Abstract
While the hominid fossil record clearly shows that brain size has rapidly expanded over the last ~2.5 M.yr., the forces driving this change remain unclear. One popular hypothesis proposes that metabolic adaptations in response to dietary shifts supported greater encephalization in humans. An increase in meat consumption distinguishes the human diet from that of other great apes. Creatine, an essential metabolite for energy homeostasis in muscle and brain tissue, is abundant in meat and was likely ingested in higher quantities during human origins. Five phosphocreatine circuit proteins help regulate creatine utilization within energy demanding cells. We compared the expression of all five phosphocreatine circuit genes in cerebral cortex, cerebellum, and skeletal muscle tissue for humans, chimpanzees, and rhesus macaques. Strikingly, SLC6A8 and CKB transcript levels are higher in the human brain, which should increase energy availability and turnover compared to non-human primates. Combined with other well-documented differences between humans and non-human primates, this allocation of energy to the cerebral cortex and cerebellum may be important in supporting the increased metabolic demands of the human brain.
Keywords: primate comparison, phosphocreatine circuit, gene expression, brain metabolism
Introduction
The rapid expansion of brain size that began ~2.5 Ma in the lineage leading to modern humans (Schoenemann, 2006; Tattersall, 2008) required substantial metabolic support (Aiello and Wheeler, 1995; Leigh, 2004; Dunbar and Shultz, 2007; Isler and van Schaik, 2009). Comparisons of both DNA sequence (Haygood et al., 2007) and mRNA abundance (Uddin et al., 2004; Khaitovich et al., 2006a; Blekhman et al., 2008; Babbitt et al., 2010) indicate that extensive changes in the regulation of metabolic associated genes help distinguish humans from chimpanzees. A dietary shift toward increased meat consumption by early hominids (Stanford, 1999; Stanford and Bunn, 2001; Ungar et al., 2006) may have contributed to some of the bioenergetic modifications necessary to support the human brain (Milton, 1987; Leonard and Robertson, 1992, 1994; Milton, 1999, 2003; Leonard et al., 2007). Comparative studies of primate genetics and molecular function provide powerful tools for identifying specific molecular changes associated with a human diet (Luca et al., 2010), an important step in understanding how changes in physiology allowed for the dramatic expansion of our brains.
Creatine, an abundant metabolite of red meat (Williams, 2007), occurs at higher concentrations in the plasma of humans who consume meat (Delanghe et al., 1989; Shomrat et al., 2000) and was likely present at higher quantities in the diet of human ancestors. Interestingly, creatine metabolism has been shown to positively correlate with brain activity (Sauter and Rudin, 1993; Du et al., 2008) and creatine supplements may improve mental performance in humans (Rae et al., 2003; McMorris et al., 2007). Thus, increased meat consumption along the hominin lineage may have influenced brain metabolism by providing additional creatine. The importance of creatine in helping maintain brain energy homeostasis (Wyss and Kaddurah-Daouk, 2000; Brosnan and Brosnan, 2007; Tachikawa et al., 2007) makes the phosphocreatine circuit a particularly attractive candidate for beginning to uncover the molecular changes associated with increased encephalization.
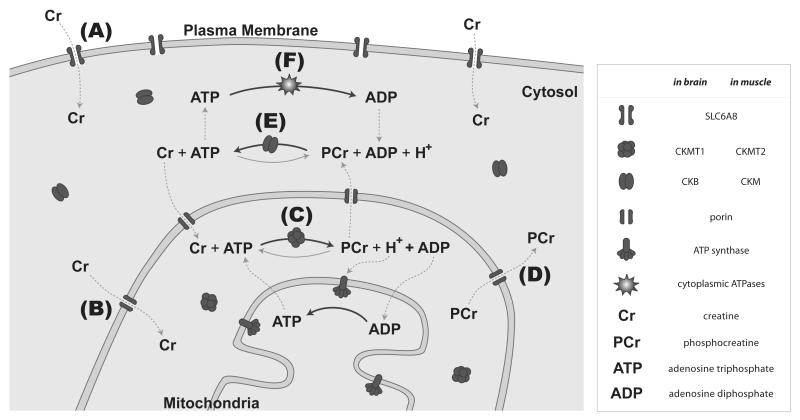
The phosphocreatine circuit begins when creatine diffuses across the plasma membrane, through the cytosol, and into the mitochondria (Figure 1A-B), where it is converted to phosphocreatine (Figure 1C), a high-energy compound (Wallimann et al., 1992). Phosphocreatine serves several functions within the brain, skeletal muscle, and other energetically demanding tissues (Brosnan and Brosnan, 2007) (Figure 1). First, phosphocreatine assists the cell in performing energy expensive processes in the cytosol (Figure 1E-F), such as contracting muscle and maintaining membrane potentials (Wyss and Kaddurah-Daouk, 2000). Second, the phosphocreatine circuit serves as an energy shuttle to cellular sites with high ATP utilization (Figure 1D) (Wyss and Kaddurah-Daouk, 2000). Phosphocreatine is able to diffuse across the cytosol faster than ATP, allowing for efficient support of biological processes farther from the mitochondria (Ellington, 2001). Third, by donating a phosphate to ADP, the phosphocreatine circuit also acts to buffer ATP concentrations (Figure 1E) (Wyss and Kaddurah-Daouk, 2000). Without ATP buffering, cellular processes regulated by ATP concentrations would be adversely affected by energy fluctuations during times of rapid energy utilization, leading to cell toxicity (Matthews et al., 1999). These functions are particularly important in the brain.
Figure 1. Schematic representation of the phosphocreatine circuit.
A. Creatine enters cells through the membrane transporter SLC6A8. B. Creatine moves across the outer mitochondrial membrane through porin. C. Creatine is phosphorylated within the outer mitochondrial space by CKMT1 or CKMT2. D. Phosphocreatine moves through porin back into the cytosol where it can diffuse to site with high ATPase activity. E. Phosphocreatine interacts with either CKB or CKM to generate ATP. F. The resulting ATP is then available as a source of energy for cytoplasmic ATPases and creatine returns to the mitochondria. ATPases, such as the sodium-potassium pump, are proteins that typically utilize energy from ATP to perform a specific cellular function. SLC6A8: creatine transporter, CKMT1: creatine kinase mitochondrial 1, CKMT2: creatine kinase mitochondrial 2, CKM: creatine kinase muscle, CKB: creatine kinase brain
While skeletal muscle contains large amounts of glycogen for energy storage (Fisher et al., 2002), the brain has few mechanisms for storing energy (Peters et al., 2004), making the phosphocreatine circuit particularly important for maintaining proper brain energy homeostasis (Wallimann and Hemmer, 1994). Given that the phosphocreatine circuit is responsible for critical cellular functions and the association between a meat-rich diet and elevated creatine levels, we hypothesize that gene regulatory changes occurred during human evolution to increase phosphocreatine circuit gene expression specifically in the brain as a way of increasing ATP energy availability and turnover. This study compares the expression of the genes that encode the phosphocreatine circuit in humans, chimpanzees, and rhesus macaques. The goals are to better understand the role of this circuit in these primates and to infer the functional significance of these differences in the context of human evolution.
