Abstract
Prediction of cancer progression after radical prostatectomy (RP) is one of the most challenging problems in the management of prostate cancer. Gene-expression profiling is widely used to identify genes associated with such progression. Usually candidate genes are identified according to a gene-by-gene comparison of expression. Recent reports suggested that relative expression of a gene pair more efficiently predicts cancer progression than single-gene analysis does. The top-scoring pair (TSP) algorithm classifies phenotypes according to the relative expression of a pair of genes. We applied the TSP approach to predict which patients would experience systemic tumor progression after RP. Relative expression of TPD52L2/SQLE and CEACAM1/BRCA1 gene pairs identified those patients, with more than 99% specificity but relatively low sensitivity (~10%). These two gene pairs were validated in three independent datasets. Additionally, combining two pairs of genes improved sensitivity without compromising specificity. Functional annotation of the TSP genes demonstrated that they cluster by a limited number of biologic functions and pathways, suggesting that relatively lower expression of genes from specific pathways can predict cancer progression. In conclusion, comparative analysis of the expression of two genes may be a simple and effective classifier for prediction of prostate cancer progression. The TSP approach can be used to identify patients whose prostate cancer will progress after they undergo radical prostatectomy. Two gene pairs can predict which men would experience progression to the metastatic form of the disease. However, because our analysis was based on a relatively small number of genes, a larger study will be needed to identify the best predictors of disease outcome overall.
Keywords: prostate cancer, gene co-expression, top-scoring pairs of genes, metastasis, cancer progression
Introduction
Screening for prostate cancer according to serum prostate-specific antigen (PSA) level has improved early detection of the disease, resulting in increased identification of patients with localized disease that is still curable with surgery and radiotherapy. However, 20–30% of treated patients experience a relapse.1,2 Thus, from the clinical perspective, it is important to be able to predict which patients will experience a relapse.3–6
Understanding the biology of prostate cancer progression is essential for enabling the development of prognostic markers and effective therapeutic targets. A number of studies have been conducted to characterize the dynamics of gene expression in prostate cancer progression by using DNA microarrays.5,7–12 In some studies, tumor-expression signatures associated with clinical parameters and outcome have been identified.13–16
Normally, researchers use one gene at a time to find any association between gene expression and phenotype. The results of recent studies, however, suggest that assessing the expression of more than one gene (i.e., co-expression analysis) yields a better prediction of tumor progression than the analysis of individual genes does.5,6 Motivated by these findings, we analyzed whether co-expression patterns can be useful in predicting prostate cancer progression by using the paired-gene approach of the top-scoring pairs (TSP) algorithm as described by Geman et al. in 2004.17 However, we modified the algorithm by controlling the specificity and sensitivity of the TSPs. This may be particularly important when the results of classification influence the selection of treatment modality or follow-up procedures that can be associated with serious side effects. For our purpose, we took this approach to analyze gene-expression data from a recently reported large case-control microarray study.5
Materials and methods
Datasets
To overcome the disadvantageous pattern of microarray data (small sample size and large number of variables), we searched the Gene Expression Omnibus (GEO) database and selected four studies of prostate cancer progression that each had at least 10 samples per phenotype. We used one as the discovery dataset and the other three as validation datasets (Table 1). Three pairs of phenotypes were compared in the four datasets: (1) systemic progression (SYS; development of distant metastasis after radical prostatectomy [RP]) vs no evidence of disease (NED) (the GSE10645 dataset),5 (2) primary prostate tumors vs distant metastases (the GSE6752 and GSE6919 datasets),7,11 and (3) hormone-sensitive vs hormone-refractory prostate cancer (the GSE6811 dataset).10
Table 1.
Gene-expression datasets used in our analysis
| GEO ID | PMID | Phenotypes | Number of samples |
Platform | Number of probes |
|---|---|---|---|---|---|
| GSE10645 | 18846227 | NED vs SYS | 213 vs 213 | GPL5858 and GPL5873 | 1028 |
| GSE6752 | 17430594 | Primary prostate tumor vs prostate tumor metastases | 10 vs 21 | CodeLink UniSet Human 20K I Bioarray (GPL2891) | 23 572 |
| GSE6811 | 17545589 | HSPCs vs HRPCs | 10 vs 25 | YN Human 36K (sets 1–8) (GPL4747) | 36 864 |
| GSE6919 | 15254046 | Primary prostate tumor vs prostate tumor metastases | 196 vs 75 | Affymetrix Human Genome U95 Version 2, U95C, and U95B Array (GPL8300, GPL93, and GPL92) | 12 625 12 646 12 620 |
Abbreviations: NED, no evidence of disease; SYS, systemic prostate cancer progression; HSPC, hormone-sensitive prostate cancers; HRPC, hormone-refractory prostate cancers.
For our discovery set, we used gene-expression data from the study by Nakagawa et al.5 (i.e., dataset 1 as described above). Those authors analyzed the association between gene expression and outcome after initial therapy, comparing data from a group of 213 patients who had NED during the 7 years after undergoing retropubic RP (RRP) with those from a group of 213 patients who experienced SYS during the 5 years after the initial rise in PSA level. The authors analyzed the expression of 1028 genes for which previous evidence had implicated their involvement in prostate cancer progression.
For our validation studies, we did not analyze exactly the same phenotypes that we analyzed in the discovery dataset. As counterparts for the NED data, we used primary tumor data from the GSE6752 and GSE6919 datasets and hormone-sensitive prostate cancer data from the GSE6811 dataset. Our counterparts for the SYS phenotype were data on distant metastases from GSE6752 and GSE6919 and hormone-refractory prostate cancer from GSE6811.
