Heparin’s role in biological processes is commonly mediated by its interaction with proteins [1]. During the past decade, our research group has relied on surface plasmon resonance (SPR)1 to monitor heparin–protein interactions [2–8]. Heparin is typically immobilized on the chip, and protein flows over the heparinized surface. Unfortunately, nonspecific interaction between analyte and the chip surface is frequently encountered. Although physical adsorption of the protein can be minimized using surfaces coated with hydrophilic polymers such as dextran, we have recently encountered a number of heparin-binding proteins that interact with dextran. Poly(ethylene glycol) (PEG)-based sensor chips might represent an alternative to the dextran-based sensor chips, for the study of heparin–protein interactions, because surfaces grafted with PEG typically show reduced protein adsorption [9]. In the current work, we describe the application of a PEG-based SPR sensor chip to acquire kinetic and affinity data on the interaction of complement protein factor P with heparin, an that could not be studied with commercially available sensor chips.
Gold (Au) and CM5 sensor chips, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), ethanolamine–HCl, and HBS-EP buffer (0.01 M Hepes (pH 7.4), 0.15 M NaCl, 3 mM ethylenediamine tetraacetic acid (EDTA), 0.005% surfactant P20) were obtained from BIAcore (Uppsala, Sweden). Phosphate-buffered saline (PBS) consisted of a 50-mM sodium phosphate solution (pH 7.2) containing 100 mM NaCl. Sulfo-N-hydroxysuccinimide-long chain-biotin (sulfo-NHS-LC-biotin) and neutravidin were obtained from Pierce (Rockford, IL, USA). Porcine intestinal heparin (sodium salt) and 3,3′-dithio-bis(propionic acid N-hydroxysuccinimide ester) (DTSP-NHS) were obtained from Sigma (St. Louis, MO, USA). mPEG-SH (5000 MW) and HCl·H2N-PEG-COOH (5000 MW) were obtained from Nektar (Huntsville, AL, USA). Double distilled water and CH2Cl2 (HPLC grade) were used. All buffers were filtered (0.22 μm) and degassed prior to using. SPR experiments were performed on a BIAcore 3000 instrument with BIAcore 3000 control and BIAevaluation software (version 4.0.1).
A solution of DTSP-NHS in CH2Cl2 (1.2 mg in 4 ml, 0.75 mM) was added to a solution of HCl·H2N-PEG-COOH in CH2Cl2 (15 mg in 1 ml, 3 mM). The mixture was incubated for 12 h at 4 °C. The solvent was evaporated in vacuo, and the residual solid was redissolved with a minimal amount of H2O, filtered (0.22 μm), dialyzed (3500 MW cutoff), and lyophilized.
An mPEG-coated sensor chip was prepared by incubating the Au sensor chip with a solution of mPEG-SH (2 mg/ml in H2O) for 24 h at 4 °C. After incubation, the chip was rinsed exhaustively with H2O.
A chip with a PEG-COOH matrix was prepared by incubating the Au sensor chip with a solution of – (SCH2CH2CONH-PEG-COOH)2 (6 mg/ml in H2O) for 24 h at 4 °C. The chip was then rinsed exhaustively with H2O. The surface immobilized with PEG-COOH was activated by incubating the PEG-COOH sensor chip for 30 min at 4 °C with an equimolar mixture of NHS/EDC (final concentration 0.05 M, mixed immediately prior to using). The chip was rinsed with 50 mM sodium acetate buffer (pH 4.0) and then incubated for 24 h at 4 °C with a solution of neutravidin (2 mg/ml in sodium acetate buffer, pH 4.0). The chip was rinsed exhaustively with water, and the surface was incubated with ethanolamine (1.0 M) for 2 h at 4 °C, to deactivate carboxyl groups that had not reacted with neutravidin.
Heparin–biotin conjugate was prepared by reaction of sulfo-NHS-LC-biotin with the free amino groups of unsubstituted glucosamine residues in heparin [10]. A solution of heparin–biotin conjugate (1 mg/ml in HBS-EP buffer) was passed over flow cell 2 of the neutravidin-PEG sensor chip (flow rate =5 μl/min, T =25 °C, running buffer: HBS-EP). The binding of heparin–biotin was confirmed by the observation of a 100-RU response.
A solution of factor P (50 nM) in PBS or HBS-EP buffer was passed over the Au, CM-5, or mPEG sensor chip at flow rate of 5 μl/min (T =25 °C, running buffer: PBS or HBS-EP). A kinetic injection mode was used, leading the protein to flow for 3 min over each chip and to be dissociated for the next 3 min.
