Abstract
Strategies to integrate instructive biomolecular signals into a biomaterial are becoming increasingly complex and bioinspired. While a large majority of reports still use repeated treatments with soluble factors, this approach can be prohibitively costly and difficult to translate in vivo for applications where spatial control over signal presentation is necessary. Recent efforts have explored the use of covalent immobilization of biomolecules to the biomaterial, via both bulk (ubiquitous) as well as spatially-selective light-based crosslinking, as a means to both enhance stability and bioactivity. However, little is known about how processing conditions during immobilization impact the degree of unintended non-covalent interactions, or fouling, that takes place between the biomaterial and the biomolecule of interest. Here we demonstrate the impact of processing conditions for bulk carbodiimide (EDC) and photolithography-based benzophenone (BP) crosslinking on specific attachment vs. fouling of a model protein (Concanavalin A, ConA) within collagen-glycosaminoglycan (CG) scaffolds. Collagen source significantly impacts the selectivity of biomolecule immobilization. EDC crosslinking intensity and ligand concentration significantly impacted selective immobilization. For benzophenone photoimmobilization we observed that increased UV exposure time leads to increased ConA immobilization. Immobilization efficiency for both EDC and BP strategies was maximal at physiological pH. Increasing ligand concentration during immobilization process led to enhanced immobilization for EDC chemistry, no impact on BP immobilization, but significant increases in non-specific fouling. Given recent efforts to covalently immobilize biomolecules to a biomaterial surface to enhance bioactivity, improved understanding of the impact of crosslinking conditions on selective attachment versus non-specific fouling will inform the design of instructive biomaterials for applications across tissue engineering.
A. Introduction
The design of biomaterial scaffolds for a range of in vitro and in vivo tissue engineering applications often requires selective optimization of structural, mechanical, and biomolecular properties.1 Scaffold developed from naturally-derived polymers represent an important group of constructs currently under development. Decellularized tissues such as small intestinal submucosa (SIS) offer one important class of naturally-derived biomaterials with demonstrated promise for clinical application.2–5 Additionally, naturally derived polymers such as collagen, fibrin, and silk have also been increasingly used to generate porous biomaterials via a range of processing approaches (e.g., freeze-drying, electrospinning, salt-leaching)1, 6–10. Collagen-glycosaminoglycan (CG) scaffolds are another class of such materials which have been optimized for a range of tissue engineering applications such as peripheral nerve, skin, and increasingly musculoskeletal tissues.11–19 Fabricated via freeze-drying, CG scaffolds maintain an open-cell foam (interconnected) pore architecture. Our laboratory and others have been responsible for developing fabrication strategies to tailor scaffold biophysical properties via pore size and shape,12–14, 20, 21 mechanical properties,22–24 and degradation rate14, 25 in order to optimize cell bioactivity. However, like with many biomaterial platforms, it has become apparent that biomolecular signals added to culture media can enhance or help direct cellular responses.26–29
While soluble supplementation of the growth media with biomolecules of interest is feasible for in vitro assays, repeated replacement of factor supplemented media is both expensive and prone to rapid diffusive loss when used in vivo.1, 30, 31 Examining processes associated with biomolecular signaling within the native ECM provides motivation for alternative supplementation strategies. Primary components of the ECM such as GAGs and proteoglycans (PGs) are responsible for sequestering factors within the matrix, a dynamic process that can lead to improved stability of biomolecular signals as well as enhanced bioactivity of the factor.1, 32, 33 Design of biomaterial platforms to mimic such interactions can induce a significantly more nuanced response of cells to local biomolecular signals. For example, heparin sulfate decorated biomaterials have alternatively been shown to upregulate cell response to FGF-2 and VEGF while down regulating cell response to PDGF.1, 32, 33 Recently, a variety of techniques have attempted to exploit concepts of transient sequestration of biomolecules within three-dimensional biomaterials via incorporation of GAGs, PGs, binding domains to proteins of interest, or their synthetic mimics to regulate biomolecule attachment.1, 32, 34–36 Notably, while GAG content had previously been shown to impact the regenerative capacity of CG scaffolds in full thickness skin wounds,14, 37 we recently demonstrated that selective alteration of the GAG content can also be used to impact transient sequestration of growth factors within the matrix as well as resultant cell bioactivity.38
Beyond transient sequestration inspired approaches, a wide range of covalent tethering methods have also been described. Carbodiimide (EDC) chemistry is commonly used to crosslink collagen biomaterials to improve their enzymatic stability and mechanical properties22, 39, 40 as well as cell response.12, 41–43 EDC has also been widely used to covalently bind a wide range of biomolecules, from matrix proteins to growth factors, to the biomaterial scaffold.26, 44–47 This approach leads to ubiquitous immobilization of the biomolecule of interest to the scaffold and does not allow decoupling of crosslinking and biomolecule attachment. Recently, we have described a benzophenone (BP) photolithography approach to create spatial patterns of factors covalently attached to the CG scaffold.48 However, given that non-covalent factor sequestration can significantly impact cell response, it is important to also consider the magnitude of unintended non-covalent interactions, biomolecule fouling on the scaffold, that arise as a result of the covalent attachment processes. Relatively little is known about how processing conditions during covalent biomolecules attachment (e.g., EDC, BP) impact non-covalent fouling. The objective of this study therefore was to examine specific attachment versus fouling of Concanavalin A (ConA), a carbohydrate-binding protein often used as a model system for covalent immobilization strategies48, 49 that retains the capacity to non-covalently bind to the ECM,50 as a function of EDC and BP processing conditions. Elucidating the relative contribution of covalent attachment versus non-specific fouling of biomolecular signals within a model CG scaffold will inform the selection of processing conditions used to add instructive biomolecular signals into CG scaffolds for a range of tissue engineering applications.