Top-scoring pairs algorithm
The original TSP classifier algorithm was described by Geman et al.17 Here we give a simplified description of the method. Given one expression matrix X = [xgn] of G genes and N samples, xgn represents the gth gene-expression value from the nth sample. Assume that every sample can be labeled as either 1 or 2. For example, the N1 samples from 1 to N1 (N1 < N) are labeled class 1 (e.g., NED), and N2 samples from (N1 + 1) to N are labeled class 2 (e.g., SYS), in which N1 + N2 = N. The focus is on detecting “marker gene pairs” (i, j) for which there is a significant difference in the probability of observing Xi < Xj from class 1 to class 2. The conditional probabilities of observing Xi < Xj in each class are defined as follows:
in which I(xin < xjn) is the indicator function, defined as
The typical TSP method is based on maximizing the scores of (i,j) Δij = |pij(1) − pij(2)|. This approach provides superior performance to that of support vector machines and other sophisticated methods for classifying cancer samples.18,19 Although maximizing the delta value identifies the best classifier, that classifier may be associated with relatively low sensitivity and specificity, which is a concern when the classifier is used in selecting a treatment. Because of this, we included an assessment of specificity and sensitivity in our analysis, keeping at least 99% specificity and trying to maximize the sensitivity.
The statistical significance of classifiers can be assessed by randomly permuting the class labels maintaining the sample sizes N1 and N2. From this permutation analysis, we estimated P values associated with a given conditional probability. This P value can be interpreted as the probability that the gene pair is not informative for the classification.
Functional annotation
For functional annotation, we applied Ingenuity Pathways Analysis (Ingenuity Systems, Inc, Redwood City, CA, USA; http://www.ingenuity.com/), which evaluates the distribution of the top-ranked genes by both pathway and gene ontology categories, testing the null hypothesis that the genes are uniformly distributed. P values characterize the statistical evidence for the clustering of the genes by pathway or functional categories: the lower the P value, the stronger the statistical evidence that the top-ranking genes belong to a specific pathway or functional category.
Results
Top-scoring pairs
Table 2 lists the TSPs we identified as useful for predicting which patients will develop SYS and which will have NED after they undergo RRP. These TSPs were selected on the basis of a delta value of at least 0.1 and on the condition that the specificity was ≥ 99%. We identified 22 TSPs for predicting SYS but only two for predicting NED.
Table 2.
Top-scoring pairs of genes for prediction of systemic progression (SYS) and no evidence for disease (NED). The last column is the estimated P value for pair genes by 106 permutation testing
| Predict | Gene1 | Gene2 | P(X1<X2| NED) |
P(X1<X2| SYS) |
Delta = |P1−P2| |
Sensitivity | Specificity | RR | P value |
|---|---|---|---|---|---|---|---|---|---|
| NED | F5 | ESR2 | 0.108 | 0.000 | 0.108 | 0.108 | 1.000 | 0.000 | <10E-6 |
| NED | COL1A1 | CDKN1B | 0.149 | 0.010 | 0.139 | 0.149 | 0.990 | 0.119 | <10E-6 |
| SYS | PAGE4 | ANXA2 | 0.000 | 0.130 | 0.130 | 0.130 | 1.000 | 2.115 | <10E-6 |
| SYS | HLF | BLM | 0.000 | 0.105 | 0.105 | 0.105 | 1.000 | 2.081 | <10E-6 |
| SYS | PASK | SQLE | 0.000 | 0.105 | 0.105 | 0.105 | 1.000 | 2.055 | 10E-6 |
| SYS | MYBPC1 | TMEM65 | 0.005 | 0.175 | 0.170 | 0.175 | 0.995 | 2.121 | <10E-6 |
| SYS | SNTB2 | SQLE | 0.005 | 0.165 | 0.160 | 0.165 | 0.995 | 2.046 | <10E-6 |
| SYS | JUN | MKI67 | 0.005 | 0.155 | 0.150 | 0.155 | 0.995 | 2.037 | 10E-6 |
| SYS | PAGE4 | TAF2 | 0.005 | 0.140 | 0.135 | 0.140 | 0.995 | 2.037 | 5 × 10E-6 |
| SYS | PTN | IMMT | 0.005 | 0.135 | 0.130 | 0.135 | 0.995 | 2.027 | <10E-6 |
| SYS | EFS | WDR67 | 0.005 | 0.130 | 0.125 | 0.130 | 0.995 | 2.018 | 3 × 10E-6 |
| SYS | EDG7 | LGR4 | 0.005 | 0.125 | 0.120 | 0.125 | 0.995 | 2.018 | <10E-6 |
| SYS | TCF7L2 | BIRC5 | 0.005 | 0.120 | 0.115 | 0.120 | 0.995 | 2.096 | <10E-6 |
| SYS | PTEN | DVL3 | 0.005 | 0.120 | 0.115 | 0.120 | 0.995 | 2.009 | <10E-6 |
| SYS | GDF15 | MKI67 | 0.005 | 0.120 | 0.115 | 0.120 | 0.995 | 2.009 | <10E-6 |
| SYS | SLC45A3 | ENY2 | 0.005 | 0.115 | 0.110 | 0.115 | 0.995 | 2.009 | 2 × 10E-6 |
| SYS | MADH2 | NOTCH2 | 0.005 | 0.115 | 0.110 | 0.115 | 0.995 | 2.009 | <10E-6 |
| SYS | PXN | TK1 | 0.005 | 0.115 | 0.110 | 0.115 | 0.995 | 2.089 | <10E-6 |
| SYS | CEACAM1 | BRCA1 | 0.005 | 0.110 | 0.105 | 0.110 | 0.995 | 1.999 | <10E-6 |
| SYS | PAGE4 | LGR4 | 0.005 | 0.110 | 0.105 | 0.110 | 0.995 | 1.999 | 10E-6 |
| SYS | TPD52L2 | SQLE | 0.005 | 0.110 | 0.105 | 0.110 | 0.995 | 1.999 | <10E-6 |
| SYS | PAGE4 | ST3GAL1 | 0.005 | 0.110 | 0.105 | 0.110 | 0.995 | 1.999 | <10E-6 |
| SYS | TMEM71 | TPX2 | 0.005 | 0.110 | 0.105 | 0.110 | 0.995 | 1.999 | 10E-6 |
Abbreviation: RR, relative risk.
