Abstract
Background
The pulmonary function measures of forced expiratory volume in one second (FEV1) and its ratio to forced vital capacity (FVC) are used in the diagnosis and monitoring of lung diseases and predict cardiovascular mortality in the general population. Genome wide association studies (GWAS) have identified numerous loci associated with FEV1 and FEV1/FVC but the causal variants remain uncertain. We hypothesized that novel or rare variants poorly tagged by GWAS may explain the significant associations between FEV1/FVC and two genes: ADAM19 and HTR4.
Methods and Results
We sequenced ADAM19 and its promoter region along with the approximately 21 kb portion of HTR4 harboring GWAS SNPs for pulmonary function and analyzed associations with FEV1/FVC among 3,983 participants of European ancestry from Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE). Meta-analysis of common variants in each region identified statistically significant associations (316 tests, P < 1.58×10−4) with FEV1/FVC for 14 ADAM19 SNPs and 24 HTR4 SNPs. After conditioning on the sentinel GWAS hit in each gene [ADAM19 rs1422795, minor allele frequency (MAF)=0.33 and HTR4 rs11168048, MAF=0.40] one SNP remained statistically significant (ADAM19 rs13155908, MAF = 0.12, P = 1.56×10−4). Analysis of rare variants (MAF < 1%) using Sequence Kernel Association Test did not identify associations with either region.
Conclusions
Sequencing identified one common variant associated with FEV1/FVC independently of the sentinel ADAM19 GWAS hit and supports the original HTR4 GWAS findings. Rare variants do not appear to underlie GWAS associations with pulmonary function for common variants in ADAM19 and HTR4.
Keywords: genetic polymorphism, lung, population studies, DNA sequencing, Genome Wide Association Study
Spirometric measures of pulmonary function are easily obtainable and reproducible indices of the physiologic state of the lung and airways commonly used in clinical medicine. Two major spirometric measures in clinical practice are the forced expiratory volume in one second (FEV1) and the ratio of FEV1 to the forced vital capacity (FVC). The FEV1 reflects airflow obstruction and lung size. Reduction in FEV1 out of proportion to the FVC leads to a reduced FEV1/FVC ratio. The FEV1/FVC provides an index of airflow obstruction that is relatively independent of lung size and is the primary criterion for the diagnosis of airway obstruction and chronic obstructive pulmonary disease (COPD). The FEV1 is used to assess the severity of the airflow obstruction and monitor the progression of lung diseases including COPD, asthma, and cystic fibrosis. In addition to its essential role in the diagnosis and monitoring of respiratory disease, lower pulmonary function has been shown in numerous studies to be related to increased cardiovascular morbidity and mortality in the general population, including among nonsmokers without lung disease, and independently of standard risk factors.1–6
Cross-sectional measures of FEV1 and FEV1/FVC in adults reflect the maximal values attained at the conclusion of growth and the inevitable decline with age thereafter. Environmental factors, most notably smoking, influence both maximal growth and the rate of decline. However, genetics also influence pulmonary function; over 40% of the variability in pulmonary function has been attributed to genetic factors.7 For decades, the role of the uncommon genetic deficiency of alpha-1 antitrypsin in reduced pulmonary function has been appreciated.8 However, other genes involved in pulmonary function remained elusive prior to the era of genome wide association studies (GWAS). Recent GWAS have identified common genetic variants related to FEV1/FVC or FEV1 in at least 27 loci.9–12 Most of these are novel loci not previously implicated in lung pathology. Two of the novel genes identified for FEV1/FVC are 5-hydroxytryptamine (serotonin) receptor 4 (HTR4) and A Disintegrin And Metallopeptidase Domain 19 (ADAM19).10 Both were subsequently associated with the clinical phenotypes of airflow obstruction and COPD in GWAS.13 Interestingly, ADAM19 also plays an essential role in cardiac development14 and copy number variants have recently been identified in patients with congenital heart disease.15
While segregation analyses suggest that genetics contribute a substantial portion of the variability in pulmonary function7, the combined effects of all GWAS-identified loci appear to explain less than eight percent of the predicted genetic variance.12 The issue of unexplained heritability in GWAS has been the subject of much recent interest. One of the explanations invoked is that rare functional variants linked to the common variants identified by GWAS platforms may be important.16 To this end, in-depth re-sequencing efforts to systematically follow-up GWAS hits were undertaken in the CHARGE Targeted Sequencing Study.
We analyzed deep sequencing data for ADAM19 and HTR4 in relation to FEV1/FVC and FEV1. The goal was to identify whether variants, common or rare, that were not included in previous GWAS datasets, might underlie the observed SNP associations from earlier GWAS of these outcomes.
Methods
Study samples
The CHARGE Targeted Sequencing Study includes participants enrolled in three cohorts: the Atherosclerosis Risk in Communities Study (ARIC), the Cardiovascular Health Study (CHS), and the Framingham Heart Study (FHS).17 All of the study participants were of European ancestry and had been included in previous GWAS meta-analysis of the pulmonary function parameters FEV1 and FEV1/FVC.10 The participants included those randomly selected from the cohort (the Cohort Random Sample) and those selected for several different extreme phenotypes (the Phenotype Groups). The Cohort Random Sample contains approximately 2,000 unrelated individuals representing the distribution of phenotypes in the general population. The Phenotype Groups contain individuals with extreme values for at least one of 14 phenotypes; each group has approximately 200 participants (Lin H, et al. Circ Cardiovasc Genet, submitted). The Phenotype Group for FEV1 and FEV1/FVC included participants selected from the ARIC cohort based on meeting the following criteria at both visits 1 and 2: FEV1 < 65% predicted and FEV1/FVC < the lower limit of normal based on NHANES III prediction equations.18 Institutional review boards at participating centers approved the study and participants provided informed consent.
