Significance
Calorie restriction (CR) has been shown to extend the lifespans of various organisms. Consequently, a considerable amount of research has been performed to elucidate its mechanisms, especially in the yeast Saccharomyces cerevisiae. Here, we show that due to small sample sizes, large variation exists between measurements. In addition, the effect of CR on lifespan has been routinely overestimated in yeast due to the use of short-lived experimental controls, which together may explain why contradictory mechanisms were found to mediate CR-induced lifespan extension. Moreover, we did not observe any lifespan-enhancing effect of CR using an alternative measurement technique. The inability of CR to robustly extend lifespan suggests that calories alone do not modulate the lifespan of this important model organism.
Abstract
Calorie restriction (CR) is often described as the most robust manner to extend lifespan in a large variety of organisms. Hence, considerable research effort is directed toward understanding the mechanisms underlying CR, especially in the yeast Saccharomyces cerevisiae. However, the effect of CR on lifespan has never been systematically reviewed in this organism. Here, we performed a meta-analysis of replicative lifespan (RLS) data published in more than 40 different papers. Our analysis revealed that there is significant variation in the reported RLS data, which appears to be mainly due to the low number of cells analyzed per experiment. Furthermore, we found that the RLS measured at 2% (wt/vol) glucose in CR experiments is partly biased toward shorter lifespans compared with identical lifespan measurements from other studies. Excluding the 2% (wt/vol) glucose experiments from CR experiments, we determined that the average RLS of the yeast strains BY4741 and BY4742 is 25.9 buds at 2% (wt/vol) glucose and 30.2 buds under CR conditions. RLS measurements with a microfluidic dissection platform produced identical RLS data at 2% (wt/vol) glucose. However, CR conditions did not induce lifespan extension. As we excluded obvious methodological differences, such as temperature and medium, as causes, we conclude that subtle method-specific factors are crucial to induce lifespan extension under CR conditions in S. cerevisiae.
In the last decades, a nutritious diet low in calories, commonly referred to as calorie restriction (CR), has been reported to extend the lifespan of a broad range of organisms, such as yeast, worms, fruit flies, mice, rats, and monkeys (1–6). This seemingly evolutionary conserved effect of CR on aging has sparked intense investigation into its underlying molecular mechanisms, especially in the budding yeast Saccharomyces cerevisiae (7–10). Some studies suggest that nutrient-responsive pathways, such as target of rapamycin (TOR), protein kinase A (PKA), and Sch9 (11), mediate CR-induced lifespan extension, whereas others hypothesize that CR increases the activity of sirtuin deacetylases (e.g., SIR2) extending lifespan by suppressing ribosomal DNA recombination (9, 12, 13).
In S. cerevisiae, lifespan can either be defined by the time cells remain viable in absence of nutrients, called chronological lifespan, or by the number of daughter cells a yeast cell produces during its life, which is referred to as replicative lifespan (RLS) (14). Although RLS can vary greatly between individual yeast cells, the average RLS (i.e., across a population of cells) is assumed to be a genotypic trait (15–19). However, besides CR, other environmental conditions, such as pH and carbon source, are known to have a strong impact on RLS (20, 21).
Here, through a systematic analysis of RLS data of S. cerevisiae using data from more than 27,000 cells, we obtained indication that small sample sizes might be the cause for the often observed significant variation between RLS data. Further, we identified that partly biased data exists. After removing such biased data, we determined the literature-wide average RLS of BY4741 and BY4742 at 2% (wt/vol) glucose and under CR conditions. As small sample sizes are inherent to the laborious microdissection technique most commonly used to measure RLS, we performed additional aging experiments with a recently developed microfluidic dissection platform that allows acquisition of data for large numbers of cells in a fully automated manner (22). Although identical RLSs were measured at 2% (wt/vol) glucose with the microfluidic dissection platform as with the classical microdissection method, CR failed to extend lifespan in the microfluidic dissection platform. After excluding factors, such as temperature and medium composition, we conclude that, in yeast, lifespan extension under CR conditions requires subtle method-specific factors.
Results
RLS Data Harbor Significant Variation.
