Abstract
RNA is arguably the most functionally diverse biological macromolecule. In some cases a single discrete RNA sequence performs multiple roles and this can be conferred by a complex three-dimensional structure. This multifunctionality can also be driven or enhanced by the ability of a given RNA to assume different conformational (and therefore functional) states1. Despite its biological importance, a detailed structural understanding of the paradigm of RNA structure-driven multifunctionality is lacking. Examples to address this gap are found in single-stranded positive-sense RNA viruses, a prototype being the tRNA-like structure (TLS) found at the 3′ end of the Turnip Yellow Mosaic Virus (TYMV). This TLS not only acts like a tRNA to drive aminoacylation of the viral genomic RNA (gRNA)2-4, but also interacts with other structures in the gRNA's 3′ untranslated region5, contains the promoter for negative strand synthesis, and influences several infection-critical processes6. This TLS RNA can provide a glimpse into the structural basis of RNA multifunctionality and plasticity, but for decades its high-resolution structure has remained elusive. Here, we present the crystal structure of the complete TYMV TLS to 2.0 Å resolution. Globally, the RNA adopts a shape that mimics tRNA, but it uses a very different set of intramolecular interactions to achieve this shape. These interactions also allow the TLS to readily switch conformations. In addition, the TLS structure is ‘two-faced’: one ‘face’ closely mimics tRNA and drives aminoacylation, the other ‘face’ diverges from tRNA and enables additional functionality. The TLS is thus structured to perform several functions and interact with diverse binding partners, and we demonstrate its ability to specifically bind to ribosomes.
Keywords: turnip yellow mosaic virus (TYMV), RNA structure, x-ray crystallography, ribosome binding, pseudoknot
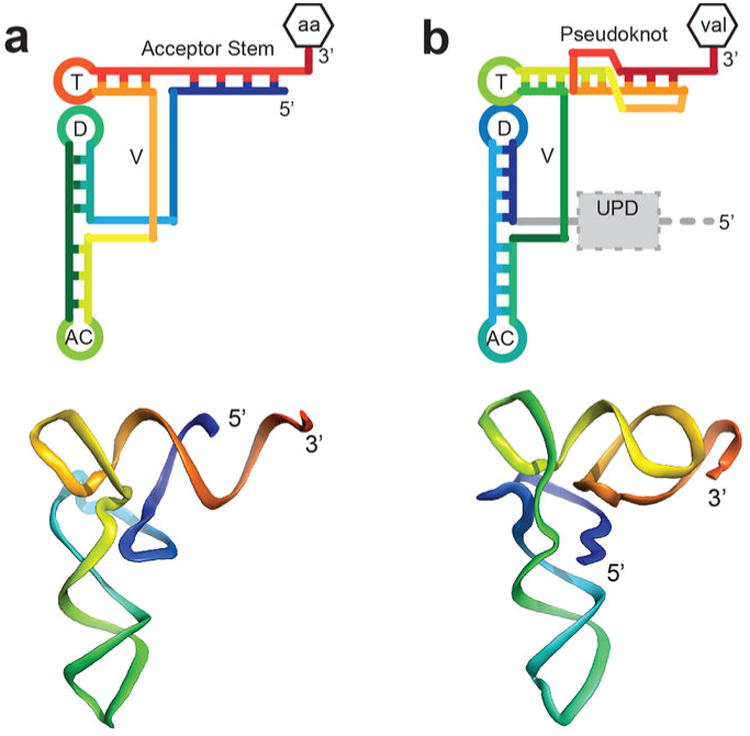
The TYMV TLS RNA (‘the TLS’) is a tRNA mimic, a subject of broad biological and evolutionary importance7, underscored by examples linked to disease8-10. Like tRNA, the aminoacylated TLS binds to eukaryotic elongation factor 1A (eEF1A) and is a substrate for tRNA-modifying enzymes6. These activities and other data suggest a tRNA-like structure11-16. However, the TLS' topology differs from tRNA, mandated by its location on the 3′ end of the gRNA (Fig. 1b, Extended Data Fig. 1). In addition to affecting many viral processes17-19, the TLS may regulate the activities of ribosomes and replicases on the gRNA6,20. This function could be conferred by the TLS' ability to readily transition between folded and unfolded states. Simple tRNA mimicry is insufficient to explain these phenomena; while tRNAs flex while transiting through the ribosome they do not unfold and refold. To explore the paradigms of tRNA mimicry and RNA structural and functional plasticity, we solved the structure of the TYMV TLS RNA by X-ray crystallography to 2.0 Å resolution (Fig. 1c, Extended Data Fig. 2), comparable to the highest-resolution structure of free tRNA, used here for comparison (1.93 Å)21.
Figure 1. Function and structure of the TYMV TLS.
