Significance
Bacterial pathogens manipulate host cellular processes using their own proteins. These proteins, so-called effectors, are injected into host cells using specialized nanomachines assembled on the bacterial cell surface. One such nanomachine, the type IV secretion system (T4SS), is evolutionarily related to bacterial conjugation systems. We describe the structure of the T4SS core complex required for effector translocation and virulence in the human pathogen Legionella pneumophila. The core complex of this T4SS is larger and more intricate than that of the pKM101 conjugation system. Analysis of premature complexes gives insights into a mechanism for core complex assembly whereby channel components dock in an outer-membrane subcomplex. Our data describing nanomachine assembly will aid toward development of drug therapies for bacterial pathogens.
Keywords: nanomachine, assembly pathway, membrane proteins, effector translocation
Abstract
Bacterial type IV secretion systems are evolutionarily related to conjugation systems and play a pivotal role in infection by delivering numerous virulence factors into host cells. Using transmission electron microscopy, we report the native molecular structure of the core complex of the Dot/Icm type IV secretion system encoded by Legionella pneumophila, an intracellular human pathogen. The biochemically isolated core complex, composed of at least five proteins—DotC, DotD, DotF, DotG, and DotH—has a ring-shaped structure. Intriguingly, morphologically distinct premature complexes are formed in the absence of DotG or DotF. Our data suggest that DotG forms a central channel spanning inner and outer membranes. DotF, a component dispensable for type IV secretion, plays a role in efficient embedment of DotG into the functional core complex. These results highlight a common scheme for the biogenesis of transport machinery.
Many bacterial pathogens transport proteins into the cytoplasm of eukaryotic host cells using specialized nanomachines assembled on the bacterial surface. The bacterial proteins being transported are called effector proteins. Effector proteins interact with host factors to modulate or hijack host cellular processes, leading to the establishment of infection. Thus, the effector transport machinery plays a critical role in bacterial pathogenesis. The type IV secretion system (T4SS) is one of bacterial nanomachines capable of transporting effector proteins directly into host cell cytoplasm (1–3). T4SSs are evolutionarily related to bacterial conjugation systems. An archetypal T4SS is the Agrobacterium tumefaciens VirB system. The VirB system is composed of 12 proteins (VirB1–VirB11 and VirD4). A. tumefaciens transports transfer (T)-DNA and several effector proteins into plant cells using the VirB system. The human pathogen Helicobacter pylori translocates the effector protein CagA using the T4SS encoded by the cag pathogenicity island, which is critical for gastric carcinogenesis (4). Many intracellular bacterial pathogens including Legionella pneumophila and Coxiella burnetii have T4SSs essential for pathogenesis.
The human pathogen L. pneumophila encodes a T4SS called the Dot/Icm system, which is essential for L. pneumophila virulence (5, 6). Using this system, L. pneumophila translocates ∼300 bacterial effector proteins into the host cell cytoplasm, where they direct a variety of virulence traits such as the establishment of a replicative niche (7–9). The Dot/Icm system, encoded by ∼30 dot/icm genes, is closely related to the conjugation systems of IncI plasmids (10, 11). The composition of the Dot/Icm system appears to be more complex than the Agrobacterium VirB system and the conjugation system of IncN plasmid pKM101, a system closely related to the VirB system (12, 13). Indeed sequence-level similarities between component proteins of the Dot/Icm system and the pKM101 conjugation system are largely undetectable, however there are a few exceptions, such as the similarity between the carboxyl-terminal domains of DotG and VirB10pKM101/TraF (see Results).
Regarding the pKM101 conjugation system, electron microscopic structures of the core complex and the crystal structure of an outer-membrane complex generated by partial digestion of the core complex were reported (14–16). The core complex spans both bacterial inner and outer membranes and consists of 14 copies each of three proteins: VirB7pKM101/TraN, VirB9pKM101/TraO, and VirB10pKM101/TraF. The core complex has 14-fold rotational symmetry along the central axis perpendicular to bacterial membranes. The view along the axis of the core complex is ring-shaped with an external diameter of ∼20 nm. Despite the limited sequence similarities between the Dot/Icm system and the pKM101 conjugation system, the ring structure of the pKM101 core complex prompted us to investigate whether L. pneumophila has a ring-shaped core complex on its surface.