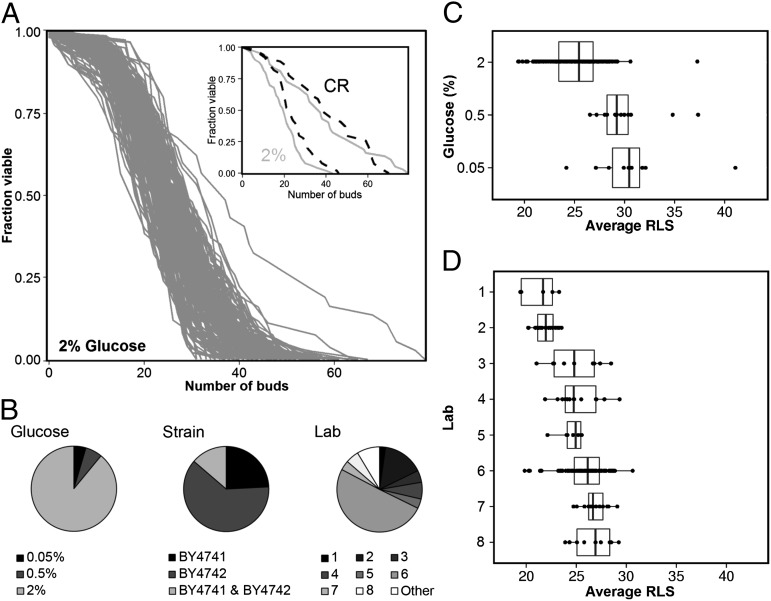
We gathered RLS data for the yeast strains BY4741 and BY4742 because of the considerable amount of lifespan data that have been published for these relatively long-lived haploid yeast strains. In total, 220 lifespan measurements from 41 papers were collected (Dataset S1), with an average RLS between 19.4 and 41.1 buds (Fig. 1A). Because the lifespan measurements were performed in various laboratories using different glucose percentages and strains with different mating types (Fig. 1B), we determined how these variables individually affected RLS using a linear mixed model (23) (for details about the model, see SI Text and Tables S1 and S2).
Fig. 1.
Replicative lifespan measurements harbor significant variation. (A) Overview of all lifespan curves at 2% (wt/vol) glucose gathered from 41 different papers for BY4741 and BY4742. (Inset) The RLS curve with the lowest and highest reported RLS for 2% (wt/vol) glucose (gray lines) and CR conditions (dashed lines). (B) Subdivision of the gathered lifespan data over variables, such as glucose percentage [2%, 0.5%, and 0.05% (wt/vol) glucose], strain (BY4741, BY4742, or an aggregate of BY4741 and BY4742), and laboratory (see Dataset S1 for details about laboratory). (C and D) Overview of the RLS published for different glucose percentages (C) and individual laboratories [D, only 2% (wt/vol) glucose]. Median RLS is indicated with a line inside the box plot. The box represents the first and third quartiles and the whiskers 1.5 times the interquartile range. Each RLS measurement is indicated by a dot.
The results of the linear mixed model showed that a reduction in glucose percentage from 2% (wt/vol) to 0.5% or 0.05% resulted in an increase in RLS (Fig. 1C; linear mixed model: P < 0.001). However, the RLS determined at 0.5% and 0.05% glucose was identical (linear mixed model: P = 0.476), contradictory to earlier studies (6, 24). Although there was no effect of mating type (BY4741 or BY4742) on lifespan (linear mixed model: P = 0.1; Fig. S1), we found that the variable “laboratory” had a strong impact on RLS (Fig. 1D; e.g., laboratory 1 vs. laboratory 8, t test: P < 0.001), suggesting that ambiguous differences between lifespan measurements (e.g., incubation times and preculture methods) may have a strong impact on RLS.
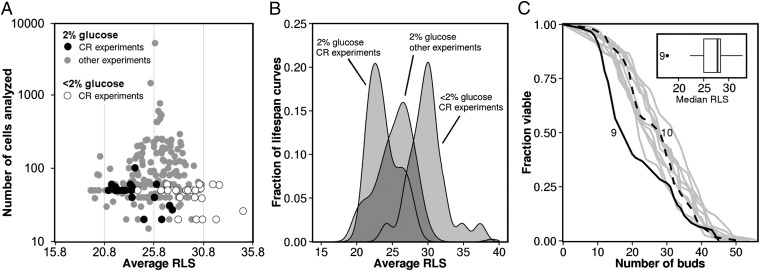
We observed that variation in RLS was not only present between laboratories, but to a varying degree also within laboratories (Fig. S2). We hypothesized that the number of cells analyzed per experiment might have caused this variation. To understand the effect of sample size on experimental variation, we used a modified funnel plot, in which sample size is plotted against the measured RLS (Fig. 2A). This plot shows an inverse relationship between sample size and the average RLS determined from all literature data. Measurements with a small sample size are scattered over a broad range of lifespans at the bottom of the graph, whereas measurements with a larger sample size converge toward the average RLS of all lifespan curves. Apparently, high numbers of cells are necessary to reliably estimate RLS. In fact, we determined using sample size estimation (25) that in order to reliably measure the effect of CR on replicative lifespan at least 100 cells need to be analysed per lifespan curve. However, in most studies, only a limited number of cells are analyzed per experiment (median of 59 cells per experiment), which means that most studies are too small to reliably determine the effect of environmental and genetic perturbations on RLS.
