation in the tailbud region. The use of the dnXolloid construct to probe GDF11 activation was limited to posterior structures, because dnXolloid expression in the prospective anterior region disrupts head development through GDF11 independent effects on anterior patterning through the chordin/BMP/Smad1 signaling axis (Piccolo et al., 1997). These data do establish, however, that xGDF11 dependent activation of p-Smad2 in the tailbud can be blocked by inhibition of endogenous BMP-1/Tolloid activity. In conjunction with the demonstration that a conserved aspartate at the BMP-1/Tolloid protease cleavage site in the xGDF11 prodomain is necessary for xGDF11 ligand activity (Fig. 6A), these data indicate that the local regulation of pSmad2 activation by endogenous xGDF11 in the tailbud is controlled by endogenous Xolloid in the tailbud.
3. Discussion
3.1 xGDF11 generates p-Smad2 signaling pattern in the tailbud-stage embryo
Several independent lines of evidence in our work suggests that the ligand xGDF11 is required for the large majority, if not all, of the observed p-Smad2 at tailbud stages. First and foremost, two independent morpholino knockdowns of xGDF11 result in abrogation of p-Smad2 as assessed by both Western blotting and immunostaining (Fig. 4). p-Smad1 was not affected (Fig. 4C), indicating that xGDF11 morpholino injection does not cause a non-specific global reduction in signaling. Second, the phenotype generated by xGDF11 MO injection is strikingly similar to the phenotype of embryos treated with SB-431542, which blocks the type I receptor and thus eliminates all p-Smad2 regardless of ligand (Fig. 5). Third, the expression pattern of xGDF11 neatly corresponds to regions of activation of p-Smad2 (Figs.1, 2, 3). Fourth, injection of dnXld, which eliminates BMP-1/Tolloid-mediated activation of GDF11 but has no effect on other ligands, results in a loss of p-Smad2 in the embryo (Fig. 6). Our data do not exclude, however, the possibility that some portion of the effect of xGDF11 on endogenous p-Smad2 occurs indirectly through the GDF11-dependent expression of other Smad2 activating ligands. While expression of only one other potential Smad-2 activating ligand, TGFß-5, has been reported in the late tailbud embryo (Kondaiah et al., 2000), the TGFß superfamily is now large enough that it is difficult to exclude the possibility that other ligands may be regulated by GDF11 signaling.
In this work we have not directly examined the potential regulation of Smad3 by xGDF11, because available antibodies are inadequate to reliably monitor endogenous phospho-Smad3 levels. While in some developmental contexts, Smad2 and Smad3 appear to be functionally interchangeable (Dunn et al., 2004), in others they likely have distinctive roles in the regulation of downstream responses. Interestingly, xSmad3 is expressed at high levels in the chordoneural hinge region of the developing tailbud, as well as in several head structures that coincide with high levels of xGDF11 expression (Howell et al., 2001). Further exploration of distinctive contributions of Smad2 and Smad3 to tailbud patterning downstream of ligands such as GDF11 will be an interesting area for further investigation.
3.2 Regulation of GDF11 signaling by BMP-1/Tolloid family proteases
We have used the frog embryo to provide the first evidence that endogenous GDF11 is regulated by BMP1/tolloid family protease activity. In Xenopus embryos, xBMP-1 is expressed throughout the head whereas Xolloid (Xld) and Xolloid-related (Xlr) are strongest in the tail (Dale et al., 2002; Goodman et al., 1998). These expression patterns suggest that xBMP-1 may be responsible for regulating the activation of GDF11 in the head, while Xld, Xlr, or both fulfill this role in the tail. Blocking these endogenous activities in the tail results in a loss of p-Smad2 signaling in the tailbud stage embryo (Fig.6D), indicating that cleavage by BMP-1/Tolloid is important for regulating GDF11 in vivo.
