Abstract
To understand the underlying mechanisms of bacterial infections, researchers have for long addressed the molecular interactions occurring when the bacterium interacts with host target cells. In these studies, primarily based on in vitro systems, molecular details have been revealed along with increased knowledge regarding the general infection process. With the recent advancements in in vivo imaging techniques, we are now in a position to bridge a transition from classical minimalistic in vitro approaches to allow infections to be studied in its native complexity—the live organ. Techniques such as multiphoton microscopy (MPM) allow cellular-level visualization of the dynamic infection process in real time within the living host. Studies in which all interplaying factors, such as the influences of the immune, lymphatic, and vascular systems can be accounted for, are likely to provide new insights to our current understanding of the infection process. MPM imaging becomes extra powerful when combined with advanced surgical procedure, allowing studies of the illusive early hours of infection. In this chapter, our intention is to provide a general view on how to design and carry out intravital imaging of a bacterial infection. While exemplifying this using a spatiotemporally well-controlled uropathogenic Escherichia coli (UPEC) infection in rat kidneys, we hope to provide the reader with general considerations that can be adapted to other bacterial infections in organs other than the kidney.
1. Introduction
Interactions occurring when pathogens encounter their host are characterized by a dynamic and complex interplay. Classical approaches to probing the process of infection has relied heavily on in vitro cell culture models. While this method has yielded substantial valuable findings, and has essentially molded our current understanding of host–pathogen interactions, the native state of infection in vivo is inherently much more complex than what any in vitro model can simulate. Host tissues are heterogeneous organizations of a diverse population of cell types whose composition changes during the time-course of infection. As tissue homeostasis is dynamically altered during infection, bacteria must adapt simultaneously to cope with the changing microenvironment. The physiology of the bacterial population is thus likely to be altered during the infection process. With these dynamic, multifactorial processes in mind, the infection process is preferentially studied in real-time within the live animal. Recent advances in imaging platforms and data analysis tools have been beneficial in greatly improving the data quality of live visualization of an infection as it progresses within the organ of an animal (Dunn et al., 2002, 2007; Månsson et al., 2007a,b). Noninvasive multiphoton imaging is one such tool.
MPM is a noninvasive technique developed by biophysicists (Goeppert-Mayer, 1931; Weissman et al., 2007) and adapted by biologists to study dynamic processes within living organs. In essence, MPM enables both high resolution and high-sensitivity fluorescence microscopy of intact tissues over time. Since its initial application by Denk et al. (1990), it has been adapted to explore a wide variety of research fields, including studies of Ca2+ fluctuations in individual synapses (Svoboda and Yasuda, 2006; Tian et al., 2006), the role of astrocytes in the brain (Tian et al., 2006), tumor vascularization (Brown et al., 2001), embryonic development (Squirrell et al., 1999), kidney physiology (Molitoris and Sandoval, 2005), and bacterial infections (Månsson et al., 2007a,b; Melican and Richter-Dahlfors, 2009a,b).
The following sections will cover both concepts and considerations behind the design of a model for real-time imaging of bacterial infections in vivo. Host–pathogen interactions when in vivo can be highly unpredictable. Depending on the anatomy and physiology of the organ, along with its relation to the vasculature and other organs, there will be a wide variation in the dynamics of pathogen clearance, persistence, or dissemination. That being so, all possible tissue compartments that may be involved in the host response or that the pathogen may access to have to be sampled. In our in vivo model, a wealth of information at a range of complexity is generated. This implies that one might not fully comprehend the significance of the data when it is encountered. Therefore, clear definition of the pathology of disease and physiology of the organ of interest must be fed into the design of the selected animal model.
Our group has primarily employed intravital imaging on UPEC infection of the upper urinary tract in live rats. The kidney is an organ containing a good variety of microenvironments for the study of host–pathogen interactions and is, in addition, well suited for surgical procedures and MPM studies using an inverted microscope setup (Dunn et al., 2003, 2007). The structure of the nephron, such as the close proximity between the tubular epithelium and peritubular capillaries, allows the bacterial infection to be initiated in the mucosal lining, while monitoring the onset of the inflammatory response. The dynamic nature of the model enables us to perform kinetic studies of bacterial colonization and host responses, as well as monitor any accompanying effects infection may have on vascular and renal flow. Thus, this model can provide new data not possible to achieve in static end-point analysis of infected tissues (Melican and Richter-Dahlfors, 2009a,b).
2. Selection of Reporters for a Complex Site of Infection
To follow the progression of infection in vivo, bacterial and host components need to be visible under the microscope. Under fluorescence-based imaging, the ironclad rule is that only fluorophore-associated molecules are visible. Fluorophores with appropriate spectral features must thus be selected and applied to the experiment using the appropriate vectors.
2.1. Visualizing bacteria
Bacteria are commonly visualized by methods such as specific dyes, antibody-based systems, and genetic approaches. While all these work well for in vitro studies, the live setting provides further challenges. There is a shortage of dyes for prokaryotes to be used in intravital studies. Use of antibodies is also limited due to the risk of them being immunogenic and the difficulty to predict their biodistribution after systemic delivery. As an alternative, bacteria can be genetically engineered to express a fluorescent protein, from either plasmid-borne or chromosomally inserted genes. Plasmid-borne reporter systems while feasible in vitro are not fully compatible with in vivo models, as the necessary absence of antibiotics in the animal may lead to a variation in plasmid copy number. By inserting a single-copy gene onto the bacterial chromosome, drawbacks of plasmid-based systems can be circumvented, including the risk of expressing unnecessarily high levels of GFP that may exert cytotoxic effects. The chromosomal site for gene insertion must be selected with care to avoid any polar effects. Similarly, the gene must be cloned under a promoter whose activity is not affected by the changing tissue microenvironment. Thus, promoters regulating ribosomal protein gene expression should be avoided as bacteria rapidly adjust ribosome numbers according to nutrient availability.
2.1.1. Required materials
Bacterial strain
UPEC strain CFT073, serotype O6:K2:H1
Oligonucleotides constructing UPEC strains with chromosomal encoding gfp
PLtetO-1: TTCGTCTTC ACCTCGAGTC CCTATCAGTG ATAGAGATTG ACATCCCTAT CAGTGATAGA GATACTGAGC ACATCAGCAG GACGCACTGA CGAATTCATT AAAGAGGAG AAAGGTACCC ATGGG
Ptet_F1: CATGCGACCC GGGTTCGTCT TCACCTCGAG TCC
Ptet_R1: GTCGCCATT CTAGACCCAT GGGTACCTTT CTCCT
P1cob: gtgacgggag gcgcacggag cgggaagagt cgccacgcaG CCTGGGGTAA TGACTCTCTA GC
P2cob: cagcgtatc gcccgtttgc ccgcccagcg tacgtttgag aCGTCATTTC TGCCATTCAT CC
Kits for genetic manipulation
Techniques and kits for genetic manipulation of bacteria vary significantly between research groups and are thus not listed here. This section will only give a brief overview of the major steps to generate a bacterium with chromosomally encoded GFP+.
2.1.2. Construction of UPEC strains with chromosomal encoding of GFP+
GFP+-producing bacteria are constructed by site-specific integration of the gene-encoding enhanced green fluorescent protein (gfp+) into the cobS site on the chromosome of uropathogenic Escherichia coli (UPEC) strain CFT073 according to procedures described by Hautefort et al. (2003). The tetracycline promoter PLtetO-1 was used in the absence of the Tet repressor, resulting in constitutive activation of the promoter.
To begin cloning, amplify the tetracycline promoter PLtetO-1 using primers Ptet_R1 and Ptet_F1 by PCR. Digest both amplified PLtetO-1 and the gfp+-cmr-containing plasmid pZEP08 (Hautefort et al., 2003) with Xbal and Smal (Sigma, Germany). Purify and ligate the digestion products to give the plasmid pKM001 (PLtetO-1-gfp+-cmr). Amplify pKM001 (PLtetO-1-gfp+-cmr) by PCR amplification with promoters P1cob and P2cob. These promoters contain a 40 nt 5′ region homologous to the cobS locus at 44 min on the CFT073 chromosome, and a 21–23 nt sequence-binding site to the gfp+-cmr-containing plasmid. Perform chromosomal recombination of PCR products by λ Red recombination (Datsenko and Wanner, 2000), deleting 1143 bp (positions 2319458–2320600). Verify accurate clones by sequencing. Perform a bacterial growth assay to ensure that gfp+ is successfully expressed and does not alter bacterial physiology.
Before applying a GFP+-producing bacterial strain to the intravital model, it would be prudent to first conduct a preliminary test of gfp+ expression in vivo. This is because bacteria will experience different microenvironments during the infection, and it is important to ensure that the GFP+ promoter activity is independent of bacterial location in the tissue. For UPEC infections, perform a traditional, ascending UPEC infection (see Section 3.4.3), sacrifice the rat 4 days postinfection, and isolate the kidneys. After fixation, use cryosections to compare the proportion of GFP+-expressing UPEC in the tissue against the total bacteria population as detected by fluorophore-conjugated (Rhodamin or Texas red) antibody staining for lipopolysaccharide (LPS). Optimally, there should be a 100% overlap.
