Abstract
Background
Metastases are the primary cause of cancer treatment failure and death, yet metastatic mechanisms remain incompletely understood.
Methods
We studied the molecular basis of head and neck cancer metastasis by transcriptionally profiling 70 samples from 27 patients—matching normal adjacent tissue, primary tumor, and cervical lymph node metastases.
Results
We identified tumor-associated expression signatures common to both primary tumors and metastases. Use of matching metastases revealed an additional 46 dysregulated genes associated solely with head and neck cancer metastasis. However, despite being metastasis-specific in our sample set, these 46 genes are concordant with genes previously discovered in primary tumors that metastasized.
Conclusions
Although our data and related studies show that most of the metastatic potential appears to be inherent to the primary tumor, they are also consistent with the notion that a limited number of additional clonal changes are necessary to yield the final metastatic cell(s), albeit in a variable temporal order.
Keywords: head and neck cancer, expression profiling, metastasis, gene signatures, oncogenomics
For most solid tumors, tumorigenesis occurs as a multistep process including both genetic and epi-genetic alterations, which together provide cancer cells with a selective growth advantage. Transformation of normal cells into malignant cells is characterized by a limited set of acquired capabilities, which are likely shared among different tumor types.1 However, metastases to distant organ sites are ultimately responsible for most cancer deaths.2 Despite much progress, important questions about metastatic progression remain to be answered.2–4 With respect to metastatic origins, 2 conflicting views have been proposed. The “rare metastatic variants” model holds that rare metastatic cells preexist within the primary tumor and subsequently progress to metastases by the process of selection.5,6 This model has recently been challenged by the results of global gene expression profiling in various cancer types. These studies have provided evidence for the independent evolution of 2 classes of primary tumors, those that are predisposed to metastasize and those that are not (“primary tumor predisposition” model).7–11 We have attempted to address the mechanism(s) of metastasis using expression profiling of genetically matched primary tumors, lymph node metastases, and adjacent normal tissue.
In this study, we focused on squamous cell carcinoma (SCC) of the head and neck (HNSCC) as a prototypic epithelial solid tumor.12–14 Several molecular alterations associated with HNSCC have been identified, but a complete understanding of the complex molecular events underlying tumori-genesis and metastasis is still lacking.13,15,16 Cervical lymph node metastases are the single most adverse independent prognostic indicator for local recurrence and/or distant metastasis.17 A reliable method to identify those tumors most likely to recur or to produce distant metastases would significantly enhance the choice of effective treatment options and, therefore, outcomes for the patients.
Several groups have attempted to predict lymph node metastases retrospectively by identifying a metastatic signature in primary tumor tissues. However, in the majority of these studies, the predictive signatures have not been compared with the expression signature that is actually present in the metastases themselves. To identify the genetic changes underlying HNSCC tumor initiation, progression, and metastasis directly, we generated comprehensive expression profiles for 70 samples, including primary tumors, genetically matched normal adjacent mucosae and genetically matched cervical lymph node metastases. Rigorous statistical analysis revealed unique expression profiles differentiating normal mucosae from primary tumor and metastatic lesions. Genetically matched primary tumor/metastasis samples (ie, from the same patients) showed highly correlated individual profiles, consistent with the notion that the metastatic potential is already encoded in the primary tumor. However, we also identified 46 genes whose dysregulated expression was specific for head and neck cancer metastasis, suggesting that the acquisition of an additional limited number of clonal changes results in the final metastatic cell(s). Findings are compared with those of other published studies, and the implications for the metastatic process in solid tumors and the utility for clinical prognostication based on gene expression in primary tumors are discussed.
PATIENTS AND METHODS
Patient Biopsy Samples
Informed consent was obtained from all patients prior to sample collection in accordance with the guidelines set by the Institutional Review Board of The Ohio State University (OSU). Biopsy samples from surgical resections were collected and banked through the OSU Head and Neck Cancer Tissue Bank. For each patient, the primary tumor, matching metastatic lymph node, and adjacent normal mucosa were collected at the same time and processed concurrently to minimize confounding clinical or technical variability. Primary tumor tissue and tissue from meta-static lymph nodes were subject to examination by a board-certified pathologist to confirm tissue histology and cellular representation. Clinically appearing tumor tissue was taken distant from the tumor margin and was macro-dissected to provide tissue that contains >70% tumor cells. Normal tissue was clinically appearing normal adjacent tissue taken at least 3 cm from the tumor margin, usually contralateral. All samples were immediately placed on ice and, after removal of portions needed for pathological diagnosis, were snap-frozen in liquid nitrogen within 30 minutes of devascularization. Snap-frozen samples were held at –80°C for long-term storage. We used samples from 5 representative anatomical locations (8 larynx, 8 tongue, 6 pharynx, 5 tonsil, 1 palate) from 27 patients. For 1 patient, we had tumor samples before (N13, P13) and after treatment (N3, P3, M3). Supplementary Table S1 summarizes the clinical and demographic features of the patients.
Sample Preparation
In all, we profiled 70 samples from 27 patients—28 primary tumors, 28 matching normal adjacent mucosae, and 14 matching lymph node metastases. RNA was extracted from snap-frozen tissue samples using the TriZol Rea-gent (Invitrogen, Carlsbad, CA) according to the manufacturer's suggestions. RNA was further purified using the RNeasy kit (Qiagen, Valencia, CA). For hybridization to the HuGeneFL and U95Av2 GeneChip arrays (Affymetrix, Santa Clara, CA) cRNA target preparation was according to the Affymetrix protocol. Thirty-five samples (12 normal adjacent mucosae, 12 primary tumors, and 11 metastatic lymph nodes from the same patients) were profiled on the HuGeneFL platform, and 35 samples (16 normal adjacent mucosae, 16 primary tumors, and 3 metastatic lymph nodes from the same patients) were profiled using the U95Av2 platform, interrogating approximately 6600 and 12,500 genes, respectively.