Sequencing data
In the CHARGE Targeted Sequencing Study, a total of 77 target regions were sequenced to follow-up selected GWAS findings across multiple phenotypes. Two of these target regions were selected based on GWAS findings for FEV1/FVC: ADAM19 and HTR4. We analyzed only these two target regions. For ADAM19 (chr 5), the following regions were submitted for sequencing based on NCBI build 36: regulatory region (CTCF binding site) between chromosomal locations 156831095 and 156832298 (hg18) and the gene region +/− 1 kb (locations 156835890 to 156936346). For HTR4 (chr 5), we submitted for sequencing the 21 kb block containing all of the high signal SNPs from our previous GWAS plus 1 kb up and downstream (locations 147815802 to 147837526).
The methods of the CHARGE Targeted Sequencing Study have been described fully in a separate manuscript (Lin H, et al. Circ Cardiovasc Genet, submitted). Briefly, approximately 2Mb of target regions were captured by a customized NimbleGen Capture array and sequenced using the ABI SOLiD V4.0 platform. The raw short reads were aligned to the reference human genome (NCBI Genome Build 36, hg18) by BFAST.19 SAMtools20 was used to pile up aligned reads and call variants with quality filters. The resulting data were then subjected to quality control procedures. Variants were categorized as known or novel by comparison with the dbSNP database and the 1000 Genomes project. The functional impact of identified variants on the encoded proteins was predicted by the ANNOVAR software package.21
Statistical analysis
Common variants
Due to the study design of CHARGE Targeted Sequencing Study, participants with extreme phenotypes were over-represented in the sequenced samples compared with those selected for the random cohort. In order to account for this sampling bias, we performed analyses with individuals weighted by the inverse of their sampling probabilities to obtain population-based effect estimates (Lin H, et al. Circ Cardiovasc Genet, submitted). We tested each common SNP for association with FEV1 and FEV1/FVC using un-weighted analyses with linear regression models with robust standard error estimates in ARIC and CHS and linear mixed effects models in FHS to account for family relatedness. All analyses assumed an additive effect of the alternate allele, and were adjusted for the same factors as in the original discovery GWAS10: age, sex, standing height, smoking status (current, past or never-smoker) and pack-years of smoking. Additional study-specific covariates included recruitment cohort (FHS), recruitment center (ARIC and CHS) and principal component eigenvalues for population stratification adjustments (ARIC and FHS). For the weighted association analysis, we used a weighted linear regression in ARIC and CHS, and weighted linear mixed model in FHS. In order to account for known GWAS loci, we compared analyses with and without conditioning on the sentinel SNPs in each GWAS locus from our earlier discovery GWAS of FEV1/FVC: HTR4 rs11168048 and ADAM19 rs1422795.10 HTR4 rs11168048 gave the smallest P value among SNPs at this locus in the discovery GWAS.10 ADAM19 rs1422795 was chosen as the sentinel SNP for conditional analyses because among the several highly correlated (r2 > 0.95) genome-wide significant SNPs in the discovery GWAS, it is a non-synonymous missense SNP. Regression models were used to adjust trait values for all covariates in addition to the genotype of the sentinel SNP. Residuals from these linear models were then used as the independent (outcome) variable in the conditional analysis.
The summary statistics (Beta, SE) from each cohort were then meta-analyzed using an inverse variance meta-analysis approach. We report the p-values from the un-weighted analysis, and the magnitude of effects from the weighted analysis (Lumley T, Dupuis J, Rice KM, Barbalic M, Bis JC, Cupples LA, et al. http://stattech.wordpress.fos.auckland.ac.nz/files/2012/05/design-paper.pdf). In order to account for multiple comparisons, we applied a Bonferroni correction for the number of common variants (N=316 with MAF>1%) analyzed. We consider common variants with association p-values less than 1.6×10−4 (0.05/316) as statistically significant.
Rare variants
Single-marker based association analysis has low power for rare variants. Therefore, we jointly analyzed all rare variants (MAF< 1%, 2166 in ADAM19 and 454 in HTR4) occurring in each of the two target regions. We tested association of rare variants with FEV1 and FEV1/FVC using the Sequence Kernel Association Test (SKAT).22 Single variant summary statistics and genotype covariance matrices were pooled for meta-analysis (Lumley T, Brody J, Dupuis J, Cupples LA http://stattech.wordpress.fos.auckland.ac.nz/files/2012/11/skat-meta-paper.pdf). We regarded statistical significance for either of the two regions tested based on P=0.05/2 = 0.025.
Predicted functional variants
Because the power of SKAT can be sensitive to the inclusion of non-functional variants, we performed additional analyses restricted to those rare variants predicted to be functional. In the ADAM19 gene region, variants were restricted to nonsynonymous and splice site SNPs (61 missense, 1 nonsense and 5 splice site). The region of sequencing around the HTR4 top GWAS hits fell in a largely intronic region. The HTR4 rare variants most likely to be functional were selected by utilizing non-coding annotation from ENCODE and TransFac tracks from the UCSC browser.23 In addition to the four rare exonic variants, SNPs in ENCODE regions annotated as DNAse hypersensitivity sites or CHiP-Seq transcription factor binding sites, and variants falling in conserved transcription factor binding motifs were selected. Applying these criteria, the 454 rare variants in the HTR4 region were refined to a set of 122 potentially functional variants.
Results
After quality control, valid sequencing data for ADAM19 and HTR4 as well as data on pulmonary function data were available for 3,983 participants from the three cohorts. This included 186 selected for severe airflow obstruction, 1,830 selected as a random sample of cohort participants and 1,967 selected because of extreme values for non-pulmonary phenotypes. Pulmonary function parameters, age, sex and smoking history of participants are shown by cohort in Table 1.
Table 1.