In the mouse, examination of this question is complicated by the partially redundant function of multiple BMP1/tolloid family members, as well as their role in other extracellular matrix processing important for normal embryogenesis. The dominant negative Xld construct used here is a general inhibitor of BMP-1/tolloid family proteases (Piccolo et al., 1997), which helps to overcome the problem of functional redundancy among family members. The specificity of action of this dominant negative construct in the context in which we are using it is confirmed by the demonstration that it has no effect on the activation of a GDF11 construct in which the BMP1/tolloid cleaved prodomain is replaced with a prodomain from a different TGFß superfamily ligand (Fig. 6A). The application of dnXolloid as a probe of the role of GDF11 cleavage in embryonic patterning is limited, however, by the additional role of BMP-1/Tolloid proteases in regulation of the BMP2/4/7 signaling axis through cleavage of the BMP inhibitor chordin, which is also critical for anterior-posterior patterning at gastrulation. The effect on chordin, particularly in the head region, produces phenotypic complications that effectively mask later BMP-1 dependent events. Further characterization of the role of the BMP-1/tolloid proteases in distinct patterning events will require additional tools to distinguish protease regulation of the GDF11 signaling pathway from the ongoing role of these proteases in the control of BMP2/4/7 signaling.
While the co-expression of xGDF11, xolloid, and xlr in the developing tailbud is consistent with the local activation of xGDF11 by this protease family, the role of this regulatory step in patterning by GDF11 remains to be defined. Extracellular activation of latent, prodomain bound TGFß superfamily ligands has been shown or proposed to be important in a variety of physiological contexts (Anderson et al., 2008; Rifkin, 2005). Extracellular activation of xGDF11 by cleavage of its pro-domain provides an additional layer of regulation of local pSmad2 activation in the tailbud, but how this regulatory step might be used in the control of patterning is not known. A complex network of extracellular regulators, including BMP1/tolloid proteases, has been shown to be critical for the establishment of morphogen gradients of BMP signaling, and it will be interesting to determine the spatial and temporal control of local pSmad2 activation in the tailbud provided by proteolytic control of GDF11 activity. Whether or not a latent pool of prodomain-bound GDF11 is available for rapid activation by BMP-1/Tolloid proteases during development and later events is also a question of interest.
3.3 Implications for the role of GDF11 in axial patterning
Studies in mouse have shown that GDF11 is important for generating correct anterior-posterior segmental identity in the developing vertebrae and spinal cord (Liu, 2006; McPherron et al., 1999). We find in the frog that GDF11 knockdown, in addition to significantly shortening the tail, results in a slight but statistically significant elongation of the trunk (Fig. 5 and Table 1). This trunk elongation may represent the same sort of anterior homeotic transformation seen in the mouse mutant. Since A-P markers of segmental identity typically used in the trunk/tail regions of the mouse do not reliably distinguish A-P segments in the pre-metamorphic frog embryo (data not shown), however, we have not been able to test this hypothesis directly. In mice, expression of GDF11 in the tailbud is complemented by somatic expression of the closely related ligand GDF8 (myostatin). Interestingly, examination of the recently completed X.tropicalis genome indicates that Xenopus, unlike most vertebrates, lacks a GDF8/myostatin orthologue, and therefore partially redundant roles for GDF11 and GDF8/myostatin in mammals may depend entirely on GDF11 in Xenopus embryos.
The shortening of the tail in xGDF11 MO embryos cannot be entirely attributed to anterior transformation of tail segments, since the total length of the embryos was also decreased (Table 1). Mouse GDF11 mutants likewise have shortened or sometimes missing tails in addition to homeotic vertebral transformation (McPherron et al., 1999), but the former defect has not been additionally characterized. GDF11 may therefore have a role in establishing tail length as well as in tail-trunk patterning; whether these two roles reflect distinct functions or are inherently linked is not resolvable in these. It is interesting to note that GDF11 is expressed in the prospective tail-forming region by the end of gastrulation (stage 13) and prior to the onset of somitogenesis, so every somite that condenses from the tailbud is likely to see GDF11 at some point. Whether GDF11 might interact with the cycling somitic clock, by regulating proliferation of tail tissues, or by some other mechanism is at present unclear. Notably, in embryos lacking GDF11 or pSmad2, the tailbud itself appears to be present and essentially functional based on the proper expression of tailbud specification genes (data not shown) and the fact that these embryos have severely shortened but not absent tails that contain recognizable structures; the defects appear rather to be primarily related to regulation of size and/or growth. Examination of the role of GDF11 in maintaining proper tail length will be interesting, as it appears to function as a positive regulator of organ size in this case. In contrast, GDF11 appears to be a negative regulator of neurogenesis in the mouse retina and olfactory epithelium (Kim et al., 2005; Wu et al., 2003), and GDF8/myostatin is a negative regulator of muscle mass (McPherron et al., 1997). These data suggest that the activity of xGDF11 in different tissues is likely to be highly context-dependent.