Data Analysis Across Platforms
All original GeneChip.cel files are available at Array Express (accession number: E-MEXP-44; www.ebi.ac.uk/ arrayexpress/). The .cel files were processed and quantified with the dChip analysis package using the Li–Wong full model (Supplementary Data S1 and S2).18,19 This choice was based on our previous finding that this method produced the most reliable results for the analysis of oligonucleotide arrays.20 All statistical analyses were performed with MATLAB (MathWorks, Natick, MA). We analyzed the 2 datasets generated on the HuGeneFL and U95Av2 platforms using standard exploratory techniques, including principal component analysis (PCA), pairwise distance matrix (PDM), and hierarchical cluster (HC) analyses. To contrast the different sample types (normal adjacent tissue, primary tumor, and lymph node metastasis), we combined the 2 datasets to generate a list of genes shared between the 2 GeneChip platforms (Supplementary Data S3).
To combine data from multiple probe sets across both platforms, we used a “pseudo” probe set approach (Supplementary Data S4).21 Briefly, the probe sequence information supplied by Affymetrix was used to identify individual probes with identical sequences present on both GeneChip platforms. Common probes were matched against the latest build of UniGene to assemble pseudo probe sets consisting of all common probes mapping to a given UniGene cluster. Because HuGeneFL and U95Av2 interrogate 20 and 16 probes per probe set/gene, respectively, the number of common probes in a pseudo probeset can vary. Therefore, computations were performed only on pseudo probe sets with at least 3 probes, interrogating a total of 4509 unique genes with 7871 pseudo probe sets. We used the robust multichip average (RMA) methodology22 for quantification of pseudo probe set intensities. RMA enables correction of spatial effects on individual chips and matching of values across chip types using quantile normalization. After combining the data using this approach, quantifications for the 2 chip types are on the same scale.
To analyze combined results for primary tumor and normal adjacent tissue samples for the genes shared between array types, we used nonpara-metric tests, which do not require the gene quantification values to be the same across array types, including a sign test that counts the number of times the gene expression in primary tumor is higher or lower than the gene expression in the paired normal adjacent tissue sample. We focused on those genes for which the sign of the difference was the same in at least 25 of 28 pairs (90%). This cutoff is conservative in that the chance of a given gene being higher in 25/28 primary tumor samples is 1.3720 e–05. Adjusting for multiple comparisons, the number of differentially expressed genes to be expected by chance is 0.1, whereas we actually see several hundred genes. A parallel argument can be made for the genes found to be higher in normal adjacent tissue than primary tumor samples. The sign test exploits the paired nature of the data by using just the paired differences as inputs. In this respect, the test is more specific than PCA, which does not take into consideration the patient-to-patient heterogeneity. We also used approaches that did not exploit the paired nature of the samples, such as Wilcoxon rank sum tests used to compare normal adjacent tissue and primary tumor samples within the same array type and then combined the p values.
For the primary tumor versus lymph node metastasis comparison, we first used a sign test that exploited the paired nature of the data similar to the test used for the normal adjacent tissue versus primary tumor comparison. For comparisons within a chip type, we also applied signed rank tests to exploit the magnitude order. Applying the sign test, we first focused on genes found to be higher in metastases in at least 12 of 14 cases (86%). With this cutoff, the chance of a given gene being higher in 12/14 lymph node metastasis samples is 6.4697 e–03. A Wilcoxon rank sum test that ignores the paired nature was also used to compare normal adjacent tissue versus lymph node metastasis samples.
RESULTS
Normal Adjacent Tissue is Distinct from Primary Tumor and Metastasis
