SUMMARY
Genetically unstable tetraploid cells can promote tumorigenesis. Recent estimates suggest that ~37% of human tumors have undergone a genome-doubling event during their development. This potentially oncogenic effect of tetraploidy is countered by a p53-dependent barrier to proliferation. However, the cellular defects and corresponding signaling pathways that trigger growth suppression in tetraploid cells are not known. Here we combine RNAi screening and in vitro evolution approaches to demonstrate that cytokinesis failure activates the Hippo tumor suppressor pathway in cultured cells as well as in naturally occurring tetraploid cells in vivo. Induction of the Hippo pathway is triggered in part by extra centrosomes, which alter small G-protein signaling and activate LATS2 kinase. LATS2 in turn stabilizes p53 and inhibits the transcriptional regulators YAP and TAZ. These findings define an important tumor suppression mechanism and uncover adaptive mechanisms potentially available to nascent tumor cells that bypass this inhibitory regulation.
INTRODUCTION
Proliferating tetraploid cells are genetically unstable and can promote tumorigenesis (Davoli and de Lange, 2011, 2012; Fujiwara et al., 2005; Ganem et al., 2007). Accumulating evidence points to a significant contribution of tetraploid intermediates in shaping the composition of cancer genomes: ~20% of all solid tumors exhibit tetraploid or near-tetraploid karyotypes, and computational analysis of human exome sequences from ~4000 human cancers reveals that ~37% of all tumors, even those with a near-diploid karyotype, have undergone at least one whole genome-doubling event at some point in their evolution (Dewhurst et al., 2014; Zack et al., 2013).
Potentially oncogenic tetraploid cells arise spontaneously, through a variety of different cell division errors. Defects in mitosis and cytokinesis are thought to be the most common routes; however, tetraploid cells also develop as a consequence of viral-induced cell fusion, chromosome endoreduplication, oncogene activation, chronic inflammation, entosis, and telomere-erosion (Davoli and de Lange, 2011; Ganem et al., 2007). Importantly, tetraploid cells are capable of promoting transformed growth irrespective of the mechanism by which they were initially generated (Davoli and de Lange, 2012; Fujiwara et al., 2005).
Given the potentially oncogenic consequences of tetraploidy, it is not surprising that tumor suppression mechanisms have evolved that limit the proliferation of these cells. Indeed, the restrained growth of tetraploid cells has long been recognized: It was first demonstrated in 1967 that inhibition of cytokinesis in non-transformed cells severely impairs the proliferation of the resulting binucleated tetraploids (Carter, 1967). Subsequently, it became clear that p53 is the key mediator of this arrest (Andreassen et al., 2001; Ganem and Pellman, 2007; Kuffer et al., 2013; Wright and Hayflick, 1972). However, the defect(s) that triggers this stress response, and the downstream signaling pathways that activate p53, remain key unresolved questions in cancer biology. Consequently, the mechanisms by which tetraploid cells might bypass this response to drive tumor development are not known.
We took two approaches to understand the mechanism of p53 activation in tetraploid cells. First, we performed RNAi screens to identify genes that are required to activate or maintain cell cycle arrest in tetraploid cells, but not cells with DNA damage. Second, we carried out in vitro evolution experiments to identify spontaneous adaptations that enable sustained proliferation by tetraploid cells. Our experiments demonstrate that the impaired proliferation of tetraploid cells is due to activation of the Hippo tumor suppressor pathway, both in vitro and in vivo.
The conserved Hippo tumor suppressor pathway regulates cell proliferation by negatively regulating the oncogenic transcriptional co-activators YAP and TAZ (Pan, 2010; Yu and Guan, 2013). This is primarily accomplished through activation of the kinases LATS1 and LATS2, which phosphorylate and inactivate YAP/TAZ. Active LATS2 also binds to and inhibits the E3 ubiquitin ligase MDM2, which normally targets p53 for destruction (Aylon et al., 2006). Thus, activation of the Hippo pathway can limit cellular proliferation in at least two ways: inactivating YAP/TAZ and stabilizing p53.
Defining the inputs that initiate Hippo signaling is central to understanding how the Hippo pathway restrains tumorigenesis. Recent studies have demonstrated that the Hippo pathway is regulated by complex inputs that monitor cell-cell adhesion, cell-matrix adhesion, and contractile tension from the actin cytoskeleton. The molecular details of these upstream signals, especially those from the actin cytoskeleton, are poorly understood (Halder et al., 2012). Here we demonstrate that cytokinesis failure, generating tetraploid cells, is another physiological activator of the Hippo pathway, and we provide mechanistic insight into this important tumor suppression mechanism.
RESULTS
Tetraploid cells activate the p53-pathway
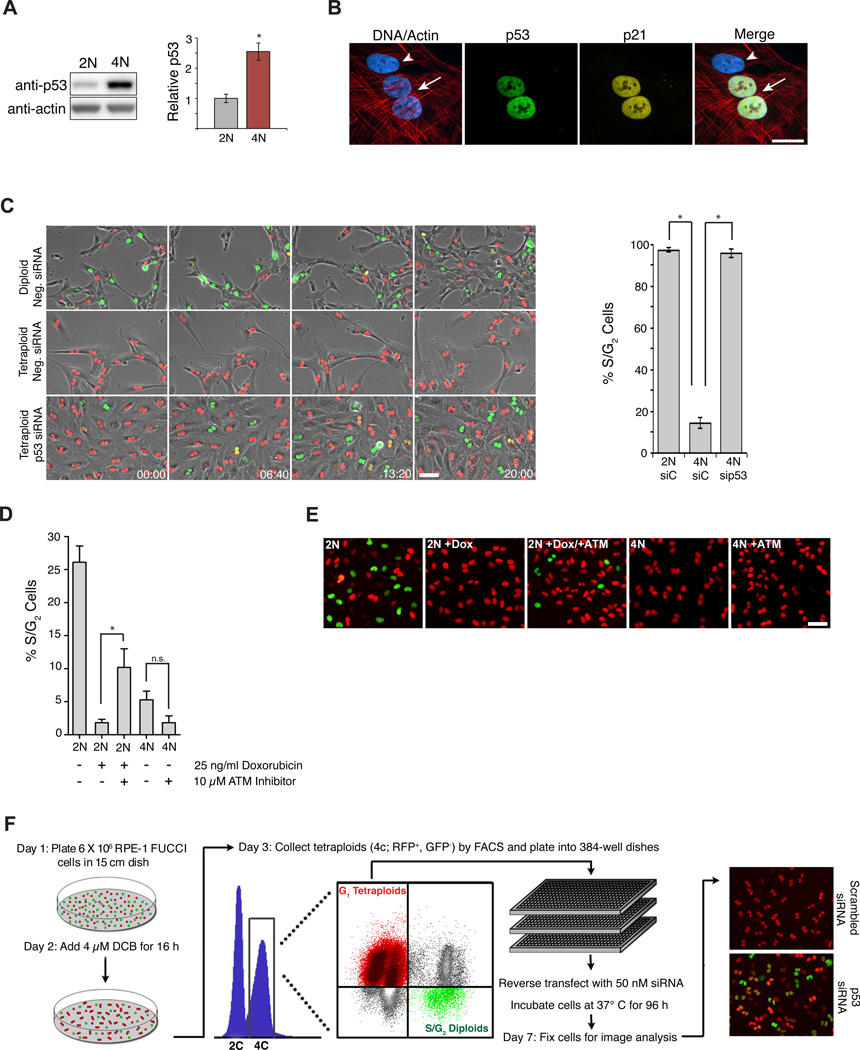
We developed a method to purify tetraploid cells as a prerequisite for an RNAi screen to identify the genes required for the p53-dependent G1 cell cycle arrest following cytokinesis failure. We used RPE-1 cells, which are diploid non-transformed human epithelial cells that maintain a normal p53-response. To generate tetraploid cells, RPE-1 cells were treated with dihydrocytochalasin B (DCB), an inhibitor of actin polymerization that prevents cytokinesis. However, separating the resulting binucleated tetraploids from the remaining diploids poses a technical challenge because the two populations cannot be distinguished by DNA content alone: diploid cells in G2/M have the same DNA content as tetraploid cells arrested G1. We overcame this difficulty by combining DNA content analysis with the FUCCI fluorescent cell cycle reporter system. FUCCI consists of two fluorescent proteins whose expression alternates based on cell cycle position: hCdt1-mCherry is expressed in G1, while hGem-Azami Green is expressed in S/G2/M (Sakaue-Sawano et al., 2008). Using this approach, we could efficiently separate G2/M diploids (green) from G1 tetraploids (red), and were able to isolate ~95% pure G1 tetraploid cells.