Materials and methods
Primate sample collection
Samples were obtained from the Kathleen Price Bryan Brain Bank at Duke University (Homo sapiens, frozen tissue), BioChain Institute Incorporated (Homo sapiens, total RNA), Southwest National Primate Research Center (Pan troglodytes and Macaca mulatta, frozen tissue), New England Regional Primate Research Center (Macaca mulatta, frozen tissue), and Yerkes National Primate Research Center (Macaca mulatta, frozen tissue) (Table 1).
Table 1.
Primate samples used in this study
| Sample | identifier | Tissue | Obtained From | |
|
| ||||
| HUMANS | Hsap1 | 1412 | Cerebral Cortex | Kathleen Price Bryan Brain Bank |
| Hsap2 | 1320 | Cerebral Cortex | Kathleen Price Bryan Brain Bank | |
| Hsap3 | 99 | Cerebral Cortex | Kathleen Price Bryan Brain Bank | |
| Hsap4 | A803148 | Cerebral Cortex | BioChain Institute Incorporated | |
| Hsap5 | A803146 | Cerebral Cortex | BioChain Institute Incorporated | |
| Hsap6 | A803159 | Cerebral Cortex | BioChain Institute Incorporated | |
| Hsap7 | A507293 | Cerebral Cortex | BioChain Institute Incorporated | |
| Hsap8 | A509243 | Cerebral Cortex | BioChain Institute Incorporated | |
| Hsap9 | A508112 | Cerebral Cortex | BioChain Institute Incorporated | |
| Hsap10 | A508285 | Cerebellum | BioChain Institute Incorporated | |
| Hsap11 | A510131 | Cerebellum | BioChain Institute Incorporated | |
| Hsap12 | A611366 | Cerebellum | BioChain Institute Incorporated | |
| Hsap13 | B104083 | Skeletal Muscle | BioChain Institute Incorporated | |
| Hsap14 | A508352 | Skeletal Muscle | BioChain Institute Incorporated | |
| Hsap15 | B104053 | Skeletal Muscle | BioChain Institute Incorporated | |
| Hsap16 | A811244 | Skeletal Muscle | BioChain Institute Incorporated | |
| Hsap17 | B207204 | Skeletal Muscle | BioChain Institute Incorporated | |
|
| ||||
| Sample | identifier | Tissue | Obtained From | |
|
| ||||
| CHIMPANZEES | Ptro1 | 4X0327 | Cerebral Cortex Cerebellum Skeletal Muscle |
Southwest National Primate Research Center |
| Ptro2 | 4X0391 | Cerebral Cortex Cerebellum Skeletal Muscle |
Southwest National Primate Research Center |
|
| Ptro3 | 4X0505 | Cerebral Cortex Cerebellum Skeletal Muscle |
Southwest National Primate Research Center |
|
| Ptro5 | 4X0519 | Cerebral Cortex Cerebellum Skeletal Muscle |
Southwest National Primate Research Center |
|
| Ptro5 | 4X0523 | Cerebral Cortex Cerebellum Skeletal Muscle |
Southwest National Primate Research Center |
|
|
| ||||
| Sample | identifier | Tissue | Obtained From | |
|
| ||||
| MACAQUES | Mmul1 | A08-296 | Cerebral Cortex Cerebellum Skeletal Muscle |
New England Regional Primate Research Center |
| Mmul2 | A08-298 | Cerebral Cortex Cerebellum Skeletal Muscle |
New England Regional Primate Research Center |
|
| Mmul3 | 19222 | Cerebral Cortex Cerebellum Skeletal Muscle |
Southwest National Primate Research Center |
|
| Mmul4 | RFc8 | Cerebral Cortex Cerebellum Skeletal Muscle |
Yerkes National Primate Research Center |
|
Tissue preparation, total RNA isolation, and cDNA synthesis
All RNA extractions from collected tissue were performed at Duke University’s Regional Biocontainment Laboratory (RBL). Special care was taken to section each tissue consistently within and between species. Whole brain tissues maintained natural form in the freezer, allowing for anatomic landmarks, such as major sulci and gyri, to be used for proper brain region identification. Cerebral cortex tissue sections were specifically taken from the frontal lobe, while skeletal muscle tissue samples were taken from the vastus lateralis. All tissue sections were superficially taken with a width of ~4mm, a depth of ~4mm. Tissues were homogenized in QIAzol lysis buffer (Qiagen) using a Tissuelyser II (Qiagen). Total RNA samples were purified using an RNeasy lipid tissue kit (Qiagen) in conjunction with an RNase-free, DNase set (Qiagen). RNA concentration and quality were determined using a Nanodrop spectrophotometer (Thermo Scientific) and an Experion system (BioRad), respectively. Total RNA was converted to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). All samples were PCR tested for genomic DNA contamination using a SDHA primer pair designed to amplify an intronic region (Supplemental Online Material Figure 1).
Primer design
Gene sequences for SLC6A8, CKMT1A, CKMT1B, CKMT2, CKB, and CKM were obtained from the Ensembl genome browser using the Homo sapiens 36.3, Pan troglodytes 2.1, and Macaca mulatta 1.1 builds. PCR primers (Sigma-Aldrich) were designed within completely conserved exonic regions among all transcript isoforms and species (Table 2). Because CKMT1A and CKMT1B encode for identical proteins (Ensembl), PCR primers could not be designed to specifically amplify one gene and not the other. As such, a single PCR primer was designed to simultaneously amplify both genes and is referred to as CKMT1. Special care was taken to ensure that the amplified exonic region was found in all known transcript isoforms for each gene. This is important because transcript isoforms may be differentially expressed between species and would complicate our interpretation of the data. This potential confounding factor is controlled by ensuring that all isoforms are captured simultaneously in our expression measurements. Primers were selected using Primer3 Input v0.4.0 (Rozen and Skaletsky, 2000). The primer sequences were blasted to all three species’ genomes using Ensembl BLAST and a test PCR was performed on human cDNA to ensure only one product for each primer pair (Supplemental Online Material Figure 2).