Results
L. pneumophila Has Ring-Shaped Structures on Its Surface.
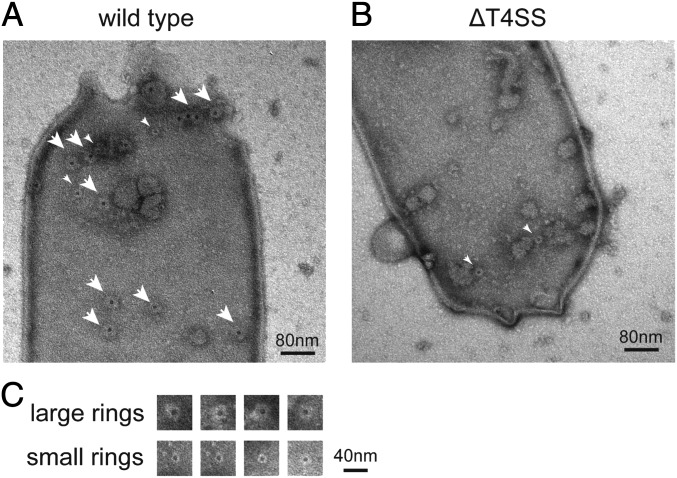
To avoid heavy electron density associated with cytoplasmic material inside bacterial cells, and to visualize bacterial envelop-associated structures, osmotically shocked L. pneumophila cells were negatively stained and examined by transmission electron microscopy (Fig. 1). At least two ring-shaped structures, with distinct diameters of ∼40 and 20 nm, were visualized (Fig. 1 A and C). Both the ring structures have a central pore. To clarify whether the formation of these ring structures requires components of the Dot/Icm system, we examined the surface of an isogenic mutant strain lacking all dot/icm genes (ΔT4SS) in the same way. The larger ring of ∼40 nm in diameter was totally absent on the cell surface of the mutant strain, whereas the smaller ring was found on the cell surface of both the wild-type and the mutant strains (Fig. 1B). These results indicate that there is a ring-shaped structure of ∼40 nm in diameter on the L. pneumophila surface, the formation of which is dependent on one or more dot/icm genes.
Fig. 1.
Ring-shaped structures on the surface of L. pneumophila strain Lp01. Osmotically shocked L. pneumophila cells were mounted on discharged carbon grids and stained with 1% uranyl acetate. Transmission electron micrographs of a wild-type cell (A) and an isogenic deletion mutant (ΔT4SS) cell lacking all dot/icm genes (B) are shown. Large and small rings are denoted by large and small white arrowheads, respectively. (Scale bar, 200 nm.) (C) Large and small ring structures found on the wild-type L. pneumophila surface. (Scale bar, 40 nm.)
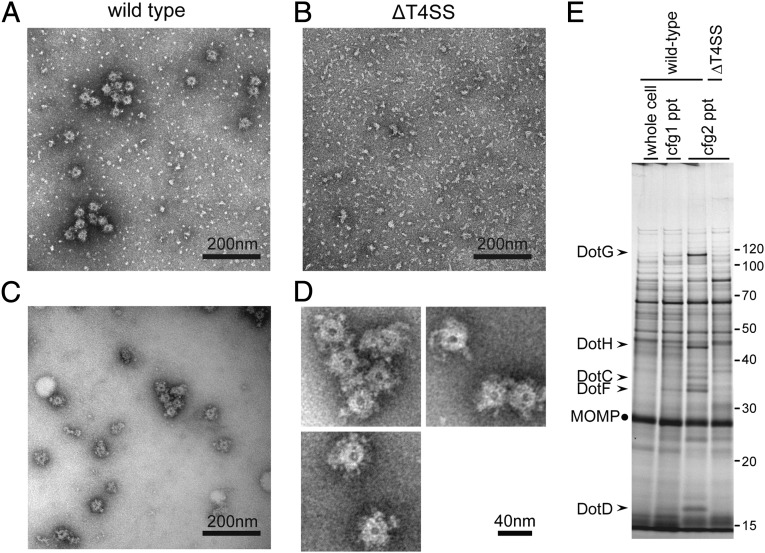
To characterize the ∼40-nm–diameter rings found on the cell surface, we isolated the ring structure from L. pneumophila. The ring structure was enriched by consecutive ultracentrifugation of detergent lysates of spheroplasts (Materials and Methods and Fig. 2A). The ring structure was absent in the fraction isolated in the same way from the ΔT4SS mutant strain (Fig. 2B). To reduce background specks observed in these electron micrographs, the enriched fraction was subjected to Superose 6 size-exclusion column chromatography (Fig. 2C). The isolated structure is ring-shaped, having a central pore and amorphous frills (Fig. 2D). Its external diameter (disregarding frills) is 38.2 ± 0.4 nm, and the diameter of the central pore is 8.0 ± 0.1 nm (n = 130).
Fig. 2.
Isolation of ring structures with a diameter of ∼40 nm. Ring structures were enriched as described in Materials and Methods, mounted on grids, and stained with 2% (wt/vol) PTA pH 7.0. Transmission electron micrographs of fractions isolated from wild-type cells (A and C) and isogenic deletion mutant (ΔT4SS) cells lacking all dot/icm genes (B) are shown. (Scale bar, 200 nm.) (C) The fraction shown in A was further purified by a size exclusion column chromatography to remove small specks observed. (D) Isolated ring structures from wild-type cells. (Scale bar, 40 nm.) (E) Proteins visualized by silver staining of fractions during the isolation procedure. The procedure involves two consecutive ultracentrifugation processes (Materials and Methods). Starting lysate (whole cell) and pellet fractions after ultracentrifugations (cfg, 1/2 ppt) were analyzed. Five Dot/Icm proteins (arrowheads) were consistently enriched during the procedure. MOMP, the major outer membrane protein of L. pneumophila, is a major contaminant protein in this procedure and is used as an internal control for comparison.