Using nonparametric tests to identify differentially expressed genes and comparing the primary tumors with their genetically matched normal adjacent mucosae, we identified 414 probe sets, representing 345 unique genes, which showed significant gene expression differences in the same direction in at least 25 of 28 samples (p value = 1.372 × 10–25) (Supplementary Table S2). We also applied the rank filter to the pseudo probe set data and identified 338 distinct Unigene clusters (genes). Of these 338 genes, 249 were also on the 345-gene list, using the Entrez Gene ID as the common identifier (Supplementary Table S2). Among these were several genes previously implicated in HNSCC carcinogenesis, including EGFR, CCNB1, and STAT1.14,23–25
Genetically Matched Primary Tumors and Metastases Share a Common Expression Profile
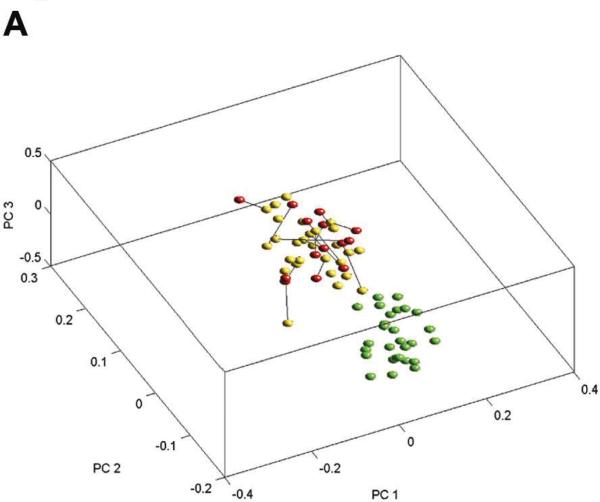
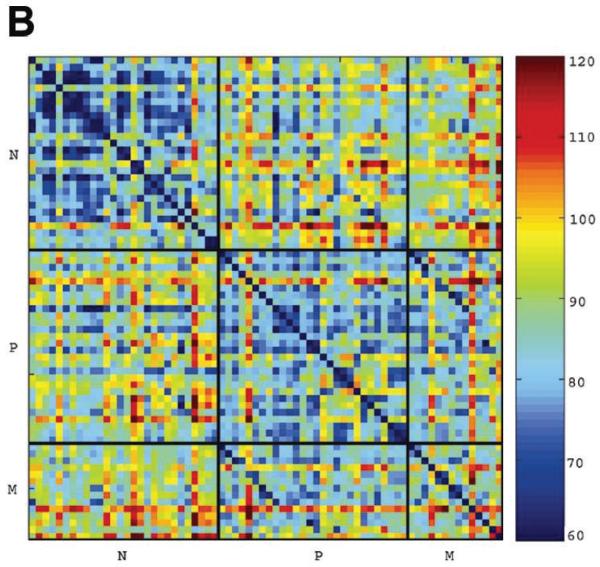
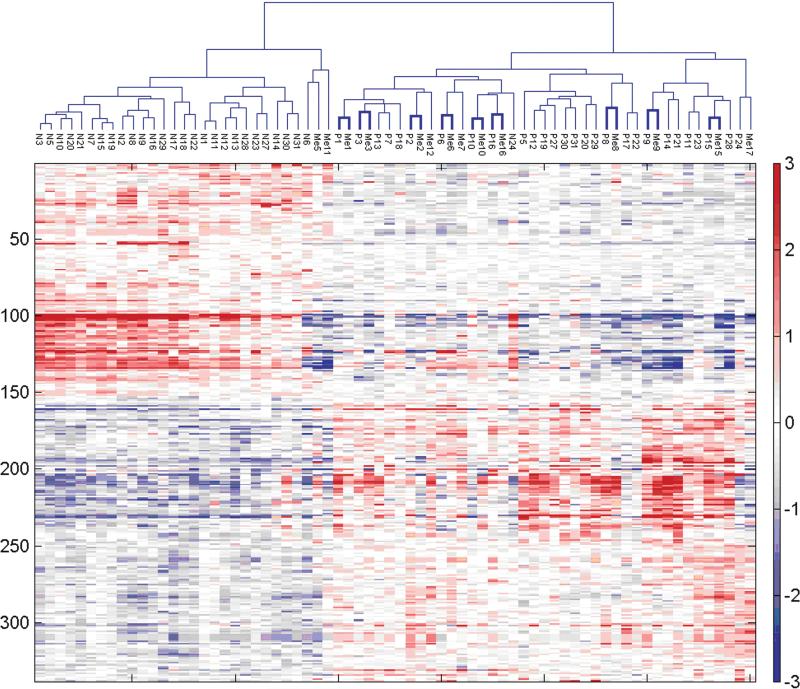
To determine the overall relationships between samples, we performed PCA (Figure 1A) and paired distance matrix (PDM, Figure 1B) analysis using all genes from the pseudo probe set approach (4509) and all 70 samples. PCA revealed specific gene signatures that distinguished primary tumors and metastases from normal adjacent tissue. PDM analysis showed more variation among the primary tumors and the metastases than among the genetically matched normal adjacent tissue. Yet, at the same time, high correlation between primary tumor and metastasis from the same patients was clearly apparent (reflected in 2 diagonal lines of blue squares comparing lymph node metastasis and primary tumor in Figure 1B). Two-way hierarchical cluster analysis based on the 338 tumor-specific genes obtained from the pseudo probe set approach confirmed the primary tumor/metastasis pairing for the majority of samples (9 of 14). Interestingly, 2 primary tumors extracted from the same patient before and after treatment (P13 and P3) clustered closely as well (Figure 2). When we performed hierarchical clustering by chip type using the 345 unique genes obtained using a rank test, sample clustering was consistent with that obtained for all samples combined; 10 of 14 primary tumors were paired with their corresponding metastases (data not shown) indicating the robustness across platforms. In fact, this degree of clustering (10/14 or 71%) is nearly as high as that obtained by comparing 2 separate biopsies of the same primary tumor by expression profile clustering (80% to 90%).26
FIGURE 1.
Analyses of expression profiles generated from the 4509 pseudo probe set genes. (A) Principal component analysis (PCA) showing all 70 samples plotted for the first 3 principal components derived using the expression matrix. The expression values for each gene were log-transformed and centered before computing the principal components. Green dots are normals, yellows are primary tumors, and reds are metastases; lines also join matched primary tumors and metastases. There is a clear separation between the genetically matching normal adjacent samples and tumor samples (primary tumors and metastases). Most metastases are closer to their corresponding primary tumors than to other metastasis profiles. (B) Pairwise distance matrix (PDM) between all 70 samples, using Euclidean distance applied to robust multichip average (RMA) quantifications of all 4509 pseudo probe set genes. The order of normals (N), primary tumors (P), and metastases (M) is the same within blocks. Distances between normals and tumors of all types are greater than distances between normals and other normals, or between tumors and other tumors. The comparison of primary tumors with metastases provided evidence for a tumor-specific signature indicated by shorter distances from primary tumors to their corresponding metastases compared with other metastases. This closer relatedness is visible as a darker blue diagonal in the primary tumor versus lymph node metastasis comparison in the upper left corner of the block of distances between primary tumor and lymph node metastasis.
FIGURE 2.
Two-way hierarchical cluster analysis of gene expression profiles generated on pseudo probe set gene expression data. Hierarchical clustering of all 70 samples (cell lines to the side) from both chip types using Euclidean distance and complete linkage applied to robust multichip average (RMA) quantifications of the 338 pseudo-probe sets passing the rank filter. Normals (N) cluster to the left, tumors to the right. Of particular interest is that most primary tumors (P) and lymph node metastases (M) from the same patient show pairing (9 of 14, 64% highlighted by thicker lines). Two primary tumor samples obtained from the same patient before and after treatment (P13 and P3) are also very near in the hierarchical samples cluster, suggesting a patient-specific tumor signature.