Table 2.
Primer sequences for quantitative RT-PCR
| Forward Primer 5′ to 3′ | Reverse Primer 5′ to 3′ | Efficiency | R2 | |
|---|---|---|---|---|
| EEF2 | AGAAGCTGTGGGGTGACAG | GATCAGCTGGCAGAAGGTG | 97.80% | 0.996 |
| SDHA | TGGGAACAAGAGGGCATCTG | CCACCACTGCATCAAATTCATG | 98.50% | 0.998 |
| SLC6A8 | GGGACCCAGATTTTCTTTTCTTAC | CCACCTTGGAGATGTGCAC | 97.10% | 0.991 |
| CKMT1 | AGAGTCAGAACTGGCCGAAG | CTGCTGTTCAGCCTCTGTCA | 98.29% | 0.991 |
| CKB | CCTTCTCCAACAGCCACAAC | CAGCTCCGCGTACAGCTC | 94.90% | 0.990 |
| CKMT2 | ATCACCCAAGGGCAGTTC | GTGATGGCCACGTTCTC | 96.60% | 0.999 |
| CKM | CAAGCCTGAGGAGGAGTACC | GGGTTGTCCACTCCTGTCTG | 98.13% | 0.996 |
Quantitative RT-PCR data collection
Quantitative RT-PCR measurements were conducted on a Mastercycler ep realplex machine (Eppendorf) in 10 μL reactions: 5.0 μl 2X QuantiFast SYBR Green PCR Kit (Qiagen), 0.25 μl for each primer (10 μM), 0.5 μl of cDNA template, and 4.0 μl PCR quality water. The following PCR program was used for all reactions: 95 °C for 5 minutes, 40 cycles of 95 °C for 15 seconds and 60 °C for 30 seconds, followed by a melt curve from 60 to 95 °C. A single peak was detected on all melt curves, ensuring a single amplification product size for all species. Ct values were determined using the CalQPlex setting with a baseline drift correction. For each primer pair, a standard curve was setup on human brain or skeletal muscle cDNA over a twelve point, factor of two dilution series to determine the efficiency and working Ct range of each primer set. All primer sets had an efficiency between 94 and 100 percent with r2 values greater than 0.99 (Table 2).
Data were collected by running each experimental and control sample in technical triplicate. For genes with medium to high expression (as defined by a mean Ct<33 PCR cycles), only measurements with low standard deviation across replicates (Std Dev <0.4 Ct) were used in the expression analysis (Karlen et al., 2007). All tissue specific genes fell into this category of medium to high expression (e.g., CKB in both brain regions). For genes with low expression (as defined by a mean Ct>33 PCR cycles), a higher standard deviation threshold was implemented (Std Dev <1.0 Ct) as a result of increased variation of measurements in this Ct range (Karlen et al., 2007). All genes that fell into the category of low expression were genes being expressed in their non-dominant tissue (e.g., CKB in skeletal muscle). Within plates, expression was normalized with two control genes (SDHA and EEF2), selected based on their performance in a geNorm™ analysis on brain and skeletal muscle tissue for humans and chimpanzees, as well as having a similar expression level to the genes of interest (Vandesompele et al., 2002; Pattyn et al., 2003; Fedrigo et al., 2010). Between plates, an inter-run calibration was conducted by running the control gene, EEF2, on IMR-32 cell cDNA (Hellemans et al., 2007). To convert the raw Ct expression into normalized relative expression, we used a modified delta-delta Ct method (our code is available at: http://www.biology.duke.edu/wraylab/wraylab/Resources.html) (Vandesompele et al., 2002; Hellemans et al., 2007; Fedrigo et al., 2010). Raw and normalized data can be found in the supplemental materials (Supplemental Online Material Table 1 and Supplemental Online Material Table 2, respectively).
Gene expression comparison
Although no generally accepted method exists for overlaying gene expression profiles onto a phylogenetic tree, a few studies have put forth alternatives for identifying natural selection from expression (Gilad et al., 2006; Khaitovich et al., 2006b). We focused on identifying gene expression differences between species (Figure 2) using a Mann-Whitney test to calculate statistical significance as this approach does not assume a normal data distribution and it works well with smaller sample sets (Table 1). Even though the Mann-Whitney test works well with small sample sets, larger sample sets have more statistical power to identify significant expression differences and have the potential to give lower p-values. This should be taken into account when comparing p-values across tissues with a higher number of samples (cerebral cortex) to those with a smaller sample set (cerebellum). The motivation behind this research is to identify human-specific expression patterns, as we believe those traits are more likely associated with human-specific phenotypes. As such, two sets of expression comparisons were performed, human versus chimpanzee and human versus rhesus macaque (Table 3).
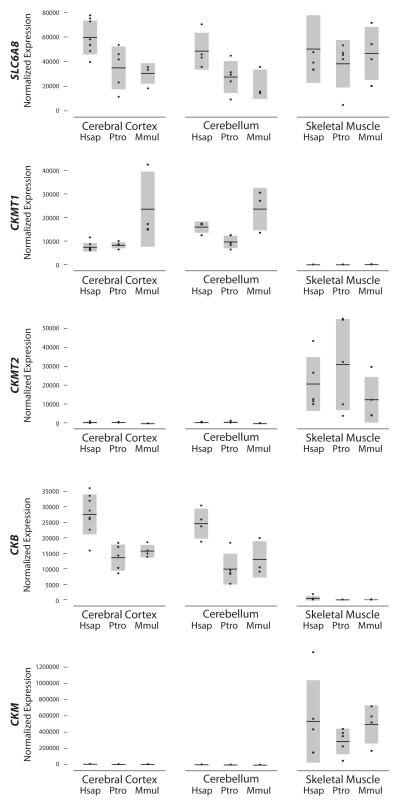
Figure 2. Phosphocreatine circuit gene expression comparisons among species.
Quantitative PCR measurements for the creatine transporter and kinases in humans, chimpanzees, and rhesus macaques. Individuals are each represented by a point, the horizontal bar is the mean, and the spread of the bar from the mean represents one standard deviation.