The Ring Structure of ∼40 nm in Diameter Is the Dot/Icm T4SS Core Complex.
Toward identification of the component proteins of the ring structure, we examined the fractions isolated from wild-type and ΔT4SS mutant strains by SDS–polyacrylamide gels and visualized the proteins by silver staining. Five major protein species of apparently 100, 45, 37, 32, and 16 kDa, found only in the fractions isolated from the wild-type strain, were reproducibly and consistently enriched during the isolation procedure (Fig. 2E). These five proteins were identified by mass spectrometry as DotG, DotH, DotC, DotF, and DotD, respectively (Fig. 2E, arrowheads). The identity of these proteins was further confirmed by Western immunoblotting analysis using specific antibodies against these proteins. We further conducted proteomic analysis of isolated fractions using liquid chromatography tandem-mass spectrometry (Fig. S1 and Table S1). The result supports the notion that these five proteins are the major Dot/Icm proteins enriched in the fraction from the wild-type strain. In addition, we found several Dot/Icm proteins—namely, DotK, IcmX, DotL, and DotA—that are less abundant but overrepresented in the fraction isolated from the wild-type strain compared with the fraction from the ∆T4SS mutant. These Dot/Icm proteins may interact with the core complex as minor components. Alternatively, these may form distinct complexes and be enriched by the procedure due to ultracentrifugation.
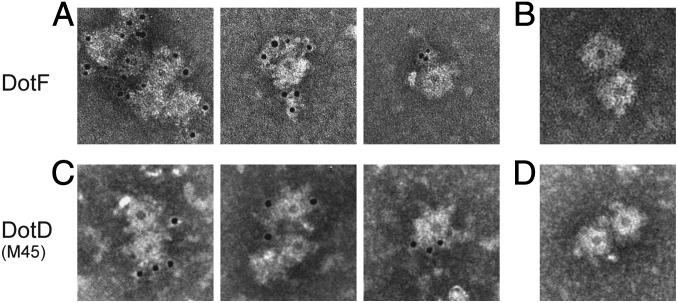
A previous systematic study of the cellular localization of Dot/Icm proteins suggested the existence of a complex composed of the same five Dot/Icm proteins: three outer-membrane proteins—DotC, DotD, and DotH—and two inner-membrane proteins—DotF and DotG (17). The complex is proposed to span both inner and outer membranes, forming the core machinery of the transport apparatus. To investigate whether these Dot/Icm proteins are components of the ring structure, we conducted immunoelectron microscopy analysis (Fig. 3). The ring structure was isolated from the wild-type strain, labeled with an affinity-purified polyclonal antibody against DotF and a gold-conjugated secondary antibody, and examined by transmission electron microscopy. Gold particles decorated the ring structures isolated from the wild-type strain (Fig. 3A). In contrast, gold particles did not accumulate around the ring structures labeled with only the secondary antibody (Fig. 3B). We further constructed an L. pneumophila strain producing M45 epitope-tagged DotD at its carboxy-terminus (DotD–M45). This protein fully complemented the intracellular growth defect of a dotD deletion mutant, suggesting the epitope did not adversely affect the formation of the secretion apparatus and its activity (Fig. S2). The ring structure was isolated from the strain producing DotD–M45, labeled with an affinity-purified antibody against the epitope tag and a gold-conjugated secondary antibody, and examined by transmission electron microscopy. Again, gold particles decorated the ring structures isolated from the strain producing DotD–M45 (Fig. 3C). In contrast, gold particles did not accumulate around the ring structures isolated from a strain producing wild-type DotD (Fig. 3D). These data demonstrate that both an inner-membrane protein DotF and an outer-membrane protein DotD are components of the ring structure.
Fig. 3.
Immunoelectron microscopy of the ring structures. (A and B) The ring structures isolated from the wild-type strain were stained with (A) affinity-purified anti-DotF antibody followed by a gold-conjugated secondary antibody or (B) the gold-conjugated secondary antibody only. (C and D) The ring structures isolated from (C) a strain producing DotD–M45 or (D) the isogenic strain producing wild-type DotD were stained with affinity-purified anti-M45 epitope antibody followed by a gold-conjugated secondary antibody. Structures were stained with 2% (wt/vol) PTA pH 7.0 and observed by transmission electron microscope.
DotC, DotD, and DotH Are Essential for Core Complex Formation.