Numbers of subjects and characteristics by cohort
| Cohort N | FEV1 – liters mean(SD) |
FEV1/FVC -% mean(SD) |
Age at exam mean(SD) |
% Female | % Ever Smoked | Pack-years Ever Smokers mean(SD) |
|
|---|---|---|---|---|---|---|---|
| ARIC | 1914 | 2.81(0.83) | 71.8 (9.9) | 54.8(5.7) | 48.8 | 64.4 | 32.1(23.0) |
| CHS | 1080 | 2.13(0.66) | 70.0(10.9) | 72.5(5.5) | 53.7 | 55.3 | 36.6(29.3) |
| FHS | 989 | 2.68(0.81) | 72.5(8.2) | 59.8(11.0) | 51.4 | 62.9 | 28.7(23.7) |
ARIC indicates Atherosclerosis Risk in Communities Study; CHS indicates Cardiovascular Health Study; FHS indicates Framingham Heart Study. Age at exam is the age at which the FEV1 and FEV1/FVC values used in this analysis were measured. This is the baseline exam for ARIC and CHS and the latest exam with acceptable pulmonary function for FHS.
For HTR4, sequencing identified a total of 2,630 SNPs including 207 coding SNPs and 2,046 that are novel defined as not present in 1000 Genomes Phase I. For ADAM19, we identified 3,494 SNPs, including 52 coding SNPs and 2,662 novel SNPs (from Table 2a in Lin H, Wang M, Brody JA, Bis JC, Dupuis J, Lumley T, et al. Methods manuscript submitted to Circ Cardiovasc Genet along with this manuscript).
Common variants
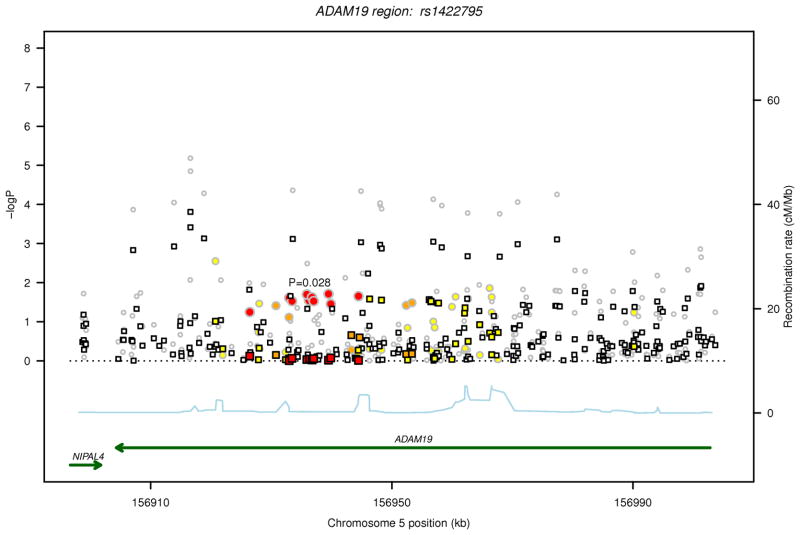
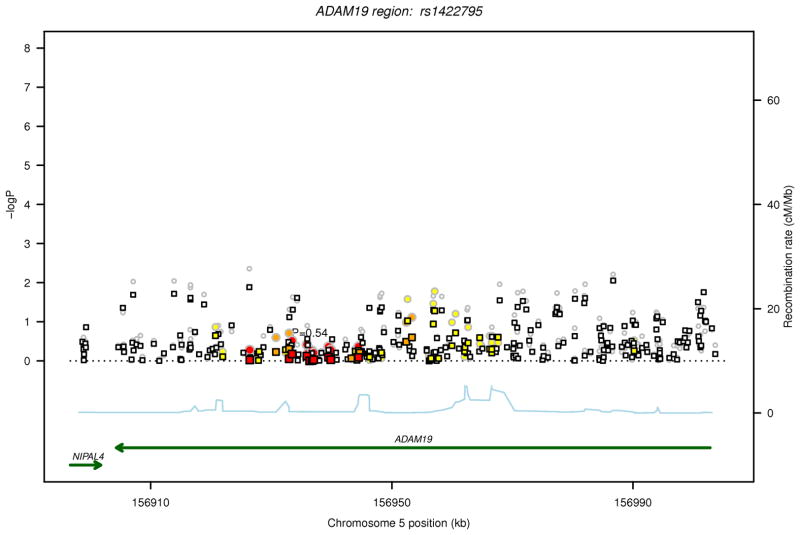
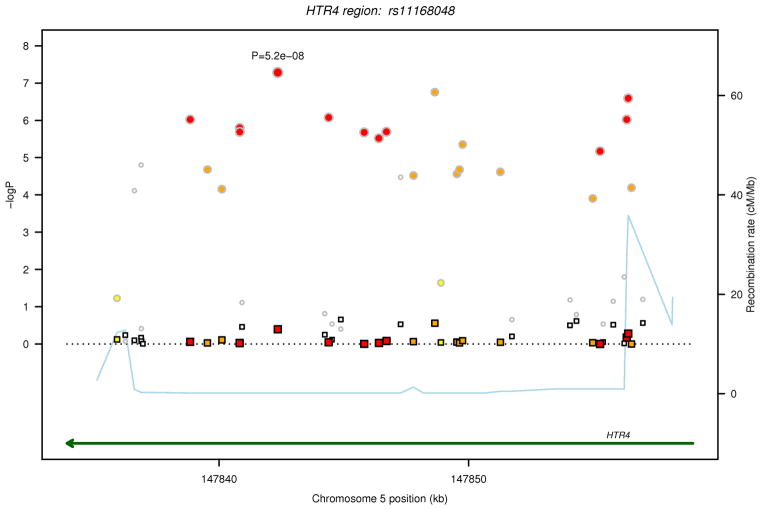
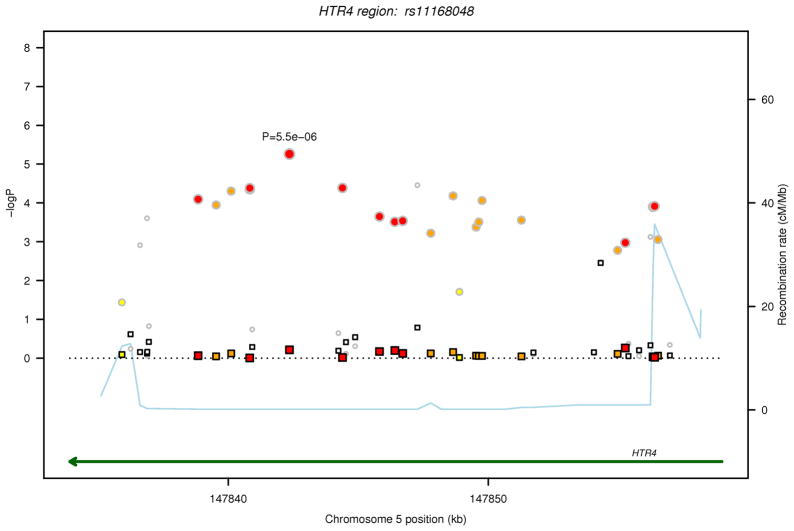
For each of the two regions (ADAM19 and HRT4), in Figure 1, the P values are plotted for FEV1/FVC and FEV1in relation to the 316 common variants (MAF>1%) before and after conditioning on the sentinel GWAS SNP from our earlier discovery analysis in each of the two regions (ADAM19 rs1422795 and HTR4 rs11168048).10 After Bonferroni correction for 316 tests, analysis of individual SNPs identified statistically significant (P<1.58 ×10−4) associations with FEV1/FVC for 14 SNPs in ADAM19 and 24 SNPs in HTR4, and with FEV1 for 12 HTR4 SNPs (Table 2). Among the statistically significant SNPs, 11 in HTR4 and 7 in ADAM19 were not included in the original GWAS discovery dataset.10 After conditioning on the sentinel GWAS SNP in each gene (ADAM19 rs1422795, HTR4 rs11168048), only one SNP surpassed the statistical significance threshold for association with FEV1/FVC (ADAM19 rs13155908, P = 1.56×10−4). The MAFs for this SNP rs13155908 (0.12), as well as the other linked SNPs that were significant in the unconditional analysis (range 0.10–0.17), are much lower than that of the sentinel GWAS SNP rs1422795 in ADAM19 (0.33). SNP rs13155908 is not in high LD with the sentinel SNP (r2=0.07) suggesting an independent signal. SNP rs13155908 did not give genome-wide statistically significant association with FEV1/FVC in the original GWAS discovery dataset (P= 1.53×10−5).
Figure 1.