Identification of Metastasis-Specific Genes Through Paired Analysis
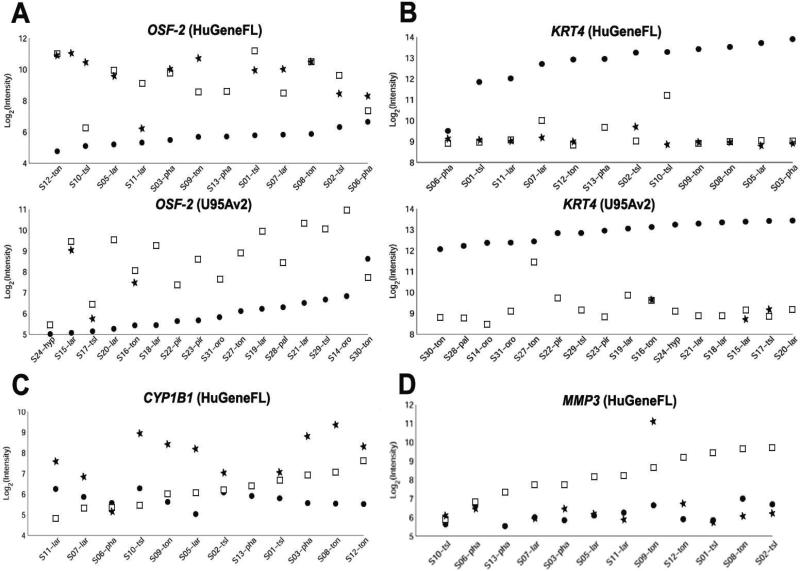
To identify genes specifically associated with the metastatic process, we compared the 14 available cervical lymph node metastases to their genetically matched primary tumors. Because of patient-to-patient heterogeneity in gene expression levels, the use of genetically unmatched primary tumors and metastases or retrospective analysis of primaries alone can be confounding. Figure 3 shows examples of variation in expression levels across individual samples for several representative genes. For each patient sample set, the general trend of dysregulation was the same. However, the range of actual levels for primary tumor and lymph node metastasis across all sets of sample overlaps, resulting in a loss of sensitivity if the paired nature of the data is ignored. To determine the power of a nonpaired relative to a paired analysis in identifying dysregulated genes, we evaluated 3 statistical tests— the Wilcoxon rank sum test, which uses a non-paired approach, and the signed rank test and sign test, both of which take advantage of the paired nature of our sample set. Both of the latter tests, which utilize the paired design, perform statistically better than the Wilcoxon rank sum test, which ignores the paired design (data not shown).
FIGURE 3.
Expression levels for specific genes in sets of genetically matching normal adjacent mucosae (●), primary tumors (□), and metastatic lymph nodes (*). The data plotted for different sample sets (S) show considerable variation in expression levels across individual sets for the same gene in the same tissue types. For each patient sample set, the general trend of dysregulation (overexpression or underexpression) is in the same direction, whereas levels across all patient sets overlap. The plots highlight the utility of the matched-based analysis approach, which improves the sensitivity of the analysis. Representative genes (A) overexpressed (OSF-2) or (B) underexpressed (KER4) in primary tumor and lymph node metastasis relative to their genetically matched normal adjacent tissue sample, in ascending order of expression in the normal tissue. Representative genes (C) overexpressed (CYP1B) or (D) under-expressed (MMP3) in lymph node metastasis relative to their genetically matched primary tumor sample, in ascending order of expression in the primary tumor.
To combine data for all 14 lymph node metastases generated on the 2 different platforms, we used the sign test to perform the paired analysis. We identified genes that showed statistically significant differences in at least 11 of 14 samples (p value = .0287) between primary tumors and matching metastases (Supplementary Table S3). Because many of the genes that were different by the sign test had absolute differences that were relatively small, an additional filter of a mean-fold change of ≥ 1.5 was applied. This resulted in a list of 46 metastasis genes—16 upregulated and 30 downregulated (Table 1). Ten of the 46 genes were represented by at least 2 probe sets. Several of these genes are known to be involved in the metastatic process of other tumor types, including ENPP2 and CXCR4, both of which were upregulated, and IL24, which was downregulated.27–29 Interestingly, of the 46 metastasis-specific genes, 9 (20%) are muscle-related genes, the highest single dysregulated functional category. This is possibly associated with the increased locomotory and invasive phenotypes of metastatic cells.30
Table 1.
HNSCC metastasis genes.