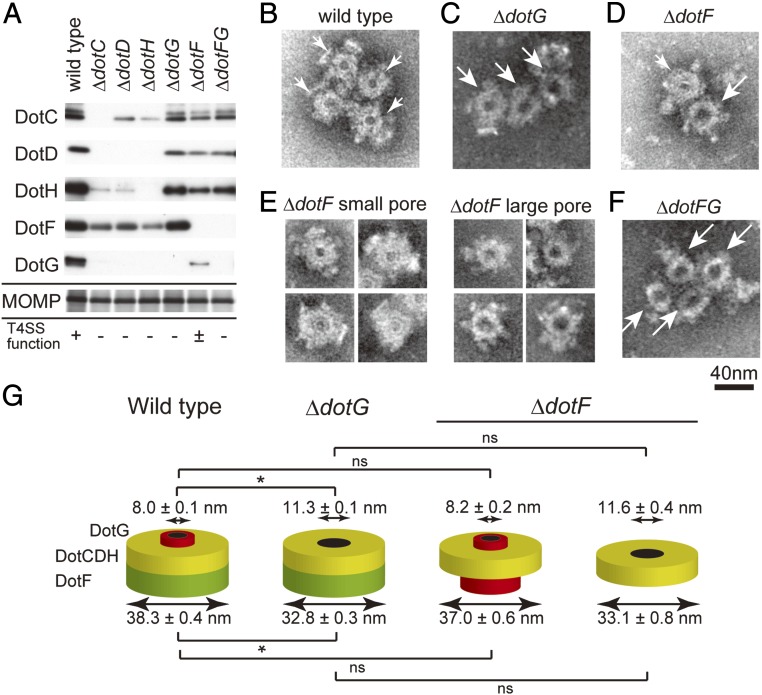
To further confirm that the ring structure is the core complex of the Dot/Icm T4SS, we isolated fractions in the same way from null mutant strains lacking DotC, DotD, DotF, DotG, or DotH. Western immunoblotting analysis of the fractions isolated from null mutants lacking outer-membrane proteins DotC, DotD, or DotH showed that levels of the other proteins were remarkably reduced (Fig. 4A, ΔdotC, ΔdotC, and ΔdotH). In those fractions, we were not able to detect the ring structure by transmission electron microscopy. These results indicate that DotC, DotD, and DotH are essential components for the formation of the ring structure. Moreover, the results are in perfect agreement with the previously reported DotC-, DotD-, and DotH-dependent outer-membrane localization of DotF and DotG (17).
Fig. 4.
Analysis of enriched fractions isolated from null mutant strains lacking indicated component protein(s). (A) Fractions isolated from designated L. pneumophila strains were analyzed by SDS–polyacrylamide gels. Core components DotC, DotD, DotH, DotF, and DotG were detected by Western immunoblotting using specific antibodies against indicated core component proteins. MOMP was detected by silver staining and used as an internal control for isolation yield. (B–F) Ring structures visualized by transmission electron microscopy analysis isolated from (B) wild-type, (C) ΔdotG, (D and E) ΔdotF, and (F) ΔdotF ΔdotG strains. In B–D and F, small and large white arrows denote the wild-type and the ΔdotG-type ring structures with small and large pores, respectively. In E, two distinct kinds of complexes isolated from the ΔdotF strain are shown. (G) Schematic cartoons illustrating the observed ring structures isolated from wild-type, ΔdotG, and ΔdotF strains. Internal and external diameters of the complexes are presented as the mean ± SEM. Asterisks denote significant difference (P < 0.001) by z-test or Student t test. ns, not significant.
An Inactive Subcomplex Lacking DotG.
In the fraction isolated from a null mutant strain lacking DotG, the proteins DotC, DotD, DotH, and DotF were recovered at comparable levels to the fraction isolated from wild-type cells (Fig. 4A, ΔdotG). Electron microscopy analysis of this fraction showed a ring structure; however, its shape is clearly distinct from the ring structure isolated from wild-type cells (Fig. 4 B versus C). The structure isolated from the dotG null mutant (Fig. 4C) has a significantly larger pore (11.3 ± 0.1 nm in diameter; n = 161; z-test P < 0.001) than the wild-type ring structure. The difference in external diameter of wild-type and ΔdotG-type ring structures is subtle but statistically significant (32.8 ± 0.3 nm for ΔdotG-type; z-test P < 0.001). This complex is inactive in effector translocation, because DotG is essential for intracellular growth within human monocyte-like U937 and human promyelocytic leukemia (HL-60) cells (6, 18). As described above, the carboxyl-terminal domain of DotG shares sequence-level similarity with that of VirB10pKM101/TraF, a component of the pKM101 core complex (13.8% identity over the 160 amino acid segment). The crystal structure of the pKM101 outer-membrane core complex demonstrated that the homologous VirB10pKM101 domain is situated adjacent to the central pore of the complex (14). Taking these previous findings into account, our data suggest that DotG is a component of the core complex located adjacent to the central pore (Fig. 4G, wild type). In the absence of DotG, a stable subcomplex lacking DotG is formed (Fig. 4G, ΔdotG). The absence of DotG results in a larger central pore, which may also result in a conformational change of the remaining part, leading to the subtle reduction of its exernal diameter.
DotF Facilitates Functional Complex Formation Containing DotG.