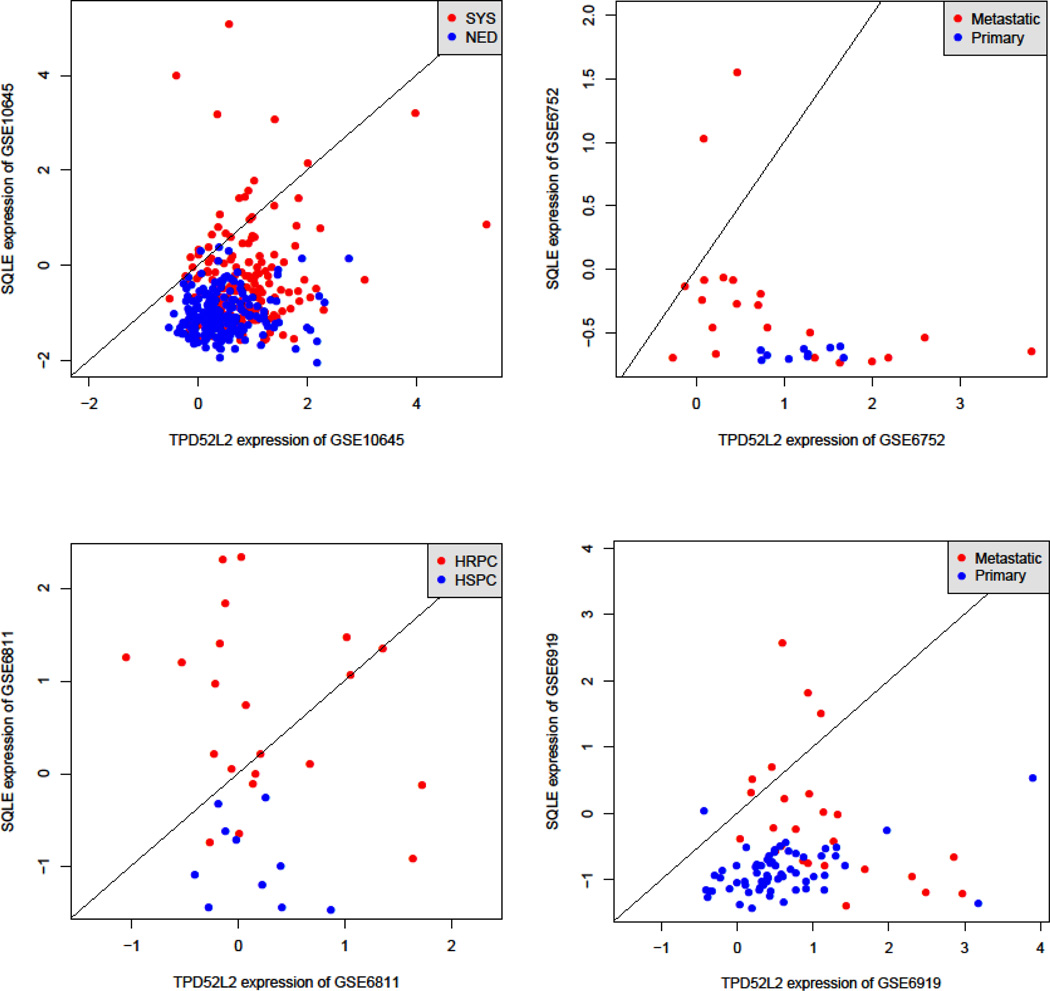
We validated the top 22 TSPs by using the three independent validation datasets (Table 3). To estimate the overall consistency between the discovery and validation sets, we looked at the correlation between proportions of positive decisions in the two sets. The discovery and validation sets tended to have a similar proportion of positive classifying decisions (i.e., Xi < Xj): the correlation coefficient was 0.44 (N = 104, P = 0.000003). We identified two pairs of genes—TPD52L2/SQLE and CEACAM1/BRCA1—that were significant in all three validation datasets. Analysis of the interactions between genes by using Pathways Studio (Ariadne, Inc, Rockville, MD, USA; http://www.ariadnegenomics.com/products/pathway-studio/) identified no interactions between the genes, which suggests that TPD52L2, SQLE, CEACAM1, and BRCA1 act independently. Figure 1 shows the classification plots for comparing the expression values of the gene pairs TPD52L2 and SQLE in the discovery and validation sets. Similar patterns can also be plotted with gene pair of CEACAM1 and BRCA1.
Table 3.
