Significance
The field of xenon magnetic resonance imaging (MRI) is moving closer to the development of targeted xenon biosensors for in vivo applications. It is motivated by a ca. 108-fold improved sensitivity compared with conventional proton MRI. This has been enabled by significant improvements to hardware (xenon polarizer design) and sensitivity (through the hyperpolarized 129Xe chemical exchange saturation transfer technique). In this paper, we capitalize on these improvements by demonstrating targeted xenon imaging on cells using a modular xenon biosensor. With this method, we can detect target cells with as little as 20 nM of our xenon contrast agent. Imaging of such low levels of cell-specific xenon hosts is unprecedented and reinforces the potential of xenon–cryptophane biosensors for molecular imaging applications.
Keywords: spin hyperpolarization, targeted imaging
Abstract
Magnetic resonance imaging (MRI) is seriously limited when aiming for visualization of targeted contrast agents. Images are reconstructed from the weak diamagnetic properties of the sample and require an abundant molecule like water as the reporter. Micromolar to millimolar concentrations of conventional contrast agents are needed to generate image contrast, thus excluding many molecular markers as potential targets. To address this limitation, we developed and characterized a functional xenon NMR biosensor that can identify a specific cell surface marker by targeted 129Xe MRI. Cells expressing the cell surface protein CD14 can be spatially distinguished from control cells with incorporation of as little as 20 nM of the xenon MRI readout unit, cryptophane-A. Cryptophane-A serves as a chemical host for hyperpolarized nuclei and facilitates the sensitivity enhancement achieved by xenon MRI. Although this paper describes the application of a CD14-specific biosensor, the construct has been designed in a versatile, modular fashion. This allows for quick and easy adaptation of the biosensor to any cell surface target for which there is a specific antibody. In addition, the modular design facilitates the creation of a multifunctional probe that incorporates readout modules for different detection methods, such as fluorescence, to complement the primary MRI readout. This modular antibody-based approach not only offers a practical technique with which to screen targets, but one which can be readily applied as the xenon MRI field moves closer to molecular imaging applications in vivo.
When referring to xenon magnetic resonance imaging (MRI), the best-known application is the use of the isotope 129Xe for facilitating the direct visualization of the airspaces and function of the lung (1–4). However, in the past few years, the scope for using 129Xe for targeted biosensor imaging has notably increased (5). The motivation to develop xenon-based biosensors is twofold. Xenon can be hyperpolarized, enhancing its signal by up to five orders of magnitude and enabling it to be detected at extremely low concentrations (5). In addition, the exquisite sensitivity of xenon to its local chemical environment, as indicated by well-resolved chemical shift changes, brings with it the potential to distinguish not only localization but also specific functional events (6–10). This sensitivity may also be exploited in the development of different xenon MRI contrast agents which could be used to perform multiplexed molecular imaging (11), especially useful as clusters of biomarkers are often more informative than a single indicator.
The development of xenon-based contrast agents has been largely enabled by the development and application of the Hyper-CEST acquisition method (12). Hyper-CEST is a technique which combines the advantages of hyperpolarized xenon gas (hp-Xe) (13) with the chemical exchange saturation transfer (CEST) detection method (14, 15) to significantly enhance signal to noise. Primarily, this technique facilitates the detection of very low amounts of contrast agent in a sample, and thus improves upon the previous sensitivity limitation of xenon biosensor MRI. An added advantage of CEST is that the contrast can be switched on and off at will to minimize false positive results. This sidesteps the common problem of distinguishing the effects of the contrast agent from the background, as is the case for relaxivity-based MRI contrast agents. For Hyper-CEST MRI, the contrast is generated through the interaction of hp-Xe with different host molecules. These chemical hosts [of which cryptophane-A (CrA) has become a molecule of choice] transiently bind the xenon and, in so doing, generate a unique (and very large, >100 ppm) xenon chemical shift in NMR spectra. By saturating at this unique resonance frequency, CEST spectroscopy experiments can easily and unambiguously detect the presence of CrA in a sample, even at subnanomolar concentrations (16, 17). In this way, xenon and CrA together act as a perfect “molecular reporter pair,” whose interaction can be monitored and spatially resolved via xenon MRI in multiple different chemical environments simultaneously.
In response to these developments, there have been a number of recent studies describing the development of new xenon contrast agents (17–19) and biosensors (10, 20–23) for xenon NMR applications. Critically however, only recently have images of cells been achieved with xenon MRI, with a nontargeted approach using the contrast agent CrA (24). Consequently, questions remain as to whether cell-specific labeling using a targeted CrA biosensor can be detected through xenon MR imaging modalities (in addition to spectroscopy). Also, if so, what are the most suitable cellular targets and what degree of CrA labeling per cell is necessary?
To this aim, we designed an antibody-based 129Xe biosensor using CrA, for evaluating the suitability of cell-surface targets for xenon MRI applications. In this paper, we describe the development of such a biosensor for the detection of cluster of differentiation 14 (CD14)-expressing cells, although, as we will discuss, the biosensors’ modular design allows for the fast and easy adaption of this methodology to any cell surface target for which there is a specific antibody. For this proof of principle study, the key design elements were versatility, biocompatibility, and the ability to quantify the CrA payload on target cells. Our target, cell membrane-associated CD14, is a glycosylphosphatidylinositol-anchored (55 kDa) protein, present on the surface of myeloid cells (25, 26). CD14, in cooperation with Toll-like receptor 4, is a cell surface receptor for bacterial lipopolysaccharide (LPS) (27), meaning that stimulating macrophage cells with LPS in vitro (as we do in this study) and pairing them with unstimulated fibroblasts provides an ideal model system for comparing CD14 expression. As CD14 is a well-characterized macrophage marker, a xenon MRI CD14 biosensor could be used to image pathologies in which macrophage activation is important, as has been demonstrated by proton MRI for atherosclerosis (28) and tumor-associated macrophage populations (29), but with the added benefit of having switchable contrast and higher sensitivity.