Regional association plot for common SNPs identified by sequencing in loci investigated for FEV1/FVC and FEV1. The two loci are ADAM19 on 5q33.3 and HTR4 on 5q.31.1. The P values for association with the trait (FEV1/FVC or FEV1) from the unconditional analysis are represented by circles and those from the conditional analysis, accounting for the sentinel SNP in each locus, are denoted by squares. For each locus, correlations in the combined study sample between the sentinel SNP from the GWAS and other SNPs identified by sequencing in the region are depicted in red when 0.8 ≤ r2 < 1, orange when 0.5 ≤ r2< 0.8, yellow when 0.2 ≤ r2< 0.5 and white when r2< 0.2. Gene annotations are shown in green, and estimated recombination rates from HapMap are shown in light blue. The sentinel SNP (rs1422795 for ADAM19 and rs11168048 for HTR4) is annotated with its unconditional association P value for the trait.
(a) FEV1/FVC, ADAM19 locus
(b) FEV1, ADAM19 locus
(c) FEV1/FVC, HTR4 locus
(d) FEV1, HTR4 locus
Table 2.
Single Nucleotide Polymorphisms in HTR4 and ADAM19 that were statistically significant (P<1.58×10−4) for association with either FEV1 or FEV1/FVC in the unconditional analysis
| Phenotype | Target | SNP position | dbSNP | Allele1 | Allele2 | Freq1 | Unconditional Analysis * | DirectionFHS, ARIC, CHS** | Conditional Analysis P value |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P value | |||||||||
| FEV1 | HTR4 | chr5:147822546 | rs11168048 | t | c | 0.6011 | −0.0309 | 0.0140 | 5.47×10 | −−− | Sentinel-NA |
| chr5:147827466 | rs4597955 | a | g | 0.5670 | 0.0165 | 0.0133 | 3.50×10−5 | +++ | 0.1630 | ||
| chr5:147824585 | rs7735184 | t | g | 0.3962 | 0.0251 | 0.0139 | 4.11×10−5 | −++ | 0.9672 | ||
| chr5:147821023 | rs6860089 | a | t | 0.3864 | 0.0317 | 0.0138 | 4.16×10−5 | −++ | 0.9888 | ||
| chr5:147821021 | rs6860087 | a | t | 0.3914 | 0.0313 | 0.0138 | 4.45×10−5 | −++ | 0.9911 | ||
| chr5:147820305 | rs10041517 | t | c | 0.3293 | 0.0307 | 0.0141 | 4.96×10−5 | −++ | 0.7584 | ||
| chr5:147828839 | rs1989153 | t | c | 0.6767 | −0.0300 | 0.0142 | 6.59×10−5 | −−− | 0.6981 | ||
| chr5:147819037 | rs10463406 | t | c | 0.3953 | 0.0235 | 0.0139 | 8.05×10−5 | −++ | 0.8637 | ||
| chr5:147829952 | rs3995091 | a | g | 0.4292 | 0.0306 | 0.0135 | 8.66×10−5 | +++ | 0.8748 | ||
| chr5:147819730 | rs10075211 | t | c | 0.5624 | −0.0285 | 0.0135 | 1.13×10−4 | −−− | 0.895 | ||
| chr5:147836585 | rs7715901 | a | g | 0.6198 | −0.0253 | 0.0140 | 1.22×10−4 | +−− | 0.9497 | ||
| chr5:147836526 | rs7733088 | a | g | 0.3792 | 0.0273 | 0.0140 | 1.27×10−4 | −++ | 0.9234 | ||
| FEV1/FVC | HTR4 | chr5:147822546 | rs11168048 | t | c | 0.6025 | −0.0093 | 0.0022 | 5.17×10−8 | −−− | Sentinel-NA |
| chr5:147828839 | rs1989153 | t | c | 0.6717 | −0.0090 | 0.0022 | 1.75×10−7 | −−− | 0.2784 | ||
| chr5:147836585 | rs7715901 | a | g | 0.6216 | −0.0090 | 0.0021 | 2.56×10−7 | −−− | 0.5276 | ||
| chr5:147824585 | rs7735184 | t | g | 0.3942 | 0.0081 | 0.0022 | 8.38×10−7 | +++ | 0.8955 | ||
| chr5:147819037 | rs10463406 | t | c | 0.3936 | 0.0079 | 0.0022 | 9.50×10−7 | +++ | 0.8758 | ||
| chr5:147836526 | rs7733088 | a | g | 0.3772 | 0.0091 | 0.0021 | 9.51×10−7 | +++ | 0.6665 | ||
| chr5:147821021 | rs6860087 | a | t | 0.3891 | 0.0086 | 0.0021 | 1.59×10−6 | +++ | 0.9395 | ||
| chr5:147826900 | rs6889822 | a | g | 0.6306 | −0.0088 | 0.0022 | 2.02×10−6 | −−− | 0.8349 | ||
| chr5:147821023 | rs6860089 | a | t | 0.3840 | 0.0083 | 0.0021 | 2.04×10−6 | +++ | 0.9517 | ||
| chr5:147826008 | rs3995090 | a | c | 0.6030 | −0.0080 | 0.0022 | 2.10×10−6 | −−− | 0.9947 | ||