In silico validation of the 24 top-scoring pairs listed in Table 2; the two top validated pairs of genes are shown in bold
| Predicted phenotype |
Dataset 1 | Gene1 | Gene2 | P(X1<X2|NED) | P(X1<X2|SYS) |
|---|---|---|---|---|---|
| SYS | GSE10645 | TPD52L2 | SQLE | 0.005128 | 0.11 |
| Meta | GSE6752 | TPD52L2 | SQLE | 0 | 0.095238 |
| HR | GSE6811 | TPD52L2 | SQLE | 0 | 0.52 |
| Metastasis | GSE6919 | TPD52L2 | SQLE | 0.015385 | 0.24 |
| SYS | GSE10645 | CEACAM1 | BRCA1 | 0.005128 | 0.11 |
| Meta | GSE6752 | CEACAM1 | BRCA1 | 0 | 0.238095 |
| HR | GSE6811 | CEACAM1 | BRCA1 | 0.2 | 0.72 |
| Metastasis | GSE6919 | CEACAM1 | BRCA1 | 0.061538 | 0.64 |
| NED | GSE10645 | COL1A1 | CDKN1B | 0.148718 | 0.01 |
| HS | GSE6811 | COL1A1 | CDKN1B | 1 | 0.64 |
| Primary | GSE6919 | COL1A1 | CDKN1B | 0.230769 | 0.08 |
| SYS | GSE10645 | EDG7 | LGR4 | 0.005128 | 0.125 |
| Meta | GSE6752 | EDG7 | LGR4 | 0 | 0.238095 |
| HR | GSE6811 | EDG7 | LGR4 | 0.6 | 0.68 |
| SYS | GSE10645 | EFS | WDR67 | 0.005128 | 0.13 |
| Meta | GSE6752 | EFS | WDR67 | 0 | 0.047619 |
| HR | GSE6811 | EFS | WDR67 | 0.5 | 0.84 |
| Metastasis | GSE6919 | EFS | WDR67 | 0.030769 | 0.12 |
| NED | GSE10645 | F5 | ESR2 | 0.107692 | 0 |
| HR | GSE6811 | F5 | ESR2 | 0.1 | 0.36 |
| Primary | GSE6919 | F5 | ESR2 | 0.753846 | 0.68 |
| SYS | GSE10645 | GDF15 | MKI67 | 0.005128 | 0.12 |
| Meta | GSE6752 | GDF15 | MKI67 | 0 | 0.142857 |
| HR | GSE6811 | GDF15 | MKI67 | 0.4 | 0.68 |
| SYS | GSE10645 | HLF | BLM | 0 | 0.105 |
| Meta | GSE6752 | HLF | BLM | 0.8 | 1 |
| HS | GSE6811 | HLF | BLM | 0.9 | 0.84 |
| Metastasis | GSE6919 | HLF | BLM | 0.507692 | 0.72 |
| SYS | GSE10645 | JUN | MKI67 | 0.005128 | 0.155 |
| HR | GSE6811 | JUN | MKI67 | 0.8 | 0.96 |
| Metastasis | GSE6919 | JUN | MKI67 | 0.015385 | 0.04 |
| SYS | GSE10645 | PAGE4 | ANXA2 | 0 | 0.13 |
| HS | GSE6811 | PAGE4 | ANXA2 | 0.1 | 0.04 |
| Metastasis | GSE6919 | PAGE4 | ANXA2 | 0.169231 | 0.92 |
| SYS | GSE10645 | PAGE4 | LGR4 | 0.005128 | 0.11 |
| Meta | GSE6752 | PAGE4 | LGR4 | 0.2 | 0.857143 |
| HR | GSE6811 | PAGE4 | LGR4 | 0.6 | 0.68 |
| SYS | GSE10645 | PAGE4 | ST3GAL1 | 0.005128 | 0.11 |
| Meta | GSE6752 | PAGE4 | ST3GAL1 | 0.2 | 1 |
| HR | GSE6811 | PAGE4 | ST3GAL1 | 0.1 | 0.32 |
| Metastasis | GSE6919 | PAGE4 | ST3GAL1 | 0.153846 | 0.96 |
| SYS | GSE10645 | PAGE4 | TAF2 | 0.005128 | 0.14 |
| Meta | GSE6752 | PAGE4 | TAF2 | 0.1 | 1 |
| HR | GSE6811 | PAGE4 | TAF2 | 0.4 | 0.68 |
| Metastasis | GSE6919 | PAGE4 | TAF2 | 0.353846 | 1 |
| SYS | GSE10645 | PASK | SQLE | 0 | 0.105 |
| HR | GSE6811 | PASK | SQLE | 0 | 0.84 |
| Metastasis | GSE6919 | PASK | SQLE | 0.723077 | 0.92 |
| SYS | GSE10645 | PTEN | DVL3 | 0.005128 | 0.12 |
| HR | GSE6811 | PTEN | DVL3 | 0.4 | 0.48 |
| Metastasis | GSE6919 | PTEN | DVL3 | 0.984615 | 1 |
| SYS | GSE10645 | PTN | IMMT | 0.005128 | 0.135 |
| HR | GSE6811 | PTN | IMMT | 0.7 | 0.88 |
| Metastasis | GSE6919 | PTN | IMMT | 0.076923 | 0.64 |
| SYS | GSE10645 | PXN | TK1 | 0.005128 | 0.115 |
| Meta | GSE6752 | PXN | TK1 | 0.9 | 0.952381 |
| HR | GSE6811 | PXN | TK1 | 0.6 | 0.8 |
| Metastasis | GSE6919 | PXN | TK1 | 0.261538 | 0.68 |
| SYS | GSE10645 | SLC45A3 | ENY2 | 0.005128 | 0.115 |
| HR | GSE6811 | SLC45A3 | ENY2 | 0.3 | 0.52 |
| SYS | GSE10645 | SNTB2 | SQLE | 0.005128 | 0.165 |
| Meta | GSE6752 | SNTB2 | SQLE | 0 | 0.952381 |
| HR | GSE6811 | SNTB2 | SQLE | 0.2 | 0.72 |
| Metastasis | GSE6919 | SNTB2 | SQLE | 0.266317 | 0.42 |
| SYS | GSE10645 | TCF7L2 | BIRC5 | 0.005128 | 0.12 |
| Meta | GSE6752 | TCF7L2 | BIRC5 | 0.8 | 0.952381 |
| HR | GSE6811 | TCF7L2 | BIRC5 | 0.4 | 0.76 |
| Metastasis | GSE6919 | TCF7L2 | BIRC5 | 0.230769 | 0.44 |
| SYS | GSE10645 | TMEM71 | TPX2 | 0.005128 | 0.11 |
| HR | GSE6811 | TMEM71 | TPX2 | 0.4 | 0.72 |
Figure 1.
