SUMMARY
Gaucher’s disease (GD) is caused by mutations that compromise β-glucocerebrosidase (GCase) folding in the endoplasmic reticulum (ER), leading to excessive degradation instead of trafficking, which results in insufficient lysosomal function. We hypothesized that ER GCase interacting proteins play critical roles in making quality control decisions, i.e., facilitating ER-associated degradation (ERAD) instead of folding and trafficking. Utilizing GCase immunoprecipitation followed by mass spectrometry-based proteomics, we identified endogenous HeLa cell GCase protein interactors, including ERdj3, an ER resident Hsp40 not previously established to interact with GCase. Depleting ERdj3 reduced the rate of mutant GCase degradation in patient-derived fibroblasts, while increasing folding, trafficking and function by directing GCase to the pro-folding ER calnexin pathway. Inhibiting ERdj3-mediated mutant GCase degradation while simultaneously enhancing calnexin-associated folding, by way of a diltiazem-mediated increase in ER Ca2+ levels, yields a synergistic rescue of L444P GCase lysosomal function. Our findings suggest a combination therapeutic strategy for ameliorating GD.
INTRODUCTION
The maintenance of the proteome in each subcellular compartment by its protein homeostasis, or proteostasis, network is crucial for normal organismal physiology (Balch, et al., 2008; Ong and Kelly, 2011; Powers, et al., 2009). One-third of the eukaryotic proteome has to be properly folded in the endoplasmic reticulum (ER) before these proteins can be trafficked to their destination environment, including outside of the cell, or in the case of lysosomal enzymes, trafficked to the lysosome (Brodsky and Skach, 2011; Futerman and van Meer, 2004; Reczek, et al., 2007; Zhao and Grabowski, 2002). Inherited mutations in genes encoding lysosomal enzymes often lead to destabilized enzymes, resulting in excessive ER mutant enzyme misfolding and ER-associated degradation (ERAD) (Brodsky and Skach, 2011; Hebert, et al., 2010; Vembar and Brodsky, 2008). Excessive degradation can lead to loss of activity of a particular enzyme in the lysosome, resulting in substrate accumulation in the lysosomes of certain cell types, potentially giving rise to clinical pathology (Sun, et al., 2010; Vitner, et al., 2010; Zhao and Grabowski, 2002). Nearly 50 distinct lysosomal storage diseases (LSDs) of this type have been described (Futerman and van Meer, 2004; Parenti, et al., 2013).
The most common LSD is Gaucher’s disease (GD), which is characterized by the accumulation of the β-glucocerebrosidase (GCase; encoded by the GBA gene) substrates, glucosylceramide and glucosylsphingosine (Edmunds, 2010; Hruska, et al., 2008). Distinct GCase mutations are subject to varying degrees of ERAD and thus exhibit different levels of residual lysosomal GCase activity–generally, the lower the activity, the more severe the GD phenotype (Bendikov-Bar and Horowitz, 2012; Grace, et al., 1994; Ron and Horowitz, 2005; Schmitz, et al., 2005; Schueler, et al., 2004). L444P homozygotes exhibiting extremely low levels of lysosomal GCase activity present with neuronopathic GD (Grabowski, 1997; Schmitz, et al., 2005). N370S GCase, exhibiting higher residual lysosomal activity, never presents with any central nervous system symptoms, even in compound heterozygotes with one null allele (Edmunds, 2010). Instead these patients present with hepatosplenomegaly and skeletal disorders, clinical features common to most GD patients (Beutler and Gelbart, 1993; Tsuji, et al., 1988). Most mutant GCase enzymes, including L444P and N370S, have been shown to be able to retain their catalytic activity in the lysosome if folded in the ER and subsequently trafficked there, albeit with generally lowered specific activity (Mu, et al., 2008; Ong, et al., 2010; Sawkar, et al., 2006; Wang, et al., 2011). As such, strategies that increase the folded and functional concentration of mutant GCase in the ER may prove to be useful for ameliorating GD. Enzyme replacement therapy with WT GCase harboring a N-glycan that facilitates lysosomal uptake is generally an effective treatment; substrate reduction therapy is an alternative for non-responders (Parenti, et al., 2013). Therapeutic strategies under development include small molecule pharmacologic chaperones and enhancers of ER folding capacity (Mu, et al., 2008; Ong, et al., 2010; Sawkar, et al., 2006; Wang, et al., 2011). The former bind to and stabilize mutant GCase in the ER, increasing the folded concentration that can be trafficked to the lysosome.
The balance between lysosomal enzyme degradation versus folding and trafficking (i.e., quality control) is determined by the stoichiometry of the ER proteostasis network components, including more than 75 proteins making up numerous integrated or competing proteostatic pathways (Adachi, et al., 2008; Lee, et al., 2003; Yamamoto, et al., 2007). Thus, we hypothesized that proteostasis network components interacting with GCase in the ER may play a critical role in facilitating GCase folding and trafficking, or alternatively in targeting misfolded GCase for ERAD. Herein, we immunoprecipitated GCase and its interacting partners, which were then identified by a mass spectrometry-based proteomic approach referred to as Multidimensional Protein Identification Technology (MudPIT) (Washburn, et al., 2001). Cellular biochemical studies on one particular ER GCase interactor, ERdj3, revealed its importance in GCase proteostasis decision-making. Depletion of ERdj3 reduced the rate of mutant GCase degradation in patient-derived fibroblasts—increasing mutant GCase folding, trafficking and function by re-directing it to the pro-folding ER chaperone calnexin. We demonstrated that inhibiting the mutant GCase–ERdj3 interaction while simultaneously promoting the chaperoning activity of the Ca2+-dependent ER lectin chaperone calnexin (by diltiazem application) yields a synergistic rescue of L444P GCase lysosomal activity to 50% of wild type (WT) GCase activity, which has the potential to be an effective combination therapeutic strategy for GD.
RESULTS
Identification of ER proteostasis network components that interact with WT GCase
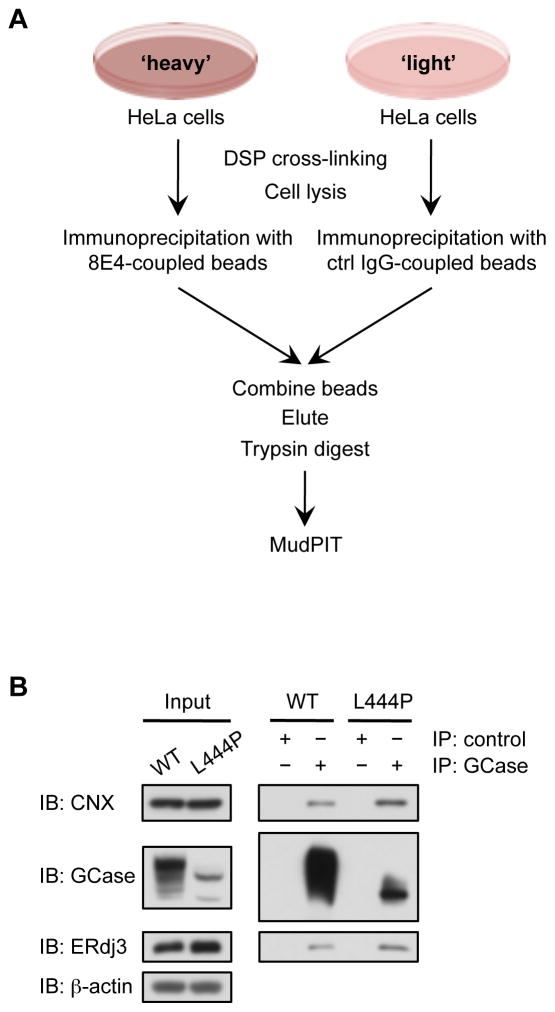
To identify the proteins that interact with GCase in the ER of HeLa cells, we utilized stable isotope labeling by amino acids in cell culture (SILAC) (Ong, et al., 2002) together with immunoprecipitation of endogenous WT GCase (Figure 1A). HeLa cells were labeled with either ‘heavy’ or ‘light’ isotopes of arginine and lysine. Prior to lysis of either ‘heavy’ or ‘light’ HeLa cells, intracellular protein complexes were cross-linked with the cell-penetrable, reversible cross-linker dithiobis(succinimidyl propionate) (DSP) in an effort to stabilize weak and/or transient interactions. The 8E4 mouse monoclonal antibody, which recognizes the extreme C-terminus of GCase (Aerts, et al., 1986; Barneveld, et al., 1983; Bleijlevens, et al., 2008), was covalently coupled to beads and used for the immunoisolation of WT GCase and its interacting proteins from the ‘heavy’ cell lysates. An irrelevant mouse antibody targeting FLAG, an epitope not present in the mammalian proteome, was covalently coupled to beads and used as a negative control in the ‘light’ population. Proteins enriched in the ‘heavy’ population were identified by MudPIT (Washburn, et al., 2001) (Table S1). We focused on GCase interacting proteins localized to the ER (Table 1), since these proteins could be important for GCase degradation or folding and trafficking.