Although an excellent candidate for xenon MRI biosensor development, previous biosensors incorporating CrA have indicated some potential limitations. Firstly, CrA is readily taken up by cells in a nonspecific manner (24). Although useful for cell labeling, this inherent hydrophobic characteristic has led to speculation that even when conjugated to a larger targeting unit, CrA may facilitate a high nonspecific binding of the biosensor to cell membranes (21). Having said that, good specificity has been reported in other constructs that carry multiple CrA units (23). Secondly, quantification of a cell-bound CrA biosensor is desirable but challenging using xenon NMR alone, as only a certain percentage of the total CrA is occupied with xenon at any one time during the experiment and this fractional occupancy can change in different microenvironments (30). Related to this fact, as there is limited information on the amount of cell-bound CrA needed for reliable xenon MRI detection, an understanding of the optimal CrA loading as well as a quantification of CrA per cell is valuable for future biosensor development as well as target selection.
Results and Discussion
The goal of this work was to demonstrate a modular, flexible biosensor design that can be used to investigate the suitability of a cellular target for xenon MRI biosensor development. The experimental design is depicted in Fig. 1.
Fig. 1.

Experimental design. The CD14 biosensor is comprised of an avidin-conjugated antibody targeting module and a set of biotin-conjugated readout modules for dual functionality (NMR/fluorescence) to selectively label high CD14-expressing cells. Multiple avidin molecules are conjugated to each targeting module, an anti-CD14 specific antibody, but for schematic simplicity, only one avidin is depicted per antibody. Step 1. The biosensor can be applied in two different ways: sequential incubation (A and B), in which cells are incubated with the targeting module, followed by the readout modules, or via incubation with the complete construct (C), in which the targeting modules and readout modules are preconnected. Step 2. Cells are washed to remove any unbound biosensor and harvested. Step 3. Cellular uptake, biosensor specificity and cellular localization can be evaluated via the fluorescence readout module. Step 4. The cell suspensions are placed into separate compartments within an NMR double phantom. The hp-Xe is bubbled through the samples for xenon MRI measurements. Step 5. Illustrative xenon MRI shows a cross-section of the NMR double phantom. The CEST effect encodes the localization of the CrA readout module in the compartment containing the high-CD14-expressing cells.
The CD14 biosensor is constructed in a two-part process and uses the high-affinity binding partners avidin and biotin to connect two functional modules (Fig. 1). The first module we refer to as the “targeting module,” and is made by conjugating multiple avidin molecules to an anti-CD14 IgG2b monoclonal antibody or to an IgG2b control antibody. Conjugation of avidin to lysine side chains, as achieved by this kit, facilitates the flexible, modular design of this method as opposed to direct conjugation of CrA to accessible side chains of a target protein (21). This module contributes a high degree of specificity of the biosensor to the target RAW 264.7 macrophage cell line, compared with the control 3T3 fibroblast cell line as confirmed by flow cytometry analysis (Fig. S1A). The second module is referred to as the “readout module.” This module can be customized to contain chemical identifiers for either 129Xe NMR or fluorescence detection. These modules are defined as either a single CrA monoacid or a single fluorescein molecule conjugated to a single biotin molecule via a polyethyleneglycol (PEG) chemical spacer (Fig. S2 A and B), and serve as readout modules for 129Xe NMR and fluorescence applications respectively. As each avidin protein can bind four biotin molecules, this modular construction results in a minimum fourfold increase in readout modules to every antibody. However, given the large number of potential avidin conjugation sites per antibody and the molecular ratios used in the avidin conjugation (4:1, avidin:antibody), it is likely there is more than one avidin per antibody and that this amplification factor is higher still. The modular approach also ensures that both targeting and readout components are equally adaptable. This includes the potential to screen different targets efficiently by making a panel of avidin-conjugated antibodies or to build new biotin-conjugated readout modules with alternative fluorophores or CrA moieties, thus reducing the time investment in design and construction of new chemical constructs for each target. Importantly, the antibody-based design should also be amenable to in vivo applications, as we will discuss later.
As one of the main aims of this modular design was to enable versatility, it was important that the biosensor system could be applied in two different ways: either by sequentially adding the targeting module and readout modules to cells in separate incubations or by preconnecting the modules and then adding the complete construct to cells in a single incubation. Sequential incubations of each module facilitate a quick customization of the readout for specific needs, e.g., changing the ratio of CrA−biotin:fluorescein−biotin in the second incubation to favor a higher CrA loading for xenon MRI experiments. This strategy relied on the equal binding affinity of the two readout modules for the avidin-labeled targeting module. To test this, a series of flow cytometry experiments were performed, which confirm that the ratio of the readout modules in the incubation media is retained in the final labeled cells (Fig. S3). This is important, as it enables a quantification of CrA on labeled cells via the relative quantification of fluorescein, as we describe later. In a parallel design strategy, we made a third readout module in which a CrA diacid molecule was linked to both a biotin and a fluorescein molecule via PEG spacers (Fig. S2C). We termed this the “branched” readout module. The goal of this construct was to facilitate a more refined quantification of the CrA on labeled cells, as the CrA:fluorescein ratio is always fixed in this construct in a 1:1 fashion.