Scatterplots of the expression of TPD52L2 and SQLE genes in the discovery set (GSE10645) and the three validation sets (GSE6752, GSE6811, and GSE6919). Each data point represents a patient: red dots are patients with systemic cancer progression (SYS) for the discovery dataset and corresponding counterparts for the validation datasets. The slanted lines are decision rules: above the decision line, the expression of TPD52L1 is lower than that of SQLE. NED, no evidence of disease; HRPC, hormone-refractory prostate cancer; HSPC, hormone-sensitive prostate cancer.
Double top-scoring pairs
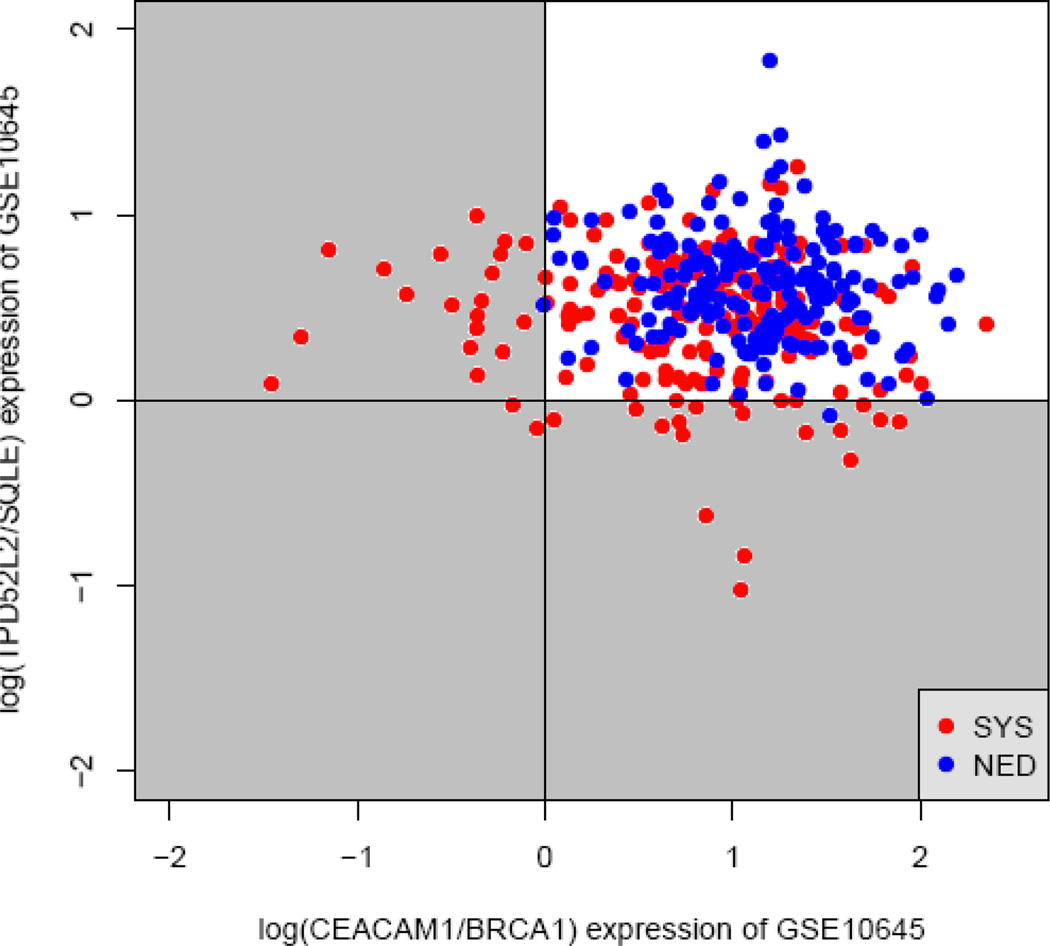
Although the specificity for predicting SYS was > 99% for the two TSPs we identified, the sensitivity was relatively low: we could make a confident prediction for only ~10% of patients. Therefore, to increase the sensitivity, we combined two TSPs, i.e., we used double TSPs (DTSPs). The decision rule for DTSP was the condition that Xi < Xj, in which Xi is the expression level of gene i and Xj is the expression level of gene j, which was met in at least one pair of genes. Figure 2 shows the scatterplots for the combination of the two pairs, TPD52L2/SQLE and CEACAM/BRCA1. The shaded quadrants in the figure represent the decision region. Combining these two pairs of genes increased the sensitivity to 21.5% without compromising the specificity, which was 99.5%.
Figure 2.
Combination of TPD52L2/SQLE and CEACAM/BRCA1 pairs for predicting prostate tumor progression. The x axis is the log-transformed expression ratio log(CEACAM/BRCA1), and the y axis is that of log(TPD52L2/SQLE). The top right quadrant is the area where the conditions CEACAM > BRCA1 and TPD52L2 > SQLE were met; the top left quadrant is where the conditions CEACAM < BRCA1 and TPD52L2 > SQLE were met; the bottom right is where the conditions TPD52L2 > SQLE and CEACAM < BRCA1 were met; and the bottom left is where the conditions TPD52L2 < SQLE and CEACAM < BRCA1 were met. According to the decision criteria of the double top-scoring pairs analysis, the three shaded quadrants comprise the decision region for predicting systemic prostate cancer progression (SYS). NED, no evidence of disease.
The statistical significance of the DTSPs was estimated by using permutation testing. Figure 3 illustrates the approach we took and shows the distribution of the conditional SYS probability for at least one (Xi < Xj) of the two gene pairs. The estimated probability value of 0.215 was not found among 10 000 permutations, suggesting that the P value for the DTSPs TPD52L2/SQLE and CEACAM/BRCA1 for predicting SYS was less than 0.0001.