SLC6A8: creatine transporter, CKMT1: creatine kinase mitochondrial 1, CKMT2: creatine kinase mitochondrial 2, CKM: creatine kinase muscle, CKB: creatine kinase brain, Hsap: Homo sapiens, Ptro: Pan troglodytes, Mmul: Macaca mulatta
Table 3.
p-values for human-specific expression comparisons (Figure 2) using a Mann-Whitney test. Significant values are highlighted (p≤0.05)
| Human to Chimpanzee | |||
|---|---|---|---|
| Cerebral Cortex | Cerebellum | Skeletal Muscle | |
| SLC6A8 | 0.0281 | 0.0500 | 0.9168 |
| CKMT1 | 0.2416 | 0.0275 | 0.9168 |
| CKB | 0.0084 | 0.0143 | 0.0163 |
| CKMT2 | 1.0000 | 0.3272 | 0.7540 |
| CKM | 0.1432 | 0.0275 | 0.4647 |
| Human to Macaque | |||
|---|---|---|---|
| Cerebral Cortex | Cerebellum | Skeletal Muscle | |
| SLC6A8 | 0.0066 | 0.0339 | 0.8065 |
| CKMT1 | 0.0066 | 0.2888 | 0.0500 |
| CKB | 0.0108 | 0.0771 | 0.1416 |
| CKMT2 | 0.0066 | 0.0339 | 0.3272 |
| CKM | 0.01742 | 0.0339 | 0.4624 |
Results
Comparative expression analysis
To our knowledge, this is the first study to comprehensively report expression measurements of the phosphocreatine circuit genes within nonhuman primates. Because we are interested in identifying which phosphocreatine circuit genes may be important for human-specific traits, we focused our attention on two comparisons: human versus chimpanzee and human versus rhesus macaque. These two comparisons allow us to identify tissue-specific expression signatures that are unique to humans among these three species.
Creatine transporter, SLC6A8
The phosphocreatine circuit begins with the active transport of creatine through a dedicated transmembrane protein, SLC6A8, into energetically expensive tissues, such as the brain and skeletal muscle (Snow and Murphy, 2001) (Figure 1A). Since creatine must be transported across the plasma membrane before the cell can harvest its energy potential (Figure 1B-F), SLC6A8 is a critical protein for fueling the underlying the phosphocreatine circuit. We therefore began by measuring transcript abundance from the SLC6A8 gene that encodes this protein.
Comparisons based on quantitative RT-PCR reveal higher mRNA transcript abundance of the creatine transporter gene SLC6A8 in humans than in chimpanzees and rhesus macaques in both the cerebral cortex (1.7-fold, p=0.028 and 2.0-fold, p=0.007, respectively) and the cerebellum (1.8-fold, p=0.05 and 2.3-fold, p=0.034, respectively) (Figure 2).
Analyzing the skeletal muscle samples reveals almost equal expression of SLC6A8 when comparing human to chimpanzee and rhesus macaque (1.3-fold, p=0.917 and 1.1-fold, p=0.807, respectively) (Figure 2).
Mitochondrial creatine kinases, CKMT1 and CKMT2
Once inside the cell, creatine primarily interacts with one family of proteins, the creatine kinases. Each of the four creatine kinase family members has tissue, intracellular, and substrate preferences, allowing for precise metabolic control (Wallimann et al., 1998) (Figure 1). Two mitochondrial kinases, creatine kinase mitochondrial 1 (CKMT1) and creatine kinase mitochondrial 2 (CKMT2), are primarily expressed in the brain and skeletal muscle, respectively. Coupled with oxidative phosphorylation, these kinases localize within the mitochondria to catalyze the production of phosphocreatine, a high-energy phosphate compound similar to ATP (Figure 1C) (Vendelin et al., 2004).
Consistent with previous reports in humans (Wyss and Kaddurah-Daouk, 2000), we found that CKMT1 and CKMT2 are also expressed in a tissue specific fashion in chimpanzees and rhesus macaques, with CKMT1 predominant in the brain regions and CKMT2 in skeletal muscle (Figure 2). Although we focus our discussion for each gene on their primary tissue of expression, all human to chimpanzee and human to rhesus macaque statistical comparisons were performed (Table 3).
When comparing human CKMT1 expression to chimpanzee in the brain regions, we observe about equal transcript abundance in the cerebral cortex (0.9-fold, p=0.242) but an increase in the cerebellum (1.7-fold, p=0.028). A more consistent pattern was measured between the human and the rhesus macaque brain samples, with humans having lower CKMT1 expression in both the cerebral cortex (0.3-fold, p=0.007) and the cerebellum (0.7-fold, p=0.289).
The other mitochondrial kinase, CKMT2, is expressed primarily in skeletal muscle. Human to chimpanzee comparisons show decreased expression (0.7-fold, p=0.754) of CKMT2 in the skeletal muscle samples, but an analysis of human to rhesus macaque shows increased expression (1.6-fold, p=0.327) (Figure 2).
Cytosolic creatine kinases, CKB and CKM
After synthesis by the mitochondrial kinases, phosphocreatine diffuses out of the mitochondria (Figure 1D) to sites with high ATPase activity (Figure 1F) where it is able to interact with cytosolic creatine kinase (Figure 1E) (Figure 1) (Wallimann et al., 1998). Two cytosolic kinases, creatine kinase brain type (CKB) and creatine kinase muscle type (CKM), are predominantly expressed in the brain and skeletal muscle, respectively. Complementary to their mitochondrial counterparts, the cytosolic kinases drive the production of ATP (Figure 1E). This reaction is coupled with cytosolic ATPases to provide energy for metabolic processes (Figure 1F), such as muscle contractions and maintaining plasma membrane potentials (Kushmerick, 1998; Wyss and Kaddurah-Daouk, 2000).
Similar to previous reports in humans (Wyss and Kaddurah-Daouk, 2000) and to the mitochondrial kinases measured in this study, expression analysis of both cytosolic creatine kinases reveals a corresponding tissue-specific expression pattern in chimpanzee and rhesus macaque (Figure 2). CKB is dominant in both brain regions of interest while CKM is dominant in skeletal muscle. As with the mitochondrial kinases, for each of these genes we concentrate on the tissue in which they are most highly expressed.
We found that CKB expression has undergone a human-specific increase in the cerebral cortex and cerebellum compared to chimpanzee (2.0-fold, p=0.008 and 2.5-fold, p=0.014 respectively) and rhesus macaque (1.8-fold, p=0.011 and 1.9-fold, p=0.077 respectively) (Figure 2).
Consistent with the other skeletal muscle genes (SLC6A8 and CKMT2), CKM lacks a species-specific expression pattern in our skeletal muscle samples. Average CKM expression in skeletal muscle shows an increase in abundance in humans compared to chimpanzee (1.9-fold, p=0.465), but about equal expression compared to rhesus macaque (1.1-fold, p=0.462).