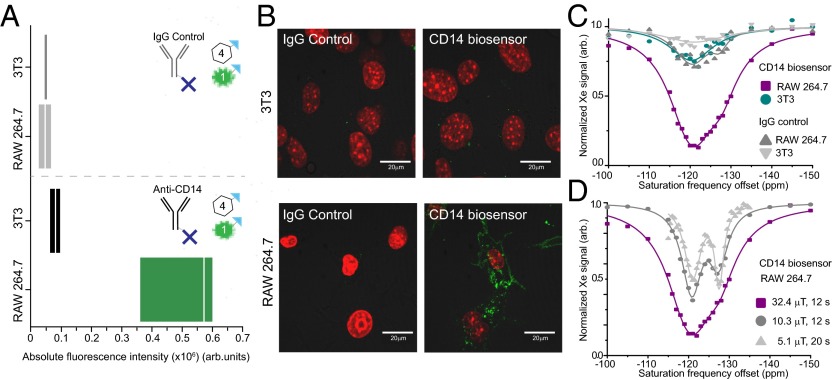
First results using the sequential incubation of the two modules (using a 4:1 CrA:fluorescein readout module ratio) confirms that the CD14 xenon MRI biosensor has good selectivity for high-CD14-expressing cells as determined by both flow cytometry (Fig. 2A) and fluorescence microscopy (Fig. 2B). As predicted for a biosensor that targets a cell membrane protein, the biosensor is predominantly localized at the surface of the target RAW 264.7 cell line (Fig. 2B). Xenon NMR spectroscopy analysis of the two cell lines also clearly demonstrates a significant response at the resonance frequency for CrA (∼120 ppm relative to free xenon in solution), in the target RAW 264.7 cell line incubated with the CD14 biosensor (Fig. 2C). By comparison, there is no significant response in either the control 3T3 fibroblast cells or in either cell line incubated with the control IgG targeting module or the readout module alone (Fig. S1B). Evaluation of the CEST spectra of the RAW 264.7 cells incubated with the CD14 biosensor at lower power (Fig. 2D) clearly indicates the presence of two CrA-associated pools. These resonances correspond to CrA within a lipidic/cellular environment (left peak) and CrA within an aqueous environment (right peak), and are consistent with results from previous cell experiments (21, 24). As the cells are thoroughly washed after the incubations with the biosensor modules, both of these CrA pools can be described as being specifically ‘cell associated’ and thus reflecting the total amount of biosensor that was attached to each cell sample. Further work would have to be done to characterize whether these represent two populations of biosensor in different local chemical environments of the cell (for example, membrane bound versus internalized biosensor). The xenon MRI detection method described in this paper employs direct bubbling of the xenon into the cell suspension (as depicted in Fig. 1). This high through-put method is the most appropriate to quickly screen the suitability of different biosensors, although it compromises cell viability in the cell lines used in this study (Fig. S4). Previous experience demonstrates that the chemical shifts detected by direct bubbling experiments are consistent with those detected in a live cell bioreactor setup (24) and that the impact of bubbling on viability can vary largely between cell lines (supporting information of ref. 23). This means that results from promising candidates in the direct bubbling experiments can be reliably translated to a more complex bioreactor experiment at a later stage.
Fig. 2.
Dual readout modules facilitate evaluation of specificity and localization of the CD14 biosensor. Cells are sequentially incubated with the biosensor modules. First, cells are incubated with the targeting module, anti-CD14 antibody, or control, IgG antibody, both of which have been previously conjugated to avidin molecules (one avidin molecule per antibody depicted for simplicity). Secondly, cells are incubated with the readout modules in a 4:1 ratio of CrA−biotin:fluorescein−biotin. (A) The absolute fluorescence of the cell populations was evaluated by flow cytometry via the fluorescein−biotin readout module and shows the biosensor has good specificity for the target RAW 264.7 cell line. Median indicated by white line; range denotes first and third quartile. (B) Localization of the biosensor bound to cells was detected via fluorescence microscopy of the fluorescein−biotin readout module (green). Cell nuclei stained with Hoechst (red). (C) Xenon CEST spectroscopy (using a saturation pulse of 32.4 μT for 12 s) of cells shows a specific response at the resonance frequency for CrA, confirming the specific binding of the CrA−biotin module to the target RAW 264.7 cell line. (D) Using low-power saturation pulses (of 32.4 μT for 12 s, 10.3 μT for 12 s, and 5.1 μT for 20 s, respectively) we can reveal two peaks that correspond to CrA within a lipidic/cellular environment (left peak) and CrA within an aqueous environment (right peak).
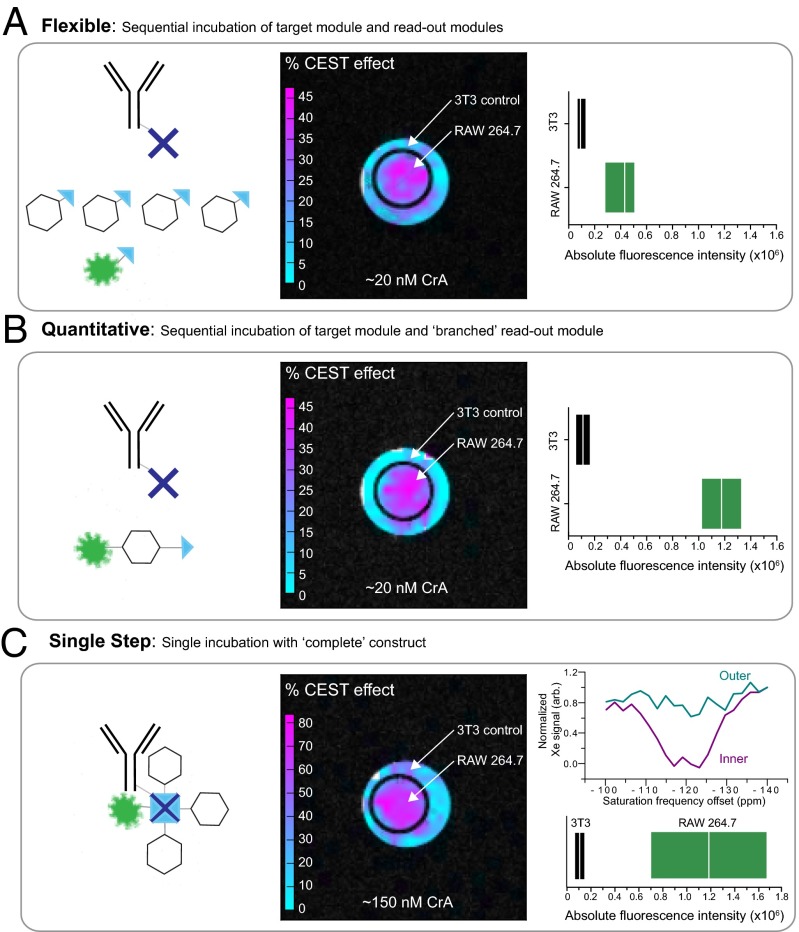
To further qualify the biosensor, we also tested its ability to distinguish the target cell line by xenon MRI. To this end, we incubated cells either sequentially with the individual reporters, as described above, or sequentially with the branched readout module. In addition, we performed a single incubation of cells with the complete biosensor construct (in which the two modules are preconnected beforehand). The combined results are shown in Fig. 3. They confirm that in all application styles, the antibody-based CD14 129Xe NMR biosensor can accurately discriminate between high-CD14-expressing RAW 264.7 macrophages and 3T3 fibroblast cells by both xenon MRI and fluorescence techniques. Flow cytometry also accurately reports the expected fourfold increase in fluorescence when using the branched readout module for the sequential incubations (comparing Fig. 3 A and B) and an improvement in labeling efficiency for the complete construct over the sequential methods. Xenon MRI results correctly localize the target cell line in the inner compartment, as visualized by overlaying the Hyper-CEST xenon MRI image over a proton MRI reference (Fig. 3 A–C, Center). In addition, localized spectroscopy can also be obtained by acquiring an image series over a range of different saturation frequencies and evaluating signal intensity in different areas in that image series (Fig. 3C, Upper Right). The quantification of cell-associated CrA in the inner compartment was determined to be ∼20 nM in the sequential incubations and ∼150 nM in cells incubated with the complete construct (Fig. 3 and Fig. S5). This low level of detection of cell-specific CrA by imaging is unprecedented and reinforces the potential of xenon−cryptophane biosensors for molecular imaging applications.