Figure 3.
Graph illustrates the permutation distribution of P (X1 < X2|SYS) for the gene pairs TPD52L2/SQLE and CEACAM/BRCA1. The arrow indicates that the estimated P value (x axis), 0.215, was not found among the 10 000 permutations (y axis).
Primary tumor vs distant metastasis
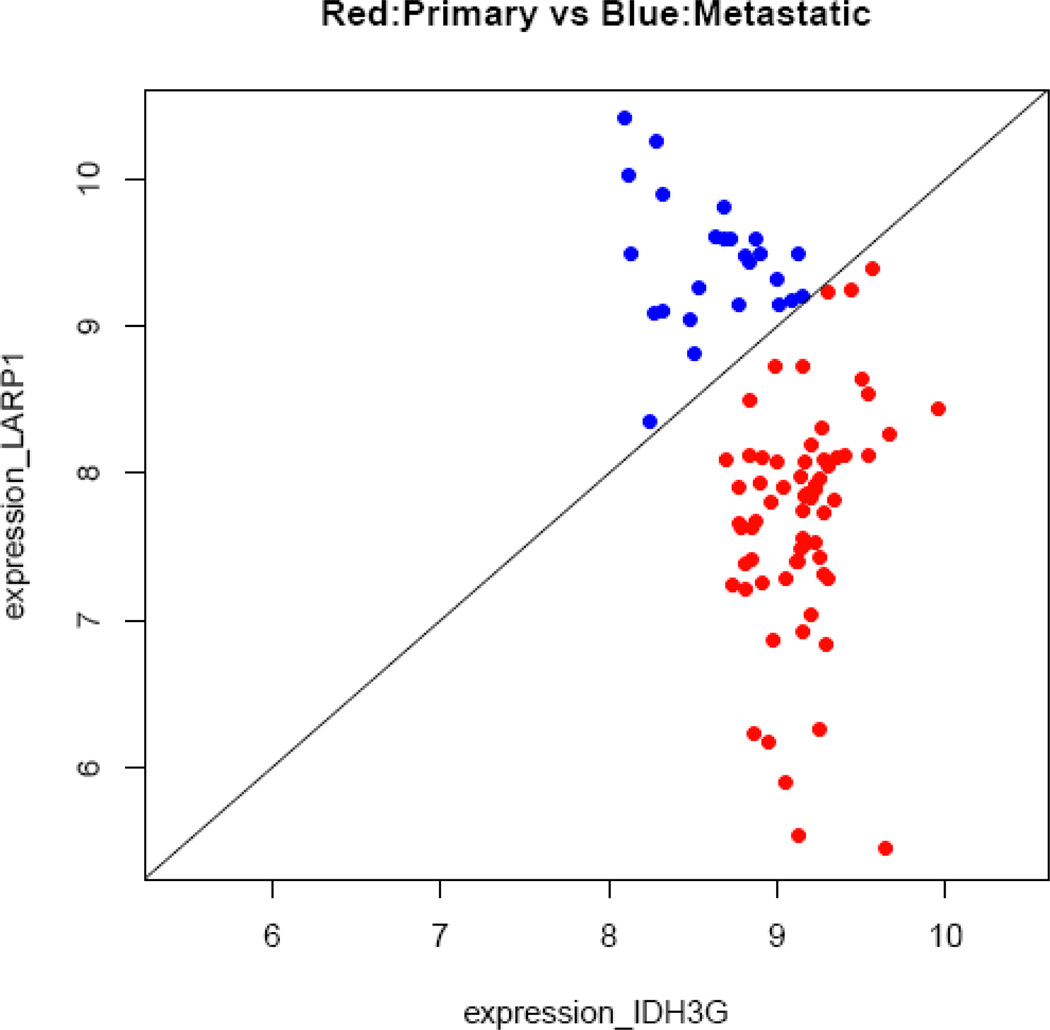
An obvious advantage of the study by Nakagawa et al.5 is its large sample size, > 200 individuals in each group. A disadvantage, though, is the limited number of genes analyzed—a total of 1028. Although those investigators selected genes that have been implicated in prostate cancer progression, it is probable that some good candidate genes were missed. The study by Chandran et al.7 was done with a smaller sample: they analyzed the gene-expression profiles of only 24 androgen-refractory metastatic samples and 64 primary tumors, but they assessed genome-wide gene expression by using Affymetrix HGU95av2, HGU95b, and HGU95c arrays (Affymetrix, Inc, Santa Clara, CA, USA). We reanalyzed the data from Chandran et al. by using all 23 572 probes from the GSE6919 dataset. The resulting TSPs for distant metastasis and primary tumors are shown in Supplementary Table S1. Three gene pairs—GRB2/ADD1, IDH3G/LARP1, and HNRNPUL1/LARP1 were perfect classifiers, with 100% sensitivity and 100% specificity. The classification plot for IDH3G/LARP1, which is representative of the three plots, is shown in Figure 4.
Figure 4.
Classification plot for discrimination between primary tumors and distant metastasis, using data from Chandran et al.5 This exemplary pair of genes, IDH3G/LARP1, has 100% sensitivity and 100% specificity. The x axis represents the log-expression value of IDH3G, and the y axis is that of LARP1.
Functional annotation
Functional annotation of the genes listed in Supplementary Table S1 was conducted using Ingenuity Pathways Analysis. We subdivided those genes into four groups by their relative expression: (1) high expression in the primary tumor, (2) low expression in the primary tumor, (3) high expression in the distant metastasis, and (4) low expression in the distant metastasis. Figure 5 shows the significant canonical pathways identified for each group of genes with top five P values. Among them, the most significant pathways include molecular mechanisms of cancer (group 2), insulin receptor signaling (group 2), integrin signaling (group 2), and regulation of actin-based motility by Rho (group 4).