Our previous work has also shown that GDF11 mRNA is upregulated during the process of Xenopus tadpole tail regeneration, concomitant with an upregulation in p-Smad2 signaling and an increase in cell proliferation in the regenerating tail tip ((Ho and Whitman, 2008) and data not shown). This result suggests that the role of GDF11 in regulating tail growth and size is recapitulated during regeneration. Further analysis of the roles of GDF11 and BMP-1/Tolloid proteases during regeneration will surely be informative.
In conclusion, we have shown that GDF11 is responsible, whether directly or indirectly, for generating the vast majority of p-Smad2 in the Xenopus embryo during tailbud stages, and that regulation of GDF11 by BMP-1/tolloid proteases is necessary for this Smad2 activation. In-depth, mechanistic studies of the effects of GDF11 and its downstream signaling in specific tissues and structures will reveal further information about the multiple roles of GDF11 during development.
4. Experimental Procedures
p-Smad2 immunostaining
Generation of a polyclonal antibody to p-Smad2 has been described elsewhere (Faure et al., 2000; Lee et al., 2001). For the experiments in this work, anti-pSmad2 was used at 1:10-1:25. Embryos were fixed in MEMFA and treated with 5 mM iodoacetamide (Sigma) to improve staining. HRP-conjugated anti-rabbit secondary antibody (Jackson Immunoresearch) was used at 1:250-1:750 and stain was developed using TrueBlue HRP substrate (KLP Laboratories) or DAB (vector laboratories). In some cases, stained embryos were dehydrated in methanol and cleared with a 1:2 solution of benzyl acetate:benzyl benzoate.
Manipulation of Xenopus embryos
Fertilization and maintenance of Xenopus embryos was as previously described (Watanabe and Whitman, 1999). Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1967). Animal cap dissections were performed at stage 8-9.
Cloning of xGDF11
We cloned xGDF11 using PCR based on available Xenopus EST sequences (GenBank IDs: BJ051697, BJ072439, and BJ034907) and 5′RACE (First Choice RLM RACE kit, Ambion). Full-length xGDF11 was subcloned into pCS4+ for mRNA injection studies. D107A mutation was made using a PCR-based mutagenesis protocol (Makarova et al., 2000) with the following primer (mutation is underlined): 5′-CAGCATGAGTTTCAGGGAGCCTCTTTCGAGCACAACACT-3′. Various epitope-tagged constructs were generated using PCR-based subcloning.
RT-PCR
Total RNA was extracted from whole embryos using the Trizol (Invitrogen) method. xGDF11 primers: Upper: 5′-CGGGACTGGGGATAGTGGAGTATA-3′, Lower: 5′-GCGAGGAGCATGGATTAAGGGA-3′. Standard primers to ODC (http://xenbase.org/xenbase/original/WWW/Marker_pages/primers.html) were used.
In situ hybridization
In situ hybridization was performed using a standard protocol (Harland, 1991). An approximately 1.1 kB antisense probe for xGDF11 was used, which contains the last 690 base pairs of the coding region and part of the 3′UTR.