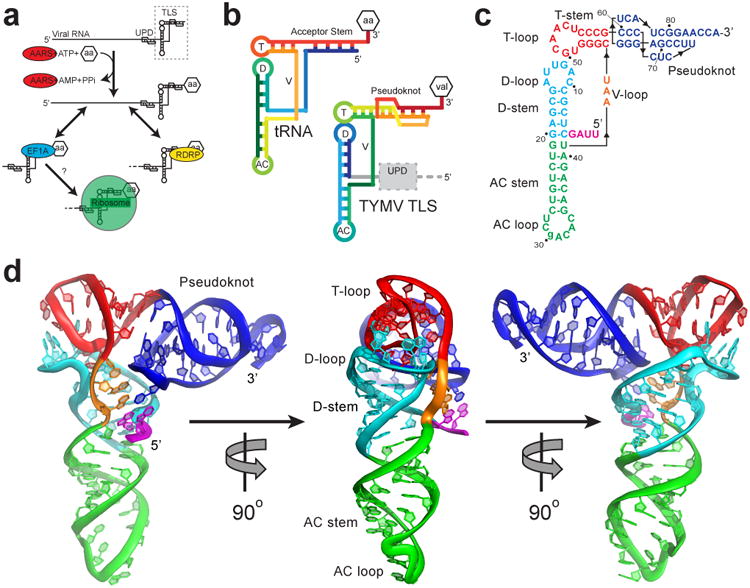
a, The TLS (dashed box) at the 3′ end of the gRNA, with the UPD upstream. AARS (red) valylates the TLS, which can interact with the RDRP (yellow) or eEF1A (blue). Ribosome binding (green) was suspected but untested. b, Topology of tRNA and the TLS in rainbow colors. 5′ ends are blue and 3′ ends are red. Attached amino acid is aa or val. T= T-loop, D= D-loop, AC= anticodon loop, V= variable loop. c, Secondary structure of the crystallized RNA. Lowercase letter indicates the single mutation. Numbering is from the 5′ end of the crystallized sequence. d, Three views of the structure, colored to match panel c. The conformation of the 3′ CCA and AC loop differ from tRNA, likely due to crystal packing (Extended Data Fig. 8).
The TLS assumes the classic L-shaped tRNA conformation (Fig. 1d), but achieves this in a way that diverges from tRNA and from predictions13,22. Compared to tRNA, the topology (Fig. 1b, Extended Data Fig. 3) and the intramolecular interactions that form the structure diverge from a tRNA's (Fig. 2a). Although the TLS pseudoknot (the first recognized RNA pseudoknot14) is in the position of the tRNA's acceptor stem and elements analogous to the D-loop, T-loop, and V-loop are positioned as in tRNA, their interactions are not tRNA-like. In the elbow region of tRNA, the V-loop interacts with the D-stem, stabilizing the L-shaped tRNA structure (Fig. 2b). In contrast, the TLS' V-loop bases point away from the D-stem to interact with the 5′ end and pseudoknot of the TLS (Fig. 2b). G4 adopts a syn conformation (Extended Data Fig. 4), forming a long-range base pair with C76 in a loop of the pseudoknot. The unexpected G4-C76 base-pair is stabilized by stacking of A3 and the V-loop base A42 on either side. V-loop bases A42-U44 continuously stack to reach from the G4-C76 pair to A15 of the D-loop, structurally linking the pair to the elbow region. These interactions explain the observation that removing 5′-UUAG sequence from the 5′ end of the TLS (including G4) destabilizes the L-shape conformation and elbow structure (Extended Data Fig. 5)5,11. Although historically not recognized as part of the minimal TLS, these 5′ nucleotides form a ‘linchpin’ interaction that stabilizes the global structure and this suggests why their presence increases aminoacylation efficiency (Fig. 2a, Extended Data Fig. 6).
Figure 2. Structural differences between tRNA and the TLS.
a, Secondary structures showing interactions that stabilize the folds of tRNA (left) and the TLS (right). Non-canonical base pairs are indicated with Leontis-Westhof symbols30, single hydrogen bonds with dashed lines. Lines with embedded arrows indicate chain connectivity. Grey nucleotides were not visible in the electron density. A grey bar indicates the long-range ‘linchpin’ interaction. b, Intramolecular interactions of the V-loop (orange) in tRNA (left) and the TLS (right). Dashed lines indicate the C76-G4 base pair. c, Conformation and interaction of the D-loop (cyan) with the T-loop (red) of tRNA (left) and the TLS (right).
The structural features of the elbow region require that the TLS D-loop be in a different conformation than tRNA's. In the TLS, the D-loop contains a tight bend that allows A15 to reach across the helix to stack on U44 in the V-loop (Fig. 2c), while U13 and U14 lie against the end of the T-loop. No analogous bases or interactions are found in tRNA. Despite this, the T-loops of the TLS and tRNA are structurally identical (Extended Data Fig. 6). D-loop bases G12 and A11 dock into the T-loop almost identically to the analogous bases of tRNA, although A11 is in a syn conformation.
The TLS' stabilizing intramolecular interactions show how it can adopt different folded states to potentially organize infection-important activities, achieving structural and functional plasticity. Disruption of the ‘linchpin’ would lead to a loss of the L-shape fold and a propagated loss of interactions extending from the V-loop to the D/T-loop interface. This effect is observed when the base pair and adjacent nucleotide that stack on and stabilize this pair are eliminated by truncating the TLS from the 5′ end5,11 (Extended Data Fig. 5). This disruption could be induced by loading of the virally-encoded RNA-dependent RNA polymerase (RDRP) at the 3′ end18,23,24. The subsequent destabilization would create a favorable template for the RDRP and effectively remove competition between the RDRP and the proteins that require the stable fold (e.g. the amino acid synthetase; AARS).