B. Materials and Methods
B.1. CG scaffold fabrication
CG scaffolds were fabricated via a previously described lyophilization process.13, 48 Briefly, a CG slurry was generated by homogenizing fibrillar collagen (Collagen Matrix, Franklin Lakes, NJ; Sigma Aldrich, St. Louis, MO) and chondroitin sulfate isolated from shark cartilage (Sigma Aldrich) in 0.05 M acetic acid (Acros Organic, NJ).14 The 0.5 wt% CG suspension was stored at 4°C and degassed prior to use. The suspension was placed in an aluminum mold and then cooled at a constant freezing rate (1 °C/min) to −40°C. Ice crystals were subsequently removed via sublimation (0°C, 200 mTorr), resulting in a dry, porous scaffold sheet. Scaffold sheets were then crosslinked and sterilized via a dehydrothermal crosslinking step (105°C, 24 hours; 0.5 mmHg).13 Experimental specimens (6 mm dia.; 3 mm thick) were cut from the sheet using a biopsy punch (Integra-Miltex, York, PA).
B.2. Immobilization of Concanavalin A within the scaffold network
B.2.1. Ubiquitous attachment via EDC crosslinking
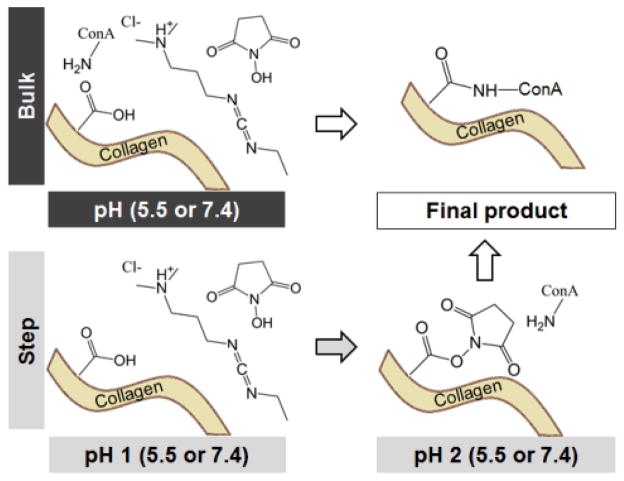
Concanavalin A AlexaFluor 647 conjugate (ConA-647; Invitrogen, Carlsbad, CA) was immobilized within the scaffold network via EDC-NHS crosslinking, utilizing 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) (Sigma Aldrich) (Figure 1). EDC-NHS crosslinking intensity was controlled by altering the molar ratio of EDC and NHS to the carboxylic acid (COOH) side chains on the CG scaffold.22, 39, 51 Here, a constant EDC: NHS ratio, but increasing solution concentrations (EDC:NHS:COOH ratios of 2.5:6.25:1, 5:12.5:1, 10:25:1) was used to generate a series of scaffold with increasing crosslinking density.
Figure 1.
Reaction schematics for bulk versus step carbodiimide crosslinking reactions with Concanavalin A (ConA).
EDC crosslinking can proceed via a bulk or step process,47 giving the opportunity to tailor solution pH during each step to improve crosslinking efficiency (Figure 1). In step EDC, the CG scaffolds were hydrated in PBS, added to a crosslinking solution of EDC and NHS dissolved in PBS, and shaken (37° C, 30 min) to activate the crosslinking process. Scaffolds were then rapidly blotted with a Kimwipe before 10 μL of a ConA-647 solution was pipetted onto each scaffold and allowed to attach (1 hr; room temperature).26 In bulk EDC, the 10 μL ConA-647 solution was added with the EDC and NHS and allowed to attach (1 hr; room temperature).47 In both cases, multiple ConA-647 solution concentrations (1 μg/mL, 750 ng/mL, 500 ng/mL, 250 ng/mL) were tested. After crosslinking, scaffolds were then blotted to remove excess protein and washed in PBS (1 hr; room temperature) on a shaker. The wash solution was then replaced and the scaffolds were left overnight on the shaker. Control scaffolds were treated with EDC: NHS solution and a PBS solution in order to generate the same scaffold crosslinking density without providing a ligand for incorporation. After the overnight wash step, scaffolds were treated with 10 μL of ConA-647 protein solution (1 hour, room temperature) prior to undergoing a second round of wash steps.
B.2.2. Selective immobilization via benzophenone photochemistry
ConA was alternatively immobilized within the CG scaffold using a previously described benzophenone photochemistry method.48 As exposure to UV light is required for BP photoimmobilization, biotinylated ConA (Vector Labs, Southfield, MI) was used for BP attachment instead of ConA-647; a fluorescent secondary antibody was added at a later step for quantification. Briefly, benzophenone isothiocyanate (Invitrogen) was covalently immobilized within the scaffold in a solution of dimethylformamide (DMF) and N-diisopropylethylamine (DIEA). Unbound BP was removed with repeated DMF, ethanol, and PBS washes. Excess moisture was blotted from the scaffolds prior to being placed on glass slides for photoimmobilization. A 10 μL solution of biotinylated ConA in PBS (5 μg/mL) was then pipetted onto each scaffold. Scaffolds were exposed to 350–365 nm UV light at 20 mW/cm2 (8, 15, 22, 30, 45, 60, 90, or 120 seconds). The scaffold was then flipped and re-exposed for the same duration. Unreacted BP was deactivated using a mild pluronic acid solution for 1 hour at room temperature prior to multiple PBS washes. Control scaffolds were generated under identical conditions, only with 10 μL of PBS added to the scaffolds prior to UV irradiation.
B.3. Analysis of biomolecular patterns generated via BP photolithography
The consistency of ConA patterns formed within the CG scaffold as a function of BP photoimmobilization was examined via fluorescence microscopy (Leica DMI4000B fluorescence microscope, Qimaging camera) as previously described.48, 52 Lateral resolution of patterning was determined for CG scaffolds fabricated from multiple collagen sources (Sigma-Aldrich, Collagen Matrix) that were patterned with 500 μm square islands of ConA. Achievable patterning depth was determined from scaffolds patterned with repeating stripes of ConA (100 μm stripes, 400 μm spacing; 2 min exposure time). Patterned scaffolds were blocked with 2% BSA solution, treated with streptavidin conjugated Alexafluor488 secondary antibody (1 hour on a shaker), then washed in PBS prior to imaging.