Fig. 2.
The average RLS at 2% (wt/vol) glucose in CR studies are biased toward shorter RLS. (A) Plot showing the measured RLS vs. the number of cells analyzed. Three outliers, which had a RLS of more than 10 buds longer than the average RLS determined for all 2% (wt/vol) glucose lifespan curves gathered in this study (25.8 ± 0.1 buds), were removed from the plot. As the sample size of the experiment increases, the reported RLS deviates less from the average RLS. (B) Density plot comparing the RLS measured under CR conditions (30.2 ± 0.3 buds) with the RLS measured at 2% (wt/vol) glucose in the accompanying CR control experiments (23.4 ± 0.3 buds) and in all other control experiments (25.9 ± 0.1 buds). (C) Overview of lifespan curves for BY4741 from Kruegel et al. (26). The lifespan curve obtained under CR conditions (curve 10, dashed black line) overlaps with most of the lifespan curves measured at 2% (wt/vol) glucose (gray lines). The CR control experiment at 2% (wt/vol) glucose (curve 9, black line) is exceptionally short-lived relative to all of the other lifespan curves. (Inset) Box-plot with the distribution of the median RLS reported for WT BY4741 in Kruegel et al. (26). Curve 9 is the single outlier.
RLS Measured at 2% Glucose Is Shorter in CR Studies Compared with Other Studies.
During our data analysis, we further noticed that the RLS reported at 2% (wt/vol) glucose in CR experiments was generally lower compared with those in other experiments in which the effect of CR on RLS was not measured (23.4 ± 0.3 vs. 25.9 ± 0.1 buds; P < 0.001; Fig. 2B; for overview of median RLS, see Dataset S1). Even within papers that studied CR, the average RLS at 2% (wt/vol) glucose was lower for controls of CR experiments than for other controls, which were not associated with CR experiments (23.4 ± 0.3 vs. 25.8 ± 0.1 buds; P < 0.001; Fig. S3). To illustrate this point, we focused on a study by Kruegel et al. (26) that reports an average RLS of 21.5 buds for 2% (wt/vol) glucose (Fig. 2C, curve 9) and 27.1 buds under CR conditions (Fig. 2C, curve 10). Although this comparison shows a substantial increase in lifespan under CR conditions (i.e., from 21.5 to 27.1 buds, P = 0.036), we discovered that the 2% (wt/vol) glucose control curve (curve 9) linked to the CR experiment (curve 10) had a significantly shorter RLS than all of the other 10 lifespan curves for 2% (wt/vol) glucose reported in the same paper (Fig. 2C, Inset). Moreover, we found that the RLS under CR conditions (curve 10) is not significantly different from the majority of the RLS measured at 2% (wt/vol) glucose (Fig. S4). Thus, the significant lifespan extension reported under CR conditions in this specific paper is solely caused by an unusually short-lived control experiment.
Because the average RLS reported for 2% (wt/vol) glucose controls in CR experiments was on average significantly shorter than the ones of other 2% (wt/vol) glucose measurements (Fig. 2B), we removed all 2% (wt/vol) glucose data originating from CR control curves from our dataset (22 of 220 lifespan curves). On the basis of the remaining literature data, we then determined that the average RLS of BY4741 and BY4742 is 25.9 ± 0.1 buds under 2% (wt/vol) glucose conditions and that CR conditions extend the average RLS by 18% to 30.2 ± 0.3 buds (P < 0.001). When we performed microdissection experiments ourselves, we measured RLS for BY4741 that were almost identical to those determined from the consolidated literature data [25.5 ± 0.9 buds at 2% (wt/vol) glucose (n = 102 cells) vs. 30.4 ± 1.0 buds at 0.5% glucose (n = 102 cells); P < 0.001; Fig. S5].