Figure 1.
Identification of GCase proteostasis network components. (A) SILAC-immunoprecipitation-MudPIT experimental scheme. (B) Western blot analysis of immunopurified GCase from WT and L444P patient-derived fibroblasts. ERdj3 and calnexin co-precipitated with WT and L444P GCase. Control experiments were performed with anti-FLAG M2 antibody. IP, immunoprecipitation; IB, immunoblot. See also Figure S1.
Table 1.
ER-localized interactors of WT GCase
| UniProt ID | Gene symbol | Protein description | H/L ratio | Spectral counts | Peptide number |
|---|---|---|---|---|---|
| P04062 | GBA | Glucocerebrosidase | 95.51 | 491 | 14 |
| P30040 | ERP29 | Endoplasmic reticulum resident protein 29 | 9.16 | 15 | 2 |
| P27824 | CANX | Calnexin | 5.05 | 11 | 2 |
| P27797 | CALR | Calreticulin | 3.61 | 11 | 4 |
| P30101 | PDIA3 | Protein disulfide-isomerase A3 (ERp57) | 1.95 | 12 | 8 |
| Q02809 | PLOD1 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | 1.69 | 3 | 2 |
| P14625 | HSP90B1 | Endoplasmin (GRP94) | 1.55 | 15 | 2 |
| Q15084 | PDIA6 | Protein disulfide-isomerase A6 | 1.55 | 13 | 6 |
| P11021 | HSPA5 | 78 kDa glucose-regulated protein (GRP78, BiP) | 1.43 | 174 | 19 |
| Q9UBS4 | DNAJB11 | DnaJ homolog subfamily B member 11 (ERdj3) | 1.41 | 5 | 2 |
| P50454 | SERPINH1 | Serpin H1 | 1.4 | 23 | 7 |
See also Table S1
Previous studies have shown that increasing calnexin levels partially restored the folding, trafficking and lysosomal function of L444P GCase (Ong, et al., 2010), establishing calnexin as an important ER chaperone for GCase folding. Calnexin, a membrane-bound lectin chaperone, and its lumenal homologue calreticulin, recognize and bind to Glc1Man9GlcNAc2 glycans on unfolded glycoproteins. The Ca2+ regulated chaperones calnexin and calreticulin assist the retention and folding of monomeric glycoproteins in the ER in collaboration with the protein disulfide isomerase ERp57 (also known as PDIA3) (Lederkremer, 2009; Maattanen, et al., 2010). All three proteostasis network components of the calnexin cycle were identified by our GCase interactome approach (Table 1), validating the potential of this immunoprecipitation/mass spectrometry-based interactome approach for discovering new GCase proteostasis network components.
ERdj3 interacts with WT and mutant GCase in patient-derived fibroblasts
The GCase interacting protein identified by our immunoprecipitation/mass spectrometry approach (Table 1) and that we focused our cellular biochemical confirmation studies on is DNAJB11 (also known as ERdj3), since its role in GCase proteostasis was unknown. ERdj3 is an ER luminal protein and is one of the seven ER J-domain-containing Hsp40 co-chaperones (Yu, et al., 2000). One established function of ERdj3 is to deliver clients to BiP, the resident Hsp70 in the ER, and stimulate its ATPase activity (Kampinga and Craig, 2010; Yu, et al., 2000). ERdj3 is able to interact directly with unfolded client proteins, recruiting them to BiP for ATP-dependent chaperoning in the Hsp70-Hsp40-nucleotide exchange factor (NEF) folding cycle. Upon stimulating the ATPase activity of BiP, ERdj3 is released from the complex (Jin, et al., 2009; Shen and Hendershot, 2005). Hsp40s like ERdj3 can also act independently of their corresponding Hsp70 (Kampinga and Craig, 2010). For example, Scj1, a yeast ER Hsp40 similar in domain architecture to ERdj3, facilitated the degradation of the epithelial sodium channel independent of BiP (Buck, et al., 2010). ERdj3 is established to be upregulated by activation of the unfolded protein response (UPR) stress-responsive signaling pathway that controls the proteostasis capacity of the secretory pathway (Shen and Hendershot, 2005).
Co-immunoprecipitates of endogenous GCase from WT and L444P GCase patient-derived fibroblasts subjected to Western blot analysis using antibodies specific for calnexin and ERdj3 (Figure 1B) corroborated our mass spectrometry findings in HeLa cells, namely that these proteostasis network components were GCase interactors. We also observed that G202R GCase interacted with calnexin and ERdj3 (Figure S1). L444P and G202R GCase appeared to interact with more calnexin and ERdj3 in comparison to WT GCase, suggesting that these misfolding-prone mutants require more assistance from ER chaperones/cochaperones for their folding and/or degradation.