The lateral pattern resolution was determined from fluorescent images generated by placing photopatterned surfaces of the scaffolds into glass coverslip bottom dishes (In Vitro Scientific, Sunnyvale, CA). The mean fluorescent intensity (MFI) of 6 randomly selected patterned and unpatterned regions were calculated using ImageJ software to determine the average signal:noise ratio of each image. Analysis was performed on 4 discrete samples for each collagen type (Sigma-Aldrich; Collagen Matrix). Achievable pattern depth was determined by first embedding striped scaffolds in Optimal Cutting Temperature (OCT) solution (Tissue-Tek, Torrance, CA) followed by flash freezing. The embedded scaffolds were then sliced longitudinally, perpendicular to the direction of the patterned stripes. Resultant sections were then rinsed in PBS and placed cut side down onto a coverslip for imaging.
B.4. Quantifying immobilized ConA within the CG scaffold
Scaffolds were subsequently digested in a papain solution (Sigma-Aldrich) in a 60° C water bath for 1 hour to recover immobilized (covalent, fouling) ConA for analysis. Total immobilized ConA within the scaffold network was determined via a F200 fluorometer (Tecan, Switzerland) from scaffold digests. For EDC-based immobilization, the inherent fluorescence of the ConA-647 was examined (620/670 nm excitation/emission). For BP-based immobilization, scaffolds containing photopatterned biotinylated ConA were rinsed in 5% Sucrose/PBS solution overnight, stained with Streptavidin-AlexaFluor488 (1:2000 dilution in 2% BSA in PBS, Invitrogen) for 1 hr then rinsed in PBS. The fluorescence of AlexaFluor488 labeled ConA within the scaffolds was examined (485/535 nm excitation/emission). Relative fluorescence between groups was used to examining covalent attachment versus fouling. For EDC crosslinking, results were compared to a standard curve of digest solutions spiked with known concentrations of soluble ConA biotin AlexaFluor 647. For BP photoimmobilization, the relative fluorescence of each specimen was compared to negative control CG scaffolds that did not contain biotinylated ConA or streptavidin-AF488 conjugates.
B.5. Statistical Methods
The effect of EDC:NHS:COOH ratio and ConA solution concentration (n = 8) was evaluated 2-way ANOVA with Bonferroni post hoc. The effects of pH on EDC immobilization methods (n = 6) were analyzed via 1-way ANOVA and Tukey HSD. The effect of BP patterning steps were evaluated using a minimum of n = 3 samples per group via 1-way ANOVA and Bonferroni post hoc testing; the correlation between UV exposure time and immobilization was tested at α = 0.05, power = 0.8 (n = 40). The effects of pH on immobilization and ConA loading (n = 9) was analyzed using a 2-way ANOVA and Bonferroni post hoc testing between pHs. The effect of one (n = 24) or two (n = 30) washes on fouling was determined via Independent Samples T-Test. The effects of wash solutions on ConA immobilization was determined by 1-way ANOVA and Dunnett’s test on a minimum of n = 5 specimens per group. The effect of sucrose treatment order on fouling was examined for a minimum of n = 3 samples per group via ANOVA and Bonferroni’s post hoc test. Significance was set at p < 0.05. Error was reported in figures as the standard error of the mean unless otherwise noted.
C. Results
C.1. Collagen source significantly impacts biomolecule patterning
The choice of collagen source significantly impacted immobilization of model biomolecules within the CG scaffold network. ConA patterns were assessed using CG scaffolds fabricated via identical lyophilization protocols from collagen-GAG suspensions generated from collagen sourced from Collagen Matrix or Sigma-Aldrich. Notably, ConA patterns produced within CG scaffolds fabricated from Collagen Matrix sourced collagen demonstrated improved definition of pattern margins (Figure 2A) as well as significantly (p < 0.01) higher pattern intensity (signal:noise ratio) (Figure 2B). All subsequent experiments were therefore performed exclusively using Collagen Matrix scaffolds.
Figure 2. The impact of collagen source on benzophenone immobilization of Concanavalin A (ConA) on CG scaffolds.
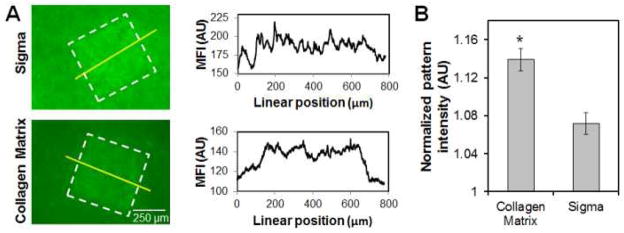
(A) Representative fluorescence image and Mean Fluorescence Intensity (MFI) of ConA signal in arbitrary units (AU) from line scan across pattern. (B) Normalized pattern:noise signal intensity for ConA immobilized on CG scaffolds. *: significant difference in signal intensity between collagen sources.
C.2. EDC immobilization efficiency depends on EDC:NHS:COOH ratio and concentration of ligand
EDC processing conditions significantly impacted total ConA immobilization within the CG scaffold network (Figure 3). Four processing parameters were identified as potential modulators of covalent versus non-covalent ConA attachment during the EDC immobilization: concentration of ConA, EDC:NHS:COOH crosslinking intensity, solution pH, and use of a bulk versus step crosslinking reaction. Not surprisingly, fouling was positively correlated with ConA concentration (data not shown). ConA solution concentration (2.5 – 10 ng, corresponding to solution concentrations of 0.25 – 1 μg/ml) and EDC:NHS:COOH crosslinking ratio (2.5:6.25:1, 5:12.5:1, and 10:25:1) both significantly (p ≤ 0.01) impacted ConA immobilization (Figure 3A). Notably, immobilization significantly increased with solution phase ConA concentration (loading) (p < 0.05 for all cases). An EDC:NHS:COOH cross-linking ratio of 5:12.5:1 let to significantly (p < 0.01) higher immobilization than the other two ratios tested. An EDC:NHS:COOH ratio of 5:12.5:1 was subsequently used for future analyses.
