Significance
The prevailing dogma since the identification of the t (11, 19) translocation gene product as a fusion of the cAMP response element binding protein (CREB)-regulated transcriptional coactivator 1 (CRTC1) and the NOTCH coactivator mastermind-like 2 (MAML2) in malignant salivary gland tumors has been that aberrant activation of CREB and/or NOTCH transcription programs drives oncogenesis. However, combined expression of the parental coactivator molecules CRTC1 and MAML2 is not sufficient to induce transformation, suggesting an added level of complexity. Here we describe gain-of-function interactions between the CRTC1/MAML2 (C1/M2) coactivator fusion and myelocytomatosis oncogene (MYC) oncoproteins that are necessary for C1/M2-driven transformation. Our findings suggest that targeting the C1/M2–MYC interface represents an attractive strategy for the development of effective and safe anticancer therapeutics in tumors harboring the t (11, 19) translocation.
Abstract
Chimeric oncoproteins created by chromosomal translocations are among the most common genetic mutations associated with tumorigenesis. Malignant mucoepidermoid salivary gland tumors, as well as a growing number of solid epithelial-derived tumors, can arise from a recurrent t (11, 19)(q21;p13.1) translocation that generates an unusual chimeric cAMP response element binding protein (CREB)-regulated transcriptional coactivator 1 (CRTC1)/mastermind-like 2 (MAML2) (C1/M2) oncoprotein comprised of two transcriptional coactivators, the CRTC1 and the NOTCH/RBPJ coactivator MAML2. Accordingly, the C1/M2 oncoprotein induces aberrant expression of CREB and NOTCH target genes. Surprisingly, here we report a gain-of-function activity of the C1/M2 oncoprotein that directs its interactions with myelocytomatosis oncogene (MYC) proteins and the activation of MYC transcription targets, including those involved in cell growth and metabolism, survival, and tumorigenesis. These results were validated in human mucoepidermoid tumor cells that harbor the t (11, 19)(q21;p13.1) translocation and express the C1/M2 oncoprotein. Notably, the C1/M2–MYC interaction is necessary for C1/M2-driven cell transformation, and the C1/M2 transcriptional signature predicts other human malignancies having combined involvement of MYC and CREB. These findings suggest that such gain-of-function properties may also be manifest in other oncoprotein fusions found in human cancer and that agents targeting the C1/M2–MYC interface represent an attractive strategy for the development of effective and safe anticancer therapeutics in tumors harboring the t (11, 19) translocation.
Gene regulatory circuits are generally controlled by transcriptional mechanisms tied to signal transduction pathways, and they allow cells to rapidly respond to environmental cues to control cell survival, growth, metabolism, and biological function. These controls are lost in cancers through various means (1–3), including chromosomal translocations that can augment the expression of oncogenes or that generate chimeric oncoproteins that are necessary and sufficient to provoke malignancy (4).
Chromosomal translocations found in epithelial tumors frequently involve the fusion of signaling molecules and regulators of transcriptional activity (5). The t (11, 19)(q21;p13.1) translocation gene product creates a unique oncoprotein fusion that is comprised of two transcriptional coactivators, the cAMP response element binding protein (CREB)-regulated transcriptional coactivator 1 (CRTC1) and the NOTCH/RBPJ coactivator mastermind-like 2 (MAML2) (6–11). The ensuing CRTC1/MAML2 (C1/M2) chimeric oncoprotein is comprised of the N-terminal 42 residues of CRTC1 encompassing a coiled-coil domain involved in CREB binding followed by the C-terminal 981 residues of MAML2, which includes a transcriptional activation domain. This C1/M2 coactivator fusion was originally identified in mucoepidermoid carcinomas (MECs) of the salivary gland and lungs, but has now also been detected in primary thyroid, breast, cervix, and skin tumors that present with MEC-like histology. As such, these are a class of epithelial cell malignancies that originate from mucous/serous glands present at different locations in the body (6, 8–15).