2.2. Visualizing tissues
Tissue autofluorescence is often regarded a problem during microscopy as it lowers the signal-to-noise ratio. During live imaging, however, it is often considered advantageous as it facilitates orientation in the tissue (Dunn et al., 2003, 2007). In addition, fluorescent probes are used to increase information obtained from the tissue. When delivered, probes are, however, constantly cycled between different bodily compartments and may not necessarily appear in the tissue of interest. Biodistribution is hence an important factor when administering probes during live imaging. Dextran is a versatile glucan polysaccharide with uses ranging from biomaterials to drugs and probes. It can be synthesized and purified to produce samples of a specific molecular mass, and it rarely exhibits toxicity at functional doses. Creative use of fluorophore-conjugated dextran allows the visualization of different tissue compartments. Store all fluorophore-conjugated dextrans wrapped in foil ≤ 1 month at 4 °C.
2.2.1. Required materials
Fluorophore-conjugated dextrans
10 kDa fluorophore-conjugated dextran in 0.9% sterile saline (20 mg/ml): A bulk probe that, when injected intravenously, is freely filtered by the glomerulus and is somewhat permeant in the vasculature. Used for assays of glomerular permeability, proximal tubule endocytosis, and vascular permeability.
500 kDa rhodamine-conjugated dextran in 0.9% sterile saline (8 mg/ml): A bulk probe that, when injected intravenously, is not filtered by the kidney but is retained in the vasculature. Used for assays of glomerular permeability, vascular flow, and vascular permeability.
Reagents and disposables
Sterile 0.9% saline solution
10,000 MWCO membrane
Surgical syringe
2.2.2. Visualizing filtrate flow in the nephron
Contents of blood are actively filtered by the glomerulus into the luminal compartment of the nephron. To visualize this, slowly inject 10 kDa fluorophore-conjugated dextran (20 mg/ml) from a surgical syringe via an access line in the femoral or jugular vein. When deciding which particular fluorophore to use, one must consider whether other fluorophores will be used in combination. With the rat positioned on the MPM stage for imaging (see Section 3.3), systemically introduced dextran first appears as a flash within the vasculature before it is rapidly filtered by the glomerulus. For detailed protocols on how to image renal filtration, readers are referred to Wang et al. (2010).
2.2.3. Visualizing the renal vasculature
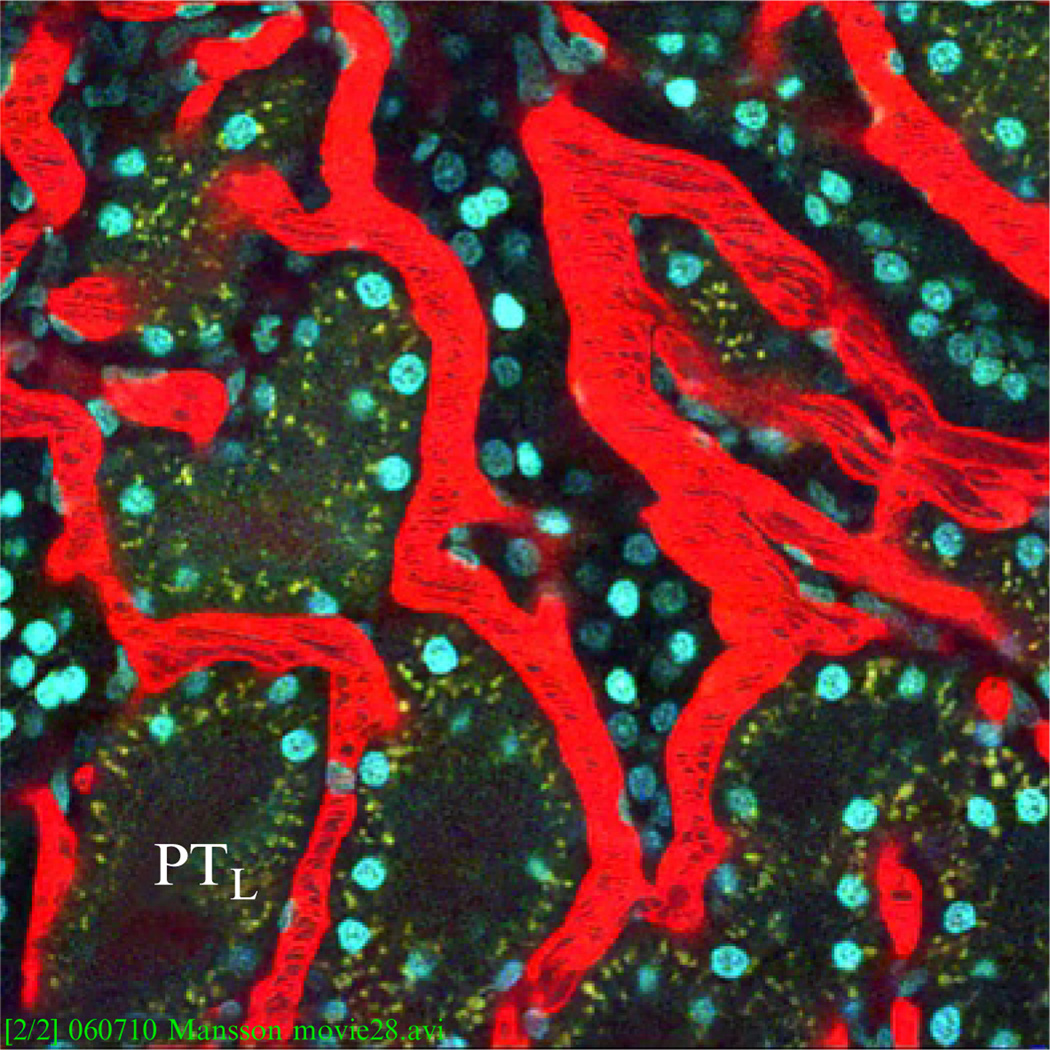
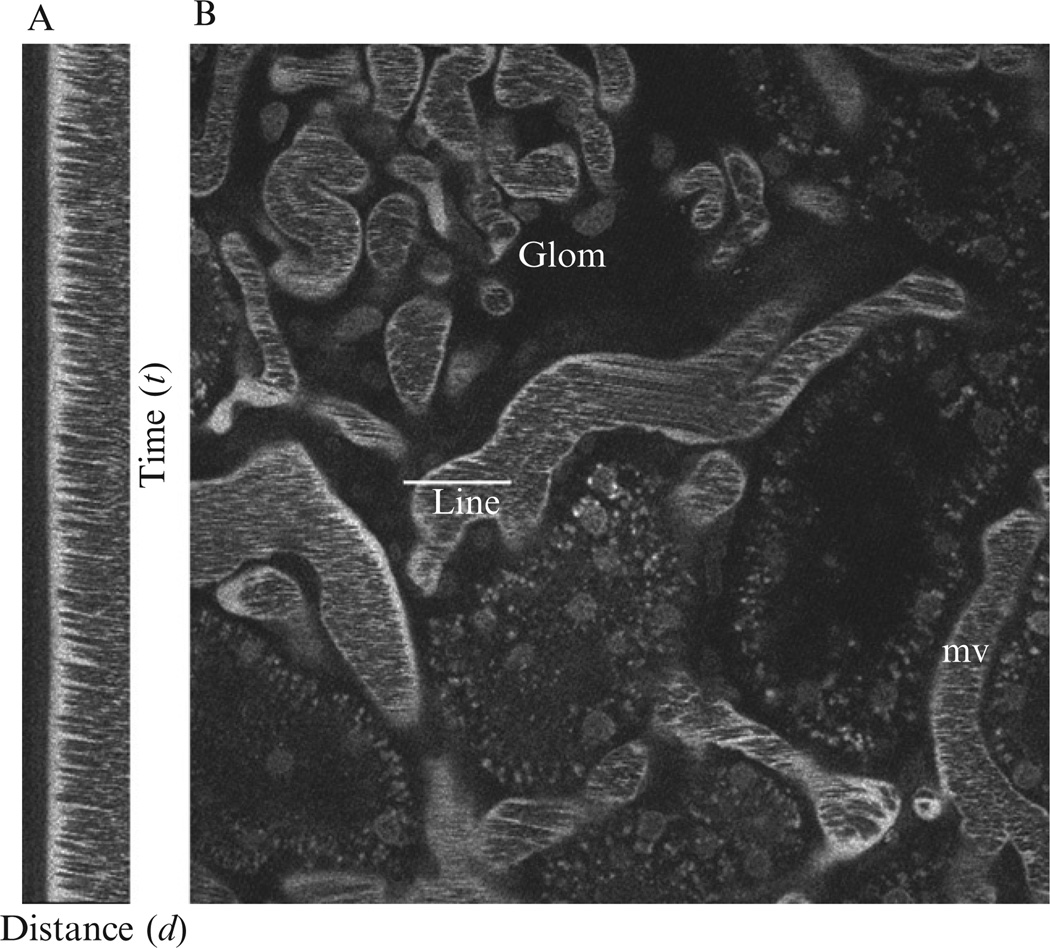
Fluorophore-conjugated molecules of molecular weight (Mw) larger than the upper glomerular filtration limit remains in the vasculature and are therefore useful to visualize the blood plasma. Prepare an 8 mg/ml sample of the rhodamine-conjugated 500 kDa dextran in 0.9% sterile saline. Dialyze 5–10 ml of this solution using a 10,000 MWCO membrane against 0.9% (w/v) sterile saline (5 l) overnight at room temperature. After systemic delivery, this fluid-phase marker remains in the vasculature for several hours (Fig. 3.1). As erythrocytes exclude the dye, these cells can be observed as black streaks within the red plasma.