In the fraction isolated from the null mutant strain lacking DotF, the DotG level was remarkably reduced compared with the levels of the proteins DotC, DotD, and DotH (Fig. 4A). Electron microscopy analysis of this fraction visualized two distinct species of ring structures (Fig. 4D). One is morphologically indistinguishable from the ring structure isolated from wild-type cells (Fig. 4E, ΔdotF small pore). The other possesses a larger pore that is similar to the ring structure isolated from the dotG null mutant (Fig. 4E, ΔdotF large pore). Indeed the external diameters of the ring structures (37.0 ± 0.6 nm and 33.1 ± 0.8 nm, respectively) and the diameters of their central pores (8.2 ± 0.2 nm and 11.6 ± 0.4 nm, respectively) are not significantly different from those of the ring structures isolated from wild-type and the dotG null strains, respectively (t test, P > 0.08). In the fraction isolated from the strain lacking both DotF and DotG, only the ring structure with the larger pore was observed (Fig. 4F). Collectively, the dotF null mutant appears to produce at least two kinds of core subcomplexes, one containing DotG and the other lacking DotG (Fig. 4G, ΔdotF).
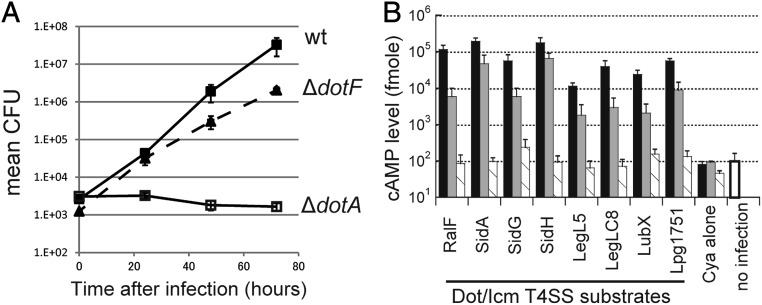
It was reported that an internal deletion/kanamycin-cassette insertion dotF mutant (JR32 icmG635::Kan) shows only a partial defect in growth within human HL-60 cells (19). Similarly our dotF null mutant shows only a partial growth defect in human U937 cells (Fig. 5A). To further investigate the DotF function in effector translocation, we measured levels of effector proteins translocated into mammalian cells via the Dot/Icm T4SS using an adenylate cyclase (cya) reporter system (20, 21). The results clearly indicate that the dotF null mutant retains the ability to translocate effectors, but the level of translocation is remarkably reduced irrespective of the effector protein tested (Fig. 5B, gray bars). Thus, the dotF null mutant produces the functional subcomplex containing DotG, at least to some extent. These results also imply that DotF is not an essential component for effector translocation, but is somehow involved in the assembly or the stability of the active core complex containing DotG.
Fig. 5.
DotF is not an essential component for the type IV secretion. (A) Intracellular growth assay of L. pneumophila wild-type (filled squares), ΔdotF (triangles; dashed line), and ΔdotA (open squares) strains within human U937 monocyte-like cells. (B) Levels of designated effector proteins translocated into Chinese hamster ovary (CHO) cells were measured by the cya reporter assay. CHO cells challenged by L. pneumophila wild-type (black bars), ΔdotF (gray bars), and ΔdotA (shaded bars) strains were extracted and cAMP levels were determined.
Discussion
Our interpretation of the data described above can be summarized by a model that predicts the ring structure of ∼40 nm in diameter, identified by electron microscopy on the surface of L. pneumophila cells, and is the core complex of the Dot/Icm T4SS. The Dot/Icm core complex contains at least five Dot/Icm proteins—DotC, DotD, DotF, DotG, and DotH—as major components. The Dot/Icm core complex of L. pneumophila has a significantly larger diameter than the core complex of the pKM101 conjugation system (18.5 nm in diameter) (15). The precise stoichiometry, nature of potential minor components, and rotational symmetry of the Dot/Icm core complex remain to be determined.
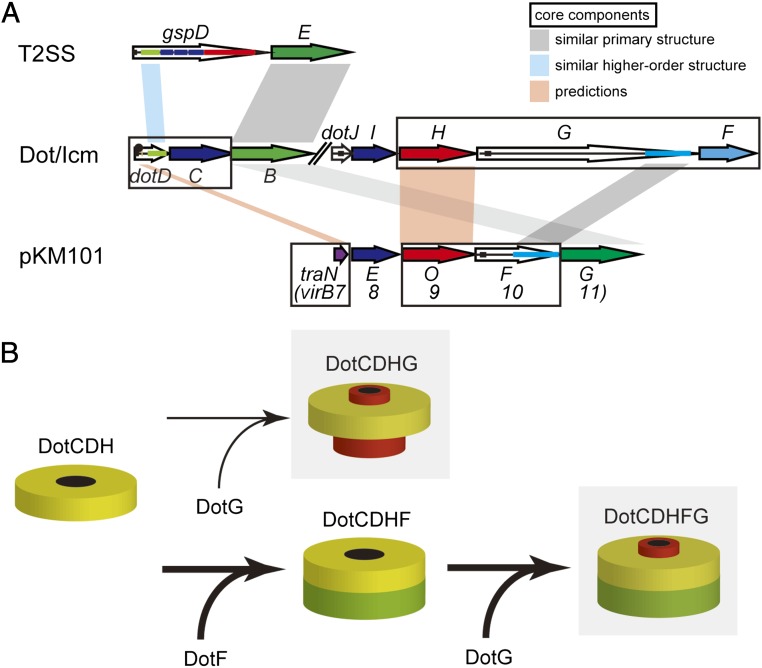
Our results demonstrated that DotC, DotD, and DotH are essential for the formation of the Dot/Icm core complex and form an outer-membrane subcomplex (Fig. 6B, DotCDH). The lipoprotein DotD shares similarity with Xanthomonas VirB7XAC2622 in domain organization; both proteins carry an amino-terminal signal sequence (for transfer across the inner membrane by a sec-dependent mechanism), followed by a lipobox (for lipidation), a disordered domain, and a carboxyl-terminal folded domain (22, 23). The folded domains share higher order structural similarity to each other and with the amino-terminal N0/T3S subdomain of secretin, which is an outer-membrane channel protein conserved in type II and type III secretion systems (Fig. 6A, gspD–dotD) (22, 23). The disordered domain of VirB7XAC2622 shares sequence-level similarity to canonical VirB7s from the Agrobacterium virB and pKM101 conjugation systems (23). VirB7pKM101/TraN is a core component situated on the surface of the pKM101 outer-membrane core complex (14), suggesting that Xanthomonas VirB7XAC2622 also forms part of an outer-membrane core complex (23). Likewise, the disordered domain of DotD may be the functional counterpart of VirB7 despite the lack of sequence-level similarity (Fig. 6A, dotD–virB7). DotD and another outer-membrane lipoprotein DotC are required for outer-membrane targeting of DotH (17). DotH and VirB9 proteins share an amino-terminal signal sequence and are recruited to outer membranes in the presence of cognate core complex components (Fig. 6A, dotH–virB9).