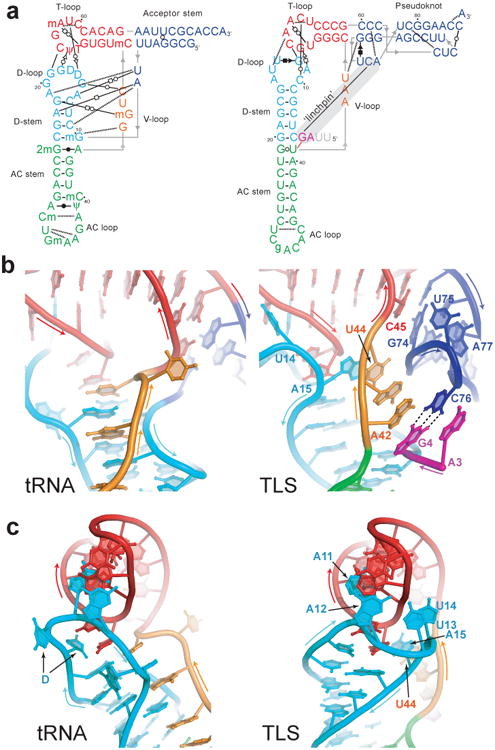
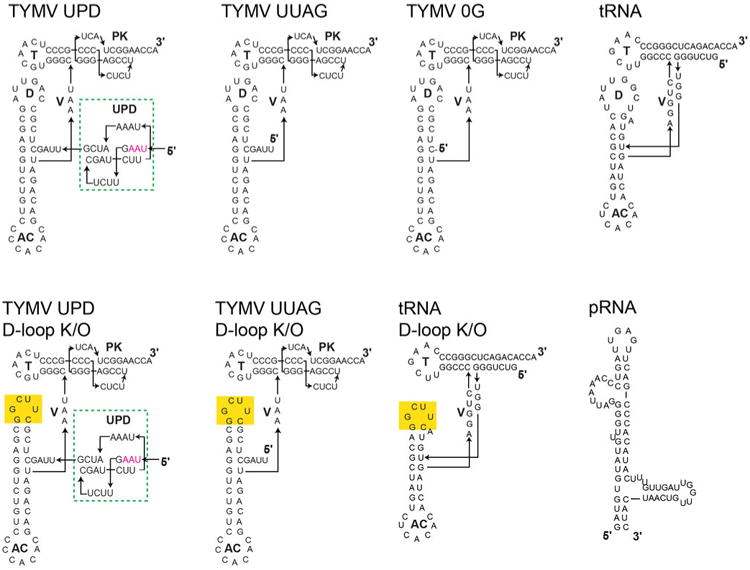
The TLS structure has two distinct ‘faces’. The tRNA-deviating features are on one side of the structure, where the UPD and the gRNA connect to the TLS (Fig. 1b, Extended Data Fig. 7). The structure reveals that the UPD is positioned to interact with the TLS' ‘divergent face’. The opposing side of the TLS, the ‘tRNA-like face’, interacts with the valine AARS when the TLS structure is modeled into a tRNAVal•AARS complex structure25 (Fig. 3a,b). The TLS structure is accommodated by the AARS, including the acceptor stem pseudoknot, which has a different structure than shown by NMR (Extended Data Fig. 6). Like tRNA, the TLS has high crystallographic B-factors in its anticodon (AC) loop and 3′ CCA, suggesting these can readily undergo structural changes (Fig. 3c,d, Extended Data Fig. 8). In the case of the AC loop, this is important to dock the valine-specifying identity elements in the AC loop onto the protein26. Modeling of the TLS structure onto an elongation factor structure also reveals an interface similar to that formed with tRNA and no obvious steric clash (Extended Data Fig. 6). Because the divergent face does not contact the AARS or eEF1A, the 5′ end of the TLS is not occluded by interaction with either protein. Thus, the UPD and viral genome do not interfere with binding (Extended Data Fig. 9), and the precise mimicry of the tRNA-like face explains how the TLS can achieve tRNA-like valylation efficiencies and eIF1A binding affinities27.
Figure 3. tRNA mimicry and AARS binding.
a, Backbone traces of superimposed tRNA (cyan) and TLS (red). The ‘tRNA-like’ face is shown. b, Superposition of the TLS onto tRNAVal bound to valine AARS (PDB: 1GAX)25. c, The AC loop of the TLS (red) must swing into position to match tRNA's (cyan). d, TLS structure colored by relative crystallographic B-factor (high=red, blue=low).
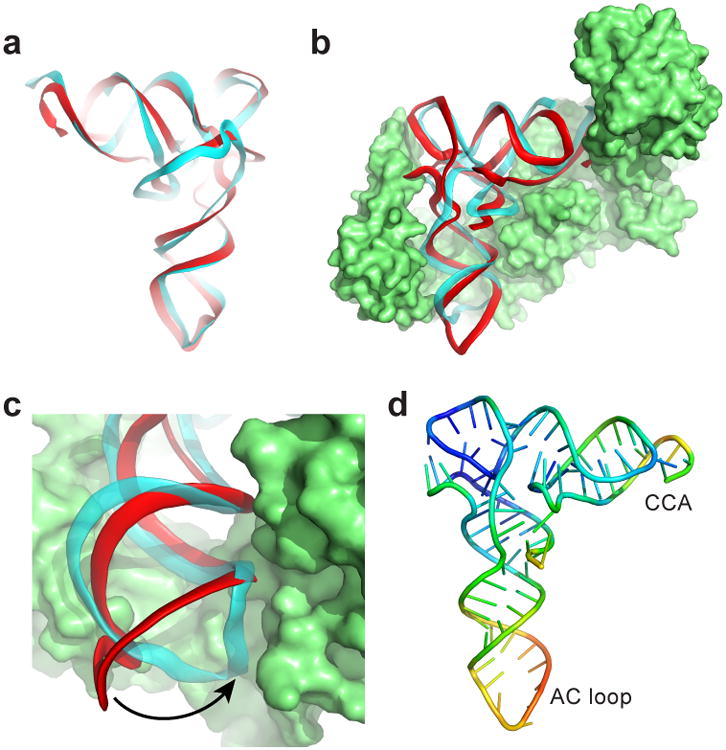
The TLS' interactions with AARS and eEF1A suggest it could bind to the ribosome as previously suggested6,28. Ribosome binding would require accommodating the entire TLS structure between the subunits, including elements that deviate from tRNA within the TYMV 3′UTR. We measured binding of TLS-containing RNAs to Thermus thermophilus 70S ribosomes, a valid model for tRNA binding assays given the interchangeability of eukaryotic and bacterial tRNAs29. In vitro transcribed A. thaliana tRNAVal bound to the 70S (Kd=0.27 +/- 0.05 nM) while a 75 nucleotide-long negative control RNA (from bacteriophage phi29 pRNA) did not (Kd >1000 nM) (Fig. 4a, Extended Data Fig. 1). Mutation of the tRNAVal D-loop to disrupt the global tRNA fold resulted in a 28-fold loss of affinity (Kd>7.6 +/- 0.8 nM) (Fig. 4b), consistent with binding being dependent on the tRNA's global conformation. A TLS RNA containing the 5′-UUAG sequence bound with tRNA-like affinity (Kd= 0.31 +/- 0.07 nM), and mutation of this RNA's D-loop decreased binding 9-fold (Kd>2.7 +/- 0.2 nM) (Fig. 4c). Likewise, truncation of the TLS' 5′ end to abrogate the ‘linchpin’ interaction reduced binding ∼3-fold (Kd=1.1 +/- 0.3 nM) (Extended Data Fig. 5). Remarkably an RNA containing the TLS, the UUAG, and the 23 nucleotide-long UPD also bound to ribosomes (TYMV UPD; Kd= 0.24 +/- 0.11 nM), and binding of this RNA was reduced 100-fold by D-loop mutation (Kd>24 +/- 8 nM) (Fig. 4d). Thus, the folded TLS can bind the ribosome even in the context of the entire 3′ UTR and binding depends on native structure. The affinity is consistent with binding to the P site, although binding to other sites is possible. The ability of the entire TYMV 3′ UTR to dock within ribosomes may relate to its functions as a regulatory switch, a translation enhancer and a means to protect the 3′ end of the genomic RNA17-19.