Figure 3.1.
Fluorescence image of live kidney tissue morphology under multiphoton. Blood plasma (red) is labeled by fluorophore-conjugated 500 kDa dextran, black streaks represent erythrocytes. Epithelial lining of the proximal tubules exhibits autofluorescence and appears dull green, PTL indicates the lumen of the proximal tubule, and cell nuclei are stained by Hoechst 33342 (blue). Image adapted from Movie S1 in Månsson et al. (2007a,b).
3. Setup of a Live Imaging Platform of Infection
Real-time live imaging of bacterial infections requires a variety of competences ranging across microbiology, renal physiology, and biophysics. A typical study would involve several phases covering bacterial cloning and cultivation; animal handling, surgery and maintenance; microscopy and data analysis. This section will provide optimum conditions and procedures covering preimaging preparations, initiation of infection, and the basics of imaging of the infected tissue.
3.1. Selecting and customizing a multiphoton imaging platform and analysis
Multiphoton imaging systems have become increasingly easy to setup, and ready-made all-in-one systems are available from several commercial suppliers, for example, Leica, Thorlabs, and Zeiss. Alternatively, “homemade” systems, which offer more freedom of expansion, can be built from scratch or by modifying confocal microscope systems (Majewska et al., 2000; Mller et al., 2003; Nguyen et al., 2001). A major decision for choosing a system is to decide between an upright and an inverted system, being that the animal preparation and use defers between the two systems.
In an inverted system, the exposed tissue, in our case the kidney, is wedged between the animal and the bottom of the dish facing the objective. This helps to reduce tissue movement as the position of the kidney is stabilized by the weight of the animal. Alternatively, for an upright system, the exposed kidney is presented and immobilized using a custom designed kidney cup.
Image capture
The quality of captured images decreases dramatically with movement of the sample. While proper surgical preparation and mounting of the animal can minimize sample movement, there is a limit to how much can be done. The chance of mortality of the animal increases with the degree of invasiveness of surgical procedures, and intrinsic pulsations of the live sample from contractions of the heart and respiration cannot be avoided. High image capture rates can accommodate sample movement by acquiring a series of clear “still” shots or a fluid video of the sample as it moves. However, such systems tend to be costly and therefore not easily available.
The system we primarily have used is limited to one frame per second for a 512-by-512 frame and one frame per 2 ms for a line scan. This has been sufficient for tracking dynamic processes of host–pathogen interactions in the kidney, as well as more rapid physiological processes such as blood flow and glomerular filtration.
Digital image analysis
Live imaging of any process in real-time generates enormous datasets/image volumes of multiples planes. Often datasets involving time series are large files stored in multiple copies and variations, which then requires massive amounts of digital real estate. Aside from the resolution capability of the imaging platform and the acquisition speed of the system, processing power and memory capacity of the data analysis platform are of equal importance. With continuous turnover of powerful personal computer systems, CPUs capable of handling most forms of imaging analysis are being increasingly easy to obtain, as are image analysis software capable of handling live imaging data. Image analysis software can be purchased commercially or obtained for free via shareware. Though sharewares may have limited capabilities, they tend to be sufficient for basic image analysis and enhancement. For image analysis, we have primarily used ImageJ (a free image-processing tool from the NIH with an extensive library of plug-ins; http://rsb.info.nih.gov/ij/) and Voxx (a voxel-based 3D imaging program; http://www.nephrology.iupui.edu/imaging/voxx/index.html), followed by Adobe Photoshop (Adobe, CA, USA) for final adjustments for presentation. In case quantitative analysis shall be performed, commercial software such as Metamorph (Molecular Devices, Sunnyvale, CA) and Amira (Visage Imaging, San Diego, CA) are better calibrated. The authors suggest beginners to first experiment and familiarize with shareware before exploring other options.
3.2. Pathogen and host
3.2.1. Required materials
Rats and bacterial strains for infection model
UPEC strain LT004 (CFT073 cobS::Φ(PLtetO-1-gfp+), cmr)
Standard Sprague-Dawley (264 ± 16 g) and Munich-Wistar (255 ± 22 g) rats
Culture medium and additional equipment
Luria–Bertani (LB) agar and LB broth
37 °C shaking incubator for liquid culture growth
37 °C incubator for agar plates
100-ml conical flask or 15-ml Falcon tube
Centrifuge
1.5-ml eppendorf tubes
Glass spreader
3.2.2. Pathogen
Our studies are primarily based on the clinical, uropathogenic E. coli isolate CFT073 (Mobley et al., 1990; Welch et al., 2002) engineered to produce GFP+. Isogenic mutants have been constructed and applied when testing the role of bacterial virulence factors in vivo. Preparation of bacterial culture for subsequent use in animal studies is relatively straightforward. Prepare a shaking overnight culture of bacteria in a 100-ml conical flask or a 15-ml Falcon tube containing LB broth at 37 °C. To ensure sufficient aeration, the volume of the culture should not exceed 20% of the total volume of the receptacle. The following morning, perform a 100-fold dilution into fresh LB broth and dispense an aliquot of suitable volume into a fresh sterile receptacle. Cultivate the culture according to above until it reaches a density of OD600 = 0.6. To minimize the volume that will be infused into the animal, concentrate the culture to an approximate density of 109 CFU/ml. In general, UPEC cultures with OD600 = 0.6 have a cell density of 108 CFU/ml. To concentrate the culture, harvest the bacteria by centrifugation at 5000×g for 5 min. Discard the supernatant and resuspend the pellet in 0.9% sterile normal saline of 10% the original volume. Add Fast Green FCF (1 mg/ml) (in order to visualize the direction and speed of fluid flow during the infusion to the nephron) and Cascade blue-conjugated 10 kDa dextran (0.2 mg/ml). The latter accumulates rapidly in endosomes/lysosomes of proximal tubule cells, thereby selectively outlining the injected tubule. This is important in perfused environments, where the bulk of bacteria are expected to be flushed out by the filtrate flow. Maintain the bacterial suspension on ice until used. After infusion, determine the actual live cell density of the bacterial culture by performing dilution plating. This involves performing serial 10-fold dilutions in 1.5-ml eppendorf tubes, and dispensing 100 µl aliquots of each dilution onto separate LB agar plates. Prepare triplicates for each dilution made. Evenly spread the 100 µl aliquot over the agar using a glass spreader and incubate plates in a 37 °C incubator overnight. The following morning, perform a visual count of all colonies on each plate. Select the dilution that gives a count of between 50 and 300 colonies per plate and determine the average colony count from all plates in the triplicate. The cell density of the original culture is estimated by multiplying this average number by the dilution factor.
3.2.3. Host
We applied Sprague-Dawley (264 ± 16 g) and Munich-Wistar (255 ± 22 g) rats in our studies. The latter has surface-localized glomeruli, which enable easy visualization of the proximal convoluted tubules. Before start of the experiment, rats were given free access to chow and water.
3.3. Anesthesia and surgical creation of retroperitoneal window
In order to perform live imaging, the animal must be anesthetized and surgically prepared for infection and orientation on an imaging platform. With the organ surgically exposed, the animal is more fragile, and vital parameters such as blood pressure, pulse rate, and breathing become prone to fluctuations. Such fluctuations not only increase the risk of mortality but also impact host responses to the induced infection. As such, much attention needs to be placed on how to optimally prepare and maintain the animal. The following section describes an optimized set of such surgical procedures.
3.3.1. Required materials
Mice strains for surgical preparation
Standard Sprague-Dawley (264 ± 16 g) and Munich-Wistar (255 ± 22 g) rats
Surgical consumables
Isoflurane/oxygen mixtures (5% (v/v) and 2% (v/v))
Pentabarbital (optional)
Buprinorphine
Germicidal soap
0.9% sterile saline, prewarmed to 37 °C
Vascular catheters (PE-60 tubing for rats and PE-50 tubing for mice; Becton Dickinson)
Surgical equipment
Anesthesia induction chamber
Homeothermic table
Rectal probe for temperature recording
Electric clippers
Kidney cup
Surgical scissors (Braintree Scientific)
Appropriate animal temperature control devices (e.g., circulating water blanket attached to a temperature-controlled circulating water bath)
Fluorescent labels and probes
See Section 2
3.3.2. Presurgery preparation
Place the animal in an anesthesia induction chamber containing a 5% isoflurane/oxygen mixture. After initial anesthesia is obtained, transfer the animal to a clean surgical area on a homeothermic table. Maintain anesthesia with a 2% isoflurane/oxygen mixture, titrate to effect. Inject 0.05 g/kg buprinorphine subcutaneously in preparation for the subsequent surgery. Shave areas to be incised. This is typically performed at the left flank area (access the kidney), neck (jugular vein and artery access line), and inner thigh (femoral vein access line) using electric clippers. Cleanse the respective areas with germicidal soap and water and then towel dry. Thoroughly remove any cut hairs. In our experience, minute presence of stray hairs can greatly interfere with imaging. Prior to surgery, insert a rectal probe for temperature monitoring.