Fig. 6.
(A) Relationship between selected components of the type II secretion system (T2SS), the Dot/Icm T4SS, and the pKM101 conjugation system. Sequence-level and higher order similarities are shown by gray and cyan shades, respectively. Core components are boxed. See text for further explanation. (B) A model describing the assembly of the Dot/Icm T4SS core complex. Shaded complexes are active in the type IV secretion. In the absence of DotF, the outer-membrane subcomplex (DotCDH) can form a functionally active subcomplex containing DotG (DotCDHG) at low efficiency. DotF facilitates the integration process of DotG into the outer-membrane subcomplex by acting as an intracomplex chaperone, resulting in robust formation of the active core complex (DotCDHFG).
Carboxyl-terminal regions of DotG and VirB10 share sequence-level similarity (5, 6), suggesting that DotG is the counterpart of VirB10 (Fig. 6A, dotG–virB10). The VirB10pKM101/TraF region homologous to DotG contains an antenna-like structure, which spans the outer membrane, forming an outer-membrane pore (14). In addition, DotG and VirB10 have an inner-membrane spanning region near their amino termini. Thus, DotG and VirB10 are core components that span both the inner and outer membranes. Our results clearly demonstrate the existence of stable subcomplexes lacking DotG (Fig. 6B, DotCDH and DotCDHF), which is surprising because DotG-like VirB10 appears to be a predominant component of the pKM101 core complex. The subcomplexes do not contain the outer-membrane spanning region of DotG and are likely anchored to the outer membrane by lipids attached to the lipoproteins DotC and DotD. The importance of DotC and DotD lipidation is supported by the observation that L. pneumophila strains producing lipidation-site mutants of DotC or DotD show partial intracellular growth defects, and the defects are additive (24).
Our results clearly indicate that DotF is not an essential component for effector translocation, but DotF is likely to facilitate formation or stability of the functional core complex containing DotG. DotF is conserved among the type IVB subset of T4SSs, which are those closely related to the L. pneumophila Dot/Icm system (13); the VirB and pKM101 systems do not have a DotF counterpart (Table S2). DotF carries an amino-terminal inner-membrane spanning region and is distributed to outer-membrane fractions in a DotC-, DotD-, and DotH-dependent manner (17). A two-hybrid interaction between DotF and DotG was reported (17), but at this moment it is not clear whether a DotF–DotG complex is actually formed and plays a role in assembly of the Dot/Icm core complex. Our results indicate the dotG null mutant produces a stable subcomplex containing DotF (Fig. 6B, DotCDHF), which is consistent with the observation that outer-membrane targeting of DotF does not require DotG (17). DotG could dock into this intermediate subcomplex to form the functional core complex (Fig. 6B, DotCDHFG).
Docking of outer- and inner-membrane complexes to form functional machinery is commonly observed in many macromolecule transport systems. One well-known example of such outer-membrane complexes is the TolC channel protein homotrimer (25). The TolC complex docks in various inner-membrane complexes, forming type I secretion systems or multidrug efflux pumps depending on the inner-membrane partners. Another example is the secretin family of outer-membrane channel proteins. Secretin serves as an outer-membrane conduit in type II and type III secretion systems, type IV pili biogenesis, and filamentous phage extrusion (26, 27). Furthermore, in a model for biogenesis of the transport apparatus of the Agrobacterium VirB system, the outer-membrane VirB7–VirB9 complex is stabilized through interaction with VirB10 (2), which is likely to result in formation of the VirB7–VirB9–VirB10 core complex. Interaction between VirB9 and VirB10 diminishes in mutant strains lacking VirB11 or VirD4 ATPase activity (28), which raises the possibility that dynamic assembly of the core complex takes place responding to ATP utilization, although the VirB7–VirB9 complex has not yet been biochemically defined. Moreover, the recently reported structure of the VirB3–10 complex of the R388 conjugation system suggests that the VirB7–VirB9–VirB10 core complex is actually a mostly outer-membrane subcomplex of the entire machinery, which docks with an inner-membrane/cytoplasmic complex containing VirB3–6 and VirB8 (29).