Figure 4. Binding of tRNA and TLS to ribosomes.
a, Binding curves of tRNAVal (positive control) and pRNA (negative control) to 70S ribosomes, fit by a Langmuir isotherm (RNA sequences in Extended Data Fig. 1). b, Binding of WT tRNA and tRNA with mutated D-loop. c, Diagram of the UUAG TLS (UUAG sequence in cyan) and binding curves of this TLS and versions with the D-loop mutated and with the UUAG removed (0G). d, Diagram of the UPD TLS (UPD shown in green) and binding curves of this UPD TLS and a D-loop mutant. Error bars are 1 s.d. from mean of 3 replicates.
Methods
General Procedures
Chemical Reagents and Synthetic DNA: General chemical reagents were all of molecular biology grade or higher. All aqueous solutions were made using diethylpyrocarbonate (DEPC)-treated milli-Q water and routinely filtered through 0.22 μm sterile filtration systems (Millipore). DNA primers were purchased from Integrated DNA Technologies and used without further purification. Nucleic acid concentrations were determined by monitoring a solution's absorbance at 260 nm using a Nanodrop UV-Vis spectrophotometer (Thermo). Iridium (III) hexammine was synthesized as described31.
RNA transcription
dsDNA templates for transcription were made by PCR using template plasmid DNA that contained the sequence of interest (plasmids made using standard mutagenesis methods). DNA from a 1 mL PCR reaction was used in a 5 mL in vitro transcription reaction with final concentrations of 30 mM Tris-HCl pH 8.0, 10 mM DTT, 0.1% Triton X-100, 0.1% Spermidine, 40 mM MgCl2, 4 mM each NTP, and T7 RNA Polymerase. The reaction was incubated at 37°C for 6 hours. Inorganic pyrophosphate was pelleted at 3000 × g for 10 minutes, followed by EtOH precipitation of the supernatant. Precipitated RNA was pelleted by centrifugation, dried, then resuspended in 8 M Urea. RNA was purified on a 10% denaturing PAGE slab gel at 40 W for 5 hours, then excised and passively eluted in DEPC-treated water overnight at 4°C. RNA was concentrated and exchanged into DEPC-treated water by ultrafiltration and stored at -20°C.
RNA crystallization and diffraction data collection
The RNA sequence used in crystallization was based on a sequence identified by in vitro selection for TYMV TLS RNAs capable of efficient valylation and contained a point mutation in the anticodon loop32. This RNA was prepared for crystallography in a solution containing 5 mg/mL RNA, 2.5 mM MgCl2, and 10 mM HEPES-KOH pH 7.5. This mixture was heated to 65°C for 3 minutes, then cooled at room temperature. After cooling, Spermidine was added to 0.5 mM. The reaction was centrifuged for 10 minutes at 13000 × g and then used in sitting-drop vapor diffusion crystallization at 4°C. 1 μL of RNA solution was combined with 2 μL of 10% MPD, 40 mM Na-Cacodylate pH 6.0, 12 mM Spermine, 80 mM NaCl, and 20 mM MgCl2. The well solution was 20-35% MPD. Crystals appeared and grew to full size over the course of 1-2 days. To obtain derivatized crystals for phasing, a solution matching the well solutions with the addition of 8 mM Iridium (III) hexamine was exchanged with the crystal growth solution. Crystals were harvested directly from the drops into nylon loops and flash-frozen by plunging into liquid nitrogen. Diffraction data were collected at Advanced Light Source Beamline 4.2.2 using “shutterless” collection at the Iridium L-III edge (1.0972 Å) at 100° K. For each crystal, multiple 180° datasets were collected with 0.1° oscillation images. Data were indexed, integrated, and scaled using XDS33,34.