3.3.3. Surgery
Small incisions are preferred when performing live imaging of the kidney, and it is easier to progressively increase the incision rather than stitch a large one. Large incisions also allow more movement of the exteriorized kidney, which is counterproductive for imaging. In our studies, a small incision (using surgical scissors) is typically made. First, make a small incision over the femoral vein and insert a venous access line (PE-60 tubing) for subsequent dye infusion. Next, expose the kidney by making a 0.5–1 cm incision (using surgical scissors) in the left flank through the retroperitoneum. We have found it beneficial to palpate the left flank for the kidney first to get an idea of its size and location. Using a pair of forceps, draw back the adipose tissues which might be covering the kidney. Lift the kidney out of the incision by holding on to the hilar fat pad. Place a Petri dish on the microscope stage and fill it with prewarmed normal saline. Move the animal to the microscope stage and position the animal such that the left kidney, immersed in saline, is facing down toward the objective. Maintain the temperature of the animal with a heating blanket and supply appropriate anesthesia.
3.4. Introducing uropathogenic E. coli to initiate infection
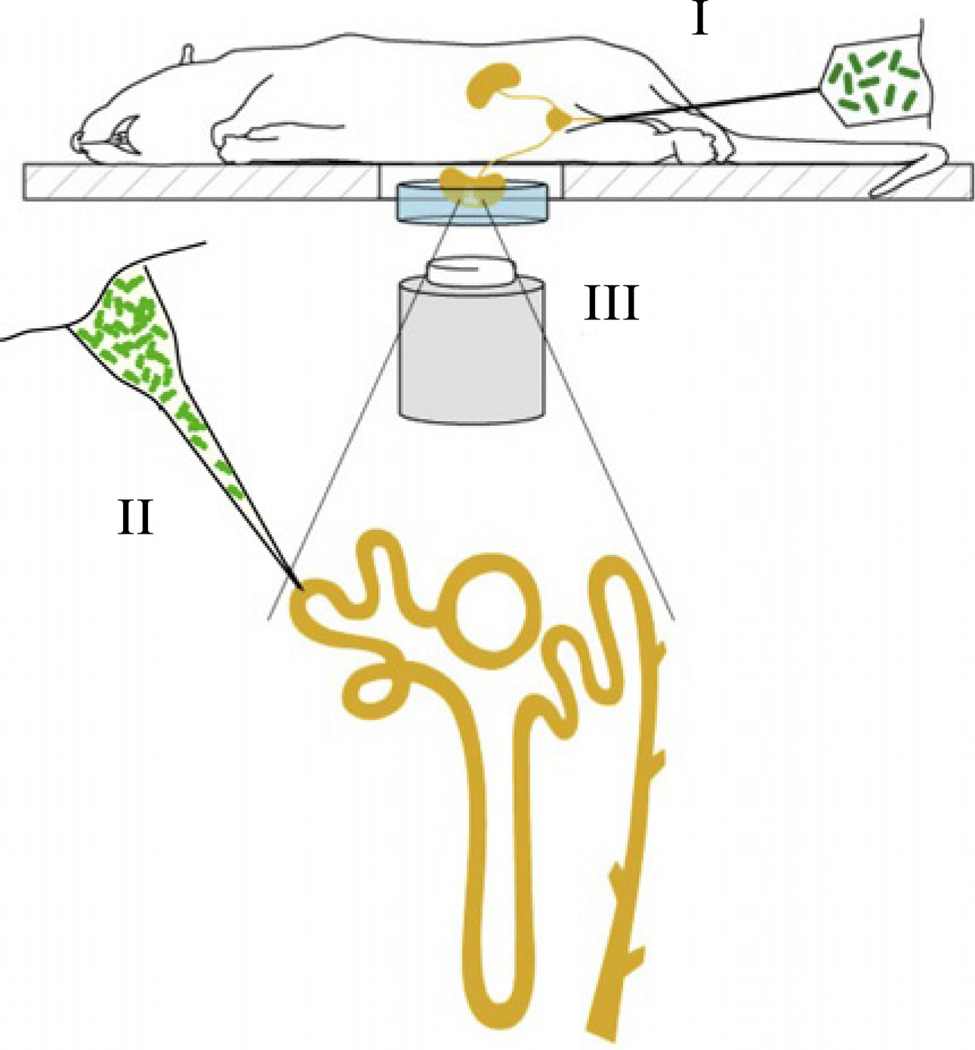
In the classical retrograde model of upper urinary tract infection, bacteria are delivered via a catheter into the bladder, from where they ascend to the kidney (Fig. 3.2). While this model resembles the natural route of infection, it suffers from drawbacks such as poor spatial and temporal control. This is because of the difficulty in predicting where and when bacteria enter into the kidney. Accordingly, this model is best suited for imaging end-stage infections when macroscopic identification of cortical abscesses directs the researcher to the site-of-interest to be imaged. To overcome these complications and generate an infection model with high spatial and temporal precision, we applied intratubular micropuncture (Fig. 3.2). Here, bacteria are slowly infused directly into the lumen of the proximal tubule in a single nephron (spatial control) at a defined time point (temporal control). For details, see below.
Figure 3.2.
Graphical depiction showing the positioning of a live rodent on an inverted multiphoton microscope. The left kidney is stabilized in a cell culture dish and immersed in isotonic saline. The microscope stage is heated, and the rat is wrapped in a heating pad (not shown). Green fluorescent bacteria can be introduced either via (I) retrograde infection where bacteria are infused into the bladder or (II) injected by micropuncture directly into the proximal tubule of one nephron (yellow) in an exposed kidney. Infection is imaged by multiphoton microscopy (III). Image adapted from Månsson et al. (2007a,b).
3.4.1. Required materials
Rats and bacterial strains for infection model and respective preparations
See Section 3.2
Consumables
Sterile PBS
Heavy mineral oil
2% isofluran/oxygen anesthesia
Halothane/oxygen anesthesia
Sudan black-stained castor oil
Appropriate fluorescent probes
1 mg/ml Fast Green Fisher
See Section 2
Infection and Imaging equipment
Sharpened micropipettes
Stereoscopic microscope
Leitz micromanipulator
Mercury leveling bulb
Imaging Platform (MPM)
3.4.2. Intratubular micropuncture
Aspirate the bacterial suspension (prepared as outlined in Section 3.2) into sharpened micropipettes (tip diameter 7–8 µm) filled with heavy mineral oil. Prepare the animal as described in Section 3.3 and position it on the stereoscopic microscope stage. Wean the animal off the isoflurane/oxygen mixture used during preparation, to halothane/oxygen anesthesia, allowing fine-tuning and recovery. Focus on the projected site of infection under observation at 96-fold magnification. Using a Leitz micromanipulator and mercury leveling bulb, inject the bacterial suspension as prepared in Section 3.2.2 into the proximal tubule. To avoid increased shear stress in the tubule during delivery, an infusion rate of approximately 50 nl/min is used for 10 min. This results in total delivery of ca. 5 × 105 CFU of bacteria. Take notice of the time point for infusion, as this marks the initiation of the infection process. We find it useful to inject one to three proximal tubules for each rat, either with bacteria or with sterile PBS. The latter serves as a control for any effects of the surgical procedure. Also inject Sudan black-stained castor oil in a nephron near each of the infection sites. This will aid in the localization of injection sites once the animal is positioned on the MPM stage.
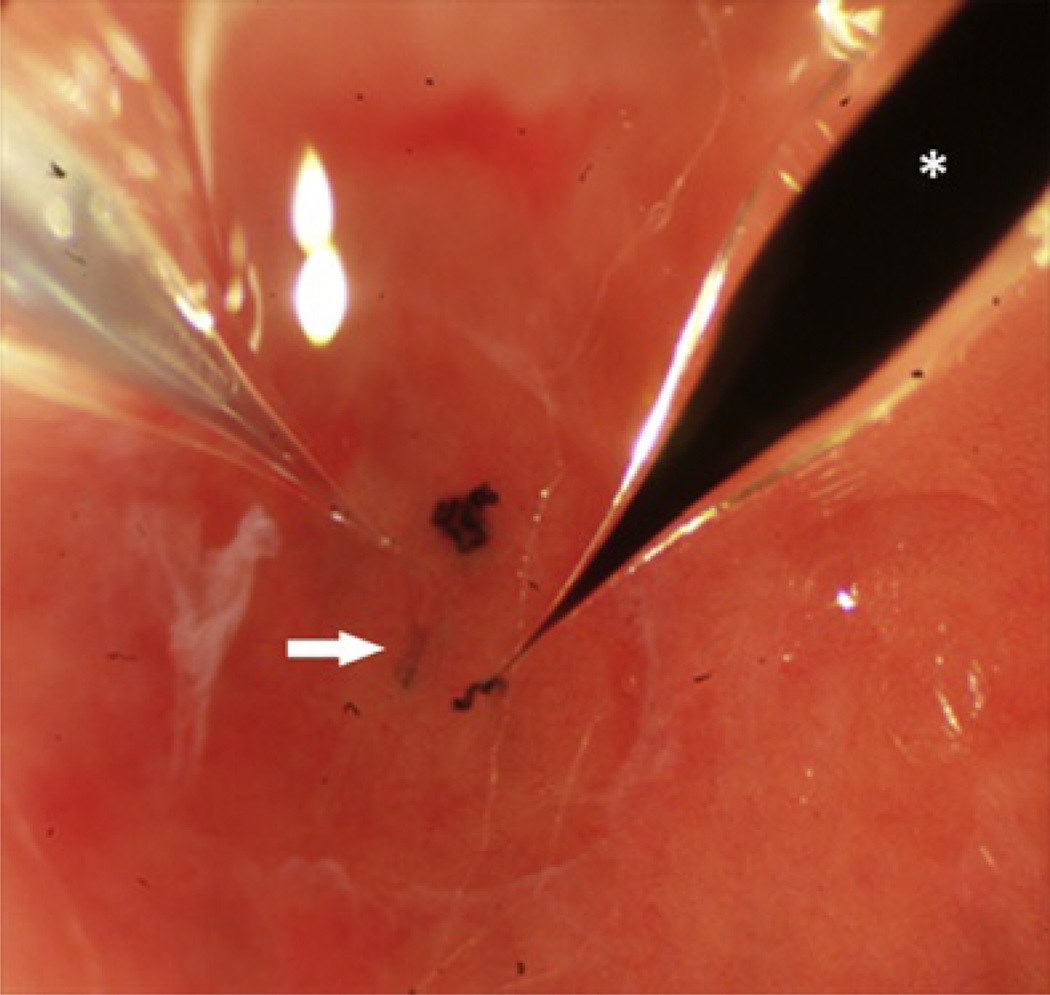
Following infusion, the animal is moved to the multiphoton microscope stage and positioned for imaging as described in Section 3.3.3. For gross identification of the infection site, use nearby tubules marked with Sudan black-stained castor oil (Fig. 3.3). To identify the bacterially injected nephron, look for the characteristic blue fluorescent outline of the tubuloepithelium, which originates from endocytosed cascade-blue-conjugated dextran coinfused with bacteria. Though the original bacterial load was ca. 5 × 105 CFU, a majority of bacteria has been flushed out by the filtrate flow, only few remain bound to the epithelium. To find these bacteria, carefully focus on the injected tubule, expect to see only few green fluorescent bacteria attached to the blue lining. In contrast, if the site is imaged at later time points, or if infection sites are used that lack perfusion, one might expect to see large masses of green fluorescent bacteria.