Figure 3. Carbodiimide crosslinking of Concanavalin A (ConA) to CG scaffold.
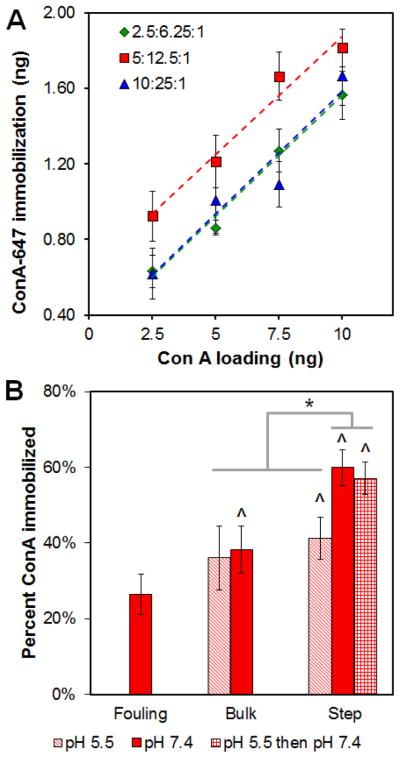
(A) Total ConA immobilization as a function of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide:N-hydroxysuccinimide:carboxylic acid ratio and total ConA added to the scaffold. (B) Impact of solution pH and the use of bulk versus step carbodiimide chemistry on total ConA immobilization within the scaffold. ^: significant increase in ConA immobilization versus non-specific fouling (ConA not present during EDC crosslinking). *: significant difference between groups.
The use of bulk versus step carbodiimide crosslinking reaction, as well as altering the pH of the crosslinking reaction (pH 5.5 vs. 7.4), were both found to significantly (p < 0.05) impact ConA immobilization (Figure 3B). Step crosslinking induced a significant increase in ConA immobilization relative to non-specific ConA fouling regardless of pH (p < 0.01). Bulk crosslinking at pH 7.4 (not at more acidic pH 5.5) resulted in a significant (p = 0.04) increase in ConA immobilization. Step EDC chemistry conducted exclusively at neutral pH (7.4) resulted in significantly higher immobilization compared to bulk crosslinking (p < 0.001). However, the increase in ConA immobilization was not seen at acidic pH (pH 5.5; p > 0.74). Together, these results indicate that carbodiimide immobilization was most efficient using the step crosslinking approach with the second step performed at neutral pH (p < 0.001); there was no impact of solution pH during the first stage of the step process (p = 0.073).
C.3. BP photoimmobilization induces significant specific and non-specific attachment
Benzophenone photoimmobilization has previously been shown via visual inspection to generate defined patterns of biomolecules within CG scaffolds.48, 52 Here, analysis of the fluorescence intensity of photoimmobilization ConA-biotin (via streptavidin-AlexaFluor 488 secondary antibody) showed a significant (p < 0.001) increase in immobilized ConA within the CG scaffold with BP photoimmobilization (Figure 4A). However, significant (p < 0.001) fouling was observed either without UV exposure (BP + ConA) or when ConA was added after UV exposure (BP + UV; ConA) (Figure 4A). However, direct BP-photoimmobilization (BP + UV + ConA) led to a significant (p < 0.001) increase in immobilized ConA compared to all non-specific (fouling) groups.
Figure 4. Benzophenone (BP) photoimmobilization of Concanavalin A (ConA) to CG scaffold.
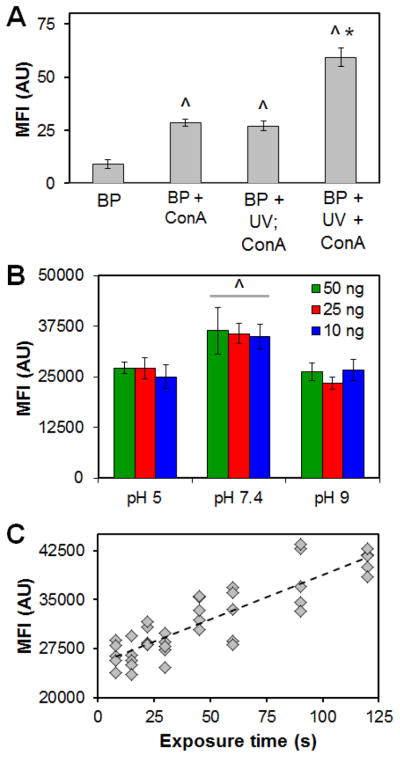
(A) Mean Fluorescence Intensity (MFI) in arbitrary units (AU) of immobilized ConA calculated from representative fluorescence images as a function of BP processing steps (50 ng ConA biotin added per scaffold) (BP: benzophenone functionalized; ConA: Concanavalin A present; UV: Ultraviolet exposure; +: concomitantly). ^: significant increase in MFI versus BP alone (fouling). *: significant increase in MFI for specific photoimmobilization of ConA versus all other treatment groups. (B) MFI of immobilized ConA as a function of solution pH and total loaded ConA. ^: significantly increased MFI versus other pH conditions. (C) Total BP photolithographic immobilization of ConA increases with increasing UV exposure time.
We subsequently examined the impact of solution pH, concentration of ConA in solution during immobilization, and UV exposure time on BP-photoimmobilization. BP photolithography was impacted by solution pH, with greatest patterning efficiency achieved at a pH of 7.4 (versus pH 5, 9; p < 0.01) (Figure 4B). While ConA immobilization did not increase (p = 0.83) with increasing ConA concentration over the range tested (Figure 4B), time of UV exposure significantly impacted immobilization (R2 = 0.77, p < 0.01) (Figure 4C).