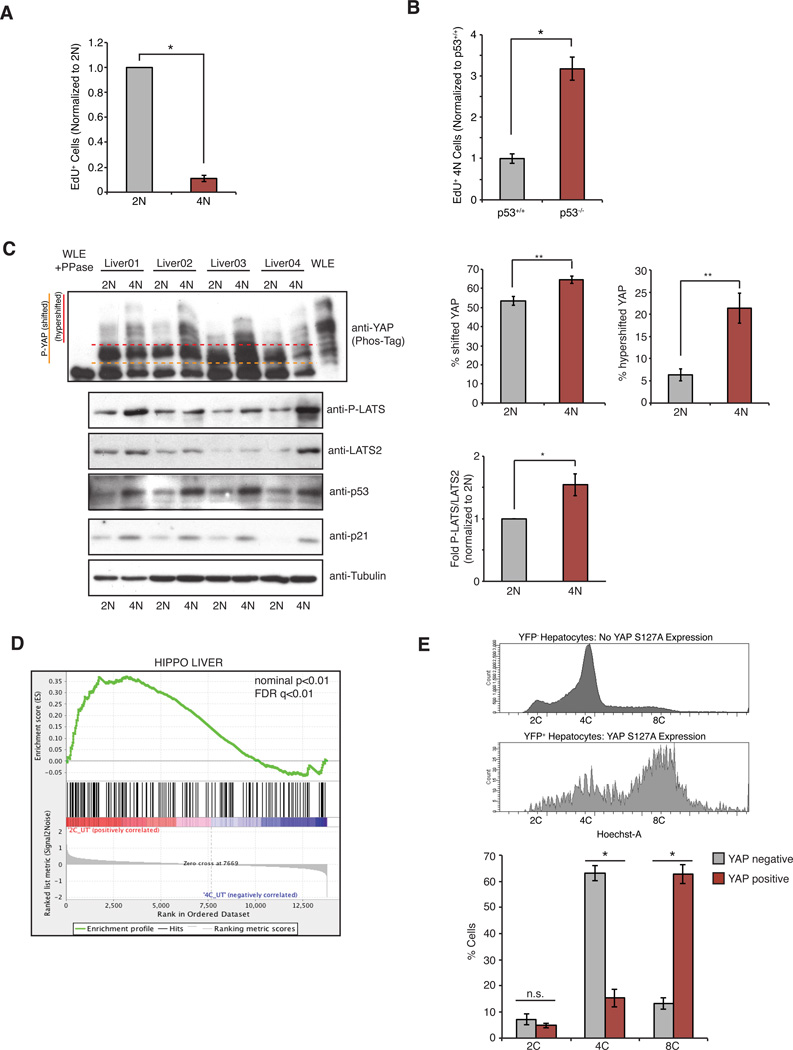
Using the RPE-FUCCI cell line, we assessed p53 levels in tetraploid cells relative to diploids. Levels of p53 (and its target p21) gradually increased in tetraploids to ~2.5-fold over diploid levels by 48 h after cytokinesis failure (Figures 1A, B and S1A, B). To examine the consequences of this p53 accumulation on cell cycle progression, live-cell imaging of diploid and tetraploid FUCCI cells was performed. Imaging revealed that 97.3% of G1 diploids entered S-phase and divided, usually within 24–36 h of re-plating (Figure 1C and Supplementary Movie S1). By contrast, only 14.3% of tetraploid cells entered S-phase within 4 days; the majority remained arrested in G1 until they became senescent (Figures 1C and S1C, Supplementary Movie S2). The rare tetraploid cells that did progress through the cell cycle and complete mitosis predominantly arrested in the following G1 phase, as previously documented (Krzywicka-Racka and Sluder, 2011; Kuffer et al., 2013). Depletion of p53 enabled 95.1% of tetraploid cells to enter S-phase (Figure 1C, Supplementary Movie S3), confirming that the tetraploid arrest is p53-dependent. Similar results were observed when cells were made tetraploid through cytokinesis failure induced by either siRNA-mediated depletion of ECT2 or by treatment with the Aurora B inhibitor Hesperadin (Figure S1E). These data suggest that tetraploidization caused by cytokinesis failure imposes a stress on cells that leads to the stabilization of p53 and eventual G1 arrest.
Figure 1. Genes that mediate cell cycle arrest of tetraploid cells.
(A) Western blot of p53 levels in diploid (2N) and tetraploid (4N) cells 48 h after sorting (n=6; *p < 0.001, unpaired t-test). (B) Diploid (arrowhead) and binucleated tetraploid (arrow) cells stained for p53 (green), p21 (yellow), actin (red), and DNA (blue). Scale bar, 25 µm. (C) Still images of sorted diploid and tetraploid RPE-FUCCI cells that were transfected with the indicated siRNAs (from a live-cell imaging experiment). The percentage of cells that progress into S-phase from 5 independent imaging experiments is shown on right (2N, n=119; 4N, n=326; 4N p53 siRNA, n=211; *p < 0.00001, unpaired t-test). Time, hrs:mins. Scale bar, 100 pm. (D) The fraction of S/G2 cells from diploid (2N) and tetraploid (4N) cells following treatment with ± 25 ng/ml doxorubicin and ± 10 µM ATM inhibitor for 24 h (n=3; *p < 0.05, unpaired t-test, n.s, not significant). (E) Representative images of FUCCI cells from the experiment in (D). Scale bar, 100 µm. (F) Protocol for genome-wide RNAi screen to identify genes necessary to activate or maintain G1 cell cycle arrest in tetraploid cells after cytokinesis failure. All error bars represent mean ± SEM.
Consistent with previous studies (Fujiwara et al., 2005; Krzywicka-Racka and Sluder, 2011), our findings exclude subtle DNA damage as the underlying cause for G1 arrest in tetraploid cells: i) Diploid cells exposed to the same cell culture conditions, drug treatments, and FACS sorting procedures as tetraploid cells did not trigger G1 arrest (Figure 1C). ii) No increase in DNA damage or reactive oxygen species was observed in tetraploid cells(Figure S1F), and the anti-oxidant N-acetylcysteine was unable to overcome tetraploid-induced arrest (Figure S1D). iii) Tetraploid cells failed to enter S-phase even after 14 days in culture, which is ample time for DNA repair (Figure S1C). iv) Tetraploid cells depleted of p53 not only progressed through G1/S, but also through G2/M, strongly arguing against the presence of persistent DNA damage which would activate p53-independent G2 arrest (as is seen in p53-depleted cells after doxorubicin-treatment; Figure 2C). v) Inhibition of ATM kinase was insufficient to overcome tetraploid-induced arrest, despite the fact that it was sufficient to overcome DNA damage-induced arrest caused by low-dose doxorubicin treatment (which elevates p53/p21 protein levels to a similar extent as tetraploidization) (Figure 1D, E). vi) Finally, below we describe genetic conditions permissive for tetraploid cells, but not cells with DNA damage, to continue proliferating (and vice versa), demonstrating that these arrest mechanisms are fundamentally distinct.
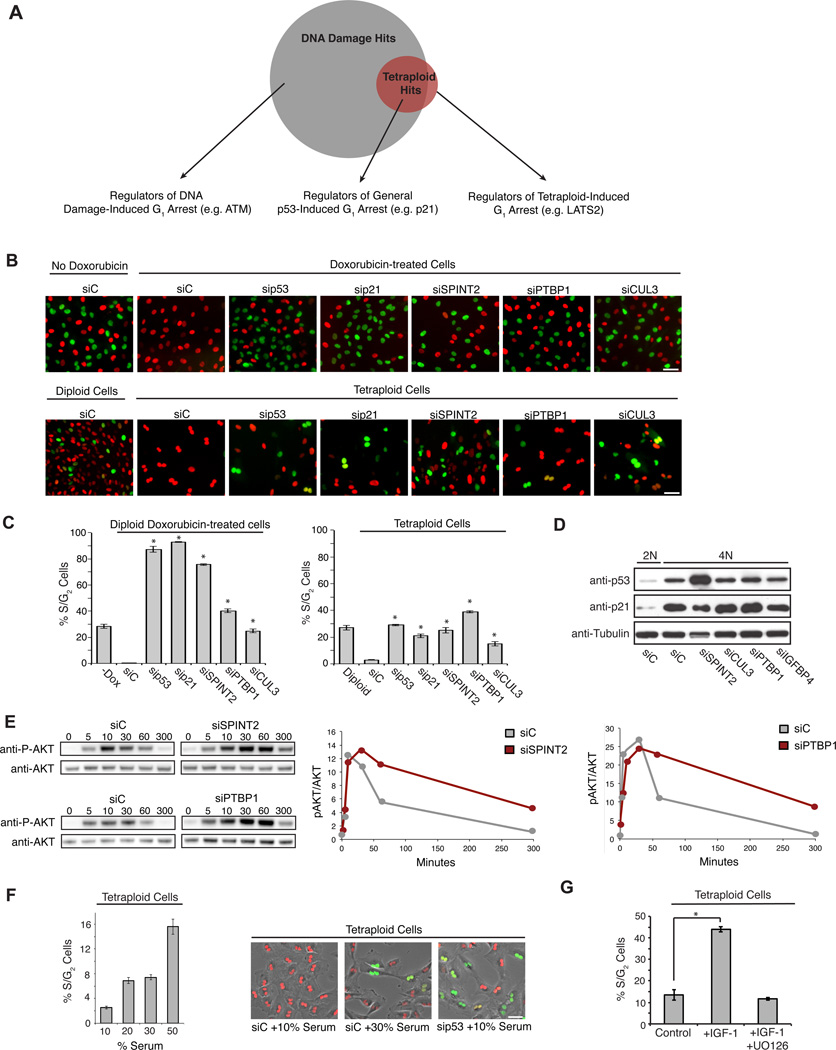
Figure 2. Enhanced growth factor signaling overcomes p53-induced G1 arrest.