Discussion
In an influential paper published in 1975, King and Wilson posited that changes in gene regulation generated many of the phenotypic differences that distinguish humans from chimpanzees (King and Wilson, 1975). Controlling the abundance of mRNA is one of the most important aspects of gene regulation, as fluctuations in the expression of specific genes are known to produce a wide variety of phenotypic consequences (Wray, 2007). Comparing gene expression between primate species provides an initial approach for understanding observed physiological differences.
In this study, we measured expression of the phosphocreatine circuit genes to determine if they are differentially expressed between primate species. Because the brain is such an energetically expensive organ, the approximately two-fold increase in cranial capacity that occurred during the past ~2 M.yr. of human evolution (Schoenemann, 2006) imposed a substantially larger metabolic demand (Aiello and Wheeler, 1995; Leonard et al., 2007). A shift toward increased meat consumption may have contributed towards meeting that increased demand (Milton, 1999; Stanford, 1999; Stanford and Bunn, 2001; Ungar et al., 2006). Knowing that creatine is an abundant nutrient in red meat (Williams, 2007) and that phosphocreatine is critical to metabolically active cells (Wyss and Kaddurah-Daouk, 2000; Brosnan and Brosnan, 2007; Tachikawa et al., 2007), we hypothesized that a brain-specific increase in of phosphocreatine circuit gene expression arose in the lineage leading to humans. Higher expression of this circuit in humans would provide additional ATP energy to brain cells by increasing ATP turnover and transport efficiency (Wyss and Kaddurah-Daouk, 2000; Snow and Murphy, 2001).
Our results show that there is higher expression of genes encoding two key components of the phosphocreatine circuit in the cerebral cortex and cerebellum of humans (Figure 2). The first of these genes, SLC6A8, encodes a protein that mediates creatine transport across the plasma membrane (Snow and Murphy, 2001). The importance of intracellular creatine for normal brain anatomy, physiology, and cognition is revealed by creatine transporter deficiency syndromes (MIM ID #300352), which involve impaired transport of creatine across the blood brain barrier and lead to serious health consequences in humans, including mental retardation, language impairment, seizures, and microcephaly (deGrauw et al., 2003; Schiaffino et al., 2005; Anselm et al., 2006). These phenotypes indicate that the transport of creatine into the brain is important within humans and suggest that differences in intracellular creatine concentrations between primate species may also be significant.
Expression comparisons reveal that SLC6A8 is expressed at about twice the level in the cerebral cortex and cerebellum of humans as it is in chimpanzees (Figure 2). Given that SLC6A8 expression is positively correlated with intracellular creatine concentrations (Wyss and Kaddurah-Daouk, 2000), the observed increase in SLC6A8 expression, combined with increased meat intake, would likely increase the transport of creatine into the brain. In contrast, SLC6A8 expression levels in skeletal muscle are not significantly different between human and chimpanzee (Figure 2). Thus, not only did more creatine become available to the body from the dietary shift toward meat consumption, but a greater proportion of this creatine is likely transported into the brain as opposed to another metabolically demanding tissue, skeletal muscle. Higher intracellular creatine concentrations in the brain would fuel the phosphocreatine circuit (Brosnan and Brosnan, 2007).
The second phosphocreatine circuit gene whose expression differs between humans and chimpanzees is CKB. This gene encodes a kinase that generates ATP from ADP using phosphocreatine as a source of high-energy phosphate in the cytosol. CKB protein plays a critical role in maintaining proper brain energy homeostasis (Wyss and Kaddurah-Daouk, 2000) and brain activity positively correlates with CKB function (Sauter and Rudin, 1993; Du et al., 2008). In rats, creatine kinase regenerates ATP twelve times faster than through oxidative phosphorylation (Wallimann et al., 1992). This rapidly available energy is important in regulating neurotransmitter release, maintaining membrane potentials, assisting growth cone migration, and restoring energy homeostasis (Wallimann et al., 1992; Wallimann and Hemmer, 1994). Further evidence for CKB’s importance comes from CKB -/- knockout mice. In addition to other neurological conditions, these mice show decreased spatial learning and decreased habituation behavior (Jost et al., 2002).
Our expression comparisons show that humans have approximately twice as much CKB mRNA in both the cerebral cortex and cerebellum compared to chimpanzees and rhesus macaques. The corresponding gene that is expressed in muscle, CKM, shows no difference in transcript abundance between these three species. Because CKB expression positively correlates with CKB protein activity (Ishikawa et al., 2005), higher CKB transcript abundance in humans may allow for more efficient ATP regeneration during energy utilization, helping support the increased metabolic demands of the human brain (Wallimann et al., 1992; Wallimann and Hemmer, 1994). The reaction catalyzed by CKB provides energy for a diverse set of enzymatic activities in the cytoplasm (Figure 1E-F), potentially providing humans with the ability to support a greater number of simultaneous enzymatic reactions in the brain than either chimpanzee or rhesus macaque.
Thus, gene expression underlying two key components of the phosphocreatine circuit are elevated in the brains of humans relative to chimpanzees and rhesus macaques. Interestingly, these observations are tissue-specific, suggesting differential allocation of a key metabolite between two metabolically demanding tissues in the body. Furthermore, the expression changes in these two genes are likely to be synergistic by transporting more creatine into cells and increasing the capacity to utilize phosphocreatine as a source of energy for ATP-dependent enzymatic reactions. These changes in gene expression are consistent with the hypothesis that humans utilize the phosphocreatine circuit more heavily than chimpanzees and rhesus macaques to support our energy demanding brain (Peters et al., 2004).
An important concern for this study and follow-up experiments is whether gene expression differences are the result of genetic changes between species or the result of environmental effects, such as different diets. It is clear that environmental factors can influence gene expression (Idaghdour et al., 2008; Somel et al., 2008; Gibson, 2008; Hodgins-Davis and Townsend, 2009). Of direct relevance to this study, creatine levels and creatine metabolism are influenced by dietary intake of creatine (Wyss and Kaddurah-Daouk, 2000; Snow and Murphy, 2001; Brosnan and Brosnan, 2007). Although it is not possible to carry out studies in chimpanzees and humans that fully control for dietary differences, these kinds of studies can readily be conducted with mice. Somel and colleagues (2008) investigated the effects of diet on gene expression in the liver and brain of mice. After feeding adult mice four different diets for eight weeks (chimpanzee diet, cafeteria food diet, McDonald’s diet, and pellet diet), the authors measured transcript abundance using microarrays (Somel et al., 2008). Importantly for the present study, they observed no significant difference in the expression levels of either SLC6A8 or CKB among the four diets (M. Somel, pers. comm.). While these data do not rule out the possibility that diet can influence the expression of these two genes, they do support the interpretation that the differences in transcription levels we observe are unlikely to be exclusively as a result of diet. An interesting question for future studies will be parsing the relative influences of genetic factors, environmental influences, or a combination of both, on creatine distribution and utilization in primates.