Fig. 3.
Cell-targeted xenon MRI. (Left) Cells were incubated either sequentially, first with the targeting module followed by the readout modules (either in a 4:1 ratio of CrA−biotin:fluorescein−biotin as in A or with the branched CrA−fluorescein−biotin readout module as in B. Alternatively, cells were incubated with the complete construct in which the targeting and readout modules were preconnected as in C. (Center) Xenon MRI scans indicate localization of the biosensor in the inner compartment containing the target cells, as determined by the high CEST effect. Images display CEST effect (false color) overlaid on proton MRI for reference (see Fig. S6 for details). Xenon CEST images using 30-μT, 26-s saturation pulse and 32 averages (A and B) or 10 averages (C) for each on- and off-resonance image. The calculated CrA concentration associated with cells in the inner compartment is shown at the bottom. A and B show similar maximum CEST responses, with C showing a much greater CEST response due to the more efficient delivery of CrA (color scale bars are different for A, B, and C). (Right) Cell specificity is confirmed by fluorescence as shown by box plots of the absolute fluorescence intensity of each cell population determined by flow cytometry, with median indicated by white line; range denotes first and third quartile. (C, Upper Right) Localized spectroscopy can also be obtained by acquiring images at different saturation frequencies and evaluating the signal from different compartments (inner and outer) throughout that image series (see Fig. S7 for details). Saturation parameters as for xenon CEST image.
Given the potential for high numbers of CrA per biosensor to mediate unspecific binding (21) and cell toxicity (9, 24), this quantification also speaks to the potential benefit of tuning the CrA load to achieve suitable detection, whether via this modular system or through other scaffolds (20, 23), while minimizing undesirable effects such as unspecific binding of the hydrophobic cage, which is prevented in our case. The higher CrA content of cells incubated with the complete construct is likely the result of a more efficient connection of the two modules, which is done before cell incubation in a phosphate buffer solution, for these samples. In the sequential incubations, the cells are incubated with the targeting module in a BSA-containing buffer, the endogenous biotin content of which may reduce the subsequent binding of readout modules. Although a limitation in this context, the sequential incubation in the presence of some endogenous biotin is an important consideration for future in vivo experiments, as we will discuss shortly.
Overall, the modular design approach of the biosensor offers several advantages. As a tool for screening different potential xenon MRI cell surface targets, the separate targeting and readout modules maximize the flexibility and minimize the cost of developing new biosensors. The ratio of the single-labeled readout modules can be selectively adapted to the needs of each experiment while maintaining an ability to reference the CrA quantification to that of the fluorescein. Alternatively, if desired, a branched readout module containing equimolar amounts of CrA and fluorescein can also be effective.
Antibody targeting in general offers high specificity, meaning that problems of nonspecific binding seen in other CrA constructs (21) may be minimized in this system, both in the case of the complete construct (which also serves to increase the solubility of the CrA moiety) and for the sequential method for which washout protocols can be used. In addition, the sequential style of these experiments offers the possibility to translate this method to in vivo settings once a suitable target is identified. Indeed, pretargeting with antibodies followed by sequential labeling with an effector molecule, including the use of the avidin/streptavidin−biotin system to connect these two modules, has already been successfully used in proton MRI (31) as well as radioimmunotherapy studies (32–34). For antibody targeted systems, there are two major advantages of separate delivery; reduced toxicity of the effector molecule and faster specific labeling. This stems from the fact that in vivo, the smaller effector molecules can diffuse more quickly to their prelabeled target (and similarly have faster clearance rates) than the larger targeting antibody modules (34). Although the avidin−biotin system is widely described, there may be some modifications that would improve the use of this system in in vivo applications. This could include exchanging the avidin conjugation with streptavidin [which has longer retention time in the blood stream and predominantly renal rather than hepatic clearance (32)], the use of more biologically stable biotin derivatives (35), or the potential to use biotin-deficient diets in mouse models (to reduce the effect of endogenous biotin binding to the targeting module) (36, 37).