(A) Venn diagram depicting genes necessary to maintain G1 arrest in response to low-level DNA damage (grey), tetraploidization (red), or both. (B) Representative images of G1-arrested diploid RPE-FUCCI cells treated ± 40 ng/ml doxorubicin (top row), and untreated tetraploid RPE-FUCCI cells (bottom row), transfected with the indicated siRNAs. Scale bar, 50 µm. (C) Quantification of the percentage of S/G2 from (B) (n=3; *p < 0.002, unpaired t-test). (D) Western blot of p53 and p21 protein levels in 2N and 4N cells transfected with the indicated siRNAs for 48 h. (E) A representative western blot (and quantitation) of phosphorylated AKT (p-AKT) relative to total AKT at various time points following serum addition to starved cells that were transfected with the indicated siRNAs. (F) The percentage of S/G2 tetraploid RPE-FUCCI cells in growth medium containing increasing concentrations of serum from one representative experiment (n=48 for each condition). Right: representative images from a live-cell experiment. Scale bar, 50 µm. (G) The percentage of S/G2 tetraploid RPE-FUCCI cells in growth medium supplemented with 50 ng/ml IGF-1 or IGF-1 and the MEK inhibitor U0126 (10 nM) (n=3; *p < 0.0005, unpaired t-test). All error bars represent mean ± SEM.
A genome-wide RNAi screen to identify regulators of tetraploid-induced cell cycle arrest
We performed an RNAi screen to identify the genes required for tetraploid-induced G1 cell cycle arrest using pooled siRNAs targeting the druggable portion of the human genome (~7300 genes). Automated image analysis measured the total number of tetraploid cells per well and the percentage that emitted green fluorescence (indicative of S/G2/M and proliferation) at 96 h post-transfection (Figure 1F).
The screen identified 98 proteins for which depletion with at least two individual siRNAs allowed tetraploid cells to escape G1 arrest (Supplementary Table 1): two of the strongest hits identified from this screen were p53 and p21, validating the overall approach. GESS analysis of all siRNA sequences that released tetraploid cells from arrest revealed no common 3’ UTR region, reducing the possibility of a common off-target gene (Sigoillot et al., 2012).
Although the primary objective of the tetraploid screen was to uncover genes that are necessary to activate or maintain tetraploid-induced cell cycle arrest, we anticipated that many of the candidates we identified would also have roles in the general maintenance of p53-induced cell cycle arrest, independent of tetraploidy. To distinguish between these two broad classes of genes, we performed a second genome-wide screen to identify genes that, when suppressed, enable cells to bypass G1 arrest in response to low-level DNA damage. This screen was similar in design to the tetraploid screen detailed above, except that instead of cells being treated with DCB to create tetraploids, they were continuously treated with 40 ng/ml of the DNA-damaging drug doxorubicin, which elevates p53 and p21 levels to a similar extent as cytokinesis failure (Figures S2A and 4C).
A comparison of the results from the tetraploid screen with the DNA damage screen identified three classes of gene knockdowns (Figure 2A – C): i) Those that allow cells with DNA damage (but not tetraploid cells) to progress into S-phase, suggesting a specific role for the encoded proteins in the DNA damage response (e.g. ATM) (Supplementary Table 2). These factors were not further characterized in this study. ii) Knockdowns that enable both tetraploid cells as well as cells with DNA damage to progress into S-phase, implying a role for the encoded proteins in the general maintenance of cell cycle arrest upon p53 activation (e.g. p21). iii) Knockdowns that allow tetraploid cells (but not cells with DNA-damage) to progress into S-phase, implying a role for the encoded proteins in activating or maintaining G1 arrest specifically in the context of tetraploidy (e.g. LATS2).
Hyperactivation of growth factor signaling is a general mechanism to bypass tetraploid-induced arrest
We expected that our screens might identify genes that are generally required to activate p53. To test this, we assessed p53/p21 levels following siRNA knockdown of several strong screen hits. Surprisingly, depletion of most genes did not reduce p53/p21 levels, demonstrating that cell proliferation can occur via mechanisms that bypass p53 activation rather than suppressing it (Figure 2D).
Interestingly, many strong hits from the screen (e.g. SPINT2) are putative negative regulators of growth factor signaling (Nakamura et al., 2011), suggesting that sustained growth factor signaling may be one route to overcome p53 cell cycle arrest. We measured the kinetics of ERK1/2 and AKT activation after serum addition to serum-starved cells and confirmed that depletion of our strongest hit, SPINT2, causes a sustained increase in growth factor signaling (Figures 2E and S2B). We also found that increasing the concentration of serum or adding recombinant IGF-1 alone was sufficient to overcome G1 arrest in tetraploid cells (Figures 2F, G). Thus, one general mechanism for bypassing p53-dependent cell cycle arrest of tetraploid cells is to activate growth factor signaling; this suggests that other hits identified from the screen might therefore represent novel negative regulators of growth factor signaling. In fact, we found that depletion of the RNA binding protein PTBP1, one of the strongest hits from the screen, similarly increased growth factor signaling (Figures 2E and S2B).
Tetraploid cells activate the Hippo pathway
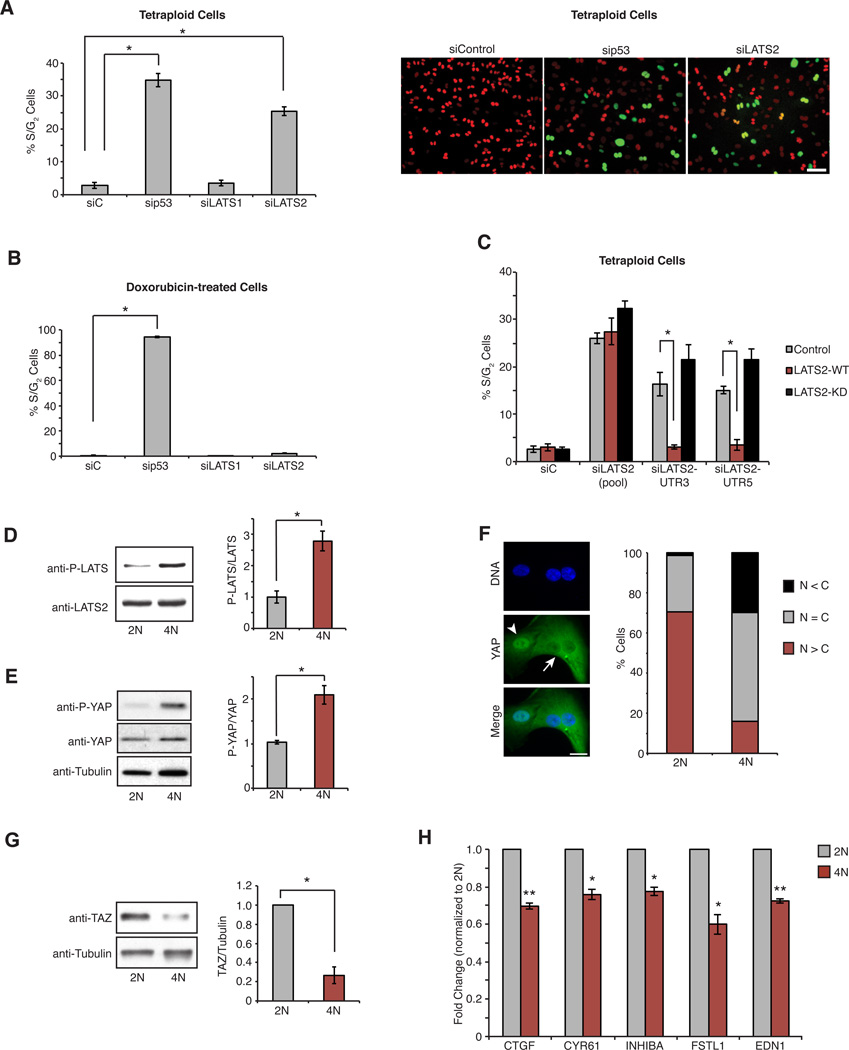
We next focused on genes that activated or maintained G1 arrest specifically in the context of tetraploidy. The strongest ploidy-specific hit was the kinase LATS2, a core component of the Hippo tumor suppressor pathway (Pan, 2010; Yu and Guan, 2013). We confirmed that depletion of LATS2 with 5 independent siRNAs, targeting both the coding and UTR regions of LATS2 mRNA, enabled the proliferation of tetraploid cells but not cells with DNA damage (Figures 3A, B and S3A–C). Depletion of LATS2 similarly promoted the proliferation of tetraploid cells generated by siRNA depletion of the cytokinesis regulators ECT2 and PRC1 (Figure S3D). Importantly, expression of siRNA-resistant wild-type LATS2 in proliferating tetraploid cells that were depleted of endogenous LATS2 was sufficient to rescue cell cycle arrest, confirming that the effect of LATS2 depletion is not due to off-target RNAi effects (Figure 3C). By contrast, expression of kinase-dead LATS2 failed to rescue, indicating that the arrest mechanism requires the kinase activity of LATS2 (Figure 3C). Depletion of LATS1 kinase, which is highly homologous to LATS2 and has many overlapping functions, was not sufficient to enable the proliferation of tetraploids (Figure 3A); however co-depletion of both LATS1 and LATS2 produced a small increase in the rate of proliferation of tetraploid cells relative to LATS2 depletion alone (Figure S3C).