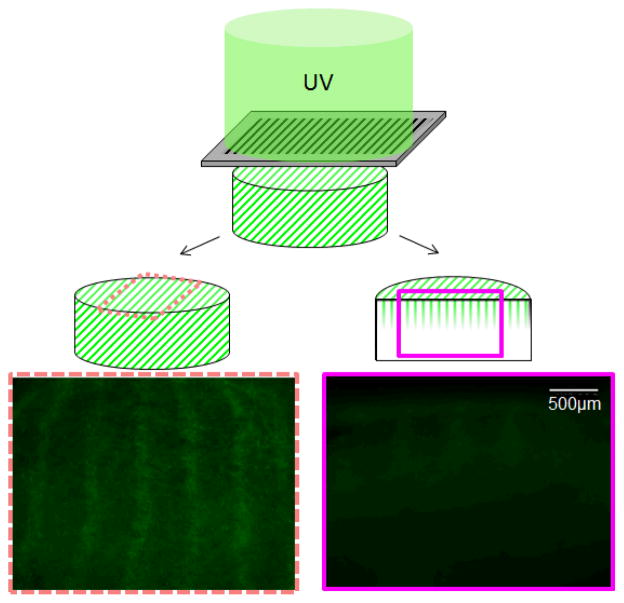
C.4. Depth of ConA pattern penetration via BP photoimmobilization
A striped pattern was used to determine the achievable patterning depth within the CG scaffold. Here, the ConA pattern was visualized in transverse cryostat sections through the patterned scaffold. BP-photopatterning was able to achieve patterning depths on the order of 500 μm into the scaffold (Figure 5), consistent with results reported for VEGF52.
Figure 5. Achievable pattern resolution via benzophenone photolithography.
Representative fluorescence images of 100 μm wide Concanavalin A stripes (400 μm spacing) patterned into CG scaffolds. Images take from longitudinal cross section (left) versus transverse section through the depth of the scaffold (right) indicate limited (500 μm) depth pattern penetration.
C.5. Impact of processing conditions on non-specific ConA immobilization
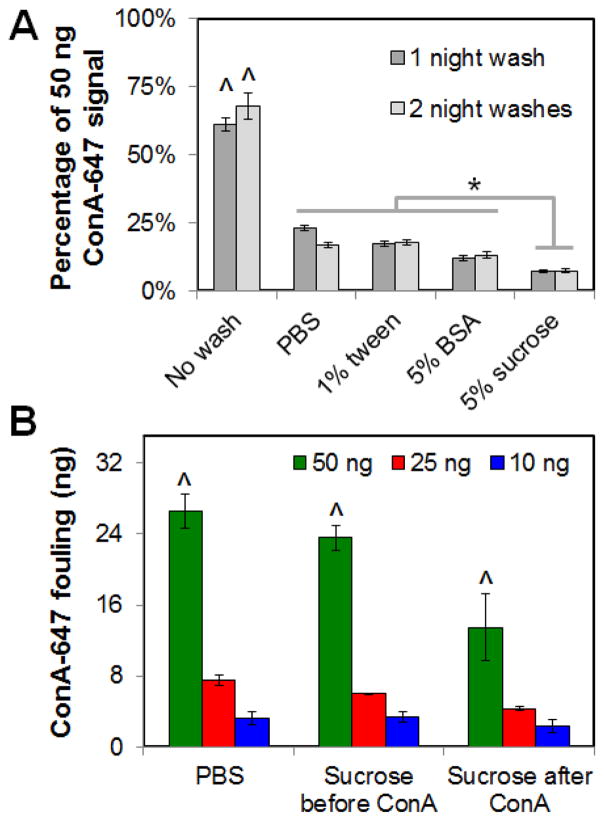
Given the presence of significant non-specific fouling during both covalent immobilization strategies tested (EDC, BP photolithography; Figures 3, 4), we explored the impact of washing steps on ligand fouling (Figure 6). Adding a 24 hour wash step after ConA exposure significantly (p < 0.001) decreased non-specific factor attachment for all wash solutions tested (PBS; PBS + 1% Tween; PBS + 5% BSA; PBS + 5% sucrose). While the addition of a second 24-hour wash step did not significantly reduce fouling for any wash solution (p = 0.69), the use of a PBS + 5% sucrose wash led to the most significant (p ≤ 0.03) reduction in ConA fouling relative to all other wash solutions (Figure 6A). The order of incorporating the sucrose wash significantly impacted ConA fouling (Figure 6B). Here, adding the sucrose wash after ConA exposure led to a significant decrease in fouling (p = 0.001) while washing the scaffolds with sucrose prior to ConA exposure had no impact (p = 0.79) Not surprisingly, longer exposure (20 vs. 5 min) to the ConA ligand led to significantly (p =0.003) increased fouling (Supp. Fig. 1A). Additionally, comparing the fraction (Supp. Fig. 1B) versus total amount (Figure 6B) of ConA fouling within the scaffold, higher ligand concentrations led to both greater total and percent fouling. However, use of a sucrose wash after ConA exposure significantly (p < 0.05) reduced total and percent fouling except at the lowest (10 ng) ligand added where the percent fouling was not significantly different from the middle (25 ng) ligand added.
Figure 6. Impact of wash steps on non-covalent fouling of Concanavalin A (ConA) within the scaffold.
(A) ConA fouling within CG scaffold scaffold as a function of wash conditions (20 minute ConA exposure). ^: significant increase in ConA fouling versus all wash groups. *: significant difference in ConA fouling between wash groups. (B) Total ConA fouling as a function of the order of sucrose wash steps (none; before ConA exposure; after ConA exposure). ^: significant increase in ConA fouling within each wash group as a function of total loaded ConA.
D. Discussion
This study examined the relationship between covalent patterning conditions sand resultant specific versus non-specific incorporation of a model ligand (ConA) within a three-dimensional CG scaffold. Previous work with CG scaffolds has shown that the incorporation of soluble and covalent biomolecules into CG scaffolds impacts cellular behavior.20, 26, 29, 53 However, these studies have focused on contributions of the total factor present without distinguishing the impacts of specific attachment and fouling individually. For this study we examined how the manipulation of treatment parameters for two covalent immobilization strategies impacted both covalent attachment and fouling.