Structure determination and refinement
Although data were collected and processed to 1.99 Å, only data to 2.5 Å were used for phasing. Fifteen Iridium (III) hexammine sites were identified and used in SAD phasing within the AUTOSOL function of PHENIX (overall FOM = 0.448)35. Scattering factors used were f′ = -11.92, f″= 10.09. Density modification using RESOLVE (solvent content set to ∼50%) led to an interpretable electron density map (Extended Data Fig. 2). Iterative rounds of model building and refinement (simulated annealing, rigid-body, B-factor refinement, phase combination using COOT36,37 and PHENIX REFINE) led to the final model. The final model contains 84 of 86 nucleotides, 2 Mg2+ ions, 12 Iridium (III) hexammine ions, one Spermine molecule and 146 water molecules. Crystal diffraction data, phasing, and refinement statistics are contained in Extended Data Table 1. For further analysis of the structure was completed using MolProbity38,39. Summary of the output: Clashscore = 12.52; Probably wrong sugar puckers: 2; Bad backbone conformations: 7; Bad bonds: 1; Bad angles: 1. Areas of concern were examined in the structure and generally fell within areas of the structure with unusual conformations, but the density and model agreed well in these regions.
Mutagenesis for ribosome binding
Mutations to the DNA templates were made using a PCR-based site-directed mutagenesis protocol (Agilent) with primers designed to modify the D-loop nucleotides. The nucleotides comprising the D-loops of tRNAVal, TYMV-UUAG TLS, and TYMV-UPD TLS were replaced with stable UUCG tetraloop sequences. For tRNA, the primer sequence was 5′-GGG TGG TGT ACT TCG GAC GCT AGT CTC-3′. The UPD primer had the sequence 5′-CTT TAA AAT CGT TAG CTC GCT TCG GCG AGG TCT GTC CCC-3′. The UUAG primer sequence was 5′-CCG TCT TAG CTC GCT TCG GCG AGG TCT GTC CCC-3′.
70S Ribosome Purification
Preparation of 70S ribosomes was done by the Noller lab (UC Santa Cruz) as described40.
Filter Binding
The filter binding protocol used was modified from previously published methods41,42. 50 μL reactions contained 25mM Tris-HCl, 50mM KCl, 10mM MgCl2, 2mM Spermine at pH 7.0, 100 counts/minute of 32P-labelled RNA. The reactions were incubated at 37°C for 30 minutes then passed through a sandwich of filters (presoaked in matching buffer) in a vacuum manifold. Filters: size exclusion (Tuffryn) filter (Pall), Nitrocellulose filter (BioRad), Hybond-N+ charged nylon filter (GE BioSciences), and filter paper (Whatman). The filters were washed 3 times with wash buffer (25mM Tris HCl, 100mM KCl, 25mM MgCl2, pH 7.5) and allowed to dry for 3 hours. Reactions were quantified by phosphorimaging and data were fit using KaleidaGraph software.
Extended Data
Extended Data Figure 1. Sequences and structures of RNAs.
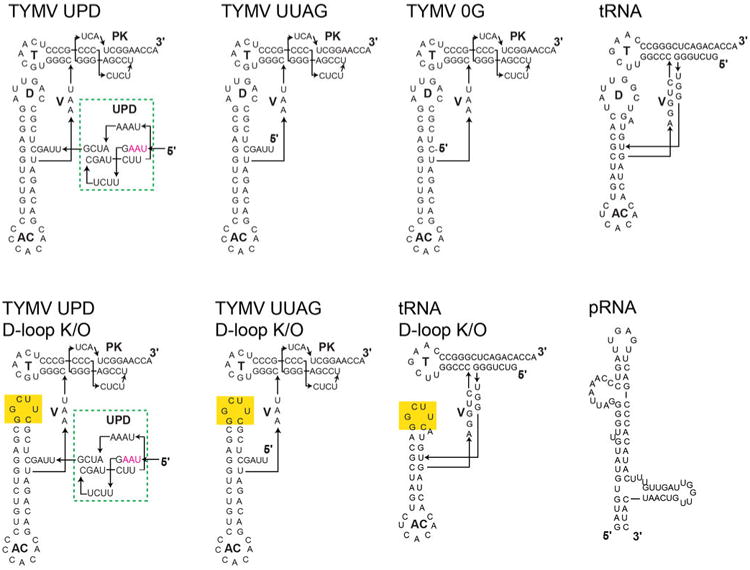
Top left: Sequence and secondary structure of the complete TYMV TLS and the Upstream Pseudoknot Domain (UPD, green dashed box). The UPD is just upstream of the UUAG sequence that is important for stabilizing the L-shaped structure and the UPD is known to be able to pack against the TLS5,11. Interestingly, the stop codon for the Coat protein is within the UPD (magenta). To the right and below: Sequences and secondary structures of all additional RNAs used in ribosome binding assays or discussed in the text. Yellow highlights indicate the location of mutation.
Extended Data Figure 2. Representative electron density and bound trivalent ions.
a, Unbiased, density-modified electron density from SAD phasing using data to 2.5 Å (grey mesh, 2σ), superimposed on the final model. The T-loop and part of the D-loop is shown. For simplicity, density and structure of water and ions is not shown. b, Final 2Fo-Fc electron density map after model building and refinement to 1.99 Å (2σ). c, Structure with the location of 12 Iridium (III) hexammine ions. Although many of these hexammine binding sites may also be Mg2+ binding sites important for stabilizing the fold, the trivalent hexammine was present at 8 mM and thus many weaker Mg2+ binding sites could have been occupied. For this reason, and because there is not a one-to-one correlation of Mg2+ binding sites and trivalent hexammine sites, we do not make conclusions about Mg2+ binding based on this structure.