ERdj3 depletion increases the lysosomal activity of mutant GCase, but not WT GCase
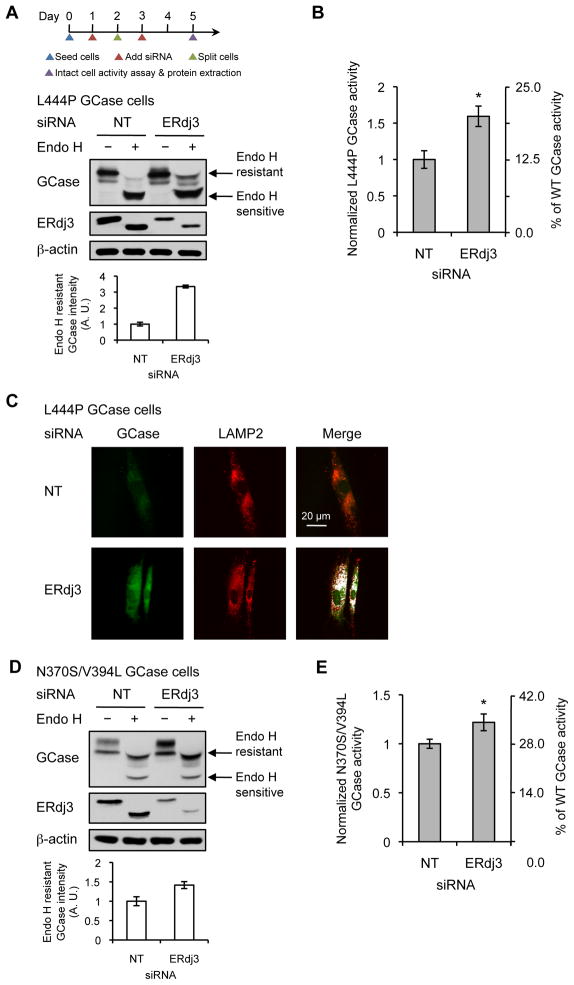
We next asked how ERdj3 affects GCase proteostasis by reducing its cellular concentration using siRNA in L444P GCase fibroblasts (>90% knockdown at the transcript level, Figure S2A, >75% knockdown at the protein level, Figure 2A, middle panel). This resulted in a 1.6-fold increase in GCase lysosomal activity (~20% of WT GCase activity), in comparison to the non-targeting (NT) siRNA control (Figure 2B), without significantly altering cellular viability (resazurin reduction assay; Figure S2B). The glucosylceramide substrate begins to accumulate when mutant GCase lysosomal activity is ≤ 11–15% of WT GCase activity (Schueler, et al., 2004), suggesting that the increase to ~20% is likely to be sufficient to ameliorate GD symptoms. To investigate if this increased activity is due to enhanced L444P GCase folding and trafficking in the ER, we performed endoglycosidase H (endo H) digestion on L444P GCase cell lysates. Endo H is able to cleave after asparaginyl-N-acetylglucosamine on glycans that have been linked to GCase in the ER, but is unable to cleave this structure once it has been enzymatically processed and remodeled into complex oligosaccharides in the Golgi. Hence, the higher molecular weight endo H-resistant band is a reporter for the properly folded and trafficked (at least to the Golgi) fraction of GCase (Ron and Horowitz, 2005; Sawkar, et al., 2006). Knockdown of ERdj3 increased the endo H-resistant glycoform band of L444P GCase (Figure 2A, top panel), indicating that L444P GCase is folded and trafficked more efficiently upon ERdj3 depletion in the ER. Furthermore, indirect immunofluorescence microscopy revealed that the knockdown of ERdj3 increased the amount of detectable L444P GCase, which is normally barely detectable due to extensive ERAD (Figure 2C, cf. left panels). Importantly, we observed an enhancement of colocalization of L444P GCase with the lysosomes as well (Figure 2C, cf. right panels, artificially colored white).
Figure 2.
ERdj3 is a proteostasis network component of mutant GCase. (A) Silencing ERdj3 increased the endo H-resistant glycoform of L444P GCase in fibroblasts. Quantification of endo H-resistant L444P GCase bands is shown below. (B) Silencing ERdj3 significantly increased L444P GCase activity in fibroblasts. (C) Silencing ERdj3 enhanced the lysosomal trafficking of L444P GCase, as assessed by indirect immunofluorescence microscopy. (D) Silencing ERdj3 increased the endo H-resistant glycoform of N370S GCase in fibroblasts. Quantification of endo H-resistant N370S GCase bands is shown below. (E) Silencing ERdj3 significantly increased N370S GCase activity in fibroblasts. The data in (A), (B), (D) and (E) are reported as mean ± SD (n = 3 for A and D, n = 8 for B and E). Statistical significance was calculated using a two-tailed Student’s t-test, * p < 0.01. See also Figures S2 and S3.
Using siRNA to reduce the ERdj3 concentration in N370S/V394L compound heterozygotic GCase fibroblasts (>75% knockdown at the protein level, Figure 2D, middle panel) resulted in a 1.2-fold increase in GCase activity in comparison to the NT control (Figure 2E). We also observed a significant increase in the endo H-resistant N370S/V394L GCase glycoform band (Figure 2D, top panel), demonstrating that more of another GCase mutant (other than L444P) is trafficked to the Golgi and then to the lysosome when the concentration of ERdj3 is reduced in the ER.
In stark contrast, depleting ERdj3 (>90% knockdown at the transcript level, Figure S2C, >75% knockdown at the protein level, Figure S3A, middle panel) did not alter WT GCase lysosomal activity (Figure S3B), or its endo H-resistant fraction (Figure S3A, top panel), even though ERdj3 interacts with WT GCase (Figures 1B and S1). We verified that ERdj3 depletion did not significantly alter mRNA levels of WT and L444P GBA using quantitative RT-PCR (Figures S2A and C). Moreover, we did not detect any significant changes in the mRNA levels of a number of established UPR markers (CHOP, spliced XBP1 and GRP78), demonstrating that stress-responsive signaling was not induced upon ERdj3 depletion (Figures S2A and C).
ERdj3 depletion slows down the degradation of L444P GCase, but not WT GCase
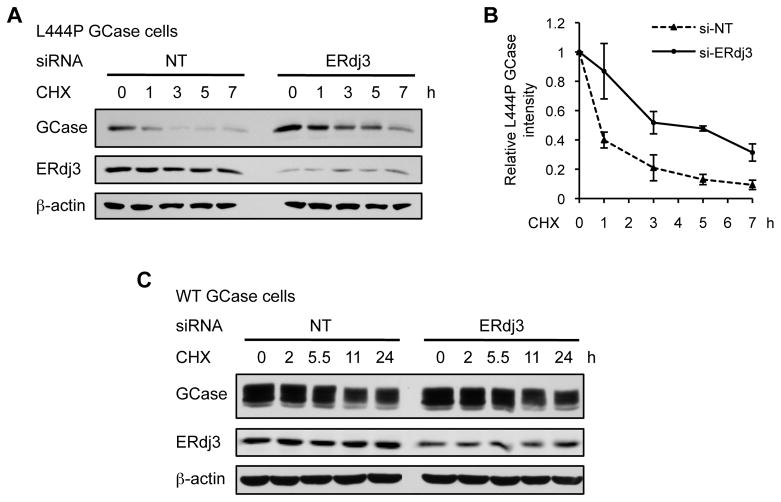
To investigate whether ERdj3 knockdown alters the rate of mutant GCase degradation, we employed cycloheximide–chase experiments in L444P GCase fibroblasts. Following NT or ERdj3 siRNA treatment, the cells were treated with cycloheximide to inhibit general protein synthesis, then harvested at the indicated timepoints after cycloheximide treatment and lysed for Western blot analysis (Figure 3A). L444P GCase typically undergoes extensive degradation over this time period, as demonstrated in the NT control. However, when the concentration of ERdj3 was reduced, the rate of degradation of L444P GCase was significantly slower (Figure 3A, quantification shown in Figure 3B). Analogous cycloheximide–chase analysis in WT GCase fibroblasts revealed no change in the degradation rate of WT GCase when ERdj3 was silenced (Figure 3C). This suggests that ERdj3 is functionally involved in the degradation pathway that targets destabilized mutant GCase, but not WT GCase, for ERAD.
Figure 3.
ERdj3 is functionally involved in the degradation of L444P GCase, but not WT GCase. (A) Silencing ERdj3 significantly reduced the rate of degradation of L444P GCase in fibroblasts, as assessed by cycloheximide–chase analysis. (B) Quantification of L444P GCase bands when NT (dashed line) and ERdj3 (solid line) siRNA were applied. (C) Silencing ERdj3 did not change the rate of degradation of WT GCase in fibroblasts, as assessed by cycloheximide–chase analysis. The data in (B) are reported as mean ± SEM (n = 4). CHX, cycloheximide. See also Figure S4.