Figure 5.
Ingenuity-defined canonical pathways for four groups of genes: (1) relatively low expression in the primary tumor (N = 463), (2) relatively high expression in the primary tumor (N = 341), (3) relatively low expression in the distant metastasis (N = 162), and (4) relatively high expression in the distant metastasis (N = 598). The numbers in parentheses in each quadrant are −log(P).
To find out whether the classifying pairs of genes form specific classifying pairs of pathways, we took the genes from each of the 17 different pathways listed in Figure 5 and identified the genes with which they made pairs. Then we looked for functional enrichment of those pairs of genes. We found that the relatively higher expression of genes associated with arginine and proline metabolism than that of genes associated with insulin receptor signaling predicts for primary tumor (P = 1.6E–7). We also found that the relatively lower expression of ILK-signaling genes than that of xenobiotic metabolism–signaling genes predicts for distant metastases. The genes in the arginine and proline metabolism, insulin receptor signaling, ILK signaling, and xenobiotic-metabolism pathways are listed in Supplementary Table S2.
Discussion
By applying the modified TSP approach to analyze the reported data from the largest retrospective study to date that has evaluated for an association between gene expression in primary tumor and disease progression after RRP, we identified two pairs of genes whose relative expression predicts such tumor progression. We validated their usefulness by using three independent datasets. The overall results of our analyses suggest that the TSP-based approach can be used to identify predictors of prostate cancer progression.
One major limitation associated with the TSP approach, however, is that a large number of tests can lead to a large number of false-positive results. That problem is expected to be most severe when the sample size is small and the number of probes is large, as they are in a typical microarray design. For example, if the number of samples in each of two groups is 10 and the number of probes is ~10 000, the expected number of perfect classifiers that can appear by chance is . To overcome this problem, we used the reported data from the largest published study conducted to date on prostate cancer progression.5 In that study, the SYS and NED groups both contained 213 individuals. This, in combination with a relatively small number of probes, contributed to the low P values estimated by our permutation testing (Table 2). For validation, we used data from studies that had much smaller sample sizes, but because we validated only the top 22 gene pairs, false-positive results were not a serious concern.
The two gene pairs we identified for use in predicting which patients will develop distant metastases of prostate cancer after undergoing RRP were TPD52L2/SQLE and CEACAM1/BRCA1. All four of those genes have some connection to prostate cancer. TPD52 reportedly regulates the migration of prostate tumor cells.20 SQLE is a gene from the 8q24 region that has been strongly associated with prostate cancer risk in several genome-wide association studies.21–23 Further, CEACAM1 plays an important role in the change from endothelial cells to an angiogenic phenotype during prostate cancer development,24 and BRCA1 is regulated by the androgen receptor and often mutates in prostate tumors.25
In the three studies from which we took our data for validation, the phenotypes were not exactly the same as those in the discovery set: Chandran et al.7 and Yu et al.11 both compared gene expression in distant metastases with that in primary tumors, and Tamura et al.10 used hormone-refractory vs hormone-sensitive prostate cancer. Despite the obvious differences in those analyzed phenotypes, we believe that it was reasonable to use them for validation because hormone-refractory tumors and distant metastases are likely to have a common gene-expression signature.
Whether the genes from the TSPs belong to specific pathways or have specific biologic functions is an interesting question. It was problematic to use genes from the study of Nakagawa et al.5 for functional annotation because those genes were selected on the basis of published evidence of their involvement in prostate cancer progression. Thus, we used the GSE6911 dataset and separately annotated four groups, as described in Results. We found that lower expression of genes from arginine and proline metabolism relative to the expression of genes from the insulin receptor signaling pathway may used to classify primary tumors vs distant metastases. Arginine is one of the most versatile amino acids in animal cells, serving as a precursor for the synthesis of proteins and nitric oxide (NO). NO has an important function in cancer, including prostate cancer, in that increased NO generation contributes to angiogenesis by up-regulating vascular endothelial growth factor.26 In addition, a high concentration of NO is associated with chronic inflammation and tumorigenesis. In prostate cancer, NO is involved in inhibiting androgen receptor activity and is associated with disease progression.27, 28 Our analyses also showed that the arginine and proline metabolism pathway may be coupled with the insulin receptor signaling pathway to classify primary prostate tumors. Insulin receptor signaling also plays an important role in prostate tumorigenesis. These two signaling pathways seem to be independent: analysis of the relationship between the genes from those pathways using all possible types of interactions identified no cross talk between them (data not shown). The pathways paired in predicting for distant metastasis were the xenobiotic metabolism and integrin-linked kinase (ILK) signaling pathways. Some experimental evidence exists that both of those pathways may also be associated with prostate cancer.29, 30
In summary, the TSP approach can be used to identify patients whose prostate cancer will progress after they undergo radical prostatectomy. Two gene pairs can predict which men would experience progression to the metastatic form of the disease. However, because our analysis was based on a relatively small number of genes, a larger study will be needed to identify the best predictors of disease outcome overall.
Supplementary Material
Acknowledgements
This study was supported by the David Koch Center for Applied Research in Genitourinary Cancer and by National Cancer Institute grant CA16672.