Morpholino oligos
All morpholino oligos were obtained from Gene Tools. Sequences of morpholino oligos are as follows: xGDF11 tMO: 5′-ACAGAGGGCACAGCTGAGACAGCAT-3′, xGDF11 scr-tMO: 5′-GAACGCAGTGACACAGGTCAGACAG-3′, xGDF11 spMO: 5′-GAAAAAACACTTACTTTCCTGTGCC-3′, xGDF11 scr-spMO: 5′-CCTAAGATCTTCAATGGCACTTCAA-3′. In some cases, a standard control morpholino obtained from Gene Tools (Cont-MO) was used. Morpholino oligos were injected marginally and bilaterally at the 2-cell stage, in a volume of 10 nl per injection. xGDF11 tMO and scr-tMO are injected at 40-60 ng per embryo and xGDF11 spMO and scr-spMO are injected at 20 ng per embryo. Cont-MO is injected at equal amount to xGDF11 MO for each experiment.
mRNA injections
mRNAs were synthesized using the mMessage mMachine SP6 kit (Ambion). Unless otherwise noted, 10 nl injections were performed animally and bilaterally at the 2-cell stage.
Western blotting and immunoprecipitation
All samples for Western blotting and immunoprecipitation were lysed in modified RIPA buffer without SDS. Anti-p-Smad2 (Faure et al., 2000) was used at 1:200-1:500. Anti-actin (AC40, Sigma), anti-Flag (M2, Sigma), and anti-HA-HRP (3F10, Roche) were used at 1:1000. For p-Smad2 blotting, embryos were dissected into regions as indicated in the figures prior to harvesting, and 2-2.5 embryo pieces or 0.2-0.25 whole embryos were loaded per lane. For animal cap experiments, 2-2.5 animal caps were loaded per lane. Anti-Flag immunoprecipitation was performed with anti-FlagM2-conjugated agarose beads (Sigma), and 3.25 embryo equivalents of immunoprecipitated material or 0.065 whole embryos (starting material) were loaded per lane.
Acknowledgments
We thank Dr. Ken Cho for the BMP-1 construct, and Dr. Leslie Dale for the dnXld construct. The human GDF11 construct was obtained from Paul Oh. As always, members of the Whitman lab contributed many hours of helpful discussion. MW was supported by Grant HD29468 from NICHD. CY was supported by Pure Basic Research Grant (070-2005-C00115) of the Korean Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson SB, Goldberg AL, Whitman M. Identification of a novel pool of extracellular pro-myostatin in skeletal muscle. J Biol Chem. 2008;283:7027–35. doi: 10.1074/jbc.M706678200. [DOI] [PubMed] [Google Scholar]
- Beck CW, Slack JM. Analysis of the developing Xenopus tail bud reveals separate phases of gene expression during determination and outgrowth. Mech Dev. 1998;72:41–52. doi: 10.1016/s0925-4773(98)00015-x. [DOI] [PubMed] [Google Scholar]
- Birsoy B, Kofron M, Schaible K, Wylie C, Heasman J. Vg 1 is an essential signaling molecule in Xenopus development. Development. 2006;133:15–20. doi: 10.1242/dev.02144. [DOI] [PubMed] [Google Scholar]
- Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- Dale L, Evans W, Goodman SA. Xolloid-related: a novel BMP1/Tolloid-related metalloprotease is expressed during early Xenopus development. Mech Dev. 2002;119:177–90. doi: 10.1016/s0925-4773(02)00359-3. [DOI] [PubMed] [Google Scholar]
- de Sousa Lopes SM, Carvalho RL, van den Driesche S, Goumans MJ, ten Dijke P, Mummery CL. Distribution of phosphorylated Smad2 identifies target tissues of TGF beta ligands in mouse development. Gene Expr Patterns. 2003;3:355–60. doi: 10.1016/s1567-133x(03)00029-2. [DOI] [PubMed] [Google Scholar]
- Dunn NR, Vincent SD, Oxburgh L, Robertson EJ, Bikoff EK. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development. 2004;131:1717–28. doi: 10.1242/dev.01072. [DOI] [PubMed] [Google Scholar]
- Faure S, Lee MA, Keller T, ten Dijke P, Whitman M. Endogenous patterns of TGFbeta superfamily signaling during early Xenopus development. Development. 2000;127:2917–31. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- Gamer LW, Wolfman NM, Celeste AJ, Hattersley G, Hewick R, Rosen V. A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer in Xenopus embryos. Dev Biol. 1999;208:222–32. doi: 10.1006/dbio.1998.9191. [DOI] [PubMed] [Google Scholar]