BiP and HYOU1 do not influence the ERdj3-mediated degradation of L444P GCase
Since ERdj3 is the Hsp40 component of the BiP(Hsp70)-Hsp40-NEF chaperone folding/degradation pathway in the ER, we wanted to investigate if any of the other components of this pathway besides ERdj3 could affect mutant GCase proteostasis. Previous work demonstrated that overexpression of BiP did not substantially alter the lysosomal activity of L444P GCase (Ong, et al., 2010; Wang, et al., 2011). Knockdown of the major NEF in the ER, HYOU1, did not increase L444P GCase activity either (Figure S4A), nor did it increase the endo H-resistant glycoform (Figure S4B). Co-knockdown of ERdj3 and HYOU1 in L444P GCase fibroblasts increased GCase activity about 1.4-fold (Figure S4A) and increased the endo H resistant band (Figure S4B) to levels that were similar to those observed after ERdj3 siRNA treatment alone. This suggests that BiP and HYOU1 do not play a significant role in the maintenance of L444P GCase proteostasis, indicating that ERdj3 may be acting independently of the BiP-Hsp40-NEF folding/degradation cycle.
Reduced interaction with ERdj3 directs L444P GCase to the calnexin pro-folding pathway
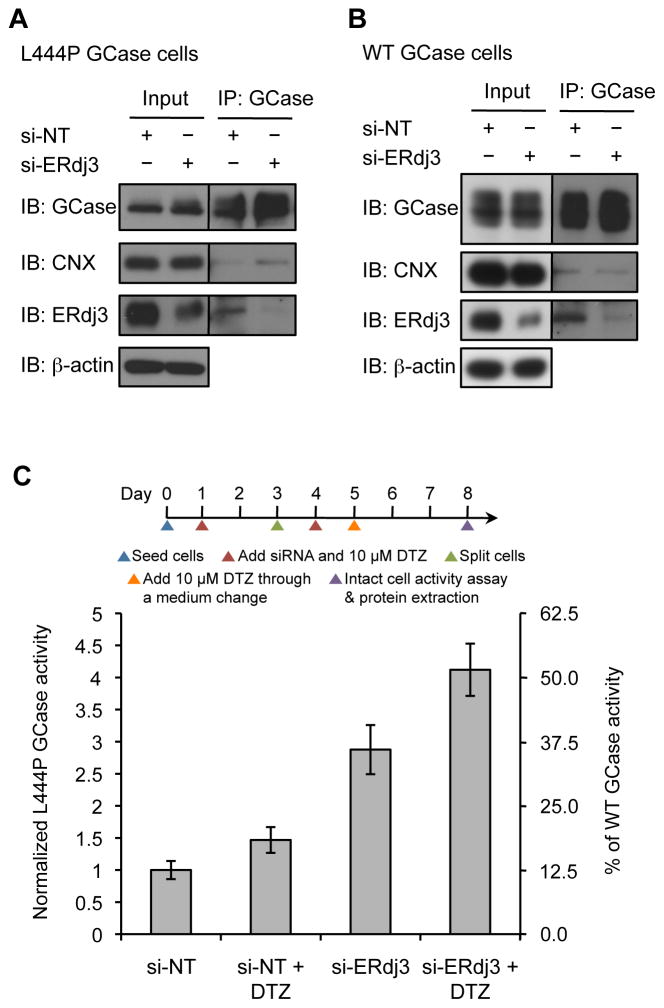
Since depletion of the pro-degradation factor ERdj3 in the ER resulted in productive folding and trafficking of L444P GCase, we hypothesized that ERdj3 competes with calnexin, an ER chaperone known to be important for GCase binding and folding (Ong, et al., 2010). When the cellular concentration of ERdj3 was reduced, more calnexin was bound to immunoprecipitated L444P GCase (Figure 4A), indicating that L444P GCase was indeed being directed towards the calnexin pro-folding pathway. In contrast, there was no significant change in the amount of calnexin bound to WT GCase when ERdj3 was silenced in WT GCase fibroblasts (Figure 4B), consistent with the finding that ERdj3 is not functionally involved in the degradation of WT GCase (Figures 3C and S3).
Figure 4.
Simultaneous inhibition of the GCase–ERdj3 interaction and enhancement of the chaperoning activity of calnexin exhibits synergy in L444P GCase fibroblasts. (A) Silencing ERdj3 enhanced the interaction between L444P GCase and calnexin in fibroblasts. (B) Silencing ERdj3 did not affect the interaction between WT GCase and calnexin in fibroblasts. IP, immunoprecipitation; IB, immunoblot. (C) Co-application of diltiazem and ERdj3 siRNA synergistically enhanced L444P GCase lysosomal activity. The data in (C) are reported as mean ± SD (n = 8). DTZ, diltiazem. See also Figure S5.
ERdj3 depletion together with diltiazem treatment synergistically enhances L444P GCase folding, trafficking and lysosomal activity
Diltiazem treatment is established to improve L444P GCase proteostasis in patient-derived fibroblasts by inhibiting the ER ryanodine receptor Ca2+ efflux channels, thereby increasing ER Ca2+ levels and enhancing the chaperoning activity of calnexin–a Ca2+-dependent chaperone (Mu, et al., 2008; Ong, et al., 2010). We hypothesized that since more L444P GCase was directed towards the calnexin pro-folding pathway in the absence of ERdj3, increasing the chaperoning activity of calnexin (by application of diltiazem) could further enhance the folding, trafficking and lysosomal function of L444P GCase.
Diltiazem and ERdj3 siRNA were co-administered to L444P GCase fibroblasts over a 7-day period (Figure 4C). Consistent with previous results (Ong, et al., 2010; Wang, et al., 2011), diltiazem treatment (10 μM) alone increased the activity of L444P GCase by 1.5-fold (or a 50 unit increase). Interestingly, application of ERdj3 siRNA over a 7-day period dramatically improved the activity of L444P GCase by 2.9-fold (or a 190 unit increase), as compared to the 1.6-fold increase observed when siRNA was applied over 4 days. This demonstrates that prolonged inhibition of the L444P GCase–ERdj3 interaction is beneficial for L444P GCase folding and trafficking, and is consistent with ERdj3 being a long-lived protein (protein half-life = 98 h, mRNA half-life = 20 h) (Schwanhausser, et al., 2011). Figure S5A depicts the linear increase in L444P GCase activity from 4 to 7 days when treated with ERdj3 siRNA. When diltiazem and ERdj3 siRNA were co-applied using the regimen shown, we observed a 4.1-fold increase (or a 310 unit increase) in L444P GCase activity (to approximately 50% of WT GCase activity; Figure 4C) after 7 days of treatment. This is significantly more than the 3.4-fold or 240 unit sum of either treatment alone, revealing a synergistic enhancement of L444P GCase activity (Figure 4C).
Endo H treatment revealed a similar trend (Figure S5B), verifying that co-administration of diltiazem and ERdj3 siRNA significantly rescued the folding, trafficking and lysosomal function of L444P GCase. We did not observe any increase in GBA transcript levels in any of the experiments depicted in Figure 4C, nor did we observe induction of the UPR based on quantitative RT-PCR (Figure S5C). Notably, cell viability was not reduced, as measured by the resazurin reduction assay (Figure S5D).