| Hu6800 probe ID | U95Av2 probe ID | Gene symbol | Title | Entrez gene | Location | Average positive fold change |
|---|---|---|---|---|---|---|
| 16 Upregulated genes | ||||||
| U03688_at | 859_at | CYP1B1 | Cytochrome P450, subfamily I (dioxin-inducible), polypeptide 1 (glaucoma 3, primary infantile) | 1545 | Chr:2p21 | 2.5564 |
| U03688_at | 40071_at | 2.5166 | ||||
| L35594_at | 41124_r_at | ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 5168 | Chr:8q24.1 | 1.6961 |
| L35594_at | 41123_s_at | 1.6668 | ||||
| J03507_at | 37394_at | C7 | Complement component 7 | 730 | Chr:5p13 | 2.1884 |
| M12529_at | 608_at | APOE | Apolipoprotein E | 348 | Chr:19q13.2 | 1.7824 |
| X13839_at | 32755_at | ACTA2 | Actin, α 2, smooth muscle, aorta | 59 | Chr:10q23.3 | 1.6111 |
| X64877_s_at | 36341_s_at | HFL3 | H factor (complement)-like 3 | 3080 | Chr:1q31–q32.1 | 1.6062 |
| Z30426_at | 37645_at | CD69 | CD69 antigen (p60, early T-cell activation antigen) | 969 | Chr:12p13–p12 | 1.5928 |
| X57809_s_at | 31459_i_at | IGL@ | Immunoglobulin lambda locus | 3535 | Chr:22q11.1–q11.2 | 1.5676 |
| X57809_s_at | 31344_at | 1.5276 | ||||
| U12535_at | 1467_at | EPS8 | Epidermal growth factor receptor pathway substrate 8 | 2059 | Chr:12q23–q24 | 1.5052 |
| U37546_s_at | 1717_s_at | BIRC3 | Baculoviral IAP repeat-containing 3 | 330 | Chr:11q22 | 1.9534 |
| U65404_at | 137_at | KLF1 | Kruppel-like factor 1 (erythroid) | 10661 | Chr:19p13.13–p13.12 | 1.9473 |
| X53331_at | 36683_at | MGP | Matrix Gla protein | 4256 | Chr:12p13.1–p12.3 | 1.8900 |
| X89101_s_at | 1441_s_at | TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 | 355 | Chr:10q24.1 | 1.6919 |
| M98539_at | 216_at | PTGDS | Prostaglandin D2 synthase 21 kDa (brain) | 5730 | Chr:9q34.2–q34.3 | 1.638 |
| U10550_at | 37279_at | GEM | GTP binding protein overexpressed in skeletal muscle | 2669 | Chr:8q13–q21 | 1.5689 |
| L06797_s_at | 649_s_at | CXCR4 | Chemokine (C–X–C motif) receptor 4 | 7852 | Chr:2q21 | 1.5148 |
| 30 Downregulated genes | ||||||
| M20642_s_at | 40157_s_at | MYL1 | Myosin, light polypeptide 1, alkali; skeletal, fast | 4632 | Chr:2q33–q34 | –4.2466 |
| M20642_s_at | 40158_r_at | –4.5890 | ||||
| V00594_s_at | 39081_at | MT2A | metallothionein 2A | 4502 | Chr:16q13 | –1.5118 |
| V00594_at | 39081_at | –1.5128 | ||||
| X90568_at | 40795_at | TTN | Titin | 7273 | Chr:2q24.3 | –3.2370 |
| S73840_at | 39101_at | MYH2 | Myosin, heavy polypeptide 2, skeletal muscle, adult | 4620 | Chr:17p13.1 | –2.9306 |
| X66141_at | 36640_at | MYL2 | Myosin, light polypeptide 2, regulatory, cardiac, slow | 4633 | Chr:12q23–q24.3 | –2.0088 |
| M24069_at | 39839_at | CSDA | Cold shock domain protein A | 8531 | Chr:12p13.1 | –1.5549 |
| M33772_s_at | 41748_at | TNNC2 | Troponin C2, fast | 7125 | Chr:20q12–q13.11 | –1.5510 |
| X53586_rna1_at | 33411_g_at | ITGA6 | Integrin, α 6 | 3655 | Chr:2q31.1 | –1.5346 |
| X53586_rna1_at | 33410_at | –1.5240 | ||||
| X64177_f_at | 39594_f_at | MT1H | Metallothionein 1H | 4496 | Chr:16q13 | –1.5339 |
| U35637_s_at | 38461_at | NEB | Nebulin | 4703 | Chr:2q22 | –2.9512 |
| X53961_at | 37149_s_at | LTF | Lactotransferrin | 4057 | Chr:3q21–q23 | –2.7714 |
| X51441_at | 33272_at | SAA1 | Serum amyloid A1 | 6288 | Chr:11p15.1 | –2.5386 |
| X51441_s_at | 33272_at | –2.3024 | ||||
| X05232_at | 437_at | MMP3 | Matrix metalloproteinase 3 (stromelysin 1, progelatinase) | 4314 | Chr:11q22.3 | –2.3736 |
| M69225_at | 40304_at | BPAG1 | Bullous pemphigoid antigen 1, 230/240kDa | 667 | Chr:6p12–p11 | –2.2511 |
| M69225_at | 32782_r_at | –2.0523 | ||||
| L20861_at | 31862_at | WNT5A | Wingless-type MMTV integration site family, member 5A | 7474 | Chr:3p21–p14 | –2.2151 |
| L20861_at | 1669_at | –1.5605 | ||||
| J00073_at | 39063_at | ACTC | Actin, alpha, cardiac muscle | 70 | Chr:15q11–q14 | –2.0703 |
| Y00787_s_at | 35372_r_at | IL8 | Interleukin 8 | 3576 | Chr:4q13–q21 | –2.0136 |
| Y07755_at | 35726_at | S100A2 | S100 calcium binding protein A2 | 6273 | Chr:1q21 | –2.0042 |
| L24564_at | 39528_at | RRAD | Ras-related associated with diabetes | 6236 | Chr:16q22 | –1.9363 |
| X06661_at | 36570_at | CALB1 | Calbindin 1, 28 kDa | 793 | Chr:8q21.3–q22.1 | –1.8359 |
| M91669_s_at | 41618_at | COL17A1 | Collagen, type XVII, alpha 1 | 1308 | Chr:10q24.3 | –1.8089 |
| U02081_at | 33894_at | NET1 | Neuroepithelial cell transforming gene 1 | 10276 | Chr:10p15 | –1.7926 |
| X06825_at | 32312_at | TPM2 | Tropomyosin 2 (beta) | 7169 | Chr:9p13.2–p13.1 | –1.7862 |
| X06825_at | 32314_g_at | –1.7145 | ||||
| U16261_at | 41848_f_at | IL24 | Interleukin 24 | 11009 | Chr:1q32 | –1.7819 |
| M93056_at | 33305_at | SERPINB1 | Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 1 | 1992 | Chr:6p25 | –1.7538 |
| M11433_at | 38634_at | RBP1 | Retinol binding protein 1, cellular | 5947 | Chr:3q23 | –1.7342 |
| X58377_at | 35464_at | IL11 | Interleukin 11 | 3589 | Chr:19q13.3–q13.4 | –1.5896 |
| U41060_at | 1798_at | SLC39A6 | Solute carrier family 39 (zinc transporter), member 6 | 25800 | Chr:18q12.1 | –1.5377 |
| M80244_at | 32186_at | SLC7A5 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 8140 | Chr:16q24.3 | –1.5284 |
| U89916_at | 39579_at | CLDN10 | Claudin 10 | 9071 | Chr:13q31–q34 | –1.5012 |
Comparison of Head and Neck Cancer Metastasis-Specific Genes with Other Published Head and Neck Cancer Metastasis Signatures