| chr5:147826596 | rs11742110 | t | c | 0.6055 | −0.0078 | 0.0022 | 3.02×10−6 | −−− | 0.9425 | ||
| chr5:147829952 | rs3995091 | a | g | 0.4278 | 0.0071 | 0.0021 | 4.44×10−6 | +++ | 0.8141 | ||
| chr5:147835457 | rs11168049 | t | c | 0.6187 | −0.0076 | 0.0022 | 6.74×10−6 | −−− | 0.997 | ||
| chr5:147817073 | rs12374521 | t | c | 0.5391 | −0.0063 | 0.0021 | 1.58×10−5 | −−− | 0.6832 | ||
| chr5:147829828 | rs1988818 | t | c | 0.4307 | 0.0064 | 0.0021 | 2.09×10−5 | +++ | 0.9314 | ||
| chr5:147819730 | rs10075211 | t | c | 0.5643 | −0.0063 | 0.0021 | 2.09×10−5 | −−− | 0.9415 | ||
| chr5:147831463 | rs6887366 | a | t | 0.5677 | −0.0067 | 0.0021 | 2.41×10−5 | −−− | 0.8996 | ||
| chr5:147829724 | rs1988819 | t | c | 0.4346 | 0.0064 | 0.0021 | 2.73×10−5 | +++ | 0.8897 | ||
| chr5:147827981 | rs1985524 | c | g | 0.4328 | 0.0065 | 0.0021 | 3.02×10−5 | +++ | 0.8684 | ||
| chr5:147827466 | rs4597955 | a | g | 0.5653 | 0.0042 | 0.0021 | 3.36×10−5 | +++ | 0.2972 | ||
| chr5:147836715 | rs7733410 | a | g | 0.4254 | 0.0069 | 0.0021 | 6.42×10−5 | +++ | 0.9967 | ||
| chr5:147820305 | rs10041517 | t | c | 0.3303 | 0.0065 | 0.0022 | 7.00×10−5 | −++ | 0.7765 | ||
| chr5:147816802 | rs13156542 | t | c | 0.5538 | −0.0059 | 0.0020 | 7.73×10−5 | −−− | 0.7984 | ||
| chr5:147835163 | rs10037493 | t | c | 0.4234 | 0.0056 | 0.0021 | 1.25×10−4 | +++ | 0.9289 | ||
| ADAM19 | chr5:156849167 | rs13155908 | a | g | 0.8769 | −0.0104 | 0.0029 | 6.56×10−6 | −−− | 1.56×10−4 | |
| chr5:156849189 | rs10058865 | a | c | 0.1108 | 0.0109 | 0.0031 | 1.41×10−5 | +++ | 3.84×10−4 | ||
| chr5:156866145 | rs13190258 | a | c | 0.8802 | −0.0097 | 0.0030 | 4.36×10−5 | −−− | 7.62×10−4 | ||
| chr5:156877473 | rs59327154 | a | g | 0.8308 | −0.0093 | 0.0027 | 4.55×10−5 | −−− | 9.26×10−4 | ||
| chr5:156851428 | rs2287749 | t | c | 0.1220 | 0.0097 | 0.0030 | 5.20×10−5 | +++ | 7.37×10−4 | ||
| chr5:156909971 | rs11748149 | a | g | 0.0980 | 0.0121 | 0.0031 | 5.55×10−5 | +++ | 7.79×10−4 | ||
| chr5:156889456 | rs72811319 | a | g | 0.1030 | 0.0123 | 0.0031 | 7.36×10−5 | +++ | 8.96×10−4 | ||
| chr5:156903456 | rs11739504 | a | c | 0.9039 | −0.0122 | 0.0032 | 8.71×10−5 | −−− | 1.03×10−3 | ||
| chr5:156846458 | rs34458213 | a | t | 0.8831 | −0.0091 | 0.0030 | 8.93×10−5 | −−− | 1.18×10−3 | ||
| chr5:156880634 | rs72811309 | a | g | 0.1020 | 0.0124 | 0.0031 | 9.21×10−5 | +++ | 1.08×10−3 | ||
| chr5:156880625 | rs72811307 | t | c | 0.8980 | −0.0122 | 0.0031 | 1.03×10−4 | −−− | 1.14×10−3 | ||
| chr5:156890868 | rs11134799 | t | c | 0.8969 | −0.0121 | 0.0031 | 1.07×10−4 | −−− | 1.24×10−3 | ||
| chr5:156880896 | rs72811310 | t | g | 0.8974 | −0.0118 | 0.0031 | 1.29×10−4 | −−− | 1.33×10−3 | ||
| chr5:156839688 | rs11466816 | t | g | 0.1126 | 0.0090 | 0.0031 | 1.36×10−4 | +++ | 1.48×10−3 | ||
Unconditional analysis includes only the SNP listed. For FEV1 the beta value is the change in liters per copy of allele 1. For FEV1/FVC the beta value is the change in FEV1/FVC scaled as a proportion (range 0–1) per copy of allele 1. Conditional analysis model includes the SNP listed plus the sentinel SNP for the specific gene target (HTR4 rs11168048, ADAM19 rs1422795) and the P value given is for the beta value for the SNP listed.