Figure 3.3.
Photograph showing the microperfusion of bacteria into a tubule with a micropipette of tip diameter 8 µm (left). (Arrow) indicates the infused tubule. (*) Identical micropipette filled with Sudan black-stained castor oil is used for injection of nearby nephron to aid in orientation of the infection site during imaging.
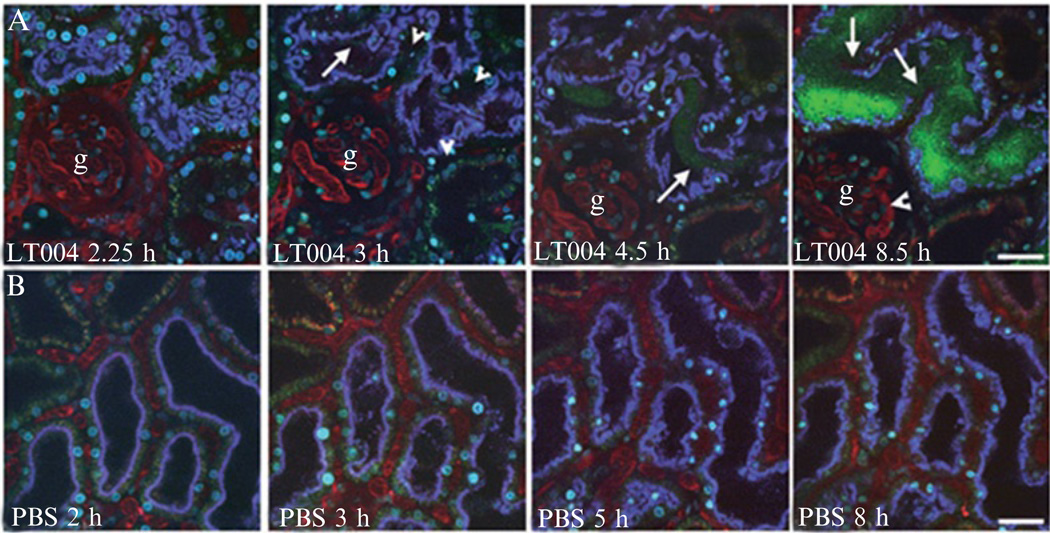
Next, introduce appropriate probes via intravenous injection, making sure to first flush dead space of catheter. In the kidney, we usually image the infection sites for 1–8 h after bacterial delivery (Fig. 3.4). While the animal is maintained on the microscope stage for image acquisition, the depth of anesthesia, core body temperature, and blood pressure (if desired) is monitored. Repeat experiments on different days, each time using freshly prepared bacterial inoculums. If desired, the rat may be taken off the stage, housed overnight, and imaged on subsequent days. Again, nephrons injected with Sudan black-stained castor oil will help in guiding the microscopist to find the infection sites.
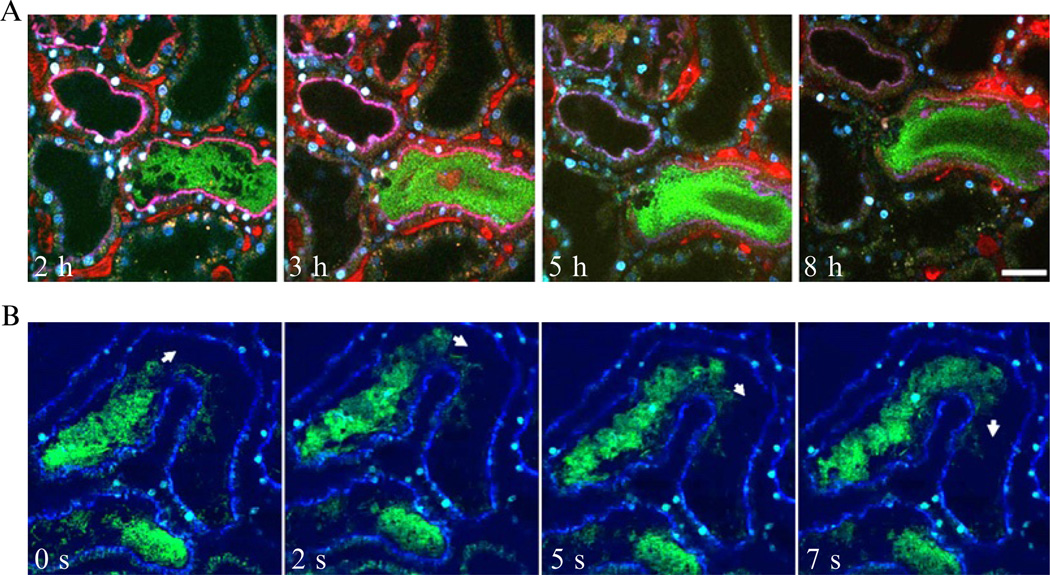
Figure 3.4.
Selected time points obtained from real-time multiphoton imaging of UPEC infected (A, top panel) and PBS control-injected (B, lower panel) proximal tubules. The injected tubules are outlined by endocytosed blue dextran while tubules in uninfected nephrons exhibit green autofluorescence. In (A), normal blood flow, visualized by infusion of large Mw fluorescently labeled dextran (red), is observed in peritubular and glomerular (g) capillaries 2.25 h after onset of infection. Erythrocytes are seen as black streaks within vessels. At 3 h, initiation of capillary collapse and altered blood flow (arrowheads) occur as a consequence of infection. At 4.5 h, bacteria fill the tubule lumen (arrow). At 8.5 h, with persistent multiplication of the pathogen, fluorescence signal of proximal tubule-specific labeling disappears indicative of epithelial linings disintegration. A single glomerular loop with slowed flow of erythrocytes is shown (arrowhead). In (B), a PBS sham-injected nephron (blue outline) shows no indication of abnormal function. Image adapted from Melican et al. (2008).
3.4.3. Imaging using the retrograde infection model of acute pyelonephritis
Prepare an overnight culture of bacteria in LB broth. The following morning, prepare a fresh 100-fold diluted culture in LB broth and cultivate it to OD600 = 0.6 as described in Section 3.2.2. Harvest the cells by centrifugation of the culture at 5000 × g. Discard the supernatant and resuspend the pellet in an equal volume of PBS to achieve a culture density of approximately 108 CFU/ml. We commonly use four to six rats in each series of experiments due to the inherited randomness of onset of pyelonephritis in the ascending model. Female Sprague-Dawley rats are anesthetized using isofluran. Via a catheter, 1 ml of the bacterial suspension is slowly infused into the bladder of each rat. Infusion of PBS is used in control rats. Allow the infection to progress for 4 days, then prepare rats for live multiphoton imaging as described in Section 3.3. Use ocular inspection to identify sites of cortical abscesses, which then are imaged on the MPM. Due to the inherited poor spatiotemporal control of this model, it lends itself best to the study of late-stage infection.
4. Studying Tissue Responses in Real-Time During Infection
In the presence of infectious agents, the host defends itself by onset of inflammatory responses, meaning that tissue homeostasis is inevitably altered during the time-course of infection. When studying these processes in real-time, a general knowledge of the organ architecture and function is required to ensure that both expected and unexpected deviations from native anatomy and physiology, occurring as result of the infection process, will be noticed. To verify whether observed effects relate to the infection process, comparative studies, controlling for the surgical procedure (PBS sham injections), should be performed. Time-course imaging is also useful to identify infection-associated effects, as such effects may become more pronounced as the infection progresses. Alternatively, infection of the animal using isogenic bacteria harboring mutations in specific virulence genes can be used to verify their role in an infection, and if the lack of such genes abrogates or alters respective contributions to pathogenesis and host responses.