DISCUSSION
Our immunoprecipitation/mass spectrometry approach to discover new GCase interactors/proteostasis network components has led to the identification and characterization of ERdj3. Reducing the concentration of ERdj3 in patient-derived fibroblasts diminishes the rate of mutant GCase degradation and enhances the folding and trafficking of mutant, but not WT GCase, despite the fact that ERdj3 interacts with both. In fact, the extent of the folding, trafficking and lysosomal activity enhancement appears to be dependent on the stability of the mutant GCase–the more unstable the GCase variant, the greater the activity enhancement observed. The L444P GCase variant is more destabilized at pH 7 in the ER compared to the N370S GCase mutant (Edmunds, 2010; Sawkar, et al., 2006; Schmitz, et al., 2005), and hence is degraded more extensively by ERAD (Ron and Horowitz, 2005). WT GCase is processed normally in the ER (Bendikov-Bar and Horowitz, 2012; Ron and Horowitz, 2005). Inhibiting ERAD through ERdj3 depletion resulted in greater enhancement of L444P GCase proteostasis than N370S GCase proteostasis–no WT GCase enhancement was observed. When these data are considered together with the observation that significantly more ERdj3 binds to mutant GCase than to WT GCase, we hypothesize that ERdj3 is able to discriminate between GCase sequences based on their extent of destabilization, consistent with ERdj3′s established ability to bind to unfolded client proteins in the ER (Jin, et al., 2009; Shen and Hendershot, 2005).
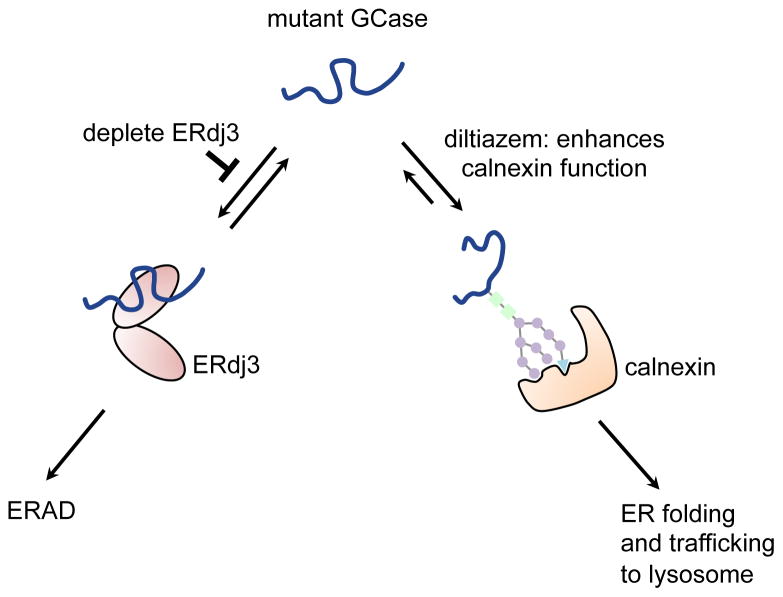
It appears that unfolded mutant GCase partitions between a calnexin-assisted folding pathway and an ERdj3-mediated ERAD pathway early in the secretory pathway (Figure 5). In our proposed model, ERdj3 and calnexin compete for the unfolded mutant GCase enzyme in the ER, resulting in its degradation or folding, respectively. Reducing the cellular concentration of ERdj3 decreases its interaction with misfolded mutant GCase, allowing pro-folding components like calnexin (and perhaps other components of this folding pathway) to compete for the unfolded enzyme, eventually resulting in its folding and subsequent trafficking to the lysosome as a functional GCase enzyme. To further support this hypothesis, we demonstrated that enhancement of the Ca2+-regulated chaperoning activity of the calnexin pathway via diltiazem treatment along with simultaneous inhibition of the ERdj3–GCase interaction synergistically rescued L444P GCase proteostasis, increasing L444P activity to 50% of WT GCase lysosomal activity. This is significantly above the GCase activity threshold at which the substrate glucosylceramide begins to accumulate in the cell (≤ 11–15% of WT GCase activity) (Schueler, et al., 2004). Despite a reduced association of WT GCase with ERdj3, we did not observe an enhanced interaction with calnexin, suggesting that reducing the binding of ERdj3 to WT GCase does not have a functional outcome.
Figure 5.
ERdj3 is a degradation versus folding partitioning factor for mutant GCase. In our proposed model, ERdj3 and calnexin compete for the unfolded mutant GCase enzyme in the ER, resulting in its degradation or folding respectively. When the ERdj3-mediated ERAD pathway is inhibited, mutant GCase partitions into the calnexin folding cycle which can be further enhanced by diltiazem treatment, thereby leading to a synergistic rescue of mutant GCase folding, trafficking and lysosomal activity.
Maintaining Ca2+ homeostasis in the ER is of particular importance, since the accumulation of glucosylceramide has been shown to cause extensive Ca2+ efflux through the ryanodine receptors in GD neurons (Korkotian, et al., 1999; Lloyd-Evans, et al., 2003; Pelled, et al., 2005). Diltiazem is an FDA-approved L-type Ca2+ channel blocker drug for the treatment of hypertension (Hockerman, et al., 1997; Massie, et al., 1984; Massie, et al., 1987). It also restores ER Ca2+ homeostasis through the inhibition of Ca2+ efflux via the ryanodine receptors (Shoshan-Barmatz, et al., 1991), potentially reversing the effect of glucosylceramide accumulation. Notably, this increase in ER Ca2+ levels has been previously shown to enhance the calnexin–mutant GCase interaction resulting in improved L444P GCase proteostasis (Ong, et al., 2010), suggesting that diltiazem treatment functions, at least partially, by increasing the calnexin chaperoning of mutant GCase. Diltiazem treatment also significantly increased GCase lysosomal activity in a number of mouse skin fibroblast lines derived from individual GCase point-mutated homozygous mice (Sun, et al., 2009). Despite mimicking the low residual GCase activity seen in human patient tissues, these mouse models do not recapitulate the phenotypic abnormalities of human patients (Farfel-Becker, et al., 2011; Xu, et al., 2003). While the effects of diltiazem in vivo appeared to be minimal when dosed in these GD mouse models (Sun, et al., 2009), it is clear from our work that the effect of diltiazem will likely be more pronounced when the ERdj3–mutant GCase interaction is also abrogated by ERdj3 depletion, since this inhibition of ERAD increases the pool of available mutant GCase in the ER which is then redirected towards the Ca2+-enhanced calnexin folding pathway (Figure 5).
In summary, our study strongly indicates that inhibition of the mutant GCase ERAD pathway by depleting ERdj3 while simultaneously enhancing the pro-folding calnexin pathway via diltiazem treatment markedly tilts the balance in favor of folding and trafficking of mutant GCase versus degradation. Since we did not observe any UPR induction or diminished cell viability with this regimen, this represents a promising therapeutic strategy for ameliorating GD, especially considering that there are a plethora of regulatory agency-approved drugs with outstanding safety records that elevate ER Ca2+ levels.
SIGNIFICANCE
The above-mentioned results show that the proteostasis network components that interact with GCase in the ER are involved in making important quality control decisions, namely, degradation versus folding and trafficking to the lysosome decisions. In particular, depletion of ERdj3 significantly diminished the rate of mutant GCase degradation, thereby enhancing its folding, trafficking and lysosomal function. Furthermore, inhibition of the ERdj3-mediated ERAD pathway along with simultaneous enhancement of the pro-folding calnexin pathway via diltiazem treatment synergistically rescues mutant GCase lysosomal activity to levels thought to be more than sufficient to ameliorate GD. These results suggest that depleting ERdj3 in combination with drugs that enhance ER Ca2+ levels (e.g., diltiazem) merits further investigation as a strategy for neuronopathic GD patients that cannot be treated by enzyme replacement therapy because the recombinant protein does not cross the blood-brain barrier.
EXPERIMENTAL PROCEDURES
Reagents
4-Methylumbelliferyl β-D-glucopyranoside (MUG) was from Sigma (St. Louis, MO). Conduritol B epoxide (CBE) was from Toronto Research Chemicals (Downsview, ON, Canada). Diltiazem hydrochloride was from Tocris Bioscience (Ellisville, MO). Cell culture media were purchased from Gibco (Grand Island, NY).