Figure 3. Tetraploid cells activate the Hippo pathway.
(A) The percentage of S/G2 tetraploid RPE-FUCCI cells following transfection with the indicated siRNAs (n=4; *p < 0.0001, unpaired t-test). Representative images from (A) are on right. Scale bar, 100 µm. (B) As in (A) except cells are treated with 40 ng/ml doxorubicin (n=3; *p < 0.0001, unpaired t-test). (C) The percentage of S/G2 tetraploid RPE-FUCCI cells stably expressing wild-type LATS2 (LATS2-WT), kinase-dead LATS2 (LATS2-KD), or empty vector control (Control), and transfected with the indicated siRNAs (n=3; *p < 0.007, unpaired t-test). (D) Western blot analysis and quantitation of LATS phosphorylation in 2N and 4N cells (n=3; *p < 0.009, unpaired t-test). Note: cells stably overexpressing LATS2 were used in this experiment to better assess LATS2 phosphorylation. (E) Western blot analysis and quantitation of YAP phosphorylation (S127) in 2N and 4N cells (n=3; *p < 0.008, unpaired t-test). (F) Diploid (arrowhead) and binucleated tetraploid (arrow) RPE-1 cells stained for YAP (green) and DNA (blue). YAP localization was quantified (n=2; N = C, YAP is evenly distributed; N < C, YAP is enriched in the cytoplasm; N > C, YAP is enriched in nucleus). Scale bar, 25 µm. (G) Western blot analysis and quantitation of TAZ levels in 2N and 4N RPE-FUCCI cells (n=4; *p < 0.005, one sample t-test). (H) qPCR analysis of YAP target gene expression in 2N and 4N cells (n=3; *p < 0.02, **p < 0.005, one sample t-test). All error bars represent mean ± SEM.
These data suggested that G1 arrest in tetraploids requires LATS2 activation. Indeed, LATS2 was phosphorylated to a significantly greater extent in tetraploid cells (Figure 3D), which led to a corresponding increase in the level of phosphorylated YAP in tetraploid cells relative to diploids (Figures 3E and S3E, H). We also found that YAP was biased to be cytoplasmic (inactive) in tetraploid cells, in contrast to its primarily nuclear (active) localization in diploid cells (Figure 3F). This finding was independently verified in tetraploid cells that were generated by use of ECT2 siRNA (Figure S3F). In addition, levels of the YAP-related transcriptional co-activator TAZ, which is proteasomally degraded upon phosphorylation by active LATS, were significantly decreased in tetraploid cells (Figure 3G). Furthermore, canonical YAP/TAZ target genes had reduced expression in tetraploid cells (Figure 3H). This differential activation of Hippo signaling between diploids and tetraploids is not due to subtle differences in cell cycle position because diploid and tetraploid cells synchronized in G1 by release from serum starvation displayed the same changes (Figure S3G and S3H). Hippo activation in tetraploids is also not a secondary consequence of p53 activation (Aylon et al., 2006), as depletion of p53 did not prevent Hippo activation in tetraploid cells (Figure S3I). Thus, LATS2-mediated Hippo signaling is selectively activated in tetraploid cells.
Next we characterized the mechanisms through which depletion of LATS2 overcomes tetraploid-induced arrest. As expected, depletion of LATS2 activated YAP, as demonstrated by a decrease in YAP phosphorylation and an accumulation of nuclear YAP (Figures 4A and S4A). Moreover, overexpression of a constitutively active mutant version of YAP (with all of the LATS phosphorylation sites mutated to alanines: YAP-S5A), released tetraploid cells into the cell cycle, irrespective of whether they were generated by DCB treatment or siRNA-mediated knockdown of ECT2 or PRC1. Importantly, expression of constitutively active YAP-S5A restored proliferation to tetraploid cells without affecting the steady-state levels of p53 (Figures 4B and S4B–D). By contrast, overexpression of WT-YAP alone was insufficient to drive tetraploid cells into the cell cycle, presumably because of the ability of endogenous LATS kinases to phosphorylate and inactivate WT-YAP.
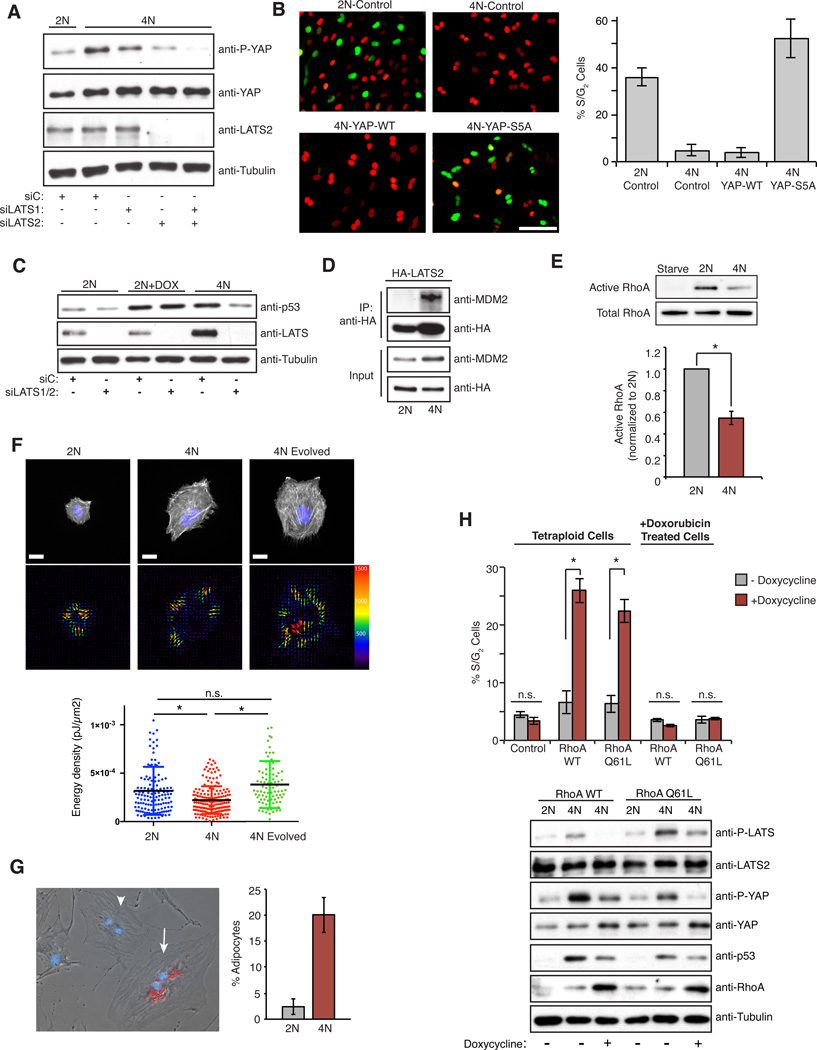
Figure 4. LATS2 inhibits the proliferation of tetraploid cells by stabilizing p53 and inactivating YAP, and is triggered by reduced RhoA activity.
(A) Western blot of p53, LATS2, and active YAP (p-S127) levels in 2N and 4N cells transfected with the indicated siRNAs. (B) Representative images of 2N and 4N RPE-FUCCI cells overexpressing either empty-vector control (Control), YAP-WT, or YAP-S5A, with quantitation of the percentage of S/G2 cells for each condition shown on right (from one representative experiment). Scale bar, 100 µm. (C) Western blot analysis of p53 and LATS2 levels in 2N, 4N, and 40 ng/ml doxorubicin-treated 2N (2N +Dox) RPE-FUCCI cells transfected with the indicated siRNAs. (D) Co-immunoprecipitation of HA-tagged LATS2-WT and endogenous MDM2 from 2N and 4N RPE-1 cells using anti-HA antibodies (one of two independent experiments). (E) Western blot analysis and quantitation of pull-down assays to detect active RhoA relative to total RhoA in serum-starved, 2N, and 4N RPE-1 cells (n=4; *p < 0.0003, one sample t-test). (F) Top Row: 2N, 4N, and Evolved 4N RPE-1 cells labeled for actin (Life-Act, white) and DNA (blue) on a PAA hydrogel. Bottom Row: Images showing the force field corresponding to the upper cells. The size and color of the arrows correspond to traction force magnitude measured (in Pascals). Below: The average energy density generated by each cell. 2N and 4N data points are from 4 independent experiments; 4N Evolved data points are from 2 independent experiments; *p < 0.0001, unpaired t-test. Scale bar, 20 µm. (G) hMSCs grown in mixed differentiation medium and stained with Oil Red. A non-differentiated diploid hMSC (arrowhead) and a tetraploid adipocyte (arrow) are highlighted. Right: The percentage of diploid and binucleated tetraploid cells that differentiate into adipocytes (n=2 independent experiments). (H) The percentage of S/G2 tetraploid or doxorubicin-treated RPE-FUCCI cells ± induction of RhoA-WT, RhoA-Q61L, or empty-vector control (Control) for 20 h with the corresponding western blot analysis of Hippo pathway activation (n=3; *p < 0.005, unpaired t-test). All error bars represent mean ± SEM.