- Ge G, Hopkins DR, Ho WB, Greenspan DS. GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol. 2005;25:5846–58. doi: 10.1128/MCB.25.14.5846-5858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gont LK, Steinbeisser H, Blumberg B, de Robertis EM. Tail formation as a continuation of gastrulation: the multiple cell populations of the Xenopus tailbud derive from the late blastopore lip. Development. 1993;119:991–1004. doi: 10.1242/dev.119.4.991. [DOI] [PubMed] [Google Scholar]
- Goodman SA, Albano R, Wardle FC, Matthews G, Tannahill D, Dale L. BMP1-related metalloproteinases promote the development of ventral mesoderm in early Xenopus embryos. Dev Biol. 1998;195:144–57. doi: 10.1006/dbio.1997.8840. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–95. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Ho DM, Whitman M. TGF-beta signaling is required for multiple processes during Xenopus tail regeneration. Dev Biol. 2008;315:203–16. doi: 10.1016/j.ydbio.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Mohun TJ, Hill CS. Xenopus Smad3 is specifically expressed in the chordoneural hinge, notochord and in the endocardium of the developing heart. Mech Dev. 2001;104:147–50. doi: 10.1016/s0925-4773(01)00365-3. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–30. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Kondaiah P, Taira M, Vempati UD, Dawid IB. Transforming growth factor-beta5 expression during early development of Xenopus laevis. Mech Dev. 2000;95:207–9. doi: 10.1016/s0925-4773(00)00326-9. [DOI] [PubMed] [Google Scholar]
- Lee MA, Heasman J, Whitman M. Timing of endogenous activin-like signals and regional specification of the Xenopus embryo. Development. 2001;128:2939–52. doi: 10.1242/dev.128.15.2939. [DOI] [PubMed] [Google Scholar]
- Liu JP. The function of growth/differentiation factor 11 (Gdf11) in rostrocaudal patterning of the developing spinal cord. Development. 2006;133:2865–74. doi: 10.1242/dev.02478. [DOI] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques. 2000;29:970–2. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet. 1999;22:260–4. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- Nakashima M, Toyono T, Akamine A, Joyner A. Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech Dev. 1999;80:185–9. doi: 10.1016/s0925-4773(98)00205-6. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) North Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- Oh SP, Yeo CY, Lee Y, Schrewe H, Whitman M, Li E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev. 2002;16:2749–54. doi: 10.1101/gad.1021802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuma Y, Takahashi S, Yokota C, Asashima M. Multiple nodal-related genes act coordinately in Xenopus embryogenesis. Dev Biol. 2002;241:94–105. doi: 10.1006/dbio.2001.0493. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Lu B, Goodman S, Dale L, De Robertis EM. Cleavage of Chordin by Xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell. 1997;91:407–16. doi: 10.1016/s0092-8674(00)80424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramis JM, Collart C, Smith JC. Xnrs and activin regulate distinct genes during Xenopus development: activin regulates cell division. PLoS One. 2007;2:e213. doi: 10.1371/journal.pone.0000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280:7409–12. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- Schohl A, Fagotto F. Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development. 2002;129:37–52. doi: 10.1242/dev.129.1.37. [DOI] [PubMed] [Google Scholar]
- Wardle FC, Welch JV, Dale L. Bone morphogenetic protein 1 regulates dorsal-ventral patterning in early Xenopus embryos by degrading chordin, a BMP4 antagonist. Mech Dev. 1999;86:75–85. doi: 10.1016/s0925-4773(99)00114-8. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Whitman M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development. 1999;126:5621–34. doi: 10.1242/dev.126.24.5621. [DOI] [PubMed] [Google Scholar]
- Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell. 2001;1:605–17. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- Wolfman NM, McPherron AC, Pappano WN, Davies MV, Song K, Tomkinson KN, Wright JF, Zhao L, Sebald SM, Greenspan DS, Lee SJ. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc Natl Acad Sci U S A. 2003;100:15842–6. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]