At this point, it is not possible to conclude that the gene expression differences we observe in the phosphocreatine circuit serve as adaptions. Comparisons of gene expression have been used to infer adaptation by concluding that interspecies differences that are significantly larger than intraspecies variation are more likely to result from positive selection than drift (Blekhman et al., 2008). To date, these techniques have been optimized and applied on a genome wide scale, making it difficult to apply these same methods to our study. However, the pattern of expression changes in both SLC6A8 and CKB loosely meet these criteria (Figure 2). The false positive rate using this test for selection is not well understood, and we do not draw any firm conclusions from it. A second line of evidence regarding adaptation comes from analysis of DNA sequences. We sought evidence of positive selection on all five phosphocreatine circuit genes by examining three regions that can house gene regulatory elements (5′ flanking region, 5′ UTR and 3′ UTR) and one that encodes the protein function (coding region) (SOM Appendix). We found no evidence of positive selection for mutations in or around any of the five genes (SOM Table 3). It is important to bear in mind, however, that these methods are generally underpowered, and are unable to identify selection on single point mutations, any other kind of mutation, or epigenetic modifications, any of which could influence gene expression. (In fact, expression of CKMT1, CKM, and CKB can be influenced by epigenetic regulation [Caretti et al., 2004; Ishikawa et al., 2005; Uzawa et al., 2006])
Although our data do not speak directly to the question of adaptation, they do focus attention on changes in specific molecular processes that may have contributed to a shift in energy allocation towards the brain during human evolution. Energy trade-off hypotheses predict that metabolic reallocation from other energetically demanding tissues to the brain allowed for greater encephalization in humans (Aiello and Wheeler, 1995; Leonard et al., 2003; Isler and van Schaik, 2009). The tissue- and species-specific differences in SLC6A8 and CKB expression we report here are consistent with these predictions. Perhaps the most convincing evidence that these expression differences are functionally important comes from genetic data showing that reducing the amount of normal SLC6A8 and CKB protein produces pathologic phenotypes related to the brain (Jost et al., 2002; deGrauw et al., 2003; Schiaffino et al., 2005; Anselm et al., 2006). The implication to our study being that elevated expression of these genes would have increased the metabolic scope of the brain.
Comparative gene expression studies in primates provide exciting opportunities to complement the extensive body of work investigating energetic trade-offs at the level of tissue mass (Aiello and Wheeler, 1995; Leonard et al., 2003; Isler and van Schaik, 2009) by giving molecular insight into the physiological underpinnings of those tissues. It seems highly unlikely that only a small set of molecular changes accounted for differential energy allocation among tissues during human evolution. Indeed, our earlier genome-wide analysis of noncoding sequences in the same three species examined here suggested that diverse genes involved in carbohydrate metabolism experienced positive selection on regulatory sequences during human origins (Haygood et al., 2007). Large-scale surveys of gene expression have begun to identify numerous genes whose expression differs among primate species (Uddin et al., 2004; Khaitovich et al., 2006a; Blekhman et al., 2008; Babbitt et al., 2010), greatly expanding our view of the specific molecular changes that accompanied the origin of humans as a distinct species. An important challenge for the coming years will be identifying which of these changes were associated with the evolution of uniquely human traits.
Conclusions
While it is well known that the anatomy and physiology of the human brain differs from other great apes in numerous regards (Deacon, 1997; Schoenemann, 2006), the underlying molecular mechanisms responsible for those differences have remained elusive. Gene expression analysis provides a rapid and powerful tool for identifying functional differences among primate species (Khaitovich et al., 2006a). Our analysis of the phosphocreatine circuit has revealed two genes, SLC6A8 and CKB, within the phosphocreatine circuit that are consistently and differentially expressed between humans, chimpanzees, and rhesus macaques specifically within the cerebral cortex and cerebellum. Given the bioenergetic importance of this circuit and its association with dietary intake, increased expression of SLC6A8 and CKB in the human brain may have a profound influence on brain energy homeostasis today and during human origins by increasing ATP energy availability and turnover.
Supplementary Material
Supplemental Online Material Figure 1: Verifying cDNA purity using SDHA control primers that amplify an intronic region. These four gels indicate that there is no genomic DNA contamination in any of cerebral cortex, cerebellum or skeletal muscle samples that could bias the expression measurements. For the last gel, C. Cortex: cerebral cortex and S. muscle: skeletal muscle. The positive control for all three gels was human genomic DNA.
Supplemental Online Material Figure 2: Primer Validation. An agarose gel ensuring the PCR amplicon size matched Ensembl BLAST predictions.
Supplemental Online Material Table 1: Raw qRT-PCR data measurements
Supplemental Online Material Table 2: Normalized qRT-PCR data measurements using a modified delta-delta Ct method
Supplemental Online Material Table 3: Test for positive selection on the human and chimpanzee phylogenetic branches
Acknowledgements
We thank the Kathleen Price Bryan Brain Bank, J. Pecotte and M.J. Aivaliotis at the Southwest National Primate Research Center (NIH-NCRR grant P51 RR013986), E. Curran at the New England Regional Primate Research Center (NIH base grant RR00168), K.L. Summerville at the Yerkes National Primate Research Center (NIH base grant RR000165) and J. Horvath at Duke University for biological materials used in this study. We would also like to thank R. Frothingham for his direction and guidance with tissue extractions in the NIAID Regional Biocontainment Laboratory at Duke University (UC6-AI058607) and J. Tung and C. Wall for helpful discussions. NSF Grant NSF-BCS-08-27552 (HOMINID) to GAW supported this study.