Next, we examined whether active LATS2 contributes to the observed increase in p53 levels in tetraploid cells. We found that depletion of LATS2 from tetraploid cells restored p53 to the basal levels observed in diploid cells (Figures 4C). By contrast, depletion of LATS1 had no effect (Figure S4E), explaining why depletion of LATS1 alone was insufficient to promote tetraploid proliferation (Figures 3A and S3A). Although depletion of LATS2 reduced p53 levels in tetraploid cells, the kinase had no effect on the accumulation of p53 in cells with DNA damage (Figure 4C). These data establish that the mechanism underlying p53 stabilization and cell cycle arrest in tetraploid cells is LATS2-dependent, but is functionally distinct from that used to activate p53 in cells with DNA damage.
Active LATS2 binds and inhibits the E3 ubiquitin ligase MDM2, which targets p53 for destruction, thereby indirectly leading to the stabilization of p53 (Aylon et al., 2006). We found that LATS2 interacted with MDM2 in tetraploid, but not diploid, cells (Figure 4D), similar to what had been previously observed in tetraploid cells generated from mitotic slippage (Aylon et al., 2006). Collectively, these findings demonstrate that activation of LATS2 in tetraploid cells inactivates YAP and stabilizes p53.
Tetraploid cells have reduced RhoA activity
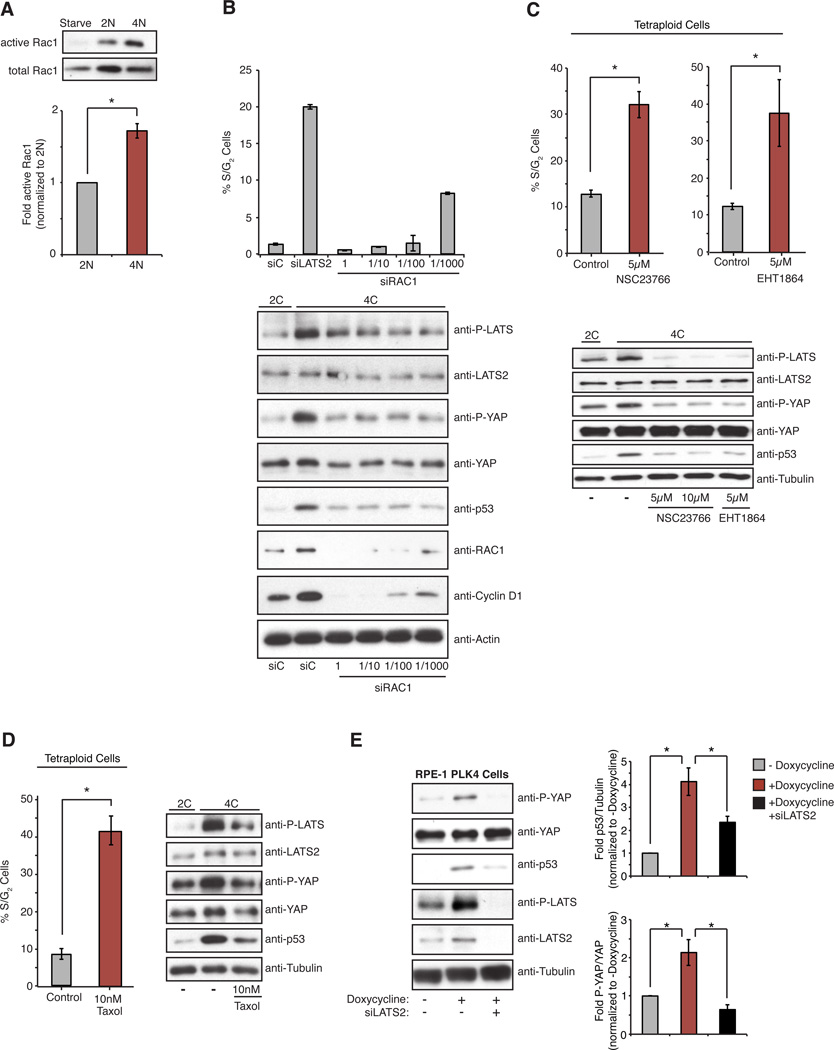
We sought to understand the mechanisms leading to LATS2 activation in tetraploid cells. Canonically, the kinases MST1 and MST2 act directly upstream of LATS2 in Hippo pathway signaling; however, we found that co-depletion of MST1/2 did not trigger the proliferation of tetraploids, demonstrating that the tetraploid-induced activation of LATS2 is MST1/2-independent (Figure S4F). Recent work shows that reduced assembly or contractility of the actin cytoskeleton, or reduced RhoA activity, can activate LATS1/2 independent of MST1/2 (Mo et al., 2012; Wada et al., 2011; Yu et al., 2012; Zhao et al., 2012). The cytoskeleton of tetraploid cells is qualitatively different from that of diploid cells: tetraploid cells are bigger, possess longer stress fibers, and have extra centrosomes that cluster together and nucleate a greater number of microtubules (Figure S4G). However, they do not exhibit obvious defects in cell attachment, migration, or cell spreading (Supplementary Movie S2). Nevertheless, we found that active RhoA was reduced by ~50% in tetraploid cells compared to diploids (Figure 4E), which correlates well with a comparable decrease in downstream phosphorylation of myosin light chain (Figure S4H).
The biological impact of this reduction of RhoA activity was assessed using two independent approaches. First, we compared the contractility of diploid and tetraploid cells using traction force microscopy. A mix of diploid and tetraploid RPE-1 cells was plated on deformable poly-acrylamide hydrogels coated with fibronectin. Gel deformation induced by the contraction of cells was then monitored to measure the forces produced. This analysis demonstrated that, relative to their size, tetraploid cells exerted significantly reduced contractile force on the underlying substrate than diploids (Figures 4F and Figure S5A, B). We also employed a commonly used indirect assay for the tensile state of cells, the differentiation of human mesenchymal stem cells (hMSCs)(McBeath et al., 2004). When hMSCs have high RhoA activity and are more contractile, which occurs when these cells are plated at low density on stiff substrates, hMSCs activate YAP and primarily differentiate into osteoblasts. By contrast, when RhoA activity is low, which occurs when cells are contact-inhibited, hMSCs exhibit less YAP activation and more frequently differentiate into adipocytes. Supporting our finding that tetraploids have less active Rho, we found that when isogenic diploid and tetraploid hMSCs were plated in mixed differentiation medium at low densities, tetraploid cells significantly more frequently differentiated into adipocytes (Figure 4G).
Finally, re-activation of RhoA was sufficient to inactivate the Hippo pathway, reduce p53 levels, and restore proliferative capacity to tetraploid cells (Figure 4H). Treatment of G1-arrested tetraploid cells with LPA or S1P (glycophospholipids known to activate RhoA) (Miller et al., 2012; Yu et al., 2012) significantly increased the fraction of tetraploid cells that entered the cell cycle (Figure S5C), as did induced expression of RhoA-WT or constitutively active RhoA-Q61L (Figure 4H). Importantly, neither of these conditions had any effect on the G1 arrest imposed by low level DNA damage (Figure 4H and Figure S5D). These data demonstrate that tetraploid cells have reduced RhoA activity, which triggers LATS2 activation and stalls the cell cycle by activating p53 and inactivating YAP.
Extra centrosomes promote activation of the Hippo pathway
We sought to define the upstream trigger for RhoA suppression and LATS2 activation in tetraploid cells. One of the most obvious differences between diploid and tetraploid cells lies in the number of centrosomes they each possess: Diploid cells have one centrosome during G1 phase, whereas tetraploid cells have two. Consequently, tetraploid cells have an increased number of centrosomal microtubules (Figures S4G). Independent work in our laboratory has revealed that extra centrosomes increase centrosomal microtubule assembly and stimulate the activity of the small G-protein Rac1 (Godinho et al., 2014). Rac1 activation by microtubules was previously described (Waterman-Storer et al., 1999) and this activation appears to require dynamic microtubules because it is suppressed by Taxol-induced microtubule stabilization. Because active Rac1 often antagonizes RhoA (Sander et al., 1999), we tested the possibility that the extra centrosomes of tetraploid cells may mediate Hippo pathway activation via microtubule-dependent hyperactivation of Rac1. We measured the relative levels of active Rac1 in diploids and tetraploids and found a nearly two-fold increase in Rac1 activity in tetraploids (Figure 5A). This increase in active Rac1 is functionally significant, as quenching hyperactivation of Rac activity in G1-arrested tetraploids through siRNA depletion or pharmacological inhibition was sufficient to inactivate Hippo signaling and reduce p53 levels (Figures 5B,C and S5E). Although complete depletion of Rac1 is known to inhibit cyclin D accumulation and halt proliferation (Figure 5B), we found that graded attenuation of Rac activity with partial siRNA knockdown or low-dose drug treatment could enable tetraploid cells to proliferate (Figures 5B, C and S5E). Strikingly, we found that Taxol-treatment of cells also inhibited Rac activation, prevented Hippo activation and p53 accumulation, and enabled cells to override tetraploid-induced G1 arrest (Figure 5D). By contrast, inactivation of Rac had no effect on G1 arrest imposed by DNA damage (Figure S5F).