CR Does Not Extend Replicative Lifespan in a Microfluidic Dissection Platform.
As the classical microdissection method is a labor intensive technique to determine RLS, especially when larger sample sizes are required, we questioned whether the recently developed microfluidic dissection methods (22, 27, 28) could generate similar lifespan data. The advantage of using a microfluidic dissection method is that RLS can be measured in a semiautomated manner with a large number of cells under constant environmental conditions, i.e., without incubation at low temperature during the night and with constant glucose concentration.
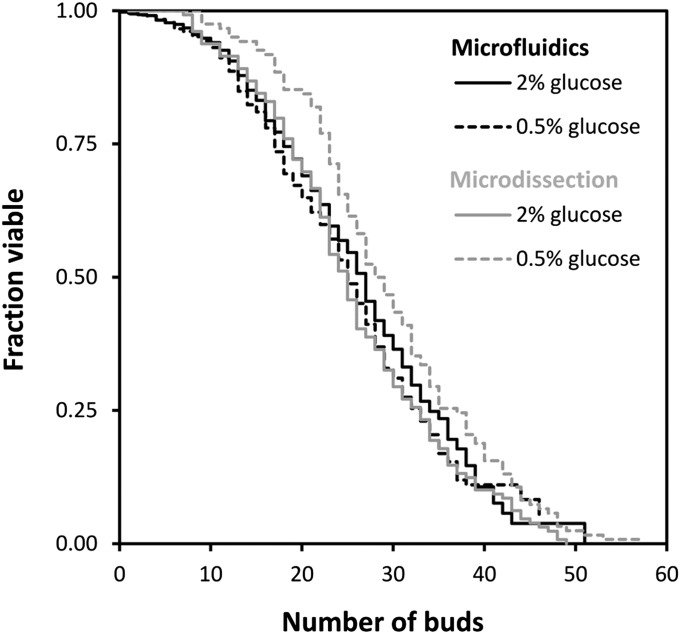
We performed a series of 10 independent RLS measurements at 2% and 0.5% (wt/vol) glucose with the microfluidic dissection platform developed by Lee et al. (22). Although at 2% (wt/vol) glucose the RLS measured with the microfluidic dissection platform was similar to the average RLS that we determined from the literature (26.5 ± 0.5 vs. 25.9 ± 0.1 buds; P = 0.8), we could not find any lifespan extending effect of CR with the microfluidic dissection platform. Instead, the RLS at 0.5% glucose was identical to the RLS measured at 2% (wt/vol) glucose (25.9 ± 0.7 vs. 26.5 ± 0.5 buds; Fig. 3; P = 0.11). Also for the longer-lived prototrophic yeast strain YSBN6 (29), we could not observe any CR-induced lifespan extension with the microfluidic dissection platform (35.3 ± 0.9 vs. 35.3 ± 0.8 buds; P = 0.42; Fig. S6).
Fig. 3.
CR does not elicit a robust extension of replicative lifespan. With the microfluidic dissection platform, we measured an average RLS of 26.5 ± 0.5 buds (n = 2,806 cells of which 2,409 were washed out before death) at 2% (wt/vol) glucose and 25.9 ± 0.7 buds at 0.5% glucose (n = 2,055 cells of which 1,716 were washed out before death). Here, CR does not extend RLS. However, with the classical dissection method RLS is extended from 25.8 ± 0.9 buds at 2% (wt/vol) glucose (n = 122 cells) to 29.6 ± 0.9 buds at 0.5% glucose (n = 129 cells) The classical dissection and microfluidic experiments were both performed with YNB medium and at a constant incubation temperature of 30 °C.
We hypothesized that a reduction in the glucose concentration of the medium may be sensed differently by the cells inside the microfluidics dissection platform compared with those on an agar plate. For instance, in the microfluidic setup, cells are completely surrounded by a flow of medium, while on an agar plate, a diffusion limitation could exist, which could generate a lower effective glucose concentration in CR experiments, leading to lifespan extension. We therefore performed additional RLS measurements with the microfluidic dissection platform with lower glucose concentrations, i.e., 0.25%, 0.1%, and 0.05% glucose. However, at these lower glucose concentrations, we obtained RLS values similar to those obtained at 2% (wt/vol) glucose [i.e., for 0.25% glucose, 25.5 ± 0.7; for 0.1% glucose, 27.9 ± 1.0; and for 0.05% glucose, 24.6 ± 0.8 vs. 26.5 ± 0.5 buds for 2% (wt/vol) glucose; Fig. S7] and therefore no significant effect of CR on RLS.