A variety of approaches have been undertaken to define a metastasis signature in HNSCC. Most groups have approached the question using primary tumors only, along with supervised analyses to identify genes that are differentially expressed in primary tumors associated with lymph node metastases, versus those without lymph node metastases, versus those without lymph node metastases.31–35 Another approach compared immortal with mortal cell lines established from HNSCC samples.36 A third approach, similar to ours, compared matched primary tumors to metastases and identified a single differentially expressed gene, MTA1.37 The specific details of these studies are compared in Supplementary Table S4. Comparing the resulting lists of differentially expressed genes from these reports with each other and with our 46-gene list yielded relatively few overlapping genes (Supplementary Figure S2). However, when cellular processes and pathways, as opposed to individual gene products, were compared between all of these studies, including ours, the results were strikingly similar. Using Ingenuity Pathway Analysis (Ingenuity Systems; www.ingenuity.com), genes involved in cellular processes including cancer, cell cycle, and cell-to-cell signaling were disproportionately overrepresented on gene lists from all 7 studies. Their degree of overrepresentation was generally similar, as indicated by the similarity in the significance level. Results of the comparison are shown in Supplementary Figure S1.
DISCUSSION
Although the primary risk factors for HNSCC (tobacco and alcohol consumption) are well recognized, and considerable progress has been made, understanding of the molecular events and mechanisms underlying the multistep carcinogenesis process in HNSCC is still incomplete and prognosis continues to be poor. The identification of genes and pathways involved in tumor initiation, progression and especially metastasis is therefore an important goal. Several groups have published distinct gene expression signatures associated with the presence of lymph node or distant metastases.31–35,38 We profiled genetically matched uninvolved adjacent tissues, primary tumors and metastatic lymph nodes from a representative set of HNSCC patients. To our knowledge, this is the largest data set yet utilizing such genetically matched samples.39–41
Genes Associated with Tumor Progression
Genes that showed concordant levels of dysregulation in the primary tumors and metastases relative to their matched normal tissues are likely involved in tumor initiation and maintenance. Tumor progression in this sample set appeared to be particularly associated with the dysregulation of genes involved in cell adhesion, cell-cycle regulation, and remodeling of the extracellular matrix (ECM) and cytoskeleton. Some of the dysregulated genes and cellular pathways in HNSCC are shared with other solid tumors, suggesting that there are a limited number of cellular functions and pathways contributing directly to tumor initiation and progression. The identification of genes previously implicated in other cancers, and HNSCC specifically, confirms the validity of our approach. In addition, we identified several genes not previously associated with HNSCC tumorigenesis, some of which constitute potential HNSCC bio-markers.
Primary Tumors are Very Similar to Their Corresponding Lymph Node Metastasis
SCC of the head and neck can be considered a prototypic solid tumor of epithelial origin that results from the accumulation of multiple genetic and epigenetic alterations.15,42 As with many other solid tumors, the presence of cervical lymph node metastasis is considered the single most important indicator for local recurrence and/or distant metastasis.17 The remarkable similarity that we observed between primary tumor and lymph node metastasis of the same patient, reflected in the paired clustering for the majority of samples, is consistent with the notion that the metastatic potential is already encoded in the bulk of the primary tumor. This finding tends to contradict the “rare metastatic variants” model and supports the “primary tumor predisposition” model.
Genes Associated with Metastasis
Despite the high similarity between primary tumors and matched metastases, incorporating genetically matched samples in our study design enabled us to overcome inter-patient tumor heterogeneity and identify 46 genes that were significantly differentially expressed in lymph node metastases of HNSCC patients. Some of the 46 genes, and the cellular pathways in which their protein products function, have been implicated in tumor progression and metastasis in previous studies. These include genes that individually have been correlated with metastatic behavior and organ-specific metastasis in other cancers of epithelial origin. Several of these genes (ENPP2, CXCR4, LTF, S100A2, and IL24) have been identified as mechanistically important in the cascade of events that drives metastasis, and many of the other genes on our list have been implicated in tumorigenesis.43–47 However, most of them were previously unrecognized as genes contributing to the development of HNSCC metastasis.
Whether our newly identified metastasis-associated genes contribute functionally to metastatic progression, not only in HNSCC but in other cancers as well, will require further study. Nonetheless, our findings are encouraging, because they suggest the involvement of a limited set of genes and pathways that might be common therapeutic targets in different solid cancers. We were able to identify these genes because of our paired design, which enabled us to overcome inter-tumor heterogeneity, a confounding factor in the analysis of unmatched samples.