Extended Data Figure 3. Topologies and three-dimensional structures of tRNA and the TYMV TLS.
a, Top: The topology of a canonical tRNA is shown in rainbow color with the 5′ end in blue and the 3′ end in red. The attached amino acid is shown (aa or val) and structural features are labeled: T= T-loop, D= D-loop, AC= anticodon loop, V= variable loop. The 5′ and 3′ ends of the RNA are shown. Bottom: Ribbon representation of the backbone of tRNAPhe roughly colored to match the cartoon diagram. b, Same as panel a, but for the TYMV TLS. The location of the UPD (grey dashed box) and gRNA (grey dashed line connected to the 5′ end) are shown on the top diagram.
Extended Data Fig. 4. Assignment of bases to the syn conformation.

a, Nucleotide G4, which forms the long-range base pair with C76 in the pseudoknot, is in a syn conformation. Top: Placement of the base into an anti conformation results in positive and negative density (green and red, respectively) in the Fo-Fc map (left, contoured at 3σ), and the 2Fo-Fc map (right) shows the base is incorrectly placed (blue density, contoured at 1.5 σ). In contrast, placement of the base into the syn conformation (bottom) results in a flat Fo-Fc map (left, contoured at 3σ) and a good fit to the 2 Fo-Fc map (right, blue density contoured at 1.5 σ). Base A11 is also in a syn conformation; the same analysis was performed to verify this (not shown). b, 2Fo-Fc map surrounding bases A3-C4. The G4′s C4′-C5′ bond is best modeled in the trans conformation.
Extended Data Figure 5. Effect of breaking the ‘linchpin’ interaction.

a, Small angle X-ray scattering (SAXS) analysis of TYMV TLS RNAs, adapted from Hammond et al. (2010)5. Left: ab initio SAXS reconstruction of the shape of the TLS when the 5′ sequence that interacts with the pseudoknot (Fig. 2) is present. The RNA overall forms an L-shape, illustrated by the black bars (stabilizing long-range interaction in grey). When these 5′ nucleotides are removed (right), the L-shape is lost and the RNA becomes more extended. b, Hydroxyl radical probing of several TYMV TLS RNAs that indicate the effect of disrupting the long-range interaction, adapted from Hammond et al. (2009 & 2010)5,11. Green and red indicate protection from cleavage by radicals and enhanced cleavage by radicals, respectively. Overall, the presence of green and red indicate tightly folded RNA. When the 5′ nucleotides that form the long-range interaction are present, the RNA stably folds (TYMV UUAG, left). Removal of the 5′ nucleotides destabilizes the fold (TYMV 0G, right). The presence of just G4 on the 5′ end partially stabilizes the RNA fold (TYMV 1G, middle), confirming its importance in folding and also indicating that nucleotides adjacent to G4 further stabilize the fold.
Extended Data Figure 6. T-loop and acceptor stems of the tRNA and TLS and elongation factor binding.
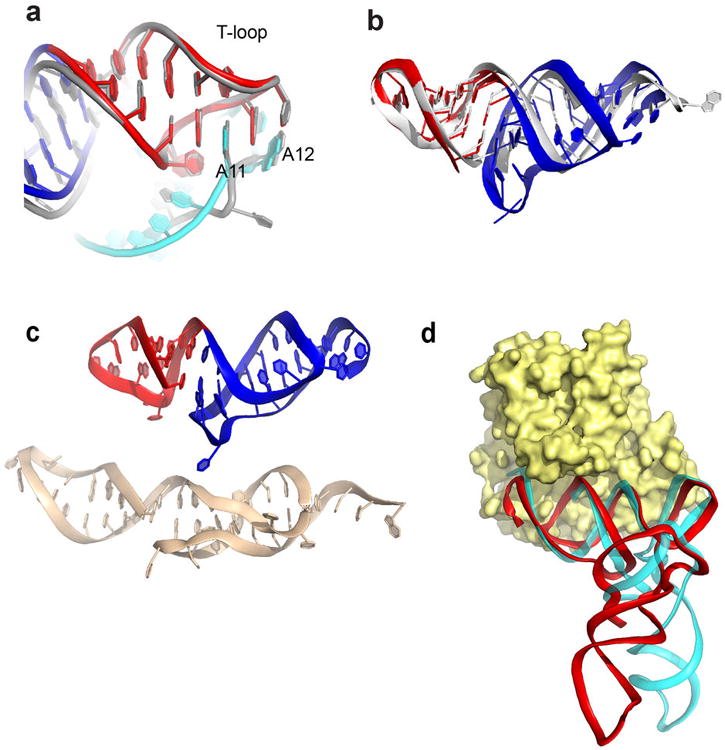
a, Superimposed structures of the TLS T-loop (red) and part of the D-loop (cyan) with the analogous structures in tRNA (grey). TLS bases A11 and A12 are shown; these bases match the interactions formed by analogous bases in tRNA. In the TLS, A11 is in a syn conformation, but the matching base in tRNA is not. This may be due to local differences in the backbone conformation. b, Superimposed structures of the TLS T-loop (red) and pseudoknot (blue) with the T-loop and acceptor stem elements in a tRNA (grey). View is from the “top” of the molecule, down the axis of the D- and AC-stems. c, Top: The structure of the T-loop (red) and acceptor stem pseudoknot (blue) in the TLS crystal structure. Bottom: structure of these elements isolated from the rest of the TLS and solved by NMR (PDB: 1A60)43. d, Superposition of the TLS structure (red) onto the tRNA (cyan) of a tRNAPhe bound to EF-Tu (yellow), the bacterial homolog of eEF1A (PDB: 1TTT)44. Binding is likely facilitated by the fact that RNA backbone conformation of the TLS pseudoknot and T-stem/loop matches that of a tRNA.