Cell cultures
Primary skin fibroblast cultures were established from Gaucher’s patients homozygous for the G202R (c.721G>A) mutation (kindly provided by Dr. K. P. Zimmer (Children’s Hospital of the University of Munster, Munster)). An apparently normal fibroblast cell line (GM00498), the Gaucher’s disease fibroblast cell line homozygous for the L444P (c.1448T>C) mutation (GM08760) and the Gaucher’s disease fibroblast cell line heterozygous for the N370S (c.1226A>G) and V394L (c. 1297G>T) mutations (GM01607) were from Coriell Cell Repositories (Camden, NJ). Fibroblasts were grown in minimal essential medium with Earle’s salts, 10% heat-inactivated fetal bovine serum and 1% glutamine Pen-Strep at 37°C in 5% CO2. Cell medium was replenished every 3 or 4 days and monolayers were passaged with TrypLE Express upon reaching confluency.
SILAC and immunoprecipitation of GCase
Two populations of HeLa cells were cultured in SILAC DMEM media containing 10% dialyzed fetal bovine serum and 1% Pen-Strep (Cambridge Isotope Laboratories, Inc, Andover, MA). The ‘light’ medium was supplemented with unlabeled L-lysine and L-arginine, and the ‘heavy’ medium was supplemented with isotopic-labeled L-lysine and L-arginine (13C6-lysine and 13C615N4-arginine). After at least five passages, the ‘light’ and ‘heavy’ HeLa cell populations were harvested and washed once with DPBS. Each population was resuspended in DPBS buffer and cross-linked with 1 mM DSP for 30 min at RT. The reaction was quenched with 100 mM Tris pH 8 for 15 min at RT. The cells were lysed with RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton-X100, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with complete protease inhibitor cocktail (Roche) on ice, clarified by centrifugation, and pre-cleared with GammaBind G Sepharose™ beads (GE Healthcare) for 1.5 h at 4 °C. The ‘heavy’ lysate was incubated with 8E4 antibody-coupled beads, and the ‘light’ lysate was incubated with FLAG M2 antibody-coupled beads overnight at 4 °C. See Supplemental Experimental Procedures for the coupling of antibodies to GammaBind G Sepharose™ beads. After overnight incubation with lysates at 4 °C, the beads were pelleted and washed three times with RIPA buffer. The beads were combined pairwise and eluted in 2% SDS, 50 mM Tris pH 7.5 for 10 min at 100 °C. GCase eluates were analyzed by mass spectrometry; see Supplemental Experimental Procedures for detailed method.
Enzyme activity assay
The intact cell GCase activity assay for GD fibroblasts using the MUG substrate has been described previously (Mu, et al., 2008). Each reported data point was evaluated at least in triplicate in each plate, and on three different days. See Supplemental Experimental Procedures for detailed method.
Western blot analysis
Cells were lysed in complete lysis-M buffer (Roche) containing complete protease inhibitor cocktail (Roche). Total protein concentration was determined using Micro BCA assay reagent (Pierce). Company specifications were followed for treatment with Endo H (New England Biolabs). Aliquots of cell lysates were separated in an 8% or 10% SDS-PAGE gel and Western blot analysis was performed using appropriate antibodies. Details of antibodies used are given in the Supplemental Experimental Procedures.
Indirect immunofluorescence microscopy
Immunofluorescence was performed as previously described (Mu, et al., 2008). The experiments were performed three times and similar results were obtained. See Supplemental Experimental Procedures for detailed method.
Transfection of siRNA
Patient-derived GD fibroblasts were transfected with siRNA as previously described (Ong, et al., 2013). Briefly, fibroblasts were seeded in 6-well plates at approximately 2 × 105 cells per well and incubated at 37 °C overnight to reach 70–80% confluency before transient transfection using HiPerfect Transfection Reagent (Qiagen), according to the manufacturer’s protocol. The small interfering RNA (siRNA) duplexes were from Dharmacon: DNAJB11/ERdj3 (J-015861-09), HYOU1 (J-003678-09) and Non-Targeting siRNA (D-001810-01-20) as control.
Quantitative RT-PCR
Relative expression levels of target genes were measured by quantitative RT-PCR using the forward and reverse primers for the genes analyzed (Table S2). Total RNA was extracted from fibroblasts using RNeasy Mini Kit (Qiagen #74104). cDNA was synthesized from 500 ng of total RNA using QuantiTect Reverse Transcription Kit (Qiagen #205311). Quantitative PCR reactions (6 min at 95 °C, then 45 cycles of 15 s at 95 °C and 60 s at 60 °C) were performed using cDNA, FastStart Universal SYBR Green Master (Roche) and corresponding primers (Table S2) in an ABI 7900HT Fast Real Time PCR machine (Applied Biosystems) and analyzed using DataAssist™ software (Applied Biosystems). Each data point was evaluated in triplicate, and measured three times.
Cycloheximide–chase assay
Fibroblasts were seeded in 6-well plates at approximately 2 × 105 cells per well and incubated at 37 °C overnight. Fibroblasts were treated with transfection complexes as indicated. To arrest protein translation, 50 μg/mL cycloheximide (Chem Service) was added to each well. Cells were then chased for the indicated period of time, harvested and lysed for Western blot analysis.
Statistical analysis
All data are presented as mean ± SEM or mean ± SD as stated, and any statistical significance was calculated using a two-tailed Student’s t-test.
Supplementary Material
Figure S1. Related to Figure 1. Western blot analysis of immunopurified GCase from WT and G202R patient-derived fibroblasts. ERdj3 and calnexin co-precipitated with WT and G202R GCase. Control experiments were performed with anti-FLAG M2 antibody. IP, immunoprecipitation; IB, immunoblot.
Figure S2. Related to Figure 2. Knockdown of ERdj3 does not significantly alter cellular viability and GBA transcription, nor activate the UPR in fibroblasts. (A) and (C) Quantitative RT-PCR analysis showed that the mRNA level of ERdj3 was significantly reduced in L444P (A) and WT (C) fibroblasts. The mRNA levels of GBA and UPR target genes, CHOP, XBP1(s) and GRP78, were not significantly altered in L444P (A) and WT (C) fibroblasts 4 d post-transfection. RiboP served as an internal control. (B) Resazurin reduction assay showed that the cellular metabolic activity of L444P GCase fibroblasts did not change upon ERdj3 siRNA treatment as compared to the NT siRNA control. The data are reported as mean ±SD (n = 3).
Figure S3. Related to Figure 2. Knockdown of ERdj3 does not alter WTGC ase proteostasis. (A) Silencing ERdj3 did not change the endo H-resistant glycoform of WT GCase in fibroblasts. Quantification of endo H-resistant WT GCase bands is shown below. (B) Silencing ERdj3 did not change WT GCase activity in fibroblasts. The data in (A) and (B) are reported as mean ± SD (n = 3 for A, n = 6 for B).
Figure S4. Related to Figure 3. HYOU1 is not involved in the ERdj3-mediated degradation of L444P GCase. Silencing HYOU1 did not change L444P GCase activity (A) and its endo H-resistant glycoform (B) in fibroblasts. Co-knockdown of HYOU1 and ERdj3 increased L444P GCase activity (A) and its endo H-resistant glycoform (B) to the same extent as knockdown of ERdj3 alone. The data in (A) are reported as mean ±SD (n = 6).