With the possibilities to adapt this system to a variety of molecular imaging targets and the ability to detect concentrations of CrA-based biosensors as low as 20 nM, this modular approach offers not only a versatile technique with which to screen targets but one which can move forward with the xenon MRI field as it moves closer to molecular imaging in vivo.
Materials and Methods
Cell Lines.
NIH/3T3 fibroblasts (ATCC CRL-1658) were grown in very low endotoxin-DMEM with stable gluatmine (Biochrom AG) supplemented with 10% (vol/vol) FBS (Biochrom AG). RAW 264.7 macrophages (Sigma-Aldrich) were grown in RPMI 1640 with stable glutamine (Biochrom AG) supplemented with 10% (vol/vol) FBS (Biochrom AG). Both cell lines were grown at 37 °C in a humidified incubator with 5% CO2.
After reaching ∼70% confluency, RAW 264.7 macrophages were stimulated for 18 h with 100 ng/mL LPS from Escherichia coli 0111:B4 (Sigma-Aldrich) in RPMI 1640 with stable glutamine supplemented with 10% (vol/vol) FBS. NIH/3T3 cells were harvested by incubation with 0.05% Trypsin-EDTA (Biochrom AG); RAW 264.7 macrophages were harvested by manual detachment of the cells by scraping.
Synthesis of the Biotinylated CrA Modules.
The CrA−biotin module was synthesized using microwave (mw)-assisted acylation to connect CrA monoacid (provided by Kangyuan Jiyi Inc.) with Biotin-PEG3-amine (ChemPrep) with a final yield of 48%. Synthesis of the branched CrA−fluorescein−biotin construct was performed through mw-assisted acylation using a “one pot” protocol (38) modified for the purpose to connect three units together: (i) Biotin-PEG3-amine, (ii) CrA diacid (provided by Kangyuan Jiyi Inc.), and (iii) PEG-FAM-amide with a final yield of 10%. PEG-FAM was synthesized as previously described (24) with the adaption of the synthesis for mw-assisted Fmoc-solid phase peptide synthesis. All constructs were purified via RP-HPLC/UV and their mass confirmed using a MALDI-TOF spectrometer. See SI Text and Figs. S2 and S8 for further details. Stock solutions of all biotin conjugates were made in DMSO.
Avidin-Antibody Biosensor Conjugates.
1 mg of both monoclonal anti-CD14 specific antibody (ABIN 1176993; antibodies-online GmbH) and control IgG2b (chain kappa) isotype antibody (ABIN 287151; antibodies-online GmbH) were conjugated with avidin using the commercially available EasyLink Avidin Conjugation Kit 1mg (ab102860; Abcam) according to manufacturer’s instructions. Avidin-antibody conjugates (molecular mass ∼160 kDa) were purified from any unbound avidin (molecular mass ∼69 kDa) by adding 10 mL sterile Dulbeco’s PBS (DPBS) to the product of the conjugation reaction and then concentrating the solution ∼fivefold through an Amicon Ultra 15, 100-kDa concentrator (Merck Millipore) with three sequential PBS additions. The final avidin-antibody conjugates were concentrated to ∼0.5 µg/µL in DPBS and stored at 4 °C. The “complete” biosensor conjugate was prepared in vitro by the incubation of 100 µg avidin-antibody conjugate with a 40-fold mole excess of biotin conjugates (readout modules) in a mole ratio of 1:4 fluorescein−biotin (Thermo Scientific) to CrA−biotin, for 30 min at room temperature. After incubation, the product was purified and concentrated fivefold through an Amicon Ultra 15, 100-kDa concentrator (Merck Millipore). This separated the antibody conjugates (molecular mass ∼160 kDa) from any unbound biotin conjugates [CrA−biotin, 1339.54 Da (see Fig. S2) and fluorescein−biotin, 732.80 Da] in the product. The final CrA−fluorescein-antibody conjugates were concentrated to 0.5 µg/µL in DPBS 1% DMSO and stored at 4 °C.
Incubation of Cells with Biosensors.
For each xenon MRI experiment, 10–20 × 106 cells were harvested and aliquoted into a 15-mL Falcon tube and pelleted by centrifugation (400 × g for 4 min at 25 °C). The cell pellet was resuspended in a 200-µL volume per million cells in DPBS (Biochrom AG) containing 3% (wt/vol) BSA and 2 µg of the avidin-labeled CD14 antibody, the avidin-labeled IgG2b control antibody, or the complete CD14 biosensor construct. The cells were incubated for 1 h in the dark at 4 °C. Following the incubation, the cells were pelleted by centrifugation (400 × g for 4 min at 25 °C) and washed twice with ice-cold DPBS containing 3% (wt/vol) BSA. A small sample was taken for cell counting and viability analysis with Trypan Blue 0.5% (Biochrom AG) using a TC20 Automated Cell Counter (Bio-Rad). All cells used in further experiments have >95% viability as assessed by Trypan Blue analysis.
Cells that were incubated with the complete biosensor constructs were then resuspended in the appropriate buffer and analyzed by either flow cytometry, xenon NMR spectroscopy, or xenon MRI.
Cells that were incubated with the avidin-labeled antibody conjugates were resuspended in 200 µL DPBS containing 3% (wt/vol) BSA per million cells with 1 µM fluorescein−biotin (Thermo Scientific) and 4 µM CrA−biotin (final DMSO concentration 1%) and incubated for 30 min in the dark at 4 °C. Cells were pelleted by centrifugation (400 × g for 4 min at 25 °C) and washed twice with ice-cold DPBS before being resuspended in the appropriate buffer and analyzed by either flow cytometry, xenon NMR spectroscopy, or xenon MRI.
Flow Cytometry.