4.1. Probing histological changes of the mucosal lining
With careful selection and introduction, fluorescent probes can be used to cleanly define respective tissue compartments. We have demonstrated that bacteria colonizing the tubule lumen can be visualized by bacterially expressed GFP+, at the same time as differently sized dextrans are used to mark the epithelial lining and the vasculature (Fig. 3.1), with no visible leakage of probes when initially introduced (Månsson et al., 2007a,b; Melican et al., 2008). As such, boundaries between mutually exclusive fluorescent signals correspond to the boundary between respective tissue compartments. By then tracking changes of the position of these target-specific fluorescent probes, changes in histology of tissues arising from the progression of UPEC infection can be detected. Several examples are described in the following section.
4.1.1. Required materials
Rats and bacterial strains for infection model and their respective preparations
See Sections 3.2 and 3.4
Fluorescent labels and probes
See Section 2
4.1.2. Integrity of the mucosal epithelium
Drawing from Månsson et al. (2007a,b) as an example, at early time points postinfection, the luminal face of the proximal tubular epithelium is seen as a continuous blue lining, arising from the internalization of coinfused small Mw cascade blue-conjugated dextran (Månsson et al., 2007a,b). Bacterium which attaches to the epithelial surface and colonize then presents as green fluorescence located directly against the blue epithelial layer (Månsson et al., 2007a,b). Within the next few hours, as bacteria multiply and their numbers escalate, green-fluorescing bacteria begin to breach the continuous blue fluorescent luminal lining of the epithelial layer, moving between individual blue units to the basal lamina (Månsson et al., 2007a,b). This illustrates how, during UPEC infection, bacteria gradually disrupt the integrity of the renal epithelium (Månsson et al., 2007a,b).
4.1.3. Exfoliation of epithelial cells
As a compound effect of infection-induced local ischemia and contact with bacterial virulence factors, exfoliation of proximal tubule epithelial lining could be observed approximately 6 h postinfusion (Melican et al., 2008). Due to the original cascade-blue staining of the epithelial lining, exfoliated cells can be identified as isolated blue fluorescent units within the lumen of the tubule with no apparent contact with the basal lamina.
4.1.4. Neutrophil Infiltration
When long-term survival of the rat is not a study objective, cytotoxic and mutagenic dyes can be introduced into the animal for short-term imaging of the tissue. In our studies, we have applied the DNA-binding fluorescent stain Hoechst 33342 to probe for the presence and location of neutrophils during UPEC infection. Neutrophils are identified based on the characteristic polymorphonuclear shape outlined in blue. As Hoechst disrupts normal cell functions by interfering with DNA replication, rat infused with Hoechst is not maintained after imaging.
4.2. Imaging vascular features such as rate, clotting, and integrity
Large Mw fluorophore-conjugated dextran injected into the vasculature remains within the systemic circulation, since glomeruli cannot filter it. This red dye, contained in the lumen of the vessels, can be used to probe several vascular features. This includes if the area under investigation has an appropriate blood supply can be visualized, the rate of the blood flow can be quantified, and the integrity of the vessel walls can be studied, as well as the formation of blood clots.
4.2.1. Imaging and estimation of vascular flow
Prepare the animal as described in Section 3. Inject rhodamine-labeled 500 kDa dextran intravenously via a jugular vein access line. Erythrocytes, which do not internalize the red dextran, remain unlabeled and appear as dark streaks rapidly moving within the vessels (Dunn et al., 2003, 2007). Using a line-scan method, the axial view of a capillary is recorded every 1–2 ms using the multiphoton microscope, for 2 s. Erythrocytes moving within the vessel are identified as diagonal black streaks in the line-scan images (Fig. 3.5) from which the velocity (µm s−1) of each erythrocyte can be determined by calculating Δd/Δt where d represents the distance (µm) and t represents time (s) (Brown et al., 2001; Kang et al., 2006; Kleinfeld et al., 1998; Ogasawara et al., 2000; Yamamoto et al., 2002). Several vessels can be analyzed to obtain an averaged estimation of the blood flow; however, it is essential to choose vessels of identical dimensions.
Figure 3.5.
Vascular blood flow as measured by MPM. Texas Red-labeled Rat Serum Albumin (TR-RSA) injected i.v. fills the plasma volume within the vasculature denoting the surrounding microvasculature (mv) and a glomerulus (Glom). A line scan shown in panel (A) was performed along the center of the vessel shown in panel (B, line). Because erythrocytes flowing throughout the systemic circulation are impermeable to TR-RSA, they appear as black streaks within the (mv). The single line show in panel B was repeatedly imaged to form the tall montage shown in (A) where the vertical axis represents time (in this case, 2 ms per line), while the horizontal axis represents distance (in this case, 330 nm per pixel). The passage of RBCs along the scan form a speed-dependent motion artifact from which velocity can be determined by using Δd/Δt; here average RBC velocity was measured to be 890 µm/s ± 134.
When analyzing the effect, an infection may have on the blood flow in nearby capillaries, line-scan data should be compared to those obtained when analyzing flow rate in capillaries located next to a PBS sham-injected tubule.
4.2.2. Imaging platelets and clots with and without anticoagulant therapy
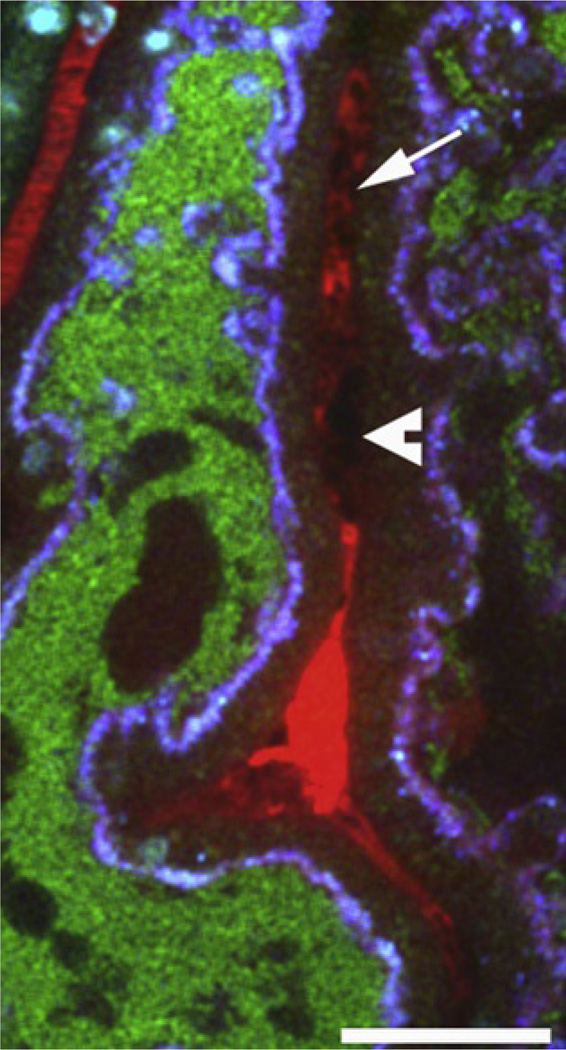
The size and shape of the black streaks seen in the red fluid-phase dextran marker varies according to cell type. Small black silhouettes, whose size indicates they are too small to be red blood cells or infiltrating immune cells, typically represent platelets. These silhouettes can be seen aggregating in capillaries, thereby forming large black nonfluorescent aggregates adhering to the vessel wall. Next to these obstructing masses, plasma, essentially devoid of red blood cells, can be identified by the intense solid red color of the fluid-phase marker. Collectively, these signs are typical for local clot formation (Fig. 3.6; Melican et al., 2008). The role of clotting, whether in blood-borne infection, or as in our model when bacteria are localized at the mucosal lining, can be analyzed by applying anticoagulant therapy.
Figure 3.6.
Images obtained during live multiphoton analysis of an UPEC (strain LT004, green) infected proximal tubule (blue) and adjacent blood vessel (red). A lack of erythrocyte movement is seen in the area. Black silhouettes within vessels are indicative of platelets (arrow). Aggregation of black masses adhering to the vessel wall (arrowhead) suggests platelet aggregates. Image adapted from Melican et al. (2008).
Immediately following bacterial infusion, rats are treated with 400 U/kg of heparin sodium via the jugular venous access line. After 4 h, a second dose (200 U/kg) is administered. The progression of infection is imaged on the multiphoton microscope as usual with the status of the animal being monitored by body temperature and blood pressure. The latter recording is important as a sudden, sharp drop in blood pressure is indicative of the onset of sepsis. If this occurs, animal should be sacrificed, and internal organs such as the liver, spleen, and heart should be aseptically removed, homogenized in PBS, and analyzed for the presence of bacteria. This is performed by serial dilution and plating, see description in Section 3.2. Blood samples shall also be taken throughout the experiment to confirm lack of clotting in animals on anticoagulant therapy as well as to analyze for the presence of bacteria (see Section 3.2).
4.2.3. Imaging the integrity of the endothelial lining
Increased endothelial permeability causing perivascular leakage of the large Mw red dextran can be observed as a local effect of infection. In case immune cells have extravasated, they can be observed as black silhouettes within the area of leakage (Melican et al., 2008).