Intriguingly, DotG proteins (typically over 100 kDa) of Legionella species and a related zoonotic pathogen C. burnetii are far larger than orthologous TraO proteins (typically 50 kDa) of IncI-plasmid conjugation systems because of an extra spacer region containing penta-peptide repeats in between inner- and outer-membrane interacting regions. Moreover, Legionella dumoffii strain Tex-KL (30) and Legionella drancourtii strain LLAP12 may encode split dotG genes: in addition to the spacer region, one containing an inner-membrane spanning region and the other containing an outer-membrane interacting region. These observations raise the possibility that the outer-membrane subcomplex and DotG are flexibly placed in some situations, with or without connection via the spacer region(s) of DotG. T4SS-associated appendage-like “pili” structures observed on the surface of H. pylori (31, 32) might be reminiscent of the flexibility allowed by DotG/VirB10-like Cag7/CagY, which is also a large and variable protein compared with canonical VirB10 (33).
In summary, the native structures of the Dot/Icm T4SS core complex and its subcomplexes reported here revealed their complexity in composition and assembly compared with the core complex of the pKM101 conjugation system. T4SSs of pathogenic bacteria specialized for effector translocation may have acquired additional intricacy to fulfill the need for efficient translocation of protein substrates into eukaryotic cells and/or to gain versatility in regulating effector translocation. Future studies of native structures of bacterial transport apparatus capable of transporting effector proteins, together with studies directed to high-resolution structures, will shed light on the molecular mechanisms of effector translocation, a central process of bacterial pathogenesis.
Materials and Methods
Bacterial Strains, Plasmids, Media, and Antibodies.
All L. pneumophila strains used in this study are Lp01 and its derivatives (34). In-frame deletion strains were constructed by allelic exchange as previously described (35). The ΔT4SS strain carries chromosomal deletions of three loci: icmX–dotA, dotB–dotD, and icmT–dotU. Plasmids for cya effector translocation assay were previously described (20, 36–38). Bacteria were grown on N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) buffered charcoal-yeast extract (CYE) agar or in liquid ACES-buffered yeast extract broth (AYE) media as previously described (39). Antibodies against Dot/Icm proteins and M45 epitope were described previously (22).
Transmission Electron Microscopy.
Samples were applied on glow-discharged carbon grids (cat. no. 649, Nisshin EM Co. Ltd.) and negatively stained with 2% (wt/vol) phosphotungstic acid (PTA) pH 7.0 or 2% (wt/vol) uranyl acetate. The samples were observed under the electron microscope (JEM-1011, JEOL). Micrographs were taken at an accelerating voltage of 80 kV.
Isolation of the Dot/Icm T4SS Core Complexes from L. pneumophila.
L. pneumophila grown on CYE plates for 2 d at 37 °C were inoculated in AYE media (OD600 = 0.2). The liquid culture was incubated for 12 h at 37 °C with rotary shaking (250 min−1). Bacteria from the total 800 mL of AYE culture were resuspended in 140 mL of 150 mM Tris⋅HCl pH 8.0 and 0.5 M sucrose containing EDTA-free Complete protease inhibitor mixture (Roche) or equivalent. To make spheroplasts, final 1 mM EDTA and 0.1 mg/mL lysozyme were added, and the suspension was incubated for 30 min at ambient temperature with gentle stirring. To lyse spheroplasts, final 1% of Triton X-100 was added, and the suspension was further incubated for 30 min. To digest released DNA, 3 mM (final) of MgSO4 and 5 μg/mL (final) DNaseI were added and incubated for 10 min. To disrupt large membrane vesicles and fragments, the pH of the lysate was adjusted to 10.0 using NaOH in the presence of final 10 mM EDTA. To remove nonlysed materials, the lysate was centrifuged at 12,000 × g for 20 min at 4 °C. To isolate large complexes including core complexes, the cleared lysate was ultracentrifuged at 100,000 × g for 30 min at 4 °C (Beckman–Coulter Type 70 Ti). Pellets were resuspended in 8 mL of TET buffer (10 mM Tris⋅HCl pH 8.0, 1 mM EDTA, 0.1% Trinton X-100). The suspension was again ultracentrifuged as above. After the second ultracentrifugation, the pellets were suspended in ∼300 μL of TET. For some electron micrographic analyses, complexes in TET buffer were further separated by Superose 6 10/300 column chromatography equilibrated with TET plus 50 mM NaCl using AKTA purifier system (GE Healthcare).