Figure 5. Increased Rac activity triggered by extra centrosomes activates the Hippo pathway.
(A) Western blot analysis and quantitation of pull-down assays to measure active Rac1 relative to total Rac1 in serum-starved, 2N, and 4N RPE-1 cells (n=5; *p < 0.003, one sample t-test). (B, C, D) The percentage of S/G2 tetraploid RPE-FUCCI cells following siRNA-mediated depletion of Rac1 (B), ± 5 µM treatment with the Rac inhibitors NSC2376 and EHT1864 (C), or 10 nM treatment with Taxol (D), along with corresponding western blot analysis of Hippo pathway activation (n ≥ 3; *p < 0.01, unpaired t-test). (E) Western blot analysis (left) and quantitation (right) of Hippo activity and p53 levels in RPE-1 cells with or without transient doxycycline-induced PLK4 overexpression and extra centrosomes. Cells were treated with control or LATS2 siRNAs prior to doxycycline treatment (n=10; *p < 0.01, unpaired t-test). Error bars represent the mean ± SEM.
These data suggested that the extra centrosome present in tetraploid cells is one trigger for Hippo pathway activation. To test this, we induced extra centrosomes in diploid RPE-1 cells by transient overexpression of PLK4 kinase (a key regulator of centriole biogenesis) and then measured Hippo pathway activity. As expected, PLK4 overexpression led to the formation of extra centrosomes in 60–80% of cells after 48 h, and these cells showed subtle activation of the Hippo pathway as judged by increases in the phosphorylation of LATS2 and YAP (Figure 5E). As previously reported, these cells also exhibited elevated levels of p53 and proliferated slowly (Figure 5E)(Holland et al., 2012). In common with what was observed in tetraploid cells, depletion of LATS2 significantly diminished Hippo pathway activation and p53 levels in PLK4 overexpressing cells (Figure 5E). These results indicate that the Hippo pathway activation observed in tetraploids is initiated, at least in part, by the effects of extra centrosomes.
Tetraploid hepatocytes activate the Hippo pathway in vivo
To determine if spontaneously arising tetraploid cells activate the Hippo pathway and exhibit reduced proliferation in vivo, we focused on the developing murine liver: hepatocytes of newborn mice are initially diploid, but become progressively tetraploid with age via programmed cytokinesis failure (Duncan, 2013). We found that the proliferation of tetraploid hepatocytes was inhibited, but not abolished, as evidenced by a significant reduction in the incorporation of EdU that was injected into 3-week old animals (Figure 6A). We also confirmed a requirement for p53 in this growth limitation: EdU incorporation in tetraploid hepatocytes was significantly increased in p53−/− mice (Figure 6B)(Kurinna et al., 2013). Furthermore, tetraploid hepatocytes showed increased levels of p53 and p21 relative to diploids (Figure 6C).
Figure 6. Tetraploid hepatocytes activate the Hippo pathway in vivo.
(A) Relative incorporation of EdU in 2N and 4N hepatocytes of wild type mice as determined by FACS (n=5; *p < 0.0005, one sample t-test). (B) Relative incorporation of EdU in 4N hepatocytes from p53+/+ and p53−/− animals (n=4; *p < 0.0005, unpaired t-test). (C) Western blot analysis of p53, p21, LATS2, and phosphorylated LATS2 protein levels in diploid (2N) and tetraploid (4N) hepatocytes isolated from the livers of 4 wild-type mice. YAP phosphorylation was analyzed using PhosTag™ gels. Whole liver extract (WLE) treated with λ-phosphatase was used as control for dephosphorylated YAP. The relative amounts of phosphorylated LATS2, and shifted (above orange line) and hypershifted (above red line) YAP are quantified on right (n=4; *p < 0.01, **p < 0.008, paired t-test). (D) Expression profiles of 2C and 4C hepatocytes, isolated from the livers of 3 different mice, were compared to a hepatocyte-specific Hippo-active gene-set signature (constructed as described in Experimental Procedures). (E) Constitutively active YAP (YAPS127A) was induced by doxycycline addition in a subset of hepatocytes in the livers of 3-week old mice. DNA content of YAP-expressing (YFP+) and YAP-non-expressing (YFP-) hepatocytes was analyzed using FACS (n=3; *p <0.0005, unpaired t-test). All error bars represent mean ± SEM.
Our experiments also indicated that primary tetraploid hepatocytes activate the Hippo pathway, as judged by increases in phosphorylated LATS and YAP in tetraploid hepatocytes relative to diploids (Figure 6C). Moreover, we performed gene expression analysis on primary diploid and tetraploid hepatocytes, and gene-set enrichment analysis (GSEA) indicated that tetraploid hepatocytes displayed significant repression of hepatocyte-specific YAP target genes relative to diploids (Figure 6D and Figure S6A, B). To address if restoring YAP activity is sufficient to promote tetraploid-hepatocyte proliferation in vivo, doxycycline-inducible YAP S127A (a constitutively active version of YAP) was expressed in the livers of 3-week old mice for 7 days. FACS analysis revealed that expression of active YAP (identified by expression of a YFP fluorescence reporter; Figure S6C) enhanced the proliferation of tetraploid hepatocytes in vivo, as evidenced by a marked increase in cell ploidy (Figure 6E). These data demonstrate that the Hippo pathway restrains the growth of tetraploid cells in vivo.
Evolution experiments identify adaptive mechanisms of proliferating tetraploid cells
Our data suggest that proliferation of tetraploid cells can be accomplished via direct inactivation the Hippo pathway, or alternatively, by hyperactivation of growth factor signaling. As an orthogonal approach to uncover pathways that can overcome the block to proliferation of tetraploids, we performed an in vitro evolution experiment: we generated a large number (~1 × 108) of tetraploid RPE-1 cells and FACS-isolated the rare tetraploid cells that were capable of re-entering the cell cycle (as indicated by an 8C DNA content). As expected, the majority of these tetraploids did not proliferate. Repeated FACS-sorting was then used to isolate a pure population of actively dividing tetraploid cells. These “evolved tetraploids” were chromosomally stable, and karyotypic analysis showed that they predominantly possessed ~92 chromosomes, with only a subset of cells carrying a gain of chromosome 12. This chromosome stability is likely due to the selection for tetraploid cells that lost their supernumerary centrosomes, and that maintain relatively balanced gene expression (Ganem et al., 2009).
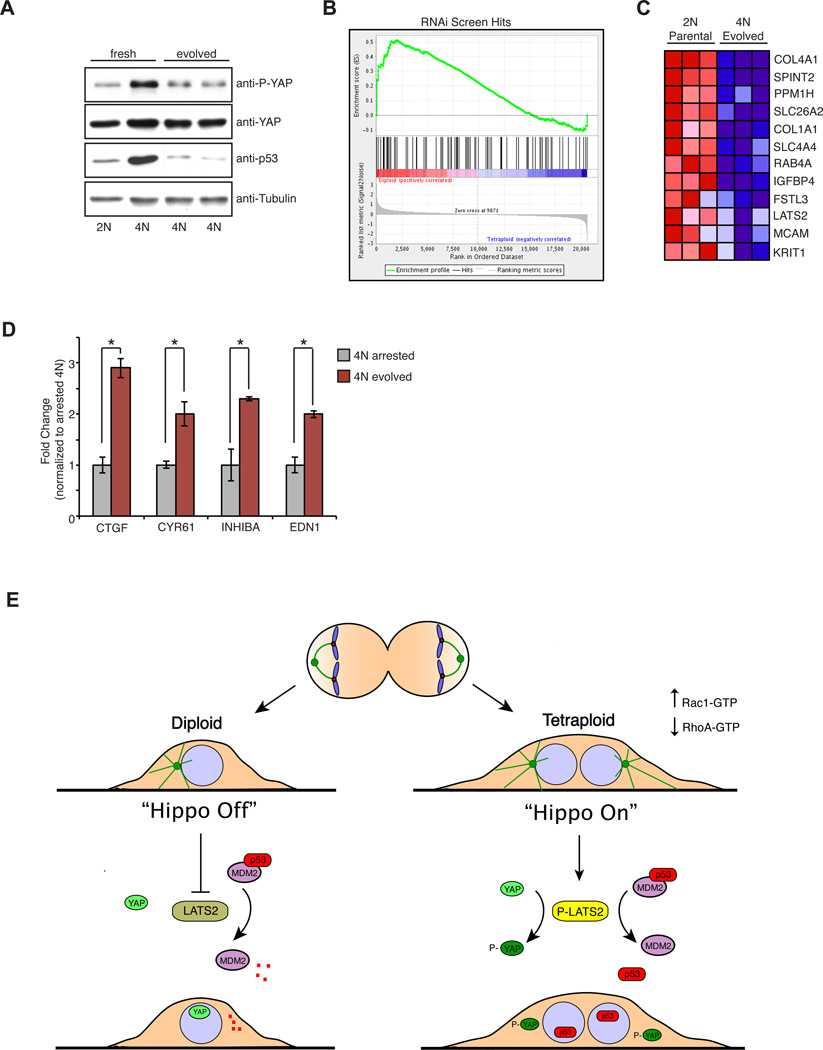
The evolved tetraploids exhibited reduced levels of p53 and p21 as compared to freshly prepared tetraploids, despite the fact that the p53 pathway remained functional in these cells (Figure 7A and S7B). Gene expression profiling was used to compare evolved tetraploids with the diploids from which they were originally derived in order to uncover adaptations that the evolved cells may have acquired to bypass the proliferative block. Remarkably, of the 98 genes identified as hits from the siRNA screen, ~20 (including LATS2) were repressed in the evolved tetraploids (Figure 7B, C). Indeed, GSEA confirmed that, as a group, hits from the RNAi screen were significantly downregulated in the evolved tetraploids (Figure 7B). Because the proliferating tetraploids arose spontaneously, plasticity in gene expression programs may enable rare cells to overcome the p53 activation triggered by tetraploidy.