Optimizing a biomaterial for a tissue engineering application requires a consideration of structural, compositional, mechanical, and biochemical cues. However, once a material such as collagen is chosen for the scaffold composition, there still remains the non-trivial consideration of source. In this study we explored the potential for fouling versus specific patterning of a model protein, ConA, on CG scaffolds fabricated using identical lyophilization conditions from CG precursor suspensions generated from two collagen sources. While both collagens evaluated were type I of bovine origin, significant differences in immobilization were detected. The increased signal:noise ratio and improved pattern margins seen with one collagen source (Figure 2) suggested that in the context of optimizing biomolecular supplementation strategies for natural-protein derived scaffolds it is critical to consider source and processing of raw ingredients.
Covalent immobilization of biomolecules within a biomaterial offers the potential to improve both the half-life and bioactivity of the factor, as well as the potential to reduce the total dose required in cases where long-term efficacy is required.1, 35, 54 Carbodiimide crosslinking represent a common approach to tether biomolecules to a biomaterial surface, as well as to crosslinking collagen-based biomaterials.22, 39, 55 In the context of crosslinking collagen-based scaffolds, previous efforts have examined the use of bulk versus step crosslinking, the impact of changing the EDC:NHS ratio, and the reaction pH. While previous efforts by Chiu et al. examined the impact of some of these conditions on VEGF immobilization within a model collagen scaffold,47 in this study we examined the impact of step versus bulk immobilization, EDC:NHS ratio, and crosslinking solution pH on covalent immobilization of a model protein (ConA) within the CG scaffold. Confirming the observations of Chiu et al., our findings demonstrate that immobilization of ConA within the CG scaffold was maximized at neutral pH using the step crosslinking reaction (Figure 3B). Additionally, we further examined the specific impact of EDC:NHS:COOH ratio (2.5:6.25:1 to 10:25:1), known to impact scaffold crosslinking,22 on ConA immobilization (Figure 3A). Not surprisingly, increasing the solution-phase concentration of ConA led to a significant increase in immobilized ConA within the scaffold for all tested EDC:NHS ratios. However, ConA immobilization efficiency did not scale with crosslinking density, known to increase with EDC:NHS:COOH ratio;22 instead, greatest immobilization was achieved with the middle EDC:NHS:COOH ratio (Figure 3A). This result may indicate a tradeoff in attachment efficiency as a function of the size of the ligand, an observation which also correlates with our finding that increasing ConA solution concentration did not lead to improved immobilization via BP photolithography (Figure 4B). Future experiments that explore crosslinking efficiency as a function of biomolecule size and concentration may help better resolve this effect.
We then considered an orthogonal covalent immobilization strategy, benzophenone photoimmobilization. Our laboratory has previously employed BP to decorate CG scaffolds with both ECM molecules (fibronectin)48 and growth factors (VEGF, PDGF),52, 56 However, the impact of patterning conditions on BP photoimmobilization has only been optimized for protein immobilization onto glass substrates,49 with degree of covalent attachment of P-selectin and mannan on glass found to be dependent on ligand concentration and time of UV exposure. Here, we explored whether both processing conditions impacted ConA immobilization with a fully three-dimensional CG scaffold. While increasing UV exposure time led to augmented ConA immobilization (Figure 4C), increasing ConA solution concentration within the scaffold during photoimmobilization surprisingly did not. Further, solution pH was found to significantly impact the BP process, with ConA immobilization efficiency maximal at physiological pH. While BP photolithography theoretically allows for patterning of biomolecules in three dimensions, such patterning requires significant penetration of the UV light into the biomaterial. Similar to observations of light-based of biomolecules within 3D hydrogels (where maximal penetration was as low at 200 μm in some cases)57–59 and for VEGF within the CG scaffold,52 effective patterning depth was found to be limited to approximately 500 μm. Such observations are likely to motivate alternative strategies for templating biomaterials at larger scales. However, the potential for non-specifically attached (fouling) biomolecules, previously estimated to range on order of 8% – 24% of total loaded factor,47 to impact cells throughout an entire construct while patterned factors only impact the first 500 μm of the construct suggest a critical need to explore processing conditions to reduce fouling. Such a need may only be magnified for studies leveraging bioinspired approaches to transiently immobilize growth factors within the biomaterial through the use of glycosaminoglycan or heparin binding peptides.32, 36, 38
In this study we pursued a strategy to assess non-specific fouling of ConA within the scaffold as a function of processing conditions and subsequent washes. In the case of EDC crosslinking (Figure 3), fouling was assessed through a separate experimental group that exposed the scaffolds to ConA after crosslinking was completed. In the case of BP photoimmobilization (Figure 4), fouling was assessed through the use of two groups. First, BP decorated scaffolds were exposed to ConA (BP + ConA) without UV exposure. Second, BP scaffolds were exposed to UV without the presence of ConA (BP + UC; ConA); after UV exposure the scaffolds were then exposed to ConA to confirm both the transience of BP-photoactivation activation as well as that UV exposure did impact the potential for fouling on the scaffold surface. In both cases, while significant fouling did occur, both EDC and BP immobilization strategies were able to incorporate significantly increased factor doses.
In an effort to combat non-specific fouling, we examined means of controlling the degree of fouling following patterning (Figure 6). Not surprisingly, adding a wash step after immobilization, regardless of the identity if the wash solution, significantly reduced fouling; however, adding multiple wash steps did not seem to further improve this effect. The most effective method to eliminate non-specific ConA attachment was by introducing a wash solution with the potential of scavenging non-covalently bound ConA: sucrose. ConA is known to bind glucose residues,50 leading to our choice of a sucrose wash solution to compare to more conventional Tween and BSA washes. The use of a 5% sucrose solution led to significant reduction in fouling; however out results suggested the sucrose solution acted most efficiently by removing ConA that had already fouled the scaffold surface (Sucrose after ConA) rather than acting to block non-specific ConA (Sucrose before ConA; Figure 6B). Although the sucrose wash solution demonstrated here was designed for the ligand of interest in this study (ConA), it is likely that appropriate choice of wash solutions (e.g., heparin solution for VEGF, PDGF; antibody tagged microspheres coupled with centrifugation) may be particularly important in the design of any biomolecule immobilization experiment in order to minimize fouling and hence maximize intended effects.