ADAM19 rs1422795 gave P = 0.02 in the unconditional analysis and is thus not listed in this table.
Direction refers to the sign of the beta coefficient by cohort in the following order: FHS (Framingham Health Study), ARIC (Atherosclerosis Risk in Communities), CHS (Cardiovascular Health Study). A “+” indicates a positive beta coefficient and a “−” indicates a negative beta coefficient
Rare variants
Meta-analysis of the cohort-specific SKAT estimates combining all variants with MAF < 1% did not provide any evidence for a role of rare variants in either ADAM19 (2166 variants) or HTR4 (454 variants) in relation to either FEV1 or FEV1/FVC (P > 0.95 for all four analyses). Because the large number of rare variants examined in ADAM19 might dilute signals from the modest expected number of associated variants, we also created 5 windows of equal size (433 in windows one to four and 434 in the fifth) and repeated the SKAT meta-analysis within those. The smallest P value in any window was 0.52.
Potential functional variants
Meta-analysis of the cohort-specific SKAT estimates combining all potential functional rare variants (62 in ADAM19 and 122 in HTR4) did not reveal any evidence for association with either FEV1 or FEV1/FVC (P > 0.68 for all four analyses).
Discussion
Spirometry is the most commonly employed assessment of lung function and FEV1/FVC and FEV1 are critical physiologic measurements in the diagnosis of airflow obstruction and monitoring of its severity and progression in clinical practice. In previous GWAS9–12, we have identified a number of novel loci containing common SNPs related to the FEV1/FVC and FEV1. Two of the novel loci were ADAM19 and HTR4. In subsequent work, we found evidence that ADAM19 and HTR4 are related to airflow obstruction and COPD.13 In the current paper, we used targeted sequencing of ADAM19 and HTR4 to address the question of whether our previous GWAS findings for FEV1/FVC were due to additional functionally relevant variants or, alternatively, due to the combined burden of rare alleles not represented in the earlier GWAS datasets.
Because HTR4 and ADAM19 were only recently identified as novel genes for pulmonary function and disease in GWAS10–13, they have not been well studied in relation to these lung phenotypes. However, within the limited published data, there is biologic plausibility for a role of both genes in lung function and lung pathobiology.
ADAM19 is a member of the “A Disintegrin And Metalloprotease” (ADAM) family of membrane tethered glycoproteins and is expressed in most tissues including the lung.24 In lung epithelial cells, TGF-β1 is a prominent mediator of the response to injury, including fibrosis. ADAM19 was found to be a key responder to stimulation by TGF- β1 in alveolar epithelial cells and a potentially critical effector of the fibrotic response to injury, an important step in the pathogenesis of pulmonary fibrosis and other lung diseases. 25 ADAM19 can potentiate pro-inflammatory activity of tumor necrosis factor alpha (TNFa), 26 a key modulator of airway inflammation. 27 ADAM19 plays a crucial role in cardiac development; Adam19−/− mice have multiple cardiac developmental defects14 and ADAM19 copy number variants have recently been identified in patients with congenital heart disease.15 Thus genetic variation and differential expression of ADAM19 are linked to both pulmonary and cardiac disease pathogenesis.
HTR4 is a member of the serotoninergic signaling cascade and is expressed in the lung.11 While serotonin (5-HT) is best studied as neurotransmitter, 5-HT signaling plays an important role in many organ systems.28 In the lung it is involved in control of breathing 28 and smooth muscle contractility29. Serotonin signaling including HTR4 is involved in human airway inflammation. 30 In a primate asthma model, ozone exposure increased HTR4 expression in the airways31 and this effect was accompanied by enhanced smooth muscle contractility.32 A recent study designed to follow-up GWAS findings for HTR4 genetic variants in lung function identified evidence for greater expression of HTR4 in fetal compared with adult human lung suggesting an important role in lung development33. The observation that HTR4 genetic variants are related to pulmonary function in both adults and children further supports a role in lung development.11, 12
Given that family history of cardiovascular disease is common among older adults in the US, our study sample included a large proportion of participants with a family history of cardiovascular disease. For example, in the participants in this analysis from the ARIC cohort, that contributed the largest number to this dataset, 49% reported that one or both biologic parents had a history of myocardial infarction. Family history of cardiovascular disease was not associated with airways obstruction [age, sex and smoking adjusted odds ratio = 1.07, 95% CI 0.79–1.42, P=0.69), a clinically relevant phenotype that showed association with both SNPs in ADAM19 and HTR413. This result is consistent with previous epidemiologic findings that reduced pulmonary function is a risk factor for mortality in the general population independent of traditional risk factors for cardiovascular disease.1–6
In analyses of all common variants (MAF>1%) in ADAM19 and HTR4 without conditioning on our sentinel GWAS SNPs, we identified a number of SNPs significantly related to either FEV1/FVC or FEV1. However these associations appeared to be explained by our previous GWAS findings because all but one (ADAM19 rs13155908, P= 1.56×10−4, cut-off P value = 1.58×10−4) were no longer significant after adjusting for the sentinel GWAS SNP at each locus.