CR-Induced Lifespan Extension Depends on Method-Specific Factors.
As the experimental conditions with the microfluidic dissection platform and the classical microdissection technique are slightly different, e.g., in the microfluidic dissection platform temperature cycles are absent and synthetic complete (SC) medium is used instead of yeast peptone dextrose (YPD) medium, we wondered whether these factors played a role in eliciting a CR-induced lifespan extension. We therefore performed microdissection experiments with BY4741 on SC medium. Further, to avoid temperature cycles, we dissected cells continuously inside a 30 °C room. Under these conditions, we obtained an RLS of 25.8 ± 0.9 buds at 2% (wt/vol) glucose (n = 129 cells) and 29.6 ± 0.9 buds at 0.5% glucose (n = 122 cells), which both correspond well with the average RLS determined from our meta-analysis [for 2% (wt/vol) glucose, 25.8 ± 0.9 buds vs. 25.9 ± 0.1 buds; P = 0.9; for 0.5% glucose, 29.6 ± 0.9 buds vs. 30.3 ± 0.4 buds; P = 0.4; Fig. 3]. Apparently, factors, such as the use of SC or YPD medium and the absence or presence of periods of lower temperature (4 °C), have no significant effect on the CR-induced extension of RLS. We therefore conclude that subtle method-specific factors, e.g., growth on a solid surface or handling with a needle, are crucial to elicit a CR-induced RLS extension.
Discussion
Although numerous studies reported lifespan extension in S. cerevisiae in response to CR, our meta-analysis uncovered several problems with the currently available RLS data, such as differences in the measured RLS between laboratories, short-lived 2% glucose CR controls, and small sample sizes. The significant variation that we observed between RLS measurements could explain several contradictions that exist about CR in literature, such as whether the histone deacetylase SIR2 (9, 30, 31) or the ability to respire are required for CR-induced lifespan extension (32, 33), whether CR is able to extend RLS in the yeast strain W303 (9, 13, 30), and whether the optimal glucose concentration for CR is at 0.5% or 0.05% glucose (24, 32). In fact, in our dataset, the RLS was identical in the presence of 0.05% and 0.5% glucose.
Based on our literature analysis of RLS data, we determined that the average RLS of the yeast strains BY4741 and BY4742 on 2% (wt/vol) glucose is 25.9 buds and 30.2 buds under CR conditions (0.5% and 0.05% glucose YPD medium). These average RLS values could serve as reference values in future yeast aging studies. In addition, we recommend that lifespan curves are generated with at least 100 cells to obtain more reliable RLS measurements. In this study, we obtained identical lifespans for BY4741 at 2% (wt/vol) glucose with the microfluidics dissection platform as with the classical dissection technique (26.5 ± 0.5 vs. 25.8 ± 0.9 buds; P = 0.3). However, we were unable to observe any CR-induced lifespan extension using the microfluidic dissection platform. This discrepancy between the two methods cannot be explained by differences in medium, the absence of temperature cycles or by a general inability of the microfluidic dissection platform to generate lifespan extension, because we previously demonstrated that the microfluidic platform generates identical RLS data for several longevity mutants as the classical dissection method [e.g., median RLS of 32 vs. 34 buds for fob1Δ and 13 vs. 13 buds for sir2Δ (22)]. Thus, apparently, CR-induced lifespan extension also depends on subtle, yet unknown, differences between the methods used to determine RLS.
In other organisms, methodology was also shown to be of key importance on the effect that CR has on lifespan. For example, although an initial study suggested that Rhesus monkeys live longer under CR conditions (3), a recent study found no evidence for CR-induced lifespan extension (34). The key difference between these studies appears to be the exact composition of the diet of the monkeys (34). Also in Drosophila (35), the effect of calorie restriction was not merely dependent on the amount of calories in the diet, but on the ratio of yeast and sugar in the diet. Overall, these recent developments suggest that lifespan extension by CR is not as robust as once believed.
Materials and Methods
Data Acquisition from the Literature.
Replicative lifespan curves from the literature were digitized using the GetData Graph Digitizer 2.25. In case there was no information provided about the number of cells used to generate the RLS curve, we inferred the cell number from the smallest step decrease in the fraction of viable cells. Dataset S1 provides a comprehensive overview of the gathered data.