Statistics.
The z-test or Student t test (where the sample size is <40) was used for analysis of electron microscopic images. Results were presented as the mean ± SEM.
Other materials and methods used are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Kazunobu Saito of the DNA-chip Development Center for Infectious Diseases (Research Institute for Microbial Diseases, Osaka University) for technical advice in mass spectrometry, Andree Hubber for discussion, and Akiko Okura and Masayo Yamada for technical assistance. This work was supported by MEXT/JSPS KAKENHI Grants 23117002, 23390105, 24659198, and 26670212 (to H.N.); 25460530 (to T.K.); and 25860317 (to M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404506111/-/DCSupplemental.
References
- 1.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73(4):775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7(10):703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein M, Ruggiero P, Rappuoli R, Bagnoli F. Helicobacter pylori CagA: From pathogenic mechanisms to its use as an anti-cancer vaccine. Front Immunol. 2013;4:328. doi: 10.3389/fimmu.2013.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95(4):1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279(5352):873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Luo ZQ. Cell biology of infection by Legionella pneumophila. Microbes Infect. 2013;15(2):157–167. doi: 10.1016/j.micinf.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton HJ, Ang DK, van Driel IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev. 2010;23(2):274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 10.Komano T, Yoshida T, Narahara K, Furuya N. The transfer region of IncI1 plasmid R64: Similarities between R64 tra and legionella icm/dot genes. Mol Microbiol. 2000;35(6):1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 11.Wilkins BM, Thomas AT. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol Microbiol. 2000;38(3):650–657. doi: 10.1046/j.1365-2958.2000.02164.x. [DOI] [PubMed] [Google Scholar]
- 12.Christie PJ, Vogel JP. Bacterial type IV secretion: Conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8(8):354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagai H, Kubori T. Type IVB secretion systems of Legionella and other gram-negative bacteria. Front Microbiol. 2011;2:136. doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandran V, et al. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462(7276):1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fronzes R, et al. Structure of a type IV secretion system core complex. Science. 2009;323(5911):266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivera-Calzada A, et al. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 2013;32(8):1195–1204. doi: 10.1038/emboj.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent CD, et al. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2006;62(5):1278–1291. doi: 10.1111/j.1365-2958.2006.05446.x. [DOI] [PubMed] [Google Scholar]
- 18.Purcell M, Shuman HA. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect Immun. 1998;66(5):2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal G, Shuman HA. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67(5):2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai H, et al. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci USA. 2005;102(3):826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14(3):583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakano N, Kubori T, Kinoshita M, Imada K, Nagai H. Crystal structure of Legionella DotD: Insights into the relationship between type IVB and type II/III secretion systems. PLoS Pathog. 2010;6(10):e1001129. doi: 10.1371/journal.ppat.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souza DP, et al. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 2011;7(5):e1002031. doi: 10.1371/journal.ppat.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yerushalmi G, Zusman T, Segal G. Additive effect on intracellular growth by Legionella pneumophila Icm/Dot proteins containing a lipobox motif. Infect Immun. 2005;73(11):7578–7587. doi: 10.1128/IAI.73.11.7578-7587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koronakis V. TolC—The bacterial exit duct for proteins and drugs. FEBS Lett. 2003;555(1):66–71. doi: 10.1016/s0014-5793(03)01125-6. [DOI] [PubMed] [Google Scholar]
- 26.Genin S, Boucher CA. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: Modular structure and specificity of the N-terminal domain. Mol Gen Genet. 1994;243(1):112–118. doi: 10.1007/BF00283883. [DOI] [PubMed] [Google Scholar]
- 27.Hardie KR, Seydel A, Guilvout I, Pugsley AP. The secretin-specific, chaperone-like protein of the general secretory pathway: Separation of proteolytic protection and piloting functions. Mol Microbiol. 1996;22(5):967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- 28.Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA. 2004;101(49):17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Low HH, et al. Structure of a type IV secretion system. Nature. 2014;508(7497):550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin T, et al. Draft genome sequences of two Legionella dumoffii strains, TEX-KL and NY-23. J Bacteriol. 2012;194(5):1251–1252. doi: 10.1128/JB.06352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49(1):219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka J, Suzuki T, Mimuro H, Sasakawa C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 2003;5(6):395–404. doi: 10.1046/j.1462-5822.2003.00286.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu G, McDaniel TK, Falkow S, Karlin S. Sequence anomalies in the Cag7 gene of the Helicobacter pylori pathogenicity island. Proc Natl Acad Sci USA. 1999;96(12):7011–7016. doi: 10.1073/pnas.96.12.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 35.Zuckman DM, Hung JB, Roy CR. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol Microbiol. 1999;32(5):990–1001. doi: 10.1046/j.1365-2958.1999.01410.x. [DOI] [PubMed] [Google Scholar]
- 36.Cambronne ED, Roy CR. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 2007;3(12):e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67(6):1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 38.Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6(12):e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feeley JC, et al. Charcoal-yeast extract agar: Primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10(4):437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.