There is functional evidence supporting the potential etiologic role of the ADAM19 sentinel GWAS SNP (rs1422795). It is a nonsynonymous coding SNP resulting in a serine to glycine substitution. This change is predicted to be “possibly damaging” in Mutation Taster (.mutationtaster.org/index.html) and PolyPhen-2 34. Evaluation of this SNP in the UCSC Genome Browser indicates that it is about 6kb upstream of the transcription start site (TSS) of an ADAM19 transcript variant suggesting that it could be part of the cis-regulatory region of this transcript. In addition, rs1422795 is close to the beginning of the translation start site (17 amino acids away) of another ADAM19 transcript variant. Because of this proximity, the amino acid change could influence the expression of this transcript. Furthermore, rs1422795 is located within a histone H3K27Ac mark—a region associated with regulatory control of gene expression.
We interrogated the HapMap3 expression Quantitative Trait Loci (eQTL) database of lymphoblastoid cell lines to assess whether ADAM19 rs13155908, that remained statistically significant in the conditional analysis, was related to gene expression.35, 36 We found a significant cis-association between rs13155908 alleles and ADAM19 expression (Spearman’s rank correlation coefficient= 0.23, P-value= 0.019) in participants of European ancestry (CEU, n = 109) but not in other ethnicities. The MAF of rs13155908 is very low in HapMap Asian populations and low in Africans. In contrast the ADAM19 sentinel GWAS SNP rs1422795 as well as the other top SNP in the original GWAS to which it is closely linked (rs2277027) were associated with ADAM19 gene expression in the CEU population (P-value = 0.001 for both SNPs) as well as several other ethnic groups. Furthermore, rs1422795 (and rs2277027) had significant cis-eQTLs in multiple other tissues including nerve (P-value = 5.3 × 10−7), adipose (P-value = 1.1 × 10−6), skeletal muscle (P-value = 3.0 × 10−5), whole blood (P-value = 6.7 × 10−6), artery (P-value = 1.2 × 10−4), and suggestive cis-eQTLs in lung (P-value = 8.0 × 10−4) based on the Genotype-Tissue Expression Portal37. We did not find other evidence in public databases supporting a functional role for rs13155908.
For HTR4, our top GWAS SNP rs11168048 is intronic and not predicted to have functional consequence for the protein. A search of transcription element binding sites (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess) identified this SNP as located within a potential binding site of the transcription factor ABF1 which is abolished when the T allele is present. In a subsequent analysis of airflow obstruction, HTR4 rs11168048 gave the smallest P value among top HTR4 SNPs identified previously by GWAS of pulmonary function10 and in an analysis limited to smokers, it gave the smallest P value overall among 75 SNP from all previous GWAS loci for pulmonary function13. Among the high signal SNPs in the current analysis, 4 fall within regulatory regions identified through overlap with fetal lung DNAse I hypersensitivity sites.38 Functional annotation of variants in high LD in this region may help inform follow-up functional studies to identify the causal variant. We acknowledge that we limited our sequencing effort to a 21 kb LD block of HTR4 that harbored all of our high signal SNPs. Thus if our top GWAS SNPs are in high LD with variants outside of this area, we would not have captured them with our sequencing effort.
SKAT analysis of possibly functional rare variants or of all rare variants did not provide any evidence for association with FEV1 or FEV1/FVC. While this suggests that our previous GWAS signals are not explained by rare variants in linkage disequilibrium with them, we acknowledge that our study and other sequencing efforts tend to be smaller than the discovery sample sizes and thus will be underpowered for rare variants.
Our analysis of targeted sequencing data for HTR4 gives support for the importance of the sentinel GWAS hit, although functional evidence in support of this SNP remains sparse. For ADAM19, the analysis conditioning on the sentinel GWAS SNP suggests the involvement of an additional SNP implying that there might be more than one causal variant at this locus.
Acknowledgments
The authors acknowledge Hong Xu, MSc and Tianyuan Wang, PhD of NIEHS for expert bioinformatics assistance and the staff and participants of the ARIC study for their important contributions.
Funding Sources: Funding support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Data for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium” was provided by Eric Boerwinkle on behalf of the Atherosclerosis Risk in Communities (ARIC) Study, L. Adrienne Cupples, principal investigator for the Framingham Heart Study, and Bruce Psaty, principal investigator for the Cardiovascular Health Study. Infrastructure for the CHARGE Consortium is supported in part by NHLBI grant HL105756.
The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN2682011000010C, HHSN2682011000011C, and HHSN2682011000012C) R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The Framingham Heart Study is conducted and supported by the NHLBI in collaboration with Boston University (Contract No. N01-HC-25195), and its contract with Affymetrix, Inc., for genome-wide genotyping services (Contract No. N02-HL-6-4278), for quality control by Framingham Heart Study investigators using genotypes in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Clusters for Genetic Analysis (LinGA) computing resources at Boston University Medical Campus. The Human Genome Sequencing Center at Baylor College of Medicine is supported by U54 HG003273. The research within CHS was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). A full list of CHS investigators and institutions can be found at http://chs-nhlbi.org/ Supported in part by the Division of Intramural Research, National Institute of Environmental Health Sciences, National Institutes of Health.
Footnotes
Conflicts of Interest Disclosures: Jemma Wilk is employed by and hold stock options in Pfizer Inc. Richard Gibbs served as a consultant to GE Clarient. All others have none.