4.3. Local tissue oxygen tension
In our UPEC infection studies, we noted an apparent cessation of blood flow in peritubular capillaries within 3–4 h of infection. As this was accompanied by typical signs of ischemia, it suggested that the tissue was suffering from a lack of vital oxygen (Melican et al., 2008). By measuring the local oxygen tension, the highly targeted and localized ischemia was demonstrated as a host defense mechanism preventing systemic dissemination of the pathogen. Measuring local-tissue oxygen tension may also provide interesting insights into the role of ROS, which is an important host innate immune response against infections. Production of reactive oxygen species is regulated by oxygen tension (Wu et al., 2007). Specifically, the rate of superoxide production by phagocytic oxygenase in leukocytes from glucose and oxygen is directly proportional to the local oxygen concentration (Hunt and Aslam, 2004). It would thus be highly appreciated if the imaging data on blood flow cessation could be complemented with actual recordings of the tissue oxygen.
4.3.1. Required materials
Equipment
Modified Clark-type microelectrode system
Stereomicroscope
Reagents
120 mg/kg Thiobutabarbital
Water saturated with Na2S2O5
Additional materials for animal preparation and infection
See Sections 3.2 and 3.4 for animal preparation and infection
Bacterial growth and enumeration
LB agar and LB broth
37 °C shaking incubator for liquid culture growth
37 °C incubator for agar plates
4.3.2. Surgical preparation of the animal
Anesthetize animals with an intraperitoneal injection of thiobutabarbital at 120 mg/kg. Transfer the animal to a temperature-controlled operating table at 37 °C. Perform a tracheotomy and insert polyethylene catheters into both femoral arteries and the right femoral vein. Use one arterial catheter to monitor blood pressure and the other for blood sampling. Maintain the fluid levels of the animal by saline infusion into the vein. Catheterize the bladder to allow urinary drainage. Expose the left kidney by a left subcostal flank incision and immobilize the kidney in a plastic cup surrounded by saline-soaked cotton wool. Cover the surface of the kidney with paraffin oil and then catheterize the left ureter for urine collection. Retain an ample amount for bacterial enumeration when required. Allow a 60-min window for the animal to stabilize. Infection can then be performed by microperfusion as described in Section 3.4.2.
4.3.3. Measurement of renal PO2
Tissue oxygen tension (PO2) is commonly measured using modified Clark-type microelectrodes (Severinghaus, 2002). Before applying the electrodes to the tissue, perform a two-point calibration in water saturated with either Na2S2O5 or air at 37 °C. Next, initiate the infection by infusing bacteria according to previous description. In parallel, PBS should be infused into a neighboring site in the same kidney to serve as internal control. While the animal still is under stereomicroscopic observation, use the micromanipulator to insert one microelectrode into each of the tubule lumens immediately after infusion of bacteria and PBS. In our setup, simultaneous recording of PO2 in both sites was conducted for a total of 240 min. At this stage, PO2 in the infected nephron had plummeted to 0 mmHg. During the whole procedure, blood pressure should be monitored either via tail cuff or via a direct arterial line with the use of a pressure transducer. By presenting recorded PO2 over time for both sites in the same graph, together with the blood pressure, a clear picture emerges whether the infection causes any local or global effects in the animal.
4.4. Systemic dissemination
In patients, bacteria sometimes find their ways from the urinary tract into the systemic circulation, thereby giving rise to urosepsis. When simulating the natural course of infection in an animal model, it is important to control different tissue compartments for the presence of the pathogen. In each experiment, blood and urine samples shall be collected from the animal at appropriate time points and analyzed for the presence of bacteria by direct enumeration of cell density by dilution plating, see Section 3.2.2. A typical sign that urosepsis has occurred is when a sharp drop in blood pressure is observed.
4.5. Imaging obstruction of a perfused environment
By infusing the vasculature with low Mw fluorophore-conjugated dextrans, the functionality of an infected nephron can be assessed. After being filtered, the fluorophore signal within the nephron is short lived as it is immediately channeled into distal reaches of the nephron. Månsson et al. observed the accumulation of a low Mw rhodamine-conjugated dextran in an UPEC infected nephron after the bacterium had established itself in the proximal tubule (Månsson et al., 2007a,b). The persistence of the otherwise transient signal here suggested that UPEC presence in the proximal tubule ultimately leads to the stoppage of renal flow.
The high flexibility inherent to our intravital imaging model then allowed the introduction of various UPEC isogenic mutants to define the root cause of the above observed obstruction. Specifically, the synergistic role of major adhesion factors Type 1 and P fimbriae in PCT obstruction was shown by applying respective knockout strains (Fig. 3.7). By imaging the site of infection and analyzing the functionality of the nephron in relation to the clearance of the intravenously infused fluorophore-conjugated low Mw dextran, the chain of events leading to the obstruction could be identified. Applying this method, Melican et al. clearly showed the synergistic role of P and Type 1 fimbriae in colonization of the luminal surface and the center of the tubule, respectively, and that the cessation of filtrate flow was the result of obstruction by large bacterial aggregate bacteria residing in the center of the proximal tubule lumen, rather than a host response to epithelium-bound colonies.
Figure 3.7.
Synergistic action of attachment organelles during colonization in presence of renal flow. (A) Mutation of the UPEC Type 1 fimbriae tip adhesion negatively impacts the ability of this strain to colonize the center of the tubule lumen. This is shown from selected time points during 8 h. The same bacterium expresses P fimbriae, which promotes normal binding to the epithelium. Scale bar = 30 µm. (B) UPEC lacking P fimbriae-mediated attachment shows an inability for epithelial attachment. Increased susceptibility to the shear stress is shown by the large bacteria aggregates that are seen “flushing” in the direction of filtrate flow (arrow) with a rate of 1 µm s−1. Image adapted from Melican et al. (2011).
5. Postimaging Analysis
While real-time study of the infection process in vivo is an attractive concept, many important techniques in the studying of host–pathogen interactions remain “endpoint” measures. By applying our spatiotemporally controllable infection model, endpoint studies can be made accurately and in parallel to the real-time setting, thereby increasing the relevance of non-real-time data in describing the natural progression of host–pathogen interactions.
5.1. Immunohistochemistry
5.1.1. Required materials
Reagents and equipment for sample preparation
Paraformaldehyde in PBS (4%)
Sucrose in sterile PBS (20%)
OCT Tissue-tek
3-methyl-butane
Microtome
Antibodies and fluorescent dyes
Phalloidin conjugated with TRITC or Texas Red
Hoechst 33342 dye
RbaCollagen IV
GtaRb-Cy3
When ending the multiphoton imaging, aseptically remove the infected kidney and fix with freshly prepared 4% paraformaldehyde in PBS pH 7.4 (Merck, Germany) for 1 h. Wash the kidney three times in PBS and immerse it overnight in 20% sucrose in PBS at 4 °C for cryoprotection of the organ. Mount pieces of the organ in OCT Tissue-tek and snap freeze the samples using 3-methyl-butane at −50 to −60 °C. In a cryostat, make 10 µm thick renal cortical slices for staining with dyes and/or antibodies and perform confocal laser scanning microscopy. In our setting, phalloidin-conjugated TRITC or Texas Red was used to visualize actin, whereas Hoechst 33342 dye stained for cell nuclei. Antibody Rb anti-Collagen IV was used to stain the basal lamina for the major extracellular matrix protein collagen IV, which was detected using the secondary antibody Gt-anti-Rb-Cy3. Bacteria were visualized using their endogenous GFP+.
6. Extending Real-Time Imaging into Molecular Analysis
Several features of the model for real-time imaging of infection have been found to be very useful in generating a molecular understanding of the infection process. Here, we summarize such features from the host as well as microbe perspectives.
6.1. Molecular view of host responses
Initiating the infection by microinfusing, one defined nephron gives this model an outstanding spatial and temporal precision. We find this very useful when supplementing the imaging data with transcriptional profiling to indicate by which molecular details the host orchestrates its tissue response. As the infection site is known, it can be precisely dissected from the kidney and used for RNA preparation. In the absence of the bulk of noninfected tissue from the entire organ, the ratio between infected and uninfected tissue enables RNA to be prepared at a quality suitable for microarray analysis. By obtaining tissue at time point of interests defined from parallel imaging experiments, and by comparing to the transcriptome obtained from noninfected tissue, a dynamic description of the host’s response to infection emerges (Boekel et al., 2011). The study can be extended by comparative tissue transcriptomics, allowing conserved trends in host responses to be identified. By doing so, we identified the importance of IFN-γ in interorgan communication already within the first hours of infection. These data illustrate that host responses at the local site of infection may result from systemic influences.
6.2. Molecular details based on bacterial genetics
Similar to the above reasoning, the spatiotemporal precision of infection provides a nice opportunity to identify bacterial gene expression in vivo. Preparing bacterial RNA from tissues dissected at selected time points, use quantitative reverse-phase RT-PCR to analyze whether bacteria express a gene-of-interest, that is, a virulence factor, during organ colonization (Månsson et al., 2007a,b). Traditionally, virulence factors have been associated to infections in specific organs. For bacterial attachment organelles, the dogma associates P fimbriae to renal infections and Type 1 fimbriae to cystitis. Using the model described herein, we tested this hypothesis and found that both fimbriae are being expressed in the kidney during colonization (Melican et al., 2011).