Flow cell 2 of the neutravidin-PEG sensor chip, with heparin–biotin attached to the surface, was used for the kinetic binding measurement. Flow cell 1, consisting of only neutravidin-PEG, was used as a control. Different concentrations of factor P (60, 50, 40, and 35 nM) in HBS-EP buffer were injected at a flow rate of 30 μl/min (T =25 °C, running buffer: HBS-EP) over flow cells 1 and 2. A kinetic injection mode was used, leading the protein to flow for 3 min and to be dissociated for the next 3 min. The surface was regenerated by the injection of 15 μl (30 s) of SDS (0.5%), followed by two 30-μl injections (60 s) of 2 M NaCl.
Two different PEG-based sensor chips were prepared. The mPEG chip, having a –S-PEG derivative but no functional group available at the end of the polymer chain, was used to determine the ability of PEG to prevent nonspecific protein binding. The PEG-COOH chip, consisting of a PEG derivative with an available carboxyl group, was used to prepare a neutravidin-PEG chip. Neutravidin, a biotin binding protein, was covalently attached to the PEG-COOH sensor chip by amide formation between the carboxyl groups of the PEG and the amino groups of the protein. Biotinylated heparin was then bound to prepare a heparin chip.
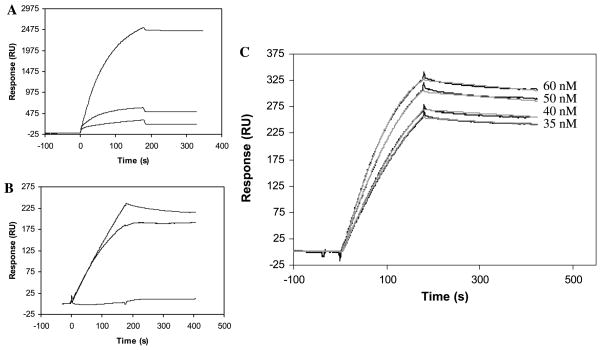
Factor P, also called properdin, functions as an enhancing regulator in the complement alternative pathway and is believed to bind to heparin [11,12]. Previous efforts to study heparin–factor P interaction using a streptavidin–dextran matrix-based sensor chip failed due to nonspecific interaction of factor P with the chip surface. The nonspecific binding of factor P was compared on an mPEG chip, an untreated gold surface (Au sensor chip), and a gold surface containing a dextran matrix (CM5 sensor chip). A solution of factor P (50 nM) in PBS was injected over all three surfaces under identical experiment conditions. The binding process on the mPEG chip gave a much lower response (315 RU) compared with RU changes from CM5 and Au chips of 2525 and 610 RU, respectively (Fig. 1A). When the experiment was repeated using HBS-EP buffer containing surfactant, the nonspecific binding of factor P with the gold surface and the dextran matrix was reduced (184 and 254 RU, respectively), and no detectable interaction was observed between factor P and PEG (Fig. 1B). By switching to PEG matrix, we removed 88% (in PBS buffer) and 100% (in HBS-EP buffer) of the nonspecific binding observed with the dextran. These results clearly demonstrate that nonspecific binding of factor P can be eliminated using a PEG-based chip.
Fig. 1.
(A and B) SPR sensorgrams of nonspecific binding of factor P in PBS (A) and HBS-EP (B) with (1) dextran (CM5 chip), (2) flat gold surface (Au sensor chip), (3) mPEG (mPEG sensor chip). (C) SPR sensorgrams (black lines) of the interaction of factor P with heparin. Gray lines represent the theoretical curves obtained from a global fitting of the sensorgrams using a Langmuir 1:1 binding model with mass transport limitation. The low density of bound heparin limits the sensitivity of SPR to 35 nM of factor P. After 1200 s of dissociation, 15% of the factor P–heparin complex is dissociated.
The interaction of factor P with heparin was studied using a neutravidin–PEG chip with immobilized heparin. At 25 °C, different concentrations (60, 50, 40, and 35 nM) of factor P in HBS-EP buffer were injected over the chip at a flow rate of 30 μl/min (running buffer: HBS-EP). The nonspecific binding of factor P protein to the material other than heparin (probably neutravidin) was less than 20% of the total response obtained. The binding curves under different concentrations were globally, and then individually, fitted with a 1:1 Langmuir binding model with mass transport limitation (Fig. 1C). The fittings showed similar kinetic and affinity data, with χ2 values less than 3.22 (Table 1). The SPR data showed that factor P binds heparin quite strongly, with a KD of 1.09 nM (global fitting).
Table 1.
Kinetic and affinity data of the interaction of factor P with heparin
| kon(×105 M−1 s−1) | koff(×10−4 s−1) | KD nM | χ2 | |
|---|---|---|---|---|
| Global fitting | 3.87 | 4.23 | 1.09 | 3.22 |
| Individual fittinga | ||||
| 60 nM | 3.52 | 3.87 | 1.10 | 1.13 |
| 50 nM | 4.25 | 4.10 | 0.97 | 0.77 |
| 40 nM | 5.66 | 4.29 | 0.76 | 0.48 |
| 35 nM | 5.79 | 4.62 | 0.80 | 0.47 |
Concentration of factor P injected over the sensor chip.