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
References
- 1.Burdick MJ, Reddy CA, Ulchaker J, Angermeier K, Altman A, Chehade N, et al. Comparison of biochemical relapse-free survival between primary Gleason score 3 and primary Gleason score 4 for biopsy Gleason score 7 prostate cancer. Int J Radiat Oncol Biol Phys. 2009;73:1439–1445. doi: 10.1016/j.ijrobp.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 2.Freedland SJ, Eastham J, Shore N. Androgen deprivation therapy and estrogen deficiency induced adverse effects in the treatment of prostate cancer. Prostate Cancer and Prostatic Diseases. 2009;12:333–338. doi: 10.1038/pcan.2009.35. [DOI] [PubMed] [Google Scholar]
- 3.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 4.Dhanasekaran SM, Dash A, Yu J, Maine IP, Laxman B, Tomlins SA, et al. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 2005;19:243–245. doi: 10.1096/fj.04-2415fje. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa T, Kollmeyer TM, Morlan BW, Anderson SK, Bergstralh EJ, Davis BJ, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3:e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 7.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 9.Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, et al. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 10.Tamura K, Furihata M, Tsunoda T, Ashida S, Takata R, Obara W, et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. doi: 10.1158/0008-5472.CAN-06-4040. [DOI] [PubMed] [Google Scholar]
- 11.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 12.Ummanni R, Teller S, Junker H, Zimmermann U, Venz S, Scharf C, et al. Altered expression of tumor protein D52 regulates apoptosis and migration of prostate cancer cells. FEBS J. 2008;275:5703–5713. doi: 10.1111/j.1742-4658.2008.06697.x. [DOI] [PubMed] [Google Scholar]
- 13.Ding GF, Xu YF, Yang ZS, Ding YL, Fang HF, Zhao HP. Coexpression of the mutated BRCA1 mRNA and p53 mRNA and its association in Chinese prostate cancer. Urol Oncol. 2009 doi: 10.1016/j.urolonc.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Schayek H, Haugk K, Sun S, True LD, Plymate SR, Werner H. Tumor suppressor BRCA1 is expressed in prostate cancer and controls insulin-like growth factor I receptor (IGF-IR) gene transcription in an androgen receptor-dependent manner. Clin Cancer Res. 2009;15:1558–1565. doi: 10.1158/1078-0432.CCR-08-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilki D, Irmak S, Oliveira-Ferrer L, Hauschild J, Miethe K, Atakaya H, et al. CEA-related cell adhesion molecule-1 is involved in angiogenic switch in prostate cancer. Oncogene. 2006;25:4965–4974. doi: 10.1038/sj.onc.1209514. [DOI] [PubMed] [Google Scholar]
- 16.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 17.Geman D, d'Avignon C, Naiman DQ, Winslow RL. Classifying gene expression profiles from pairwise mRNA comparisons. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1071. Article 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan AC, Naiman DQ, Xu L, Winslow RL, Geman D. Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics. 2005;21:3896–3904. doi: 10.1093/bioinformatics/bti631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu L, Tan AC, Naiman DQ, Geman D, Winslow RL. Robust prostate cancer marker genes emerge from direct integration of inter-study microarray data. Bioinformatics. 2005;21:3905–3911. doi: 10.1093/bioinformatics/bti647. [DOI] [PubMed] [Google Scholar]
- 20.Ummanni R, Teller S, Junker H, Zimmermann U, Venz S, Scharf C, et al. Altered expression of tumor protein D52 regulates apoptosis and migration of prostate cancer cells. FEBS J. 2008;275:5703–5713. doi: 10.1111/j.1742-4658.2008.06697.x. [DOI] [PubMed] [Google Scholar]
- 21.Cheng I, Plummer SJ, Jorgenson E, Liu X, Rybicki BA, Casey G, et al. 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooker S, Hernandez W, Chen H, Robbins C, Torres JB, Ahaghotu C, et al. Replication of prostate cancer risk loci on 8q24, 11q13, 17q12, 19q33, and Xp11 in African Americans. Prostate. 2009 doi: 10.1002/pros.21061. [DOI] [PubMed] [Google Scholar]
- 23.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 24.Antognelli C, Mearini L, Talesa VN, Giannantoni A, Mearini E. Association of CYP17, GSTP1, and PON1 polymorphisms with the risk of prostate cancer. Prostate. 2005;63:240–251. doi: 10.1002/pros.20184. [DOI] [PubMed] [Google Scholar]
- 25.Schayek H, Haugk K, Sun S, True LD, Plymate SR, Werner H. Tumor suppressor BRCA1is expressed in prostate cancer and controls insulin-like growth factor I receptor (IGF-IR) gene transcription in an androgen receptor-dependent manner. Clin Cancer Res. 2009;15:1558–1565. doi: 10.1158/1078-0432.CCR-08-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, Liu LZ, Loizidou M, Ahmed M, Charles IG. The role of nitric oxide in cancer. Cell Res. 2002;12:311–320. doi: 10.1038/sj.cr.7290133. [DOI] [PubMed] [Google Scholar]
- 27.Cronauer MV, Ince Y, Engers R, Rinnab L, Weidemann W, Suschek CV, et al. Nitric oxide-mediated inhibition of androgen receptor activity: possible implications for prostate cancer progression. Oncogene. 2007;26:1875–1884. doi: 10.1038/sj.onc.1209984. [DOI] [PubMed] [Google Scholar]
- 28.Kroncke KD, Suschek CV, Kolb-Bachofen V. Implications of inducible nitric oxide synthase expression and enzyme activity. Antioxid Redox Signal. 2000;2:585–605. doi: 10.1089/15230860050192341. [DOI] [PubMed] [Google Scholar]
- 29.Moon YJ, Wang X, Morris ME. Dietary flavonoids: effects on xenobiotic and carcinogen metabolism. Toxicol In Vitro. 2006;20:187–210. doi: 10.1016/j.tiv.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 30.Okamura M, Yamaji S, Nagashima Y, Nishikawa M, Yoshimoto N, Kido Y, et al. Prognostic value of integrin β1-ILK-pAkt signaling pathway in non–small cell lung cancer. Hum Pathol. 2007;38:1081–1091. doi: 10.1016/j.humpath.2007.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.