For flow cytometry, 0.5 × 106 cells were removed from samples after incubation with either sequential or complete biosensor modules as described in Incubation of Cells with Biosensors. For control experiments with unlabeled antibody, the cells were incubated with 2 µg CD14 antibody for 1 h at 4 °C, followed by 1:1,000 dilution of secondary goat anti-rat IgG FITC (ABIN102120, antibodies-online GmbH) for 30 min in the dark at 4 °C. All cell samples were resuspended in 500 µL DPBS and kept in the dark, on ice, before the flow cytometry measurement. The absolute fluorescence intensity of fluorescein was measured on the FL1 channel and compared for each cell line and biosensor under identical acquisition parameters.
Flow cytometry was performed on a BD FACSCalibur flow cytometer using BD CellQuest Pro Acquisition software. Data were then analyzed with FlowJo analysis software. The absolute fluorescence from triplicate experiments is presented.
Hyper-CEST Experiments.
After incubation with the biosensor (as described above), the cells were resuspended at a concentration of 5 million cells per milliliter in PBS. To reduce foam formation when bubbling xenon in the gas mix, 0.2% of Pluronic l-81 (BASF) was added just before MRI measurements. The cell suspension was transferred to an NMR double bubbling chamber, with the RAW 264.7 cell suspension in the inner compartment and the 3T3 cells in the outer compartment. This setup was then connected to the hp-Xe MRI system and placed inside the NMR magnet.
Xenon Hyperpolarization and MR.
All NMR studies were performed using a 9.4 T Bruker AV 400 wide-bore NMR spectrometer (Bruker Biospin) fitted with gradient coils for imaging. A 10-mm-inner-diameter double-resonant (129Xe and 1H) probe was used for excitation and detection. A custom-designed continuous-flow polarizer was used to produce hp-Xe by spin-exchange optical pumping (13, 39). A gas mixture of 2% xenon (26.4% 129Xe natural abundance), 10% N2, and 88% He was used. The achieved 129Xe spin polarization was ∼25%. The hp-Xe was directly bubbled into a 10-mm NMR tube containing 1.5 mL of the sample by using a spectrometer-triggered bubble dispenser (3.5 bar overpressure) via fused silica capillaries (Polymicro Technologies, Molex Incorporated). The bubbling was triggered by using spectrometer-triggered gas flow regulators (mass flow controllers, Omega Newport).
Hyper-CEST Spectroscopy and Imaging.
For each data point in the CEST spectrum, the gas mixture was directly bubbled for 12 s into the sample solution (flow rate 0.1 standard liters per minute) followed by a 4-s delay to allow bubbles to collapse. This was followed by a 12-s, 32.4-µT saturation pulse. Evaluation of the CEST spectra of the RAW 264.7 cells incubated with the CD14 biosensor at lower power (10.3 µT, 12 s and 5.1 µT, 20 s) (Fig. 2D) clearly indicates the presence of two CrA-associated pools. These resonances correspond to CrA within a lipidic/cellular environment (left peak) and CrA within an aqueous environment (right peak), and are consistent with results from our previous cell experiments (referenced to the xenon in free solution peak at 0 ppm). CEST spectra were fit to the negative exponential of a sum of Lorentzians (40) (see SI Text) in OriginPro 8.6.OG (OriginLab). For the Hyper-CEST image of the cells incubated sequentially with the biosensor modules, 32 on-resonant (−124 ppm relative to xenon in free solution at 0 ppm) and 32 off-resonant (124 ppm) scans were taken and averaged. For the Hyper-CEST image of the cells incubated with the complete biosensor, 10 on-resonant and 10 off-resonant scans were taken and averaged. For each image, the xenon gas mix was bubbled into solution for 15 s followed by a 3-s delay to allow bubbles to collapse. This was followed by a 26-s, 30-μT saturation pulse. The image was read out using a Rapid Acquisition with Relaxation Enhancement (RARE) (41) readout (slice thickness, 20 mm; matrix size, 32 × 32; in-plane resolution, 625 μm; centric k-space encoding; bandwidth, 16 kHz; echo time, 5 ms; acquisition time, 160 ms; RARE factor, 32). Image processing (see SI Text and Fig. S6) was done using Python.
Quantification of Cell-Bound CrA.
The concentration of cell-associated CrA was estimated by measuring the fluorescence intensity in cell lysates in parallel to MRI measurements. RAW 264.7 and 3T3 cells were incubated with biosensor for hp-Xe NMR measurements (as described). After washing with PBS, 1 million cells were removed from each sample and pelleted by centrifugation. Cells were lysed by adding 50 µL of cell lysis buffer [PBS containing 2% (vol/vol) SDS and 1% Triton X]. The cell lysate was diluted by the addition of 100 µL PBS containing DMSO, to give a final 150-µL volume of cell lysate in PBS, 0.5% SDS, 0.25% Triton X, 1% DMSO per sample. Two standard curves were generated by dissolving the two readout modules: CrA−biotin and the branched CrA−fluorescein−biotin in PBS containing 0.5% SDS, 0.25% Triton X, 1% DMSO final. The fluorescence of each sample in a 96-well black fluorescence plate was read at excitation 485 nm, emission 520 nm by a Tecan Safire fluorescence plate reader. The number of cell-associated CrA molecules was calculated by assuming the ratio of fluorescein to CrA in the sample is consistent with the ratio of the two readout modules used in the incubation. Therefore, for the sequential incubation, the quantification of CrA was calculated as 4 times that of the fluorescein. For the branched construct with a 1:1 ratio of fluorescein:CrA, no conversion was necessary.
The effective MRI concentration is then calculated according to the MRI sample setup with 5 million cells per milliliter for each sample. Example calculations are shown in Fig. S5.
Laser Scanning Microscopy.