Using an isogenic mutant, which is unable to produce a specific virulence factor, the role of this factor for disease progression in vivo can be tested. When knocking out expression of the UPEC-associated toxin a-hemolysin (HlyA), the kinetics of the host response, rather than the actual outcome of infection at 24 h, was altered, suggesting a role for this factor in modulating the inflammatory response (Månsson et al., 2007a,b).
Intravital imaging allows the function of bacterial proteins to be tested in contexts not previously attainable. Many tissue compartments expose bacteria to mechanical challenges, such as shear stress from the filtrate flow. By comparing the infection process using strains expressing or not expressing the above-mentioned attachment organelles, we studied the mechanisms that help bacteria maintain themselves in perfused environments. A synergistic function of the P and Type 1 fimbriae was identified, with P being important for attachment to the tubule epithelium, while Type 1 is engaged in interbacterial binding and biofilm formation (Melican et al., 2011). The latter enables bacterial colonization across the central part of the tubule lumen where there is no epithelium to hold on to (Fig. 3.7). In dynamic environments, the infectious niche can thus be defined to be as small as center or periphery of the tubule lumen, rather than bladder and kidney as previously suggested.
In conclusion, we hope this chapter illustrates that real-time intravital imaging holds great potential in studying the dynamics of host–pathogen interactions. The concerns we have raised are in principle valid no matter which organ is studied. Hopefully, this chapter can assist others in defining systems for future analysis.
ACKNOWLEDGMENTS
The research relevant to this chapter was supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, and the Swedish Royal Academy of Sciences (to A. Richter-Dahlfors), and NIH Grant DK061594 awarded to B. A. Molitoris to establish a “George M. O’Brien Center for Advanced Renal Microscopic Analysis” at the Indiana Center for Biological Microscopy, Indianapolis, Indiana.
REFERENCES
- Boekel J, Källskog O, Rydén-Aulin M, Rhen M, Richter-Dahlfors A. Comparative tissue transcriptomics reveal prompt inter-organ communication in response to local bacterial kidney infection. BMC Genomics. 2011;12:123. doi: 10.1186/1471-2164-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EB, Campbell RB, Tsuzuki Y, Xu L, Carmeliet P, Fukumura D, Jain RK. In vivo measurement of gene expression, angiogenesis and physiological function in tumors using multiphoton laser scanning microscopy. Nat. Med. 2001;7(7):864–868. doi: 10.1038/89997. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248(4951):73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Dunn KW, Sandoval RM, Kelly KJ, Dagher PC, Tanner GA, Atkinson SJ, Bacallao RL, Molitoris BA. Functional studies of the kidney of living animals using multicolor two-photon microscopy. AmJPhysiol. Cell Physiol. 2002;283(3):C905–C916. doi: 10.1152/ajpcell.00159.2002. [DOI] [PubMed] [Google Scholar]
- Dunn KW, Sandoval RM, Molitoris BA. Intravital imaging of the kidney using multiparameter multiphoton microscopy. Nephron Exp. Nephrol. 2003;94(1):e7–e11. doi: 10.1159/000070813. [DOI] [PubMed] [Google Scholar]
- Dunn K, Sutton T, Sandoval R. Live-animal imaging of renal function by multiphoton microscopy. Curr. Protoc. Cytom. 2007:12.9.1–12.9.18. doi: 10.1002/cpcy.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeppert-Mayer M. Über Elementarakte mit zwei Quantensprüngen. Ann. Phys. 1931;9(3):273–295. [Google Scholar]
- Hautefort I, Proença MJ, Hinton JCD. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 2003;69(12):7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Aslam R. Oxygen 2002: Wounds. Undersea Hyperb. Med. 2004;31:147–153. [PubMed] [Google Scholar]
- Kang JJ, Toma I, Sipos A, McCulloch F, Peti-Peterdi J. Quantitative imaging of basic functions in renal (patho)physiology. AmJPhysiol. Renal Physiol. 2006;291(2):F495–F502. doi: 10.1152/ajprenal.00521.2005. [DOI] [PubMed] [Google Scholar]
- Kleinfeld D, Mitra PP, Helmchen F, Denk W. Fluctuations and stimulus-induced changes in blood flow observed in individual capillaries in layers 2 through 4 of rat neocortex. Proc. Natl. Acad. Sci. USA. 1998;95(26):15741–15746. doi: 10.1073/pnas.95.26.15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska A, Yiu G, Yuste R. A custom-made two-photon microscope and deconvolution system. Pflugers Arch. 2000;441(2–3):398–408. doi: 10.1007/s004240000435. [DOI] [PubMed] [Google Scholar]
- Månsson LE, Melican K, Boekel J, Sandoval RM, Hautefort I, Tanner GA, Molitoris BA, Richter-Dahlfors A. Real-time studies of the progression of bacterial infections and immediate tissue responses in live animals. Cell. Microbiol. 2007a;9(2):413–424. doi: 10.1111/j.1462-5822.2006.00799.x. [DOI] [PubMed] [Google Scholar]
- Månsson LE, Melican K, Molitoris BA, Richter-Dahlfors A. Progression of bacterial infections studied in real time—Novel perspectives provided by multiphoton microscopy. Cell. Microbiol. 2007b;9(10):2334–2343. doi: 10.1111/j.1462-5822.2007.01019.x. [DOI] [PubMed] [Google Scholar]
- Melican K, Richter-Dahlfors A. Multiphoton imaging of host-pathogen interactions. Biotechnol. J. 2009a;4(6):804–811. doi: 10.1002/biot.200800347. [DOI] [PubMed] [Google Scholar]
- Melican K, Richter-Dahlfors A. Real-time live imaging to study bacterial infections in vivo. Curr. Opin. Microbiol. 2009b;12(1):31–36. doi: 10.1016/j.mib.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Melican K, Boekel J, Månsson LE, Sandoval RM, Tanner GA, Källskog O, Palm F, Molitoris BA, Richter-Dahlfors A. Bacterial infection-mediated mucosal signalling induces local renal ischaemia as a defence against sepsis. Cell. Microbiol. 2008;10(10):1987–1998. doi: 10.1111/j.1462-5822.2008.01182.x. [DOI] [PubMed] [Google Scholar]
- Melican K, Sandoval RM, Kader A, Josefsson L, Tanner GA, Molitoris BA, Richter-Dahlfors A. Uropathogenic Escherichia coli P and Type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 2011;7(2):e1001298. doi: 10.1371/journal.ppat.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mller M, Schmidt J, Mironov S, Richter D. Construction and performance of a custom-built two-photon laser scanning system. J. Phys. D Appl. Phys. 2003;36:1747. [Google Scholar]
- Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: Role of hemolysin in some strains. Infect. Immun. 1990;58(5):1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitoris BA, Sandoval RM. Intravital multiphoton microscopy of dynamic renal processes. AmJPhysiol. Renal Physiol. 2005;288(6):F1084–F1089. doi: 10.1152/ajprenal.00473.2004. [DOI] [PubMed] [Google Scholar]
- Nguyen Q, Callamaras N, Hsieh C, Parker I. Construction of a two-photon microscope for video-rate Ca2+ imaging. Cell Calcium. 2001;30(6):383–393. doi: 10.1054/ceca.2001.0246. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Takehara K, Yamamoto T, Hashimoto R, Nakamoto H, Kajiya F. Quantitative blood velocity mapping in glomerular capillaries by in vivo observation with an intravital videomicroscope. Methods Inf. Med. 2000;39(2):175–178. [PubMed] [Google Scholar]
- Severinghaus JW. The invention and development of blood gas analysis apparatus. Anesthesiology. 2002;97(1):253–256. doi: 10.1097/00000542-200207000-00031. [DOI] [PubMed] [Google Scholar]
- Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat. Biotechnol. 1999;17(8):763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50(6):823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Tian GF, Takano T, Lin JHC, Wang X, Bekar L, Nedergaard M. Imaging of cortical astrocytes using 2-photon laser scanning microscopy in the intact mouse brain. Adv. Drug Deliv. Rev. 2006;58(7):773–787. doi: 10.1016/j.addr.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wang E, Sandoval R, Campos S, Molitoris B. Rapid diagnosis and quantification of acute kidney injury using fluorescent ratio-metric determination of glomerular filtration rate in the rat. AmJPhysiol. Renal Physiol. 2010;299(5):F1048. doi: 10.1152/ajprenal.00691.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman SJ, Warren JW, Mobley HL, Donnenberg MS. Host-pathogen interactions and host defense mechanisms. Dis. Kidney Urin. Tract. 2007;1:3776. 8th edition. [Google Scholar]
- Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99(26):17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Platoshyn O, Firth AL, Yuan JXJ. Hypoxia divergently regulates production of reactive oxygen species in human pulmonary and coronary artery smooth muscle cells. AmJPhysiol. Lung Cell. Mol. Physiol. 2007;293(4):L952–L959. doi: 10.1152/ajplung.00203.2007. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Lee CE, Marcus JN, Williams TD, Overton JM, Lopez ME, Hollenberg AN, Baggio L, Saper CB, Drucker DJ, Elmquist JK. Glucagon-like peptide-1 receptor stimulation increases blood pressure and heart rate and activates autonomic regulatory neurons. J. Clin. Invest. 2002;110(1):43–52. doi: 10.1172/JCI15595. [DOI] [PMC free article] [PubMed] [Google Scholar]