Figure S5. Related to Figure 4. Co -application of ERdj3 siRNA and diltiazem is a viable strategy to rescue L444P GCase proteostasis. (A) L444P GCase activity increased linearly as a function of time when ERdj3 was silenced. L444P GCase fibroblasts were transfected with the same amount of NT and ERdj3 siRNA and analyzed 4 to 7 d post-transfection. (B) Co-application of diltiazem and ERdj3 siRNA significantly increased the endo H-resistant glycoform of L444P GCase in fibroblasts. (C) Quantitative RT-PCR analysis showed that the mRNA levels of GBA and UPR target genes, CHOP, XBP1(s) and GRP78, were not significantly altered in L444P GCase fibroblasts 7 d post-treatment. RiboP served as an internal control. (D) Resazurin reduction assay showed that the cellular metabolic activity of L444P GCase fibroblasts was not significantly altered 7 d post-treatment. The data in (C) and (D) are reported as mean ±SD (n = 3). DTZ, diltiazem.
Table S1. List of proteins that interact with endogenous WT GCase in HeLa cells as identified by SILAC-immunoprecipitation-MudPIT. See separate Excel file. Related to Table 1.
Table S2. Primer sequences for quantitative RT-PCR. Related to Experimental Procedures.
HIGHLIGHTS.
Immunoprecipitation/mass spectrometry identifies ERdj3 as a GCase interactor
ERdj3 depletion enhances mutant GCase folding, trafficking and lysosomal function
The ERdj3 degradation factor competes with pro-folding calnexin for mutant GCase
ERdj3 depletion and increased calnexin function synergize to rescue mutant GCase
Acknowledgments
This work was supported by the NIH (DK075295), the Skaggs Institute for Chemical Biology and the Lita Annenberg Hazen Foundation. Y.L. Tan is supported by a predoctoral fellowship from the Agency for Science, Technology and Research (A*STAR) Singapore. We thank M. Fukuda (Burnham Institute) for generously providing us with the rabbit anti-LAMP2, and C. Fearns for critical feedback on the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
Y.L.T., J.C.G., and J.W.K. designed research; Y.L.T., J.C.G., and S.P. performed research; J.M.F.G.A., and J.R.Y. contributed new reagents/analytic tools; Y.L.T., J.C.G., S.P., and J.W.K. analyzed data; and Y.L.T., and J.W.K. wrote the paper.
Supplemental Information contains five figures, two tables, and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Aerts JM, Donker-Koopman WE, Murray GJ, Barranger JA, Tager JM, Schram AW. A procedure for the rapid purification in high yield of human glucocerebrosidase using immunoaffinity chromatography with monoclonal antibodies. Anal Biochem. 1986;154:655–663. doi: 10.1016/0003-2697(86)90043-6. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Barneveld RA, Tegelaers FP, Ginns EI, Visser P, Laanen EA, Brady RO, Galjaard H, Barranger JA, Reuser AJ, Tager JM. Monoclonal antibodies against human beta-glucocerebrosidase. Eur J Biochem. 1983;134:585–589. doi: 10.1111/j.1432-1033.1983.tb07606.x. [DOI] [PubMed] [Google Scholar]
- Bendikov-Bar I, Horowitz M. Gaucher disease paradigm: from ERAD to comorbidity. Hum Mutat. 2012;33:1398–1407. doi: 10.1002/humu.22124. [DOI] [PubMed] [Google Scholar]
- Beutler E, Gelbart T. Gaucher disease mutations in non-Jewish patients. Br J Haematol. 1993;85:401–405. doi: 10.1111/j.1365-2141.1993.tb03185.x. [DOI] [PubMed] [Google Scholar]
- Bleijlevens B, van Breemen MJ, Donker-Koopman WE, de Koster CG, Aerts JM. Detection of mutant protein in complex biological samples: glucocerebrosidase mutations in Gaucher’s disease. Anal Biochem. 2008;372:52–61. doi: 10.1016/j.ab.2007.09.033. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell. 2010;21:1047–1058. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds T. Gaucher disease. In: Ramirez-Alvarado JWKM, Dobson CM, editors. Protein Misfolding Diseases: Current and Emerging Principles and Therapies. Hoboken, New Jersey: John Wiley & Sons, Inc; 2010. pp. 469–485. [Google Scholar]
- Farfel-Becker T, Vitner EB, Futerman AH. Animal models for Gaucher disease research. Dis Model Mech. 2011;4:746–752. doi: 10.1242/dmm.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- Grabowski GA. Gaucher disease: gene frequencies and genotype/phenotype correlations. Genet Test. 1997;1:5–12. doi: 10.1089/gte.1997.1.5. [DOI] [PubMed] [Google Scholar]
- Grace ME, Newman KM, Scheinker V, Berg-Fussman A, Grabowski GA. Analysis of human acid beta-glucosidase by site-directed mutagenesis and heterologous expression. J Biol Chem. 1994;269:2283–2291. [PubMed] [Google Scholar]
- Hebert DN, Bernasconi R, Molinari M. ERAD substrates: which way out? Semin Cell Dev Biol. 2010;21:526–532. doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Hockerman GH, Peterson BZ, Johnson BD, Catterall WA. Molecular determinants of drug binding and action on L-type calcium channels. Annu Rev Pharmacol Toxicol. 1997;37:361–396. doi: 10.1146/annurev.pharmtox.37.1.361. [DOI] [PubMed] [Google Scholar]
- Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- Jin Y, Zhuang M, Hendershot LM. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry. 2009;48:41–49. doi: 10.1021/bi8015923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkotian E, Schwarz A, Pelled D, Schwarzmann G, Segal M, Futerman AH. Elevation of intracellular glucosylceramide levels results in an increase in endoplasmic reticulum density and in functional calcium stores in cultured neurons. J Biol Chem. 1999;274:21673–21678. doi: 10.1074/jbc.274.31.21673. [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans E, Pelled D, Riebeling C, Bodennec J, de-Morgan A, Waller H, Schiffmann R, Futerman AH. Glucosylceramide and glucosylsphingosine modulate calcium mobilization from brain microsomes via different mechanisms. J Biol Chem. 2003;278:23594–23599. doi: 10.1074/jbc.M300212200. [DOI] [PubMed] [Google Scholar]
- Maattanen P, Gehring K, Bergeron JJ, Thomas DY. Protein quality control in the ER: the recognition of misfolded proteins. Semin Cell Dev Biol. 2010;21:500–511. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Massie BM, Hirsch AT, Inouye IK, Tubau JF. Calcium channel blockers as antihypertensive agents. Am J Med. 1984;77:135–142. doi: 10.1016/s0002-9343(84)80049-2. [DOI] [PubMed] [Google Scholar]
- Massie BM, Tubau JF, Szlachcic J. Comparative studies of calcium-channel blockers and beta-blockers in essential hypertension: clinical implications. Circulation. 1987;75:V163–169. [PubMed] [Google Scholar]
- Mu TW, Fowler DM, Kelly JW. Partial restoration of mutant enzyme homeostasis in three distinct lysosomal storage disease cell lines by altering calcium homeostasis. PLoS Biol. 2008;6:e26. doi: 10.1371/journal.pbio.0060026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DS, Kelly JW. Chemical and/or biological therapeutic strategies to ameliorate protein misfolding diseases. Curr Opin Cell Biol. 2011;23:231–238. doi: 10.1016/j.ceb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DS, Mu TW, Palmer AE, Kelly JW. Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. Nat Chem Biol. 2010;6:424–432. doi: 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DS, Wang YJ, Tan YL, Yates JR, 3rd, Mu TW, Kelly JW. FKBP10 depletion enhances glucocerebrosidase proteostasis in Gaucher disease fibroblasts. Chem Biol. 2013;20:403–415. doi: 10.1016/j.chembiol.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Parenti G, Pignata C, Vajro P, Salerno M. New strategies for the treatment of lysosomal storage diseases (review) Int J Mol Med. 2013;31:11–20. doi: 10.3892/ijmm.2012.1187. [DOI] [PubMed] [Google Scholar]