Conclusions
The development of biomaterials for increasingly complex tissue engineering applications is increasingly calling for approaches to either improve the efficiency of action or regionally control the presentation of biochemical factors within a three-dimensional matrix. However, like as is seen within the native ECM, non-covalent or transient immobilization of factors can significantly impact cell response. In this study we investigated the impact of processing conditions on covalent immobilization of a model protein (ConA) within a CG scaffold using methods that enable either ubiquitous (carbodiimide crosslinking) or regionally-controlled (BP photolithography) immobilization. We identified processing conditions that increased attachment efficiency, notably pH (both), ligand concentration (carbodiimide), and crosslinking intensity (both). We found that for both methods, significant non-specific fouling is observed and can be quantified. Lastly, we report post-immobilization washing steps to minimize ConA fouling. These efforts are informing ongoing efforts to generate CG scaffolds containing instructive biomolecular signals to augment scaffold bioactivity for a range of musculoskeletal repair applications.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Aurora Alsop (UIUC) for assistance with BP-photopatterning. This material is based upon work supported by the National Science Foundation under Grant No. 1105300. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers R03 AR062811. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We are grateful for additional funding for this study provided by the Chemical and Biomolecular Engineering Dept. (BAH) and the Institute for Genomic Biology (BAH) at the University of Illinois at Urbana-Champaign.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
References
- 1.Place ES, Evans ND, Stevens MM. Nat Mater. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- 2.Badylak SF, Freytes DO, Gilbert TW. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Wu S, Liu Y, Bharadwaj S, Atala A, Zhang Y. Biomaterials. 2011;32:1317–1326. doi: 10.1016/j.biomaterials.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Lin HK, Frimberger D, Epstein RB, Kropp BP. BJU Int. 2005;96:1120–1125. doi: 10.1111/j.1464-410X.2005.05741.x. [DOI] [PubMed] [Google Scholar]
- 5.Rossetto VJ, da Mota LS, Rocha NS, Miot HA, Grandi F, Brandao CV. Acta Vet Scand. 2013;55:39. doi: 10.1186/1751-0147-55-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornwell KG, Pins GD. J Biomed Mater Res A. 2007;82:104–112. doi: 10.1002/jbm.a.31057. [DOI] [PubMed] [Google Scholar]
- 7.Malafaya PB, Silva GA, Reis RL. Adv Drug Deliv Rev. 2007;59:207–233. doi: 10.1016/j.addr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Kim TG, Chung HJ, Park TG. Acta Biomater. 2008;4:1611–1619. doi: 10.1016/j.actbio.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Powell HM, Supp DM, Boyce ST. Biomaterials. 2008;29:834–843. doi: 10.1016/j.biomaterials.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Sill TJ, von Recum HA. Biomaterials. 2008;29:1989–2006. doi: 10.1016/j.biomaterials.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Soller EC, Tzeranis DS, Miu K, So PT, Yannas IV. Biomaterials. 2012;33:4783–4791. doi: 10.1016/j.biomaterials.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 12.Harley BA, Kim HD, Zaman MH, Yannas IV, Lauffenburger DA, Gibson LJ. Biophys J. 2008;95:4013–4024. doi: 10.1529/biophysj.107.122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Brien FJ, Harley BA, Yannas IV, Gibson LJ. Biomaterials. 2005;26:433–441. doi: 10.1016/j.biomaterials.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 14.Yannas IV, Lee E, Orgill DP, Skrabut EM, Murphy GF. Proc Natl Acad Sci USA. 1989;86:933–937. doi: 10.1073/pnas.86.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silver FH, Yannas IV, Salzman EW. Thromb Res. 1978;13:267–277. doi: 10.1016/0049-3848(78)90014-2. [DOI] [PubMed] [Google Scholar]
- 16.Yannas IV, Burke JF, Huang C, Gordon PL. Polym Prepr Am Chem Soc. 1975;16:209–214. [Google Scholar]
- 17.Harley BAC, Gibson LJ. Chem Eng J. 2008;137:102–121. [Google Scholar]
- 18.Getgood AMJ, Kew SJ, Brooks R, Aberman H, Simon T, Lynn AK, Rushton N. The Knee. 2012;19:422–430. doi: 10.1016/j.knee.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Yannas IV, Tzeranis DS, Harley BA, So PT. Philos Trans A Math Phys Eng Sci. 2010;368:2123–2139. doi: 10.1098/rsta.2010.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caliari SR, Harley BAC. Biomaterials. 2011;32:5330–5340. doi: 10.1016/j.biomaterials.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haugh MG, Murphy CM, O’Brien FJ. Tissue Eng Part C Methods. 2010;16:887–894. doi: 10.1089/ten.TEC.2009.0422. [DOI] [PubMed] [Google Scholar]
- 22.Harley BA, Leung JH, Silva EC, Gibson LJ. Acta Biomater. 2007;3:463–474. doi: 10.1016/j.actbio.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Vickers SM, Gotterbarm T, Spector M. J Orthop Res. 2010;28:1184–1192. doi: 10.1002/jor.21113. [DOI] [PubMed] [Google Scholar]