Abbreviations
- Cr
creatine
- PCr
phosphocreatine
- SLC6A8
creatine transporter
- CKMT1
creatine kinase mitochondrial 1
- CKMT2
creatine kinase mitochondrial 2
- CKB
creatine kinase brain
- CKM
creatine kinase muscle
- UTR
untranslated region
- ATP
adenosine triphosphate
- ADP
adenosine diphosphate
- kb
kilobase
- DNA
deoxyribonucleic acid
- mRNA
messenger ribonucleic acid
- mya
million years ago
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiello LC, Wheeler P. The expensive-tissue hypothesis: the brain and the digestive system in human and primate evolution. Curr. Anthropol. 1995;36:199–221. [Google Scholar]
- Anselm IA, Alkuraya FS, Salomons GS, Jakobs C, Fulton AB, Mazumdar M, Rivkin M, Frye R, Poussaint TY, Marsden D. X-linked creatine transporter defect: a report on two unrelated boys with a severe clinical phenotype. J. Inherit. Metab. Dis. 2006;29:214–219. doi: 10.1007/s10545-006-0123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt CC, Fedrigo O, Pfefferle AD, Boyle AP, Horvath JE, Furey TS, Wray GA. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol. Evol. 2010;2:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Gen. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan JT, Brosnan ME. Creatine: endogenous metabolite, dietary, and therapeutic supplement. A. Rev. Nutr. 2007;27:241–261. doi: 10.1146/annurev.nutr.27.061406.093621. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon TW. What makes the human brain different? Ann. Rev. Anthropol. 1997;26:337–357. [Google Scholar]
- deGrauw TJ, Cecil KM, Byars AW, Salomons GS, Ball WS, Jakobs C. The clinical syndrome of creatine transporter deficiency. Mol. Cell Biochem. 2003;244:45–48. [PubMed] [Google Scholar]
- Delanghe J, De Slypere JP, De Buyzere M, Robbrecht J, Wieme R, Vermeulen A. Normal reference values for creatine, creatinine, and carnitine are lower in vegetarians. Clin. Chem. 1989;35:1802–1803. [PubMed] [Google Scholar]
- Du F, Zhu XH, Zhang Y, Friedman M, Zhang N, Ugurbil K, Chen W. Tightly coupled brain activity and cerebral ATP metabolic rate. Proc. Natl. Acad. Sci. 2008;105:6409–6414. doi: 10.1073/pnas.0710766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar RI, Shultz S. Evolution in the social brain. Science. 2007;317:1344–1347. doi: 10.1126/science.1145463. [DOI] [PubMed] [Google Scholar]
- Ellington WR. Evolution and physiological roles of phosphagen systems. Ann. Rev. Physiol. 2001;63:289–325. doi: 10.1146/annurev.physiol.63.1.289. [DOI] [PubMed] [Google Scholar]
- Fedrigo O, Warner LR, Pfefferle AD, Babbitt CC, Cruz-Gordillo P, Wray GA. A pipeline to determine candidate qRT -PCR control genes for evolutionary studies: application to primate gene expression across multiple tissues. PLoS ONE. 2010;5:e12545. doi: 10.1371/journal.pone.0012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JS, Nolte LA, Kawanaka K, Han DH, Jones TE, Holloszy JO. Glucose transport rate and glycogen synthase activity both limit skeletal muscle glycogen accumulation. Am. J. Physiol. Endocrinol. Metab. 2002;282:E1214–E1221. doi: 10.1152/ajpendo.00254.2001. [DOI] [PubMed] [Google Scholar]
- Gibson G. The environmental contribution to gene expression profiles. Nat. Rev. Genet. 2008;9:575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Rifkin SA. Natural selection on gene expression. Trends Genet. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat. Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins-Davis A, Townsend JP. Evolving gene expression: from G to E to GxE. Trends Ecol. Evol. 2009;24:649–658. doi: 10.1016/j.tree.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idaghdour Y, Storey JD, Jadallah SJ, Gibson G. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PLoS Genet. 2008;4:e1000052. doi: 10.1371/journal.pgen.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa J, Taniguchi T, Takeshita A, Maekawa M. Increased creatine kinase BB activity and CKB mRNA expression in patients with hematologic disorders: relation to methylation status of the CKB promoter. Clin. Chim. Acta. 2005;361:135–140. doi: 10.1016/j.cccn.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Isler K, van Schaik CP. The expensive brain: a framework for explaining evolutionary changes in brain size. J. Hum. Evol. 2009;57:392–400. doi: 10.1016/j.jhevol.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Jost CR, Van Der Zee CE, In ’t Zandt HJ, Oerlemans F, Verheij M, Streijger F, Fransen J, Heerschap A, Cools AR, Wieringa B. Creatine kinase B-driven energy transfer in the brain is important for habituation and spatial learning behaviour, mossy fibre field size and determination of seizure susceptibility. Eur. J. Neurosci. 2002;15:1692–1706. doi: 10.1046/j.1460-9568.2002.02001.x. [DOI] [PubMed] [Google Scholar]
- Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N. Statistical significance of quantitative PCR. BMC Bioinformatics. 2007;8:131. doi: 10.1186/1471-2105-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006a;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Tang K, Franz H, Kelso J, Hellmann I, Enard W, Lachmann M, Paabo S. Positive selection on gene expression in the human brain. Curr. Biol. 2006b;16:R356–R358. doi: 10.1016/j.cub.2006.03.082. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kushmerick MJ. Energy balance in muscle activity: simulations of ATPase coupled to oxidative phosphorylation and to creatine kinase. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1998;120:109–123. doi: 10.1016/s0305-0491(98)00026-1. [DOI] [PubMed] [Google Scholar]
- Leigh SR. Brain growth, life history, and cognition in primate and human evolution. Am. J. Primatol. 2004;62:139–164. doi: 10.1002/ajp.20012. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Robertson ML. Nutritional requirements and human evolution: A bioenergetics model. Am. J. Hum. Biol. 1992;4:179–195. doi: 10.1002/ajhb.1310040204. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Robertson ML. Evolutionary perspectives on human nutrition: The influence of brain and body size on diet and metabolism. Am. J. Hum. Biol. 1994;6:77–88. doi: 10.1002/ajhb.1310060111. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Robertson ML, Snodgrass JJ, Kuzawa CW. Metabolic correlates of hominid brain evolution. Comp. Biochem. Physiol. A. 2003;136:5–15. doi: 10.1016/s1095-6433(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Snodgrass JJ, Robertson ML. Effects of brain evolution on human nutrition and metabolism. A. Rev. Nutr. 2007;27:311–327. doi: 10.1146/annurev.nutr.27.061406.093659. [DOI] [PubMed] [Google Scholar]