Comparison with Previously Published Studies
Given that several HNSCC metastatic signatures have now been published,31–36 it appeared worthwhile to examine the studies for overlap as a means to derive information that may have been missed in the analysis of any single study in isolation. These HNSCC metastatic signatures were derived using several different profiling platforms and analytical approaches (ie, supervised analysis of primary tumor signatures based on association with presence of lymph node metastases, comparison of immortal versus mortal HNSCC samples, and in our case, comparison of metastases with paired primary tumors). These diverse study designs can explain some of the discrepancies seen in the direction of gene expression changes and strengthen the significance of the genes that are independently discovered in multiple studies (Supplementary Figure S2). Similarities seen between all 7 studies when pathways, rather than individual gene products, were compared indicates that higher-level systems analysis will likely be essential for the understanding and development of clinical applications from primary gene expression data. Interestingly, the metastasis signatures for different solid tumors10 and ours for HNSCC, showed a similar enrichment of muscle-related and metallothionein genes. The fact that tumor progression is reflected in the remodeling of the ECM is consistent with the observations that epithelial–mesenchymal interactions are critical factors of tumor cell behavior.48–50 The theme that is emerging is one of loss of cell adhesion and acquisition of increased cell motility, which endows the cell with the ability to migrate, invade, and home to specific organs—in the case of HNSCC, first lung then bone.
Progression to Metastasis in HNSCC
Clearly, head and neck cancer tumorigenesis is a multistep process that results from the accumulation of multiple genetic and epigenetic alterations. Together, these events provide the tumor cell with a selective growth advantage, which drives the development of the primary tumor and metastasis. The striking similarity of the expression profiles between primary tumors and their genetically matched metastases, and the identification of the 46 metastasis-specific genes together support the hypothesis that, compared with the development of the primary tumor, fewer additional clonal changes are necessary to yield metastatic cells. Expression profiling of cancer cell lines selected in vivo for their metastatic potential also indicated that changes in only a limited number of genes and cellular functions underlie the acquisition of the metastatic phenotype.44,51,52 Similarity between our metastatic signature (derived from metastases themselves) and other metastatic signatures (derived from primary tumors alone) suggests that the order of these changes may be variable, because our study found these expression changes only in metastases, but others identified them as already present in the primary tumor. This “variable order” metastasis progression model would be in contrast to current models of cancer progression, in which events in tumorigenesis are proposed to occur in the same sequence every time a tumor arises.
Further studies, especially using multiple primary tumors and metastases from patients with different clinical histories, will be needed to clarify the role of specific genes and their cellular pathways to the overall metastatic potential. Our results on lymph node metastases should also be extended to distant metastases to identify genes responsible for the establishment and maintenance of metastasis.
In conclusion, our data provide experimental evidence for the model recently proposed by Hynes53 that postulates the development of meta-static variants from a metastasis-prone primary tumor cell population. Moreover, our data extend this model by indicating that the sum of the changes in pathways and processes, rather than the specific genes or the order in which changes occur, determines the final metastatic outcome.
Supplementary Material
Acknowledgments
The authors thank Erin Hertlein, Kimmo Virtaneva, James W. Clarke, and the OSU Human Cancer Genetics Program Microarray Unit for help with data generation and organization.
Contract grant sponsor: National Cancer Institute; contract grant number: P30 CA16058; contract grant sponsor: American Cancer Society; contract grant number: IRG-98-278-01; contract grant sponsor: DoD; contract grant number: W81XWH-05-2-0027; contract grant sponsors: OSU-Comprehensive Cancer Center (OSU-CCC)/Cancer Hospital and Research Institute Research Fund; OSU-CCC Molecular Biology and Cancer Genetics Program; Kleberg Foundation; State of Texas Tobacco Research Funds; Texas Tobacco Settlement Funds.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 3.Gupta PB, Mani S, Yang J, Hartwell K, Weinberg RA. The evolving portrait of cancer metastasis. Cold Spring Harb Symp Quant Biol. 2005;70:291–297. doi: 10.1101/sqb.2005.70.033. [DOI] [PubMed] [Google Scholar]
- 4.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 5.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 6.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 7.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 8.van't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 10.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 11.Buckhaults P. Gene expression determinants of clinical outcome. Curr Opin Oncol. 2006;18:57–61. doi: 10.1097/01.cco.0000198022.76476.b2. [DOI] [PubMed] [Google Scholar]
- 12.Califano J, van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 13.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Ordonez B, Beauchemin M, Jordan RC. Molecular biology of squamous cell carcinoma of the head and neck. J Clin Pathol. 2006;59:445–453. doi: 10.1136/jcp.2003.007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cells. 2004;5:311–316. doi: 10.1016/s1535-6108(04)00090-x. [DOI] [PubMed] [Google Scholar]
- 16.Lothaire P, de Azambuja E, Dequanter D, et al. Molecular markers of head and neck squamous cell carcinoma: promising signs in need of prospective evaluation. Head Neck. 2006;28:256–269. doi: 10.1002/hed.20326. [DOI] [PubMed] [Google Scholar]
- 17.Ferlito A, Rinaldo A, Devaney KO, et al. Prognostic significance of microscopic and macroscopic extracapsular spread from metastatic tumor in the cervical lymph nodes. Oral Oncol. 2002;38:747–751. doi: 10.1016/s1368-8375(02)00052-0. [DOI] [PubMed] [Google Scholar]