- 24.Caliari SR, Weisgerber DW, Ramirez MA, Kelkhoff DO, Harley BAC. J Mech Behav Biomed Mater. 2012;11:27–40. doi: 10.1016/j.jmbbm.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harley BA, Spilker MH, Wu JW, Asano K, Hsu HP, Spector M, Yannas IV. Cells Tissues Organs. 2004;176:153–165. doi: 10.1159/000075035. [DOI] [PubMed] [Google Scholar]
- 26.Caliari SR, Harley BA. Tissue Eng Part A. 2013;19:1100–1112. doi: 10.1089/ten.tea.2012.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capito RM, Spector M. Osteoarthritis Cartilage. 2006;14:1203–1213. doi: 10.1016/j.joca.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Farrell E, O’Brien FJ, Doyle P, Fischer J, Yannas I, Harley BA, O’Connell B, Prendergast PJ, Campbell VA. Tissue Eng. 2006;12:459–468. doi: 10.1089/ten.2006.12.459. [DOI] [PubMed] [Google Scholar]
- 29.Caliari SR, Harley BAC. Tissue Eng A. 2014 [Google Scholar]
- 30.Masters KS. Macromol Biosci. 2011;11:1149–1163. doi: 10.1002/mabi.201000505. [DOI] [PubMed] [Google Scholar]
- 31.Holland TA, Tessmar JKV, Tabata Y, Mikos AG. Journal of Controlled Release. 2004;94:101–114. doi: 10.1016/j.jconrel.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Hudalla GA, Murphy WL. Adv Funct Mater. 2011;21:1754–1768. doi: 10.1002/adfm.201002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz GS, Wysocki A. Wound Repair Regen. 2009;17:153–162. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 34.Hudalla GA, Kouris NA, Koepsel JT, Ogle BM, Murphy WL. Integr Biol (Camb) 2011;3:832–842. doi: 10.1039/c1ib00021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudalla GA, Koepsel JT, Murphy WL. Adv Mater. 2011;23:5415–5418. doi: 10.1002/adma.201103046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim JJ, Temenoff JS. Biomaterials. 2013;34:5007–5018. doi: 10.1016/j.biomaterials.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yannas IV, Burke JF, Gordon PL, Huang C, Rubenstein RH. J Biomed Mater Res. 1980;14:107–132. doi: 10.1002/jbm.820140203. [DOI] [PubMed] [Google Scholar]
- 38.Hortensius RA, Harley BA. Biomaterials. 2013;34:7645–7652. doi: 10.1016/j.biomaterials.2013.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olde Damink LHH, Dijkstra PJ, van Luyn MJA, Van Wachem PB, Nieuwenhuis P, Feijen J. Biomaterials. 1996;17:765–773. doi: 10.1016/0142-9612(96)81413-x. [DOI] [PubMed] [Google Scholar]
- 40.Osborne CS, Barbenel JC, Smith D, Savakis M, Grant MH. Med Biol Eng Comput. 1998;36:129–134. doi: 10.1007/BF02522870. [DOI] [PubMed] [Google Scholar]
- 41.Haugh MG, Murphy CM, McKiernan RC, Altenbuchner C, O’Brien FJ. Tissue Eng Part A. 2010 doi: 10.1089/ten.TEA.2010.0590. [DOI] [PubMed] [Google Scholar]
- 42.Lee JC, Pereira CT, Ren X, Huang W, Weisgerber DW, Yamaguchi DT, Harley BAC, Miller TA. Submitted. [Google Scholar]
- 43.Cornwell KG, Lei P, Andreadis ST, Pins GD. J Biomed Mater Res A. 2007;80:362–371. doi: 10.1002/jbm.a.30893. [DOI] [PubMed] [Google Scholar]
- 44.Wissink MJ, Beernink R, Pieper JS, Poot AA, Engbers GH, Beugeling T, van Aken WG, Feijen J. Biomaterials. 2001;22:151–163. doi: 10.1016/s0142-9612(00)00164-2. [DOI] [PubMed] [Google Scholar]
- 45.Wu JM, Xu YY, Li ZH, Yuan XY, Wang PF, Zhang XZ, Liu YQ, Guan J, Guo Y, Li RX, Zhang H. J Mater Sci Mater Med. 2011;22:107–114. doi: 10.1007/s10856-010-4176-4. [DOI] [PubMed] [Google Scholar]
- 46.Shen YH, Shoichet MS, Radisic M. Acta Biomater. 2008;4:477–489. doi: 10.1016/j.actbio.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Chiu LL, Weisel RD, Li RK, Radisic M. J Tissue Eng Regen Med. 2011;5:69–84. doi: 10.1002/term.292. [DOI] [PubMed] [Google Scholar]
- 48.Martin TA, Caliari SR, Williford PD, Harley BA, Bailey RC. Biomaterials. 2011;32:3949–3957. doi: 10.1016/j.biomaterials.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toh CR, Fraterman TA, Walker DA, Bailey RC. Langmuir. 2009 doi: 10.1021/la9019537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soderstrom KO. Histochemistry. 1987;87:557–560. doi: 10.1007/BF00492470. [DOI] [PubMed] [Google Scholar]
- 51.Pieper JS, Hafmans T, Veerkamp JH, van Kuppevelt TH. Biomaterials. 2000;21:581–593. doi: 10.1016/s0142-9612(99)00222-7. [DOI] [PubMed] [Google Scholar]
- 52.Alsop AT, Pence JC, Weisgerber DW, Harley BAC, Bailey RC. 2014 doi: 10.1016/j.actbio.2014.07.002. In revision. [DOI] [PubMed] [Google Scholar]
- 53.Caliari SR, Harley BAC. Advanced healthcare materials. 2014 doi: 10.1002/adhm.201300646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyons FG, Gleeson JP, Partap S, Coghlan K, O’Brien FJ. Clin Orthop Relat Res. 2014;472:1318–1328. doi: 10.1007/s11999-013-3438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vickers SM, Squitieri LS, Spector M. Tissue Eng. 2006;12:1345–1355. doi: 10.1089/ten.2006.12.1345. [DOI] [PubMed] [Google Scholar]
- 56.Caliari SR, Gonnerman EA, Grier WK, Weisgerber DW, Banks JM, Alsop AJ, Lee J-S, Bailey RC, Harley BAC. Advanced healthcare materials. 2014 doi: 10.1002/adhm.201400252. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hahn MS, Miller JS, West JL. Advanced Materials. 2006;18:2679. [Google Scholar]
- 59.DeForest CA, Anseth KS. Angew Chem Int Ed Engl. 2012;51:1816–1819. doi: 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.