Statistical Analysis.
Meta-analysis of lifespan data were performed using a linear mixed model (23). Survival curves were compared with each other using the log-rank test (36). For each condition the average replicative lifespan was given with the SE. Median RLS for all conditions can be found in Dataset S1. An overview of all lifespan data acquired in this study can be found in Dataset S1.
Strains, Media, and Cultivation.
Lifespan measurements were performed with BY4741 and YSBN6 [MATa FY3 ho::HphMX4 (29)], which is derived from S288c. YPD plates were prepared by adding 20 g/L agar to complete rich medium (50 g/L YPD; Formedium). Minimal medium [6.9 g/L yeast nitrogen base (YNB); Formedium] supplemented with filter-sterilized glucose (d-glucose monohydrate; Sigma) was used for liquid cultures. For BY4741, SC medium was used, which is minimal medium to which complete supplemental mixture was added (790 mg/L). A glucose concentration of 20 g/L [2% (wt/vol) glucose] was used for control experiments and 5 g/L [0.5% (wt/vol) glucose], 2.5 g/L [0.25% (wt/vol) glucose], 1 g/L [0.1% (wt/vol) glucose], and 0.5 g/L [0.05% (wt/vol) glucose] for calorie restriction experiments. One or 2 d before the experiment, cells were taken from the −80 °C freezer, streaked on YPD plates, and incubated overnight at 30 °C. A single colony was used to inoculate a 100-mL flask containing 10 mL SC medium with 2% (wt/vol) glucose. The flask was then incubated overnight at 30 °C with shaking at 300 rpm (Climo-Shaker ISF1-x, Kühner AG, Switzerland). The next morning, the culture was diluted (∼30 µL) into a new 100-mL flask containing 10 mL medium with 2% or 0.5% (wt/vol), 0.25% (wt/vol), 0.1% (wt/vol), or 0.05% (wt/vol) glucose to yield an OD600nm of 0.3 after roughly 8 h. The culture was then diluted again and incubated overnight to yield an OD600nm of about 0.3 the next morning at the start of the aging experiment. This preculture procedure ensured that the cells were growing exponentially at the start of the RLS measurements. For 0.25% or 0.05% (wt/vol) glucose, the cultures were grown to an OD600nm of 0.05–0.1 to ensure that the cells were still growing exponentially at the start of the aging experiment.
RLS Measurements with the Microfluidic Dissection Platform.
RLS was determined using a microfluidic dissection platform (22, 37) and an inverted fluorescence microscope (Eclipse Ti-E; Nikon Instruments). The microscope was placed in an incubator (Life Imaging Services) for cultivation at 30 °C. Brightfield images were taken every 10 min for ∼100–120 h using a UV blocking filter and 60× objective (CFI Plan Apo; Nikon; NA = 1.4; working distance = 0.13 mm). Replicative lifespan was determined for each cell by counting the number of buds produced during its entire life. Experimental data obtained from cells that were washed out before death were incorporated into the lifespan curve as right-censored data using Kaplan-Meier analysis (38).
RLS Measurements with the Microdissection Technique.
At the start of each experiment, cells were taken from a frozen −80 °C stock and streaked on a fresh YPD plate containing 2% (wt/vol) glucose. Cells were then grown for 2 d at 30 °C before being restreaked on a fresh YPD plate containing either 2% or 0.5% (wt/vol) glucose. The following day, the cells were streaked again on fresh YPD or SC plates containing either 2% or 0.5% (wt/vol) glucose and allowed to grow for another 12 h before being restreaked very thinly on a fresh plate. The cells were then incubated for 12 h at 30 °C. The very faint colonies present on the plate were then used to create small patches on each microdissection plate. In general, only one patch of cells was made per microdissection plate. The microdissection plates were then incubated for another 2–3 h before the start of the experiment.