Extended Data Figure 7. The ‘two-faced’ architecture of the TYMV TLS and connection with the UPD.

Several views of the TLS (red) superimposed on tRNAPhe (cyan)21 are shown, rotated 90° relative to each other. The dashed line bisects the structure into its two faces. The backbones are very similar on the tRNA-like face, but differ on the divergent face. Locations where the two structures diverge most dramatically are shaded grey. The 5′ end of the TLS, where the UPD connects, is indicated.
Extended Data Figure 8. The anticodon loop: structures and crystal packing.

a, Structure of the anticodon loop of tRNAPhe, solved to 1.93 Å21. The loop is colored to reflect relative B-factors, with red as the highest and blue as the lowest. b, Structure of the anticodon loop of the TYMV TLS, colored identically to panel a. The asterisk marks the C30 base that was mutated to G to enhance crystallization. This was the only mutation made to the TLS for crystallization and does not inhibit aminoacylation32. Overall, the loop structures are similar and both have high crystallographic B-factors compared to other parts of the structures, a common feature of tRNAs. There is no evidence that the TYMV TLS anticodon loop is post-transcriptionally modified, yet it has structural features and conformal flexibility similar to a tRNA's anticodon loop (which is often modified, Fig. 2a). c, Crystal packing involving the anticodon loop of the TYMV TLS. Two interacting copies of the RNA are shown in red and magenta, with the C30G mutation in yellow. This mutation, while not appearing to alter the overall anticodon loop structure compared to a tRNA, induces intermolecular base-pairing in the crystal (pattern shown to the right), suggesting why this mutation aided crystallization. d, Crystal packing of the TLS's 3′ CCA (red, labeled) against an adjacent molecule (magenta) likely causes the CCA to adopt a folded-back conformation.
Extended Data Figure 9. Models of protein binding to the TLS and the location of the UPD.
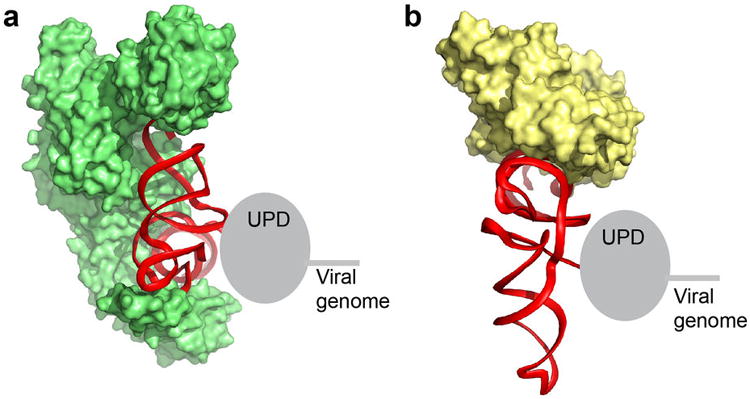
a, Model of the TLS (red, backbone ribbon shown) on the valine AARS (green; PDB: 1GAX), similar to Figure 3b, but viewed from the top and with the tRNAVal not shown. The location of the UPD directly 5′ of and against the TLS is shown as a grey oval. The viral genomic RNA is 5′ of the UPD. Note that the strategy used by the TYMV TLS to interact with this protein is likely very different from that used by the TLSs that are histidylated or tyrosylated, which are very different in terms of their secondary structure and fold6,9. b, Same as panel a, but with the TLS modeled onto the bacterial homolog of eEF1A (EF-Tu) as in Extended Data Fig. 6. tRNAPhe is not shown. In both complexes, the location of the 5′ end, the UPD, and viral genome would not interfere with protein binding. This would not be true if the TLS had a tRNA-like topology with the 5′ end paired to the 3′ end.
Extended Data Table 1.
Crystallographic data collection, phasing and refinement statistics. One crystal was used.