Figure 7. In vitro evolution to generate proliferating tetraploid cells.
(A) Western blot analysis of YAP phosphorylation and p53 levels in evolved 4N RPE-1 cells relative to freshly generated 4N cells. (B) GSEA analysis demonstrates that the expression of those genes identified from the RNAi screen are, as a whole, significantly reduced in the evolved tetraploids (nominal p-value < 0.01; FDR q-value < 0.01). (C) Expression of genes (uncovered by the RNAi screen) that are the most significantly repressed in the evolved tetraploids. Each column represents a technical replicate. Red to blue coloring represents high to low gene expression. (D) qPCR analysis of Hippo target genes in evolved tetraploids relative to freshly generated tetraploids (n=3; *p < 0.05, unpaired t-test). Error bars represent the mean ± SEM. (E) Binucleated tetraploid cells arising from cytokinesis failure have an additional centrosome that leads to microtubule-dependent hyperactivation of Rac1 and a corresponding decrease in active RhoA. Reduced RhoA activity leads to activation of LATS2 kinase, which phosphorylates and inactivates YAP, as well as stabilizes p53 through direct inhibition of MDM2 (“Hippo On”). Both p53 stabilization and YAP inactivation are necessary to prevent the proliferation of tetraploid cells. Cells that overcome this tumor suppression mechanism are genetically unstable and can promote tumorigenesis.
Finally, we found that the evolved tetraploids inactivated the Hippo pathway, as judged by a decrease in phosphorylated YAP, restoration of YAP nuclear localization, and a corresponding increase in the expression of YAP target genes (Figures 7A, D and S7A). This is likely due, at least in part, to both reduced level of LATS2 expression (Figure 7C) as well as loss of the additional centrosome. Indeed, evolved tetraploids with a normal number of centrosomes exhibited normal contractility by traction force microscopy, suggesting restoration of normal Rac and Rho function (Figures 4F and Figure S5A, B). These data provide an independent confirmation of the importance of silencing Hippo signaling to enable proliferation of tetraploid cells.
DISCUSSION
Spontaneously arising tetraploid cells that result from non-programmed mitotic failures pose a serious threat to organismal health because proliferating tetraploid cells are genomically unstable and can facilitate tumor development (Davoli and de Lange, 2011; Ganem et al., 2007). Tumor suppression mechanisms appear to have evolved to neutralize potential risks associated with tetraploidy (Ganem and Pellman, 2007; Senovilla et al., 2012). However, the mechanisms that sense tetraploidization and trigger p53 pathway activation have been poorly defined and controversial.
Early studies that used drug treatments to induce cytokinesis failure found that tetraploid, but not diploid cells within the same population displayed a near complete loss of cell proliferation, a finding that led to the proposed existence of a tetraploidy-checkpoint (Andreassen et al., 2001; Carter, 1967). However, subsequent work documented that G1 arrest is not an obligatory outcome of tetraploidization, and that when tetraploid cells are maintained under ideal tissue culture conditions, a significant fraction of cells can re-enter the cell cycle (Uetake and Sluder, 2004; Wong and Stearns, 2005). The data we present here can reconcile this apparent discrepancy: we show that tetraploidization does not impose a requisite cell cycle block, but rather initiates a gradually accumulating p53 response (Figure S1B) which only manifests as a G1 arrest once p53 levels induce sufficient p21 to cross a critical threshold that is necessary to inhibit S-phase entry. Accordingly, conditions that prolong G1 phase in tetraploid cells, and thus provide more time for p53/p21 to accumulate, are more efficient at promoting an immediate cell cycle arrest. The gradual accumulation of stress, which ultimately triggers cell cycle arrest in tetraploids, is very similar to the arrest observed in cells with extra centrosomes (Holland et al., 2012).
We demonstrate that the Hippo tumor suppressor pathway has a central role in limiting the proliferation of tetraploid cells, both in vitro and in vivo. We show that tetraploid cells activate LATS2 kinase, inactivate YAP/TAZ-dependent transcription, and stabilize p53 (Figure 7E). Our data are consistent with prior work showing that LATS2 is required for cell cycle arrest after mitotic slippage (Aylon et al., 2006). These data confirm that tetraploidy imposes stress on cells, and suggest that the development of high-ploidy tumors requires that cells adapt to overcome or bypass these inherent limitations to growth.
Defects that activate the Hippo pathway in tetraploid cells
A number of recent studies demonstrate that mechanical forces generated by the actomyosin cytoskeleton have a major role in regulating YAP/TAZ through both LATS-dependent and independent pathways (Aragona et al., 2013; Dupont et al., 2011; Halder et al., 2012; Mana-Capelli et al., 2014; Mo et al., 2012; Wada et al., 2011; Yu et al., 2012). Although the mechanisms that link the contractility of the actin cytoskeleton to YAP activity remain unknown, it is clear that RhoA plays a key role: active RhoA promotes the assembly and contraction of actin filaments within cells and leads to LATS2 inhibition and YAP/TAZ activation, whereas cells with reduced RhoA activity display LATS2 activation and inhibition of YAP/TAZ. Our data demonstrate that a major defect in tetraploid cells is a significant reduction in RhoA activity: the level of active RhoA in these cells is roughly half of that observed in diploids, and increasing RhoA activity is sufficient to rescue proliferation of tetraploid cells (Figures 4E, H).
Several distinct, but not mutually exclusive, mechanisms may explain the observed reduction of RhoA activity in tetraploid cells. Centrosome amplification, one of the most obvious differences between diploid and tetraploid cells, is one contributing factor. Centrosomes are microtubule nucleating and organizing centers in cells, and tetraploids, with their doubled centrosome content, exhibit increased microtubule mass (Figure S5G), which can have significant consequences on cellular physiology. For example, consistent with prior work demonstrating that dynamic microtubules stimulate the activity of the small G-protein Rac1 (Godinho et al., 2014; Waterman-Storer et al., 1999), we find that tetraploid cells display a nearly two-fold increase in Rac1 activity relative to diploids (Figure 5A). Because active Rac1 in many contexts can antagonize RhoA (Sander et al., 1999), increased Rac1 activity provides one molecular explanation for the observed loss of RhoA activity in tetraploids. In support of this view, we find that inhibition of Rac activation is sufficient to inhibit Hippo pathway signaling and override the cell division arrest of tetraploid cells (Figures 5B–D): this also explains our observation that centrosome amplification alone, in the absence of tetraploidy, can trigger Hippo pathway activation (Figure 5E). Recently, Holland et al. (2012) showed that centrosome amplification leads to an elevation of p53 levels and a corresponding decrease in the proliferation of non-transformed cells (Holland et al., 2012); our data that the increase in p53 levels in such cells is LATS2-dependent now provide a molecular underpinning for this phenomenon. Moreover, we find that evolved tetraploid RPE-1 cells, which adapt to silence Hippo signaling, consistently lose their extra centrosomes (Ganem et al., 2009).
In addition to extra centrosomes, other scaling/cell size effects associated with tetraploidy might contribute to reductions in RhoA activity and Hippo pathway activation. For example, tetraploid cells possess longer actin stress fibers and the impact of length changes on stress fiber function and mass are unknown. Alternatively, the reduction in RhoA activity in tetraploids may be an indirect consequence of a reduced surface area to volume ratio, which may alter signaling from the plasma membrane (such as from receptor tyrosine kinases and GPCRs), which are important for RhoA activation and silencing of Hippo signaling (Yin et al., 2013; Yu et al., 2012).