The 3T3 and RAW 264.7 cells (400,000) were seeded on 30-mm glass coverslips (pretreated with 100 μg/mL poly-l-lysine) in six-well plates and grown for 24 h. The immobilized cells were incubated sequentially with the target module and then the readout module components of the biosensor in DPBS (Biochrom AG) containing an additional 1 mM Ca2+, 3% (wt/vol) BSA for 1 h each at 37 °C in a humidified incubator with 5% CO2. Cells were then washed twice with DPBS (Biochrom AG), and fresh media were added to each well. The cells were incubated with Hoechst Stain solution (H6024; Sigma) for 15 min before each imaging. Live cells were visualized on a LSM 510 (Carl Zeiss Microimaging GmbH) microscope using an ×100 oil objective. Fluorescence signals were recorded using the following lasers: a 488-nm argon laser/ BP 505–550 nm for the fluorescein coupled biosensor and a 543-nm UV laser/LP 364 nm for Hoechst nuclear stain. Images were processed using the ZEN 2009 light Edition (Carl Zeiss Microimaging GmbH).
Supplementary Material
Acknowledgments
This work has been supported by the European Research Council (ERC) under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement 242710, the Leibniz Association (Wissenschaftsgemeinschaft Gottfried Wilhelm Leibniz e.V.; Grant SAW-2011-FMP-2), the Federal Ministry of Education and Research (BMBF) Grant 13EZ1010B, and the Human Frontier Science Program (C.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406797111/-/DCSupplemental.
References
- 1.Mugler JP, 3rd, et al. MR imaging and spectroscopy using hyperpolarized 129Xe gas: preliminary human results. Magn Reson Med. 1997;37(6):809–815. doi: 10.1002/mrm.1910370602. [DOI] [PubMed] [Google Scholar]
- 2.Driehuys B, et al. Imaging alveolar-capillary gas transfer using hyperpolarized 129Xe MRI. Proc Natl Acad Sci USA. 2006;103(48):18278–18283. doi: 10.1073/pnas.0608458103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mugler JP, 3rd, Altes TA. Hyperpolarized 129Xe MRI of the human lung. J Magn Reson Imaging. 2013;37(2):313–331. doi: 10.1002/jmri.23844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikolaou P, et al. Near-unity nuclear polarization with an open-source 129Xe hyperpolarizer for NMR and MRI. Proc Natl Acad Sci USA. 2013;110(35):14150–14155. doi: 10.1073/pnas.1306586110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schröder L. Xenon for NMR biosensing—Inert but alert. Phys Med. 2013;29(1):3–16. doi: 10.1016/j.ejmp.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Spence MM, et al. Functionalized xenon as a biosensor. Proc Natl Acad Sci USA. 2001;98(19):10654–10657. doi: 10.1073/pnas.191368398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spence MM, et al. Development of a functionalized xenon biosensor. J Am Chem Soc. 2004;126(46):15287–15294. doi: 10.1021/ja0483037. [DOI] [PubMed] [Google Scholar]
- 8.Wei Q, et al. Designing 129Xe NMR biosensors for matrix metalloproteinase detection. J Am Chem Soc. 2006;128(40):13274–13283. doi: 10.1021/ja0640501. [DOI] [PubMed] [Google Scholar]
- 9.Seward GK, Wei Q, Dmochowski IJ. Peptide-mediated cellular uptake of cryptophane. Bioconjug Chem. 2008;19(11):2129–2135. doi: 10.1021/bc8002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garimella PD, et al. Hyperpolarized xenon-based molecular sensors for label-free detection of analytes. J Am Chem Soc. 2014;136(1):164–168. doi: 10.1021/ja406760r. [DOI] [PubMed] [Google Scholar]
- 11.Kunth M, Döpfert J, Witte C, Rossella F, Schröder L. Optimized use of reversible binding for fast and selective NMR localization of caged xenon. Angew Chem Int Ed Engl. 2012;51(33):8217–8220. doi: 10.1002/anie.201202481. [DOI] [PubMed] [Google Scholar]
- 12.Schröder L, Lowery TJ, Hilty C, Wemmer DE, Pines A. Molecular imaging using a targeted magnetic resonance hyperpolarized biosensor. Science. 2006;314(5798):446–449. doi: 10.1126/science.1131847. [DOI] [PubMed] [Google Scholar]
- 13.Witte C, Kunth M, Döpfert J, Rossella F, Schröder L. Hyperpolarized xenon for NMR and MRI applications. J Vis Exp. 2012;67:4268. doi: 10.3791/4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinogradov E, Sherry AD, Lenkinski RE. CEST: From basic principles to applications, challenges and opportunities. J Magn Reson. 2013;229:155–172. doi: 10.1016/j.jmr.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terreno E, Castelli DD, Aime S. Encoding the frequency dependence in MRI contrast media: The emerging class of CEST agents. Contrast Media Mol Imaging. 2010;5(2):78–98. doi: 10.1002/cmmi.369. [DOI] [PubMed] [Google Scholar]
- 16.Bai Y, Hill PA, Dmochowski IJ. Utilizing a water-soluble cryptophane with fast xenon exchange rates for picomolar sensitivity NMR measurements. Anal Chem. 2012;84(22):9935–9941. doi: 10.1021/ac302347y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens TK, Ramirez RM, Pines A. Nanoemulsion contrast agents with sub-picomolar sensitivity for xenon NMR. J Am Chem Soc. 2013;135(26):9576–9579. doi: 10.1021/ja402885q. [DOI] [PubMed] [Google Scholar]
- 18.Huber G, et al. Water soluble cryptophanes showing unprecedented affinity for xenon: candidates as NMR-based biosensors. J Am Chem Soc. 2006;128(18):6239–6246. doi: 10.1021/ja060266r. [DOI] [PubMed] [Google Scholar]