Plates intended to be used for microdissection contained 50 mL of medium plus agar and were allowed to dry for 2 d on the laboratory bench to remove excess water before the start of the experiment. During the experiment, each individual microdissection plate was kept inside a plastic bag with a wet towel to prevent desiccation. All other plates used for initial plating before the microdissection experiment were parafilmed to prevent dessication. A SporePlay dissection microscope (Singer Instruments) equipped with a 50-μm–diameter dissection needle (Singer Instruments) was used to perform the microdissection experiments. Small groups of cells, which produced one to two buds in the 2- to 3-h incubation time before the start of the experiment, were gathered and rapidly moved to a fresh position on the agar plate for virgin daughter cell selection. It appeared crucial for overall cell viability to keep the amount of time that the cells spend in the microdissection needle to a bare minimum. Replicative lifespan measurements were then performed as described by Steffen et al. (39) with the exception that the fridge incubation steps were omitted, and cells were dissected 24 h/d in a 30 °C room during the entire lifespan measurement (roughly 5 d).
Supplementary Material
Acknowledgments
We thank Michael Chang and his laboratory for helpful suggestions and generous sharing of their laboratory and equipment with us, which allowed us to perform the microdissection experiments described in this paper. In addition, we thank Ida van der Klei for allowing us to borrow a microdissection microscope, Nate Thayer and Annina Denoth for advice on how to perform microdissection experiments, Jasper Paauwe and Bas Stringer for assistance with the analysis of the microfluidics aging movies, Georges Janssens and Jakub Radzikowksi for discussions and helpful comments on the manuscript, and Marcus de Goffau for advice about statistical analyses. This study was funded by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek-funded Systems Biology Center of Energy Metabolism and Aging.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410024111/-/DCSupplemental.
References
- 1.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95(22):13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325(5937):201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition. 1989;5(3):155–171, discussion 172. [PubMed] [Google Scholar]
- 5.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289(5487):2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 6.Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14(14):2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- 7.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447(7144):550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 8.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamming DW, et al. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309(5742):1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 10.Lin SJ, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418(6895):344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 11.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 12.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101(45):15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5(10):e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: The means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinclair D, Mills K, Guarente L. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol. 1998;52:533–560. doi: 10.1146/annurev.micro.52.1.533. [DOI] [PubMed] [Google Scholar]
- 16.Defossez PA, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3(4):447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 17.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13(19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005;126(4):491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Schleit J, et al. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12(6):1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botta G, Turn CS, Quintyne NJ, Kirchman PA. Increased iron supplied through Fet3p results in replicative life span extension of Saccharomyces cerevisiae under conditions requiring respiratory metabolism. Exp Gerontol. 2011;46(10):827–832. doi: 10.1016/j.exger.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami C, et al. pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast. Cell Cycle. 2012;11(16):3087–3096. doi: 10.4161/cc.21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SS, Avalos Vizcarra I, Huberts DH, Lee LP, Heinemann M. Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc Natl Acad Sci USA. 2012;109(13):4916–4920. doi: 10.1073/pnas.1113505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. J Am Stat Assoc. 1993;88(421):9–25. [Google Scholar]
- 24.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2(9):E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dell RB, Holleran S, Ramakrishnan R. Sample size determination. ILAR J. 2002;43(4):207–213. doi: 10.1093/ilar.43.4.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruegel U, et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7(9):e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehrmann S, et al. Aging yeast cells undergo a sharp entry into senescence unrelated to the loss of mitochondrial membrane potential. Cell Reports. 2013;5(6):1589–1599. doi: 10.1016/j.celrep.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS ONE. 2012;7(11):e48275. doi: 10.1371/journal.pone.0048275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kümmel A, et al. Differential glucose repression in common yeast strains in response to HXK2 deletion. FEMS Yeast Res. 2010;10(3):322–332. doi: 10.1111/j.1567-1364.2010.00609.x. [DOI] [PubMed] [Google Scholar]
- 30.Kaeberlein M, et al. Comment on “HST2 mediates SIR2-independent life-span extension by calorie restriction”. Science. 2006;312(5778):1312–, author reply 1312. doi: 10.1126/science.1124608. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: A skeptical perspective. Ageing Res Rev. 2007;6(2):128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Kaeberlein M, et al. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1(5):e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin SJ, Guarente L. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2006;2(3):e33–, author reply e34. doi: 10.1371/journal.pgen.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattison JA, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489(7415):318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3(7):e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50(3):163–170. [PubMed] [Google Scholar]
- 37.Huberts DH, et al. Construction and use of a microfluidic dissection platform for long-term imaging of cellular processes in budding yeast. Nat Protoc. 2013;8(6):1019–1027. doi: 10.1038/nprot.2013.060. [DOI] [PubMed] [Google Scholar]
- 38.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- 39.Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009;(28):1209. doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.