- Pelled D, Trajkovic-Bodennec S, Lloyd-Evans E, Sidransky E, Schiffmann R, Futerman AH. Enhanced calcium release in the acute neuronopathic form of Gaucher disease. Neurobiol Dis. 2005;18:83–88. doi: 10.1016/j.nbd.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Reczek D, Schwake M, Schroder J, Hughes H, Blanz J, Jin X, Brondyk W, Van Patten S, Edmunds T, Saftig P. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Ron I, Horowitz M. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum Mol Genet. 2005;14:2387–2398. doi: 10.1093/hmg/ddi240. [DOI] [PubMed] [Google Scholar]
- Sawkar AR, Schmitz M, Zimmer KP, Reczek D, Edmunds T, Balch WE, Kelly JW. Chemical chaperones and permissive temperatures alter localization of Gaucher disease associated glucocerebrosidase variants. ACS Chem Biol. 2006;1:235–251. doi: 10.1021/cb600187q. [DOI] [PubMed] [Google Scholar]
- Schmitz M, Alfalah M, Aerts JM, Naim HY, Zimmer KP. Impaired trafficking of mutants of lysosomal glucocerebrosidase in Gaucher’s disease. Int J Biochem Cell Biol. 2005;37:2310–2320. doi: 10.1016/j.biocel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Schueler UH, Kolter T, Kaneski CR, Zirzow GC, Sandhoff K, Brady RO. Correlation between enzyme activity and substrate storage in a cell culture model system for Gaucher disease. J Inherit Metab Dis. 2004;27:649–658. doi: 10.1023/b:boli.0000042959.44318.7c. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, Pressley TA, Higham S, Kraus-Friedmann N. Characterization of high-affinity ryanodine-binding sites of rat liver endoplasmic reticulum. Differences between liver and skeletal muscle. Biochem J. 1991;276 (Pt 1):41–46. doi: 10.1042/bj2760041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liou B, Quinn B, Ran H, Xu YH, Grabowski GA. In vivo and ex vivo evaluation of L-type calcium channel blockers on acid beta-glucosidase in Gaucher disease mouse models. PLoS One. 2009;4:e7320. doi: 10.1371/journal.pone.0007320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liou B, Ran H, Skelton MR, Williams MT, Vorhees CV, Kitatani K, Hannun YA, Witte DP, Xu YH, et al. Neuronopathic Gaucher disease in the mouse: viable combined selective saposin C deficiency and mutant glucocerebrosidase (V394L) mice with glucosylsphingosine and glucosylceramide accumulation and progressive neurological deficits. Hum Mol Genet. 2010;19:1088–1097. doi: 10.1093/hmg/ddp580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji S, Martin BM, Barranger JA, Stubblefield BK, LaMarca ME, Ginns EI. Genetic heterogeneity in type 1 Gaucher disease: multiple genotypes in Ashkenazic and non-Ashkenazic individuals. Proc Natl Acad Sci U S A. 1988;85:2349–2352. doi: 10.1073/pnas.85.7.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitner EB, Platt FM, Futerman AH. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Agnello G, Sotolongo N, Segatori L. Ca2+ homeostasis modulation enhances the amenability of L444P glucosylcerebrosidase to proteostasis regulation in patient-derived fibroblasts. ACS Chem Biol. 2011;6:158–168. doi: 10.1021/cb100321m. [DOI] [PubMed] [Google Scholar]
- Wang F, Chou A, Segatori L. Lacidipine remodels protein folding and Ca 2+ homeostasis in Gaucher’s disease fibroblasts: a mechanism to rescue mutant glucocerebrosidase. Chem Biol. 2011;18:766–776. doi: 10.1016/j.chembiol.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Xu YH, Quinn B, Witte D, Grabowski GA. Viable mouse models of acid beta-glucosidase deficiency: the defect in Gaucher disease. Am J Pathol. 2003;163:2093–2101. doi: 10.1016/s0002-9440(10)63566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yu M, Haslam RH, Haslam DB. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J Biol Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- Zhao H, Grabowski GA. Gaucher disease: Perspectives on a prototype lysosomal disease. Cell Mol Life Sci. 2002;59:694–707. doi: 10.1007/s00018-002-8458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1. Western blot analysis of immunopurified GCase from WT and G202R patient-derived fibroblasts. ERdj3 and calnexin co-precipitated with WT and G202R GCase. Control experiments were performed with anti-FLAG M2 antibody. IP, immunoprecipitation; IB, immunoblot.
Figure S2. Related to Figure 2. Knockdown of ERdj3 does not significantly alter cellular viability and GBA transcription, nor activate the UPR in fibroblasts. (A) and (C) Quantitative RT-PCR analysis showed that the mRNA level of ERdj3 was significantly reduced in L444P (A) and WT (C) fibroblasts. The mRNA levels of GBA and UPR target genes, CHOP, XBP1(s) and GRP78, were not significantly altered in L444P (A) and WT (C) fibroblasts 4 d post-transfection. RiboP served as an internal control. (B) Resazurin reduction assay showed that the cellular metabolic activity of L444P GCase fibroblasts did not change upon ERdj3 siRNA treatment as compared to the NT siRNA control. The data are reported as mean ±SD (n = 3).
Figure S3. Related to Figure 2. Knockdown of ERdj3 does not alter WTGC ase proteostasis. (A) Silencing ERdj3 did not change the endo H-resistant glycoform of WT GCase in fibroblasts. Quantification of endo H-resistant WT GCase bands is shown below. (B) Silencing ERdj3 did not change WT GCase activity in fibroblasts. The data in (A) and (B) are reported as mean ± SD (n = 3 for A, n = 6 for B).
Figure S4. Related to Figure 3. HYOU1 is not involved in the ERdj3-mediated degradation of L444P GCase. Silencing HYOU1 did not change L444P GCase activity (A) and its endo H-resistant glycoform (B) in fibroblasts. Co-knockdown of HYOU1 and ERdj3 increased L444P GCase activity (A) and its endo H-resistant glycoform (B) to the same extent as knockdown of ERdj3 alone. The data in (A) are reported as mean ±SD (n = 6).
Figure S5. Related to Figure 4. Co -application of ERdj3 siRNA and diltiazem is a viable strategy to rescue L444P GCase proteostasis. (A) L444P GCase activity increased linearly as a function of time when ERdj3 was silenced. L444P GCase fibroblasts were transfected with the same amount of NT and ERdj3 siRNA and analyzed 4 to 7 d post-transfection. (B) Co-application of diltiazem and ERdj3 siRNA significantly increased the endo H-resistant glycoform of L444P GCase in fibroblasts. (C) Quantitative RT-PCR analysis showed that the mRNA levels of GBA and UPR target genes, CHOP, XBP1(s) and GRP78, were not significantly altered in L444P GCase fibroblasts 7 d post-treatment. RiboP served as an internal control. (D) Resazurin reduction assay showed that the cellular metabolic activity of L444P GCase fibroblasts was not significantly altered 7 d post-treatment. The data in (C) and (D) are reported as mean ±SD (n = 3). DTZ, diltiazem.
Table S1. List of proteins that interact with endogenous WT GCase in HeLa cells as identified by SILAC-immunoprecipitation-MudPIT. See separate Excel file. Related to Table 1.
Table S2. Primer sequences for quantitative RT-PCR. Related to Experimental Procedures.