- Luca F, Perry GH, Di Rienzo A. Evolutionary adaptations to dietary changes. A. Rev. Nutr. 2010;30:291–314. doi: 10.1146/annurev-nutr-080508-141048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews RT, Ferrante RJ, Klivenyi P, Yang L, Klein AM, Mueller G, Kaddurah-Daouk R, Beal MF. Creatine and cyclocreatine attenuate MPTP neurotoxicity. Exp. Neurol. 1999;157:142–149. doi: 10.1006/exnr.1999.7049. [DOI] [PubMed] [Google Scholar]
- McMorris T, Mielcarz G, Harris RC, Swain JP, Howard A. Creatine supplementation and cognitive performance in elderly individuals. Neuropsychol. Dev. Cogn. B Aging Neuropsychol. Cogn. 2007;14:517–528. doi: 10.1080/13825580600788100. [DOI] [PubMed] [Google Scholar]
- Milton K. Primate diets and gut morphology: implications for hominid evolution. In: Harris M, Ross EB, editors. Food and Evolution: Toward a Theory of Human Food Habits. Temple University Press; Philadelphia: 1987. pp. 93–115. [Google Scholar]
- Milton K. A hypothesis to explain the role of meat-eating in human evolution. Evol. Anthropol. 1999;8:11–21. [Google Scholar]
- Milton K. The critical role played by animal source foods in human (Homo) evolution. J. Nutr. 2003;133:3886S–3892S. doi: 10.1093/jn/133.11.3886S. [DOI] [PubMed] [Google Scholar]
- Pattyn F, Speleman F, De Paepe A, Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res. 2003;31:122–123. doi: 10.1093/nar/gkg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Schweiger U, Pellerin L, Hubold C, Oltmanns KM, Conrad M, Schultes B, Born J, Fehm HL. The selfish brain: competition for energy resources. Neurosci. Biobehav. Rev. 2004;28:143–180. doi: 10.1016/j.neubiorev.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Rae C, Digney AL, McEwan SR, Bates TC. Oral creatine monohydrate supplementation improves brain performance: a double-blind, placebo-controlled, cross-over trial. Proc. Biol. Sci. 2003;270:2147–2150. doi: 10.1098/rspb.2003.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sauter A, Rudin M. Determination of creatine kinase kinetic parameters in rat brain by NMR magnetization transfer: Correlation with brain function. J. Biol. Chem. 1993;268:13166–13171. [PubMed] [Google Scholar]
- Schiaffino MC, Bellini C, Costabello L, Caruso U, Jakobs C, Salomons GS, Bonioli E. X-linked creatine transporter deficiency: clinical description of a patient with a novel SLC6A8 gene mutation. Neurogenetics. 2005;6:165–168. doi: 10.1007/s10048-005-0002-4. [DOI] [PubMed] [Google Scholar]
- Schoenemann PT. Evolution of the size and functional areas of the human brain. Ann. Rev. Anthropol. 2006;35:379–406. [Google Scholar]
- Shomrat A, Weinstein Y, Katz A. Effect of creatine feeding on maximal exercise performance in vegetarians. Eur. J. Appl. Physiol. 2000;82:321–325. doi: 10.1007/s004210000222. [DOI] [PubMed] [Google Scholar]
- Snow RJ, Murphy RM. Creatine and the creatine transporter: a review. Mol. Cell Biochem. 2001;224:169–181. doi: 10.1023/a:1011908606819. [DOI] [PubMed] [Google Scholar]
- Somel M, Creely H, Franz H, Mueller U, Lachmann M, Khaitovich P, Paabo S. Human and chimpanzee gene expression differences replicated in mice fed different diets. PLoS One. 2008;3:e1504. doi: 10.1371/journal.pone.0001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford CB. The hunting apes: meat eating and the origins of human behavior. Princeton University Press; New Jersey: 1999. [Google Scholar]
- Stanford CB, Bunn HT. Meat-eating and human evolution. Oxford University Press Inc.; New York: 2001. [Google Scholar]
- Tachikawa M, Hosoya K, Ohtsuki S, Terasaki T. A novel relationship between creatine transport at the blood-brain and blood-retinal barriers, creatine biosynthesis, and its use for brain and retinal energy homeostasis. Subcell. Biochem. 2007;46:83–98. doi: 10.1007/978-1-4020-6486-9_5. [DOI] [PubMed] [Google Scholar]
- Tattersall I. An evolutionary framework for the acquisition of symbolic cognition by Homo sapiens. Comp. Cog. Beh. Rev. 2008;3:99–114. [Google Scholar]
- Uddin M, Wildman DE, Liu G, Xu W, Johnson RM, Hof PR, Kapatos G, Grossman LI, Goodman M. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl. Acad. Sci. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar PS, Grine FE, Teaford MF. Diet in early Homo: A review of the evidence and a new model of adaptive versatility. Ann. Rev. Anthropol. 2006;35:209–228. [Google Scholar]
- Uzawa OT, Endo Y, Bukawa H, Yokoe H, Shibahara T, Tanzawa H. Ubiquitous mitochondrial creatine kinase downregulated in oral squamous cell carcinoma. Br. J. Cancer. 2006;94:698–709. doi: 10.1038/sj.bjc.6602986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendelin M, Lemba M, Saks VA. Analysis of functional coupling: mitochondrial creatine kinase and adenine nucleotide translocase. Biophys. J. 2004;87:696–713. doi: 10.1529/biophysj.103.036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallimann T, Dolder M, Schlattner U, Eder M, Hornemann T, O’Gorman E, Ruck A, Brdiczka D. Some new aspects of creatine kinase (CK): compartmentation, structure, function and regulation for cellular and mitochondrial bioenergetics and physiology. Biofactors. 1998;8:229–234. doi: 10.1002/biof.5520080310. [DOI] [PubMed] [Google Scholar]
- Wallimann T, Hemmer W. Creatine kinase in non-muscle tissues and cells. Mol. Cell Biochem. 1994;133:193–220. doi: 10.1007/BF01267955. [DOI] [PubMed] [Google Scholar]
- Wallimann T, Wyss M, Brdiczka D, Nicolay K, Eppenberger HM. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the ’phosphocreatine circuit’ for cellular energy homeostasis. Biochem. J. 1992;281(Pt 1):21–40. doi: 10.1042/bj2810021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. Nutritional composition of red meat. Nutr. Diet. 2007;64:S113–S119. [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol. Rev. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Online Material Figure 1: Verifying cDNA purity using SDHA control primers that amplify an intronic region. These four gels indicate that there is no genomic DNA contamination in any of cerebral cortex, cerebellum or skeletal muscle samples that could bias the expression measurements. For the last gel, C. Cortex: cerebral cortex and S. muscle: skeletal muscle. The positive control for all three gels was human genomic DNA.
Supplemental Online Material Figure 2: Primer Validation. An agarose gel ensuring the PCR amplicon size matched Ensembl BLAST predictions.
Supplemental Online Material Table 1: Raw qRT-PCR data measurements
Supplemental Online Material Table 2: Normalized qRT-PCR data measurements using a modified delta-delta Ct method
Supplemental Online Material Table 3: Test for positive selection on the human and chimpanzee phylogenetic branches