References
- 1.Hole DJ, Watt GC, Davey-Smith G, Hart CL, Gillis CR, Hawthorne VM. Impaired lung function and mortality risk in men and women: Findings from the renfrew and paisley prospective population study. BMJ. 1996;313:711–715. doi: 10.1136/bmj.313.7059.711. discussion 715–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroeder EB, Welch VL, Couper D, Nieto FJ, Liao D, Rosamond WD, et al. Lung function and incident coronary heart disease: The atherosclerosis risk in communities study. Am J Epidemiol. 2003;158:1171–1181. doi: 10.1093/aje/kwg276. [DOI] [PubMed] [Google Scholar]
- 3.Schunemann HJ, Dorn J, Grant BJ, Winkelstein W, Jr, Trevisan M. Pulmonary function is a long-term predictor of mortality in the general population: 29-year follow-up of the buffalo health study. Chest. 2000;118:656–664. doi: 10.1378/chest.118.3.656. [DOI] [PubMed] [Google Scholar]
- 4.Sin DD, Wu L, Man SF. The relationship between reduced lung function and cardiovascular mortality: A population-based study and a systematic review of the literature. Chest. 2005;127:1952–1959. doi: 10.1378/chest.127.6.1952. [DOI] [PubMed] [Google Scholar]
- 5.Strachan DP. Ventilatory function, height, and mortality among lifelong non-smokers. J Epidemiol Community Health. 1992;46:66–70. doi: 10.1136/jech.46.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young RP, Hopkins R, Eaton TE. Forced expiratory volume in one second: Not just a lung function test but a marker of premature death from all causes. Eur Respir J. 2007;30:616–622. doi: 10.1183/09031936.00021707. [DOI] [PubMed] [Google Scholar]
- 7.Wilk JB, Djousse L, Arnett DK, Rich SS, Province MA, Hunt SC, et al. Evidence for major genes influencing pulmonary function in the nhlbi family heart study. Genet Epidemiol. 2000;19:81–94. doi: 10.1002/1098-2272(200007)19:1<81::AID-GEPI6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Pauwels RA, Rabe KF. Burden and clinical features of chronic obstructive pulmonary disease (copd) Lancet. 2004;364:613–620. doi: 10.1016/S0140-6736(04)16855-4. [DOI] [PubMed] [Google Scholar]
- 9.Hancock DB, Artigas MS, Gharib SA, Henry A, Manichaikul A, Ramasamy A, et al. Genome-wide joint meta-analysis of snp and snp-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilk JB, Shrine NR, Loehr LR, Zhao JH, Manichaikul A, Lopez LM, et al. Genome-wide association studies identify chrna5/3 and htr4 in the development of airflow obstruction. Am J Respir Crit Care Med. 2012;186:622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou HM, Weskamp G, Chesneau V, Sahin U, Vortkamp A, Horiuchi K, et al. Essential role for adam19 in cardiovascular morphogenesis. Mol Cell Biol. 2004;24:96–104. doi: 10.1128/MCB.24.1.96-104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldmuntz E, Paluru P, Glessner J, Hakonarson H, Biegel JA, White PS, et al. Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis. 2011;6:592–602. doi: 10.1111/j.1747-0803.2011.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for heart and aging research in genomic epidemiology (charge) consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general u.S. Population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 19.Homer N, Merriman B, Nelson SF. Bfast: An alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Hakonarson H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, et al. The ucsc genome browser database: Extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei P, Zhao YG, Zhuang L, Ruben S, Sang QX. Expression and enzymatic activity of human disintegrin and metalloproteinase adam19/meltrin beta. Biochem Biophys Res Commun. 2001;280:744–755. doi: 10.1006/bbrc.2000.4200. [DOI] [PubMed] [Google Scholar]
- 25.Keating DT, Sadlier DM, Patricelli A, Smith SM, Walls D, Egan JJ, et al. Microarray identifies adam family members as key responders to tgf-beta1 in alveolar epithelial cells. Respir Res. 2006;7:114. doi: 10.1186/1465-9921-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, Saftig P, Hartmann D, Blobel C. Evaluation of the contribution of different adams to tumor necrosis factor alpha (tnfalpha) shedding and of the function of the tnfalpha ectodomain in ensuring selective stimulated shedding by the tnfalpha convertase (tace/adam17) J Biol Chem. 2004;279:42898–42906. doi: 10.1074/jbc.M403193200. [DOI] [PubMed] [Google Scholar]
- 27.Berry MA, Hargadon B, Shelley M, Parker D, Shaw DE, Green RH, et al. Evidence of a role of tumor necrosis factor alpha in refractory asthma. N Engl J Med. 2006;354:697–708. doi: 10.1056/NEJMoa050580. [DOI] [PubMed] [Google Scholar]
- 28.Berger M, Gray JA, Roth BL. The expanded biology of serotonin. Annu Rev Med. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arreola-Ramirez JL, Vargas MH, Manjarrez-Gutierrez G, Alquicira J, Gutierrez J, Cordoba G, et al. Modifications of plasma 5-ht concentrations during the allergic bronchoconstriction in guinea pigs. Exp Lung Res. 2013;39:269–274. doi: 10.3109/01902148.2013.805855. [DOI] [PubMed] [Google Scholar]
- 30.Bayer H, Muller T, Myrtek D, Sorichter S, Ziegenhagen M, Norgauer J, et al. Serotoninergic receptors on human airway epithelial cells. Am J Respir Cell Mol Biol. 2007;36:85–93. doi: 10.1165/rcmb.2006-0151OC. [DOI] [PubMed] [Google Scholar]
- 31.Murphy SR, Schelegle ES, Miller LA, Hyde DM, Van Winkle LS. Ozone exposure alters serotonin and serotonin receptor expression in the developing lung. Toxicol Sci. 2013;134:168–179. doi: 10.1093/toxsci/kft090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore BD, Hyde D, Miller L, Wong E, Frelinger J, Schelegle ES. Allergen and ozone exacerbate serotonin-induced increases in airway smooth muscle contraction in a model of childhood asthma. Respiration. 2012;83:529–542. doi: 10.1159/000336835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodge E, Nelson CP, Miller S, Billington CK, Stewart CE, Swan C, et al. Htr4 gene structure and altered expression in the developing lung. Respir Res. 2013;14:77. doi: 10.1186/1465-9921-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: A database and java application for the analysis and visualization of snp-gene associations in eqtl studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The genotype-tissue expression (gtex) project. Nat Gen. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]