- 18.Li C, Wong HW. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:1–11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemon WJ, Palatini JJ, Krahe R, Wright FA. Theoretical and experimental comparisons of gene expression indexes for oligonucleotide arrays. Bioinformatics. 2002;18:1470–1476. doi: 10.1093/bioinformatics/18.11.1470. [DOI] [PubMed] [Google Scholar]
- 21.Morris JS, Yin G, Baggerly KA, Wu C, Zhang L. Pooling information across different studies and oligonucleotide chip types to identify prognostic genes for lung cancer. In: Shoemaker JS, Lin JS, editors. Methods of microarray data analysis IV. Springer-Verlag; New York: 2005. pp. 51–66. [Google Scholar]
- 22.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 23.Dong Y, Sui L, Watanabe Y, Sugimoto K, Tokuda M. Clinical relevance of cyclin B1 overexpression in laryngeal squamous cell carcinoma. Cancer Lett. 2002;177:13–19. doi: 10.1016/s0304-3835(01)00770-4. [DOI] [PubMed] [Google Scholar]
- 24.Hassan KA, Ang KK, El-Naggar AK, et al. Cyclin B1 overexpression and resistance to radiotherapy in head and neck squamous cell carcinoma. Cancer Res. 2002;62:6414–6417. [PubMed] [Google Scholar]
- 25.Xi S, Dyer KF, Kimak M, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcino-genesis. J Natl Cancer Inst. 2006;98:181–189. doi: 10.1093/jnci/djj020. [DOI] [PubMed] [Google Scholar]
- 26.Pramana J, Pimentel N, Hofland I, et al. Heterogeneity of gene expression profiles in head and neck cancer. Head Neck. 2007:1083–1089. doi: 10.1002/hed.20621. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Medicine. 2006;3:2229–2243. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramesh R, Ito I, Gopalan B, Saito Y, Mhashilkar AM, Chada S. Ectopic production of MDA-7/IL-24 inhibits invasion and migration of human lung cancer cells. Mol Ther. 2004;9:510–518. doi: 10.1016/j.ymthe.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Zlotnik A. Chemokines and cancer. Int J Cancer. 2006;119:2026–2029. doi: 10.1002/ijc.22024. [DOI] [PubMed] [Google Scholar]
- 30.Levine MD, Liotta LA, Stracke ML. Stimulation and regulation of tumor cell motility in invasion and metastasis. EXS. 1995;74:157–179. doi: 10.1007/978-3-0348-9070-0_8. [DOI] [PubMed] [Google Scholar]
- 31.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cells. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 32.Cromer A, Carles A, Millon R, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 33.Roepman P, Wessels LF, Kettelarij N, et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet. 2005;37:182–186. doi: 10.1038/ng1502. [DOI] [PubMed] [Google Scholar]
- 34.O'Donnell RK, Kupferman M, Wei SJ, et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24:1244–1251. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- 35.Schmalbach CE, Chepeha DB, Giordano TJ, et al. Molecular profiling and the identification of genes associated with metastatic oral cavity/pharynx squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130:295–302. doi: 10.1001/archotol.130.3.295. [DOI] [PubMed] [Google Scholar]
- 36.Hunter KD, Thurlow JK, Fleming J, et al. Divergent routes to oral cancer. Cancer Res. 2006;66:7405–7413. doi: 10.1158/0008-5472.CAN-06-0186. [DOI] [PubMed] [Google Scholar]
- 37.Roepman P, de Jager A, Groot Koerkamp MJ, Kummer JA, Slootweg PJ, Holstege FC. Maintenance of head and neck tumor gene expression profiles upon lymph node metastasis. Cancer Res. 2006;66:11110–11114. doi: 10.1158/0008-5472.CAN-06-3161. [DOI] [PubMed] [Google Scholar]
- 38.Giri U, Ashorn CL, Ramdas L, et al. Molecular signatures associated with clinical outcome in patients with high-risk head-and-neck squamous cell carcinoma treated by surgery and radiation. Int J Radiat Oncol Biol Phys. 2006;64:670–677. doi: 10.1016/j.ijrobp.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 39.Sotiriou C, Lothaire P, Dequanter D, Cardoso F, Awada A. Molecular profiling of head and neck tumors. Curr Opin Oncol. 2004;16:211–214. doi: 10.1097/00001622-200405000-00003. [DOI] [PubMed] [Google Scholar]
- 40.Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci U S A. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belbin TJ, Singh B, Smith RV, et al. Molecular profiling of tumor progression in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2005;131:10–18. doi: 10.1001/archotol.131.1.10. [DOI] [PubMed] [Google Scholar]
- 42.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 43.Nam SW, Clair T, Campo CK, Lee HY, Liotta LA, Stracke ML. Autotaxin (ATX), a potent tumor motogen, augments invasive and metastatic potential of ras-transformed cells. Oncogene. 2000;19:241–247. doi: 10.1038/sj.onc.1203263. [DOI] [PubMed] [Google Scholar]
- 44.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 45.Tsuda H, Sekine K, Fujita K, Ligo M. Cancer prevention by bovine lactoferrin and underlying mechanisms—a review of experimental and clinical studies. Biochem Cell Biol. 2002;80:131–136. doi: 10.1139/o01-239. [DOI] [PubMed] [Google Scholar]
- 46.Kyriazanos ID, Tachibana M, Dhar DK, et al. Expression and prognostic significance of S100A2 protein in squamous cell carcinoma of the esophagus. Oncol Rep. 2002;9:503–510. [PubMed] [Google Scholar]
- 47.Ellerhorst JA, Prieto VG, Ekmekcioglu S, et al. Loss of MDA-7 expression with progression of melanoma. J Clin Oncol. 2002;20:1069–1074. doi: 10.1200/JCO.2002.20.4.1069. [DOI] [PubMed] [Google Scholar]
- 48.Rubin H. Selected cell and selective microenvironment in neoplastic development. Cancer Res. 2001;61:799–807. [PubMed] [Google Scholar]
- 49.Matrisian LM, Cunha GR, Mohla S. Epithelial-stromal interactions and tumor progression: meeting summary and future directions. Cancer Res. 2001;61:3844–3846. [PubMed] [Google Scholar]
- 50.Liotta LA, Kohn EC. The microenvironment of the tumour–host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 51.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z, Zhang K, Zhang X, et al. Comparison of gene expression between metastatic derivatives and their poorly metastatic parental cells implicates crucial tumor-environment interaction in metastasis of head and neck squamous cell carcinoma. Clin Exp Metastasis. 2003;20:335–342. doi: 10.1023/a:1024039802134. [DOI] [PubMed] [Google Scholar]
- 53.Hynes RO. Metastatic potential: generic predisposition of the primary tumor or rare, metastatic variants-or both? Cell. 2003;113:821–823. doi: 10.1016/s0092-8674(03)00468-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.