| Iridium (III) hexammine | |
|---|---|
| Data collection | |
| Space group | I 222 |
| Cell dimensions | |
| a, b, c (Å) | 55.3, 101.6, 111.6 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 28.87-1.99 (2.06-1.99)* |
| Rsym or Rmerge | 5.4 (82.3) |
| Rmeas# | 5.8 (89.5) |
| I|σ| | 21.71 (2.19) |
| CC(1/2)## | 99.9 (83.3) |
| Completeness (%) | 99.4 (94.7) |
| Redundancy | 7.5 (6.5) |
| Refinement | |
| Resolution (Å) | 28.9-1.99 |
| No. reflections | 308254 (18783) |
| Rwork/Rfree | 20.3 (30.2) / 24.1 (33.9) |
| No. atoms | 2038 |
| RNA | 1785 |
| Ligand/ion | 107 |
| Water | 146 |
| B-factors | 43.9 |
| Protein | 43.6 |
| Ligand/ion | 56.0 |
| Water | 37.5 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 2.43 |
Acknowledgments
We thank Harry Noller (Univ. of CA, Santa Cruz) for the gift of 70S ribosomes. We thank Ignacio Tinoco, Jr., Catherine Musselman, and Theo Dreher for critical reading of this manuscript. The UC Denver X-ray Facility is supported by UC Cancer Center Support Grant P30CA046934. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences of the U.S. Department of Energy under Contract #DE-AC02-05CH11231. J.S.K. is supported by grants GM081346 and GM097333 from the National Institutes of Health and is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Online Content: Any additional Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author contributions: J.A.H. and G.M.R. designed the crystallization RNAs and identified initial crystals. T.M.C. and D.A.C. improved and grew the crystals. Crystals were harvested by T.M.C., who also solved, built, and refined the structure. J.C.N. collected and processed synchrotron diffraction data. G.M.R. conducted the ribosome binding experiments. J.S.K. provided overall supervision and guidance, and together with T.M.C. and D.A.C. wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yot P, Pinck M, Haenni AL, Duranton HM, Chapeville F. Valine-specific tRNA-like structure in turnip yellow mosaic virus RNA. Proc Natl Acad Sci USA. 1970;67:1345–1352. doi: 10.1073/pnas.67.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinck M, Yot P, Chapeville F, Duranton HM. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature. 1970;226:954–956. doi: 10.1038/226954a0. [DOI] [PubMed] [Google Scholar]
- 4.Litvak S, Carr DS, Chapeville F. TYMV RNA As a substrate of the tRNA nucleotidyltransferase. FEBS Lett. 1970;11:316–319. doi: 10.1016/0014-5793(70)80557-9. [DOI] [PubMed] [Google Scholar]
- 5.Hammond JA, Rambo RP, Kieft JS. Multi-domain packing in the aminoacylatable 3′ end of a plant viral RNA. J Mol Biol. 2010;399:450–463. doi: 10.1016/j.jmb.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreher TW. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009;139:217–229. doi: 10.1016/j.virusres.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giege R, Frugier M, Rudinger J. tRNA mimics. Curr Opin Struct Biol. 1998;8:286–293. doi: 10.1016/s0959-440x(98)80060-2. [DOI] [PubMed] [Google Scholar]
- 8.Cliffe AR, Nash AA, Dutia BM. Selective uptake of small RNA molecules in the virion of murine gammaherpesvirus 68. J Virol. 2009;83:2321–2326. doi: 10.1128/JVI.02303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreher TW. Viral tRNAs and tRNA-like structures. Wiley Interdiscip Rev RNA. 2010;1:402–414. doi: 10.1002/wrna.42. [DOI] [PubMed] [Google Scholar]
- 10.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond JA, Rambo RP, Filbin ME, Kieft JS. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA. 2009;15:294–307. doi: 10.1261/rna.1360709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rietveld K, Linschooten K, Pleij CW, Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984;3:2613–2619. doi: 10.1002/j.1460-2075.1984.tb02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rietveld K, Pleij CW, Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rietveld K, Van Poelgeest R, Pleij CW, Van Boom JH, Bosch L. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982;10:1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florentz C, et al. The tRNA-like structure of turnip yellow mosaic virus RNA: structural organization of the last 159 nucleotides from the 3′ OH terminus. EMBO J. 1982;1:269–276. doi: 10.1002/j.1460-2075.1982.tb01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giege R, Florentz C, Dreher TW. The TYMV tRNA-like structure. Biochimie. 1993;75:569–582. doi: 10.1016/0300-9084(93)90063-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda D, Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 18.Singh RN, Dreher TW. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997;233:430–439. doi: 10.1006/viro.1997.8621. [DOI] [PubMed] [Google Scholar]
- 19.Skuzeski JM, Bozarth CS, Dreher TW. The turnip yellow mosaic virus tRNA-like structure cannot be replaced by generic tRNA-like elements or by heterologous 3′ untranslated regions known to enhance mRNA expression and stability. J Virol. 1996;70:2107–2115. doi: 10.1128/jvi.70.4.2107-2115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuda D, Yoshinari S, Dreher TW. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dumas P, et al. 3-D graphics modelling of the tRNA-like 3′-end of turnip yellow mosaic virus RNA: structural and functional implications. J Biomol Struct Dyn. 1987;4:707–728. doi: 10.1080/07391102.1987.10507674. [DOI] [PubMed] [Google Scholar]
- 23.Singh RN, Dreher TW. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and -CCR- boxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998;4:1083–1095. doi: 10.1017/s1355838298980694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deiman BA, Koenen AK, Verlaan PW, Pleij CW. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998;72:3965–3972. doi: 10.1128/jvi.72.5.3965-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukai S, et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 26.Dreher TW, Tsai CH, Florentz C, Giege R. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry. 1992;31:9183–9189. doi: 10.1021/bi00153a010. [DOI] [PubMed] [Google Scholar]
- 27.Dreher TW, Goodwin JB. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998;26:4356–4364. doi: 10.1093/nar/26.19.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barends S, Bink HH, van den Worm SH, Pleij CW, Kraal B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–129. doi: 10.1016/s0092-8674(02)01256-4. [DOI] [PubMed] [Google Scholar]
- 29.Berthelot F, Bogdanovsky D, Schapira G, Gros F. Interchangeability of factors and tRNA's in bacterial and eukaryotic translation initiation systems. Mol Cell Biochem. 1973;1:63–72. doi: 10.1007/BF01659939. [DOI] [PubMed] [Google Scholar]
- 30.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keel AY, Rambo RP, Batey RT, Kieft JS. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wientges J, Putz J, Giege R, Florentz C, Schwienhorst A. Selection of viral RNA-derived tRNA-like structures with improved valylation activities. Biochemistry. 2000;39:6207–6218. doi: 10.1021/bi992852l. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch W. Xds. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis IW, Murray LW, Richardson JS, Richardson DC. MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, et al. Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc Natl Acad Sci USA. 2011;108:1839–1844. doi: 10.1073/pnas.1018582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolk MH, et al. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science. 1998;280:434–438. doi: 10.1126/science.280.5362.434. [DOI] [PubMed] [Google Scholar]
- 44.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 45.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]