- 19.Delacour L, et al. “Clickable” hydrosoluble PEGylated cryptophane as a universal platform for 129Xe magnetic resonance imaging biosensors. Chemistry. 2013;19(19):6089–6093. doi: 10.1002/chem.201204218. [DOI] [PubMed] [Google Scholar]
- 20.Meldrum T, et al. A xenon-based molecular sensor assembled on an MS2 viral capsid scaffold. J Am Chem Soc. 2010;132(17):5936–5937. doi: 10.1021/ja100319f. [DOI] [PubMed] [Google Scholar]
- 21.Boutin C, et al. Cell uptake of a biosensor detected by hyperpolarized 129Xe NMR: The transferrin case. Bioorg Med Chem. 2011;19(13):4135–4143. doi: 10.1016/j.bmc.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Sloniec J, et al. Biomembrane interactions of functionalized cryptophane-A: Combined fluorescence and 129Xe NMR studies of a bimodal contrast agent. Chemistry. 2013;19(9):3110–3118. doi: 10.1002/chem.201203773. [DOI] [PubMed] [Google Scholar]
- 23.Palaniappan KK, et al. Molecular imaging of cancer cells using a bacteriophage-based 129Xe NMR biosensor. Angew Chem Int Ed Engl. 2013;52(18):4849–4853. doi: 10.1002/anie.201300170. [DOI] [PubMed] [Google Scholar]
- 24.Klippel S, et al. Cell tracking with caged xenon: Using cryptophanes as MRI reporters upon cellular internalization. Angew Chem Int Ed Engl. 2014;53(2):493–496. doi: 10.1002/anie.201307290. [DOI] [PubMed] [Google Scholar]
- 25.Wright SD. CD14 and innate recognition of bacteria. J Immunol. 1995;155(1):6–8. [PubMed] [Google Scholar]
- 26.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249(4975):1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 27.Schütt C, Schilling T, Krüger C. sCD14 prevents endotoxin inducible oxidative burst response of human monocytes. Allerg Immunol (Leipz) 1991;37(3-4):159–164. [PubMed] [Google Scholar]
- 28.Te Boekhorst BC, van Tilborg GA, Strijkers GJ, Nicolay K. Molecular MRI of inflammation in atherosclerosis. Curr Cardiovasc Imaging Rep. 2012;5(1):60–68. doi: 10.1007/s12410-011-9114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daldrup-Link HE, et al. MRI of tumor-associated macrophages with clinically applicable iron oxide nanoparticles. Clin Cancer Res. 2011;17(17):5695–5704. doi: 10.1158/1078-0432.CCR-10-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnurr M, Witte C, Schröder L. Functionalized 129Xe as a potential biosensor for membrane fluidity. Phys Chem Chem Phys. 2013;15(34):14178–14181. doi: 10.1039/c3cp51227d. [DOI] [PubMed] [Google Scholar]
- 31.Sano K, et al. A pre-targeting strategy for MR imaging of functional molecules using dendritic Gd-based contrast agents. Mol Imaging Biol. 2011;13(6):1196–1203. doi: 10.1007/s11307-010-0463-1. [DOI] [PubMed] [Google Scholar]
- 32.Sakahara H, Saga T. Avidin–biotin system for delivery of diagnostic agents. Adv Drug Deliv Rev. 1999;37(1-3):89–101. doi: 10.1016/s0169-409x(98)00101-x. [DOI] [PubMed] [Google Scholar]
- 33.Sung C, van Osdol WW. Pharmacokinetic comparison of direct antibody targeting with pretargeting protocols based on streptavidin-biotin binding. J Nucl Med. 1995;36(5):867–876. [PubMed] [Google Scholar]
- 34.Frost SH, Jensen H, Lindegren S. In vitro evaluation of avidin antibody pretargeting using 211At-labeled and biotinylated poly-L-lysine as effector molecule. Cancer. 2010;116(4) Suppl:1101–1110. doi: 10.1002/cncr.24798. [DOI] [PubMed] [Google Scholar]
- 35.Foulon CF, Alston KL, Zalutsky MR. Astatine-211-labeled biotin conjugates resistant to biotinidase for use in pretargeted radioimmunotherapy. Nucl Med Biol. 1998;25(2):81–88. doi: 10.1016/s0969-8051(97)00166-2. [DOI] [PubMed] [Google Scholar]
- 36.Hamblett KJ, et al. A streptavidin-biotin binding system that minimizes blocking by endogenous biotin. Bioconjug Chem. 2002;13(3):588–598. doi: 10.1021/bc010087t. [DOI] [PubMed] [Google Scholar]
- 37.Rusckowski M, Fogarasi M, Fritz B, Hnatowich DJ. Effect of endogenous biotin on the applications of streptavidin and biotin in mice. Nucl Med Biol. 1997;24(3):263–268. doi: 10.1016/s0969-8051(97)00061-9. [DOI] [PubMed] [Google Scholar]
- 38.Carnaroglio D, et al. One-pot sequential synthesis of isocyanates and urea derivatives via a microwave-assisted Staudinger-aza-Wittig reaction. Beilstein J Org Chem. 2013;9:2378–2386. doi: 10.3762/bjoc.9.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witte C, Kunth M, Rossella F, Schröder L. Observing and preventing rubidium runaway in a direct-infusion xenon-spin hyperpolarizer optimized for high-resolution hyper-CEST (chemical exchange saturation transfer using hyperpolarized nuclei) NMR. J Chem Phys. 2014;140(8):084203. doi: 10.1063/1.4865944. [DOI] [PubMed] [Google Scholar]
- 40.Zaiss M, Schnurr M, Bachert P. Analytical solution for the depolarization of hyperpolarized nuclei by chemical exchange saturation transfer between free and encapsulated xenon (HyperCEST) J Chem Phys. 2012;136(14):144106. doi: 10.1063/1.3701178. [DOI] [PubMed] [Google Scholar]
- 41.Hennig J, Nauerth A, Friedburg H. RARE imaging: A fast imaging method for clinical MR. Magn Reson Med. 1986;3(6):823–833. doi: 10.1002/mrm.1910030602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.