Hippo pathway inactivation in the development of high-ploidy tumors
An inference that can be drawn from this work is that the development of high-ploidy tumors from spontaneously arising tetraploid cells might commonly require inactivation of Hippo pathway signaling. We previously demonstrated that tetraploid p53−/− mouse mammary epithelial cells that were transplanted in nude mice rapidly developed into invasive cancers, whereas isogenic diploids did not (Fujiwara et al., 2005). Array-CGH analysis performed on 10 of these tetraploid-derived tumors revealed that all 10 (derived from 5 independent experiments) showed double minute chromosomes with a recurrent amplicon of chromosome 9. A similar amplicon has been identified in mouse and human hepatocellular carcinomas, and YAP was identified as the major oncogenic driver on this amplicon (Overholtzer et al., 2006; Zender et al., 2006). We confirmed that all of the tetraploid-derived tumors overexpressed YAP (Figure S7C). This suggests that, although loss of p53 is sufficient to enable the initial proliferation of tetraploid cells, Hippo pathway silencing and YAP activation is likely required to sustain proliferation and promote tumorigenesis. Indeed, analysis of cell lines from the Cancer Cell Line Encyclopedia shows that cancers of high-ploidy are significantly more likely to amplify YAP and/or delete LATS1/2 than are near-diploid cancers (Figure S7D–F).
YAP hyperactivation, p53 loss, and other tumor-promoting mutations can arise from the massive genomic instability that occurs in proliferating tetraploid cells. However, this raises a paradox: how do tetraploid cells, which activate p53 through Hippo pathway signaling and arrest the cell cycle, proliferate initially, to acquire the necessary oncogenic mutations required for tumor development? Our data demonstrate that this can be accomplished by subtly activating growth factor signaling, which is sufficient to overcome tetraploid-induced cell cycle arrest. We have identified many known (IGFBP4) and perhaps novel (PTBP1) negative regulators of growth factor signaling, which represent potential tumor suppressors. One such regulator, the serine protease inhibitor SPINT2, is a candidate tumor suppressor that is silenced through DNA methylation in many human cancers (Nakamura et al., 2011). Bypassing physiologically relevant low-level activation of the p53 pathway through repression of such genes may be of broad general relevance to tumor initiation, because numerous events that accompany oncogenesis (e.g. oxidative stress, whole chromosome aneuploidy, hypoxia) are known to initially trigger p53.
EXPERIMENTAL PROCEDURES
Cell Culture, siRNA, and Imaging
RPE-1 cells were grown in DMEM:F12 media containing 10% FBS, 100 IU/ml penicillin, and 100 Hg/ml streptomycin. Cells were maintained at 37°C with 5% CO2 atmosphere. All siRNA transfections were performed using 50 nM siRNA with Lipofectamine RNAi MAX. Fixed and live-cell imaging were performed as described (Ganem et al., 2009). Complete details can be found in the Extended Experimental Procedures.
Genome-wide RNAi Screening
Tetraploid RPE-1 FUCCI cells were generated by 16 h of treatment with DCB, isolated by FACS, and plated into 384-well screening dishes. Cells were reverse-transfected with pooled siRNAs targeting the human druggable genome (Dharmacon). Fluorescent images were acquired after 96 h to determine the fraction of proliferating tetraploid cells. The DNA damage screen was similar in design, only cells were continuously treated with 40 ng/ml doxorubicin. Complete screening details can be found in the Extended Experimental Procedures.
Protein Extraction, Immunoprecipitation and Immunoblotting
Cells were lysed in RIPA buffer and protein was resolved using SDS-PAGE. For immunoprecipitations, cells were lysed in NP-40 buffer, pre-cleared with Protein A-sepharose and rabbit control antibody, and then immunoprecipitated with anti-HA sepharose for 4 h at 4° C. Phos-Tag gels were prepared and run according to the manufacturer’s instructions (NARD).
RhoA and Rac1 Activation Assays
Diploid and tetraploid RPE-FUCCI cells were purified by FACS as described, serum starved for 24 h, and then stimulated with media containing 10% serum for 6 h. GTP-bound RhoA and Rac1 were immunoprecipitated from diploid and tetraploid cells according to the manufacturers’ protocol (Cytoskeleton, Inc.).
hMSC Differentiation Assay
Diploid and tetraploid hMSCs (generated by 16 h DCB treatment) were plated at low densities on plastic dishes containing a 50:50 mix of adipocyte and osteogenic differentiation media. After 9–14 days, cells were fixed with paraformaldehyde and stained for adipocytes (Oil Red) and DNA (Hoechst).
Traction Force Microscopy
Diploid and tetraploid RPE-1 cells were plated on deformable poly-acrylamide (PAA) hydrogels coated with fibronectin and gel deformation was monitored to measure the contractile forces produced. See Extended Experimental Procedures for details.
Primary Hepatocyte Isolation and Hippo Inactivation In Vivo
Hepatocytes were collected from the livers of three week-old male C57BL/6 mice (see Extended Experimental Procedures for details). For gene expression profiling, hepatocytes were stained with Hoechst33342 (for DNA content) and sorted by DNA content. To induce YAP S127A expression in vivo, three-week-old tetracycline-inducible YAP S127A mice containing Rosa26-lox-STOP-lox-rtTA and Rosa26-lox-STOP-lox-EYFP alleles were injected with AAV8-TBG-Cre, and then doxycycline (1 mg/ml) was provided in the drinking water after 7 days. Following 7 days of doxycycline treatment, hepatocytes were isolated and analyzed for DNA content and YFP expression by FACS.
Antibodies, Plasmids and siRNAs
A complete list of all antibodies, plasmids, qPCR and siRNA sequences used in this study can be found in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Tetraploid cells activate the Hippo tumor suppressor pathway in vitro and in vivo
Extra centrosomes and altered G-protein signaling trigger LATS2 activation
Active LATS2 stabilizes p53 and inactivates YAP/TAZ in tetraploid cells
Evolution experiments identify adaptations that bypass Hippo pathway activation
ACKNOWLEDGEMENTS
We would like to thank S. Jhaveri-Schneider and A. Salic for comments on the manuscript; members of the ICCB screening facility; K. Lunquist for technical assistance; H. Li, K. Ross and K. Stegmaier for assistance with microarray analysis; R. Beroukhim for assistance with ploidy analysis of the CCLE collection; S. Godinho for reagents; P. Nagaruri Gonchi and D. Williams for discussions; and T. Vignaud, F. Senger and J.-L. Martiel for advice on trsction force microscopy. NIH grant 1S10RR026582-01 to C. Shamu supported the IsoCyte/Velos laser scanning cytometer. N. Ganem is supported by an NCI K99 award (K99CA154531-01), and is a member of the Shamim and Ashraf Dahod Breast Cancer Research Laboratories; H. Cornils is a fellow of the Leukemia and Lymphoma Society; M. Théry is supported by ERC grant 31047; D. Yimlamai is supported by a BCH Career Development Award and is a George Ferry Young Investigator of NASPGHAN; F. Camargo is supported by the Manton Center for Orphan Disease Research and NIH grant DK099559; D. Pellman is a HHMI investigator and is supported by NIH grant GM083299-1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
N.G, H.C., and D.P. designed the experiments and wrote the manuscript. N.G. and H.C. conducted and performed data analysis for most experiments. S.C. performed the experiments in Figs. 6A, B, and E, and isolated murine hepatocytes for gene expression analysis. K.P.O. assisted with the RNAi screens, and contributed data for figures 2F and 7B. D.Y and F.C assisted with the YAP transgenic mouse experiments in Fig. 6E. J.A. and M.T performed the traction-force microscopy in Figs. 4F and S5A, B.
References
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SB. Effects of cytochalasins on mammalian cells. Nature. 1967;213:261–264. doi: 10.1038/213261a0. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Gronroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer discovery. 2014;4:175–185. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol. 2013;24:347–356. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007;131:437–440. doi: 10.1016/j.cell.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer; Current opinion in genetics &. development. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Thery M, Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014 doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nature Reviews Molecular Cell Biology. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 2012;26:2684–2689. doi: 10.1101/gad.207027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywicka-Racka A, Sluder G. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J Cell Biol. 2011;194:199–207. doi: 10.1083/jcb.201101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffer C, Kuznetsova AY, Storchova Z. Abnormal mitosis triggers p53-dependent cell cycle arrest in human tetraploid cells. Chromosoma. 2013;122:305–318. doi: 10.1007/s00412-013-0414-0. [DOI] [PubMed] [Google Scholar]
- Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW, Barton MC. p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology. 2013;57:2004–2013. doi: 10.1002/hep.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell. 2014 doi: 10.1091/mbc.E13-11-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Miller E, Yang J, Deran M, Wu C, Su AI, Bonamy GMC, Liu J, Peters EC, Wu X. Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP; Chemistry &. Biology. 2012;19:955–962. doi: 10.1016/j.chembiol.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Mo J-S, Yu F-X, Gong R, Brown JH, Guan K-L. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Hongo A, Kodama J, Hiramatsu Y. The role of hepatocyte growth factor activator inhibitor (HAI)-1 and HAI-2 in endometrial cancer. Int J Cancer. 2011;128:2613–2624. doi: 10.1002/ijc.25606. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng C-X, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science. 2012;337:1678–1684. doi: 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K-I, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999;1:45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Hayflick L. Formation of anucleate and multinucleate cells in normal and SV 40 transformed WI-38 by cytochalasin B. Experimental cell research. 1972;74:187–194. doi: 10.1016/0014-4827(72)90496-x. [DOI] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F-X, Guan K-L. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F-X, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang C-Z, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan S-T, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang C-Y, Yu J, Guan K-L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.