We have demonstrated that PEG-based biosensor chips offer an alternative to dextran-based chips when an analyte interacts nonspecifically with a dextran matrix. PEG-based chips are easy to prepare and afford high baseline stability. We repeatably obtained excellent binding sensorgrams for the interaction between factor P and heparin using these chips. Other heparin-binding proteins examined in our laboratory have also exhibited significant nonspecific interactions with dextran matrix. Thus, we have replaced standard commercial chips with new PEG-based sensor chips to measure interactions between these proteins and heparin.
Footnotes
Abbreviations used: SPR, surface plasmon resonance; PEG, poly(ethylene glycol); Au, gold; EDC, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride; NHS, N-hydroxysuccinimide; EDTA, ethylenediamine tetraacetic acid; PBS, phosphate-buffered saline; sulfo-NHS-LC-biotin, sulfo-N-hydroxysuccinimide-long chain-biotin; DTSP-NHS, 3,3′-dithio-bis(propionic acid N-hydroxysuccinimide ester).
References
- 1.Capila I, Linhardt RJ. Heparin–protein interactions. Angew Chem Int Ed Engl. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 2.Ibrahimi OA, Zhang F, Hrstka SC, Mohammadi M, Linhardt RJ. Kinetic model for FGF, FGFR, and proteoglycan signal transduction complex assembly. Biochemistry. 2004;43:4724–4730. doi: 10.1021/bi0352320. [DOI] [PubMed] [Google Scholar]
- 3.Peterson FC, Elgin ES, Nelson TJ, Zhang F, Hoeger TJ, Linhardt RJ, Volkman BF. A glycosaminoglycan recognition element of lymphotactin essential for in vivo chemokine activity. J Biol Chem. 2004;279:12598–12604. doi: 10.1074/jbc.M311633200. [DOI] [PubMed] [Google Scholar]
- 4.Olsen SK, Ibrahimi OA, Raucci A, Zhang F, Eliseenkova AV, Yayon A, Basilico C, Linhardt RJ, Schlessinger J, Mohammadi M. Insights into the molecular basis for FGF receptor autoinhibition and ligand binding promiscuity. Proc Natl Acad Sci USA. 2004;101:935–940. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barth H, Schaefer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsaecker F, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Fath M, Marks R, Linhardt RJ. A highly stable covalent conjugated heparin biochip for heparin–protein interactions studies. Anal Biochem. 2002;304:271–273. doi: 10.1006/abio.2002.5617. [DOI] [PubMed] [Google Scholar]
- 7.Rathore D, McCutchan TF, Garboczi DN, Toida T, Hernaiz MJ, LeBrun LA, Lang SC, Linhardt RJ. Direct measurement of the interactions of glycosaminoglycans and a heparin decasaccharide with malaria circumsporozoite protein. Biochemistry. 2001;40:11518–11524. doi: 10.1021/bi0105476. [DOI] [PubMed] [Google Scholar]
- 8.Dong J, Peters-Libeu CA, Weisgraber KH, Segelke BW, Rupp B, Capila I, Hernaiz MJ, LeBrun LA, Linhardt RJ. Interaction of the N-terminal domain of apolipoprotein E4 with heparin. Biochemistry. 2001;40:2826–2834. doi: 10.1021/bi002417n. [DOI] [PubMed] [Google Scholar]
- 9.Masson JF, Battaglia TM, Kim YC, Prakash A, Beaudoin S, Booksh KS. Preparation of analyte-sensitive polymeric supports for biochemical sensors. Talanta. 2004;64:716–725. doi: 10.1016/j.talanta.2004.03.051. [DOI] [PubMed] [Google Scholar]
- 10.Hernaiz MJ, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparin sulfate on a biochip promotes its interaction with antithrombin III. Biochem Biophys Res Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 11.Holt GD, Pangburn MK, Ginsburg V. Properdin binds to sulfatide [Gal(3-SO4)β 1-1 Cer] and has a sequence homology with other proteins that bind sulfated glycoconjugates. J Biol Chem. 1990;265:2852–2855. [PubMed] [Google Scholar]
- 12.Wilson JG, Fearon DT, Stevens RL, Seno N, Austen KF. Inhibition of the function of activated properdin by squid chondroitin sulfate E glycosaminoglycan and murine bone marrow-derived mast cell chondroitin sulfate E proteoglycan. J Immunol. 1984;132:3058–3063. [PubMed] [Google Scholar]