The genesis of these adult solid tumors was thought to be due to the direct activation of both CREB and NOTCH/RBPJ, the transcription factor targets of the CRTC1 and MAML2 coactivators involved in the t (11, 19) translocation, respectively (6, 15). However, forced expression of both CRTC1 and MAML2 is not sufficient to provoke transformation, whereas ectopic and/or inducible expression of C1/M2 transforms epithelial cells. Furthermore, the domain within MAML2 required for interacting with NOTCH is absent in the C1/M2 fusion protein, and those NOTCH genes originally identified as aberrantly regulated C1/M2 targets were found to also possess CREB-responsive promoters (16, 17). However, a C1/M2 deletion mutant (C1/M2Δ48–222) that is fully capable of interacting with and activating CREB cannot drive transformation (17). Collectively, these observations point to added levels of regulation outside of simply activating CREB or NOTCH, which is further supported by clinical patient data, where the C1/M2 translocation alone is characterized by a benign phenotype, and where the full tumorigenic potential of mucoepidermoid C1/M2-positive cells requires additional cooperating signals for a full-blown malignancy (18).
Collectively, these observations hint at additional functional activities present in the C1/M2 coactivator fusion that contribute to its oncogenic potential. To define these functions, we used a mammalian cell-based screen and discovered a C1/M2 gain-of-function, where C1/M2 binds to and coopts the function of MYC oncoproteins, and show that C1/M2-driven transformation requires MYC. Furthermore, a C1/M2 gene signature identifies human tumors having combined activation of MYC and CREB pathways. These findings suggest that gain-of-function activities may be a common feature of oncoprotein fusions and that such activities represent new avenues for therapeutic intervention.
Results
C1/M2 Interacts with the MYC Oncoprotein Network.
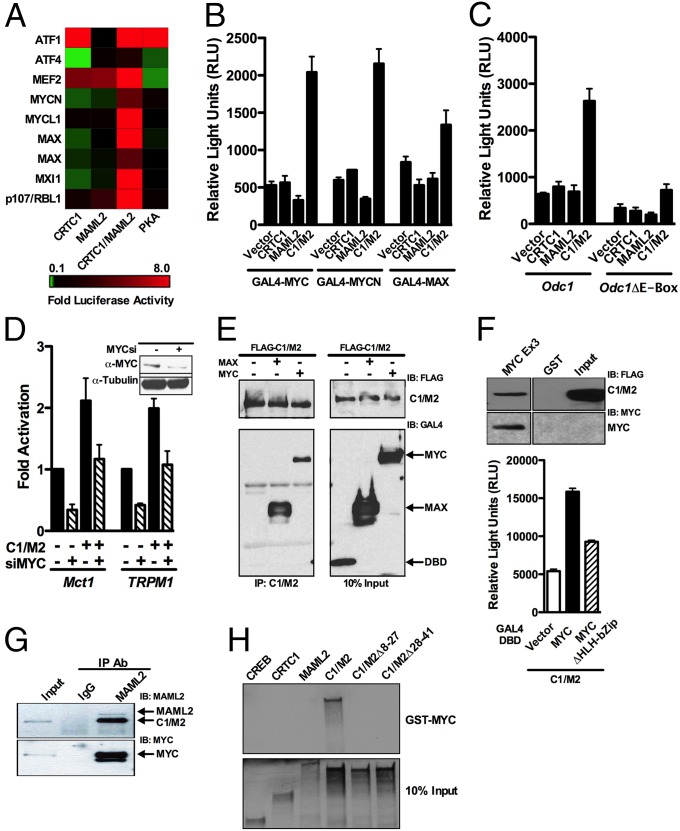
To assess if the C1/M2 coactivator fusion interacts with transcriptional regulators in addition to CREB or NOTCH/RBPJ, we used a previously developed and validated cell-based functional screen coined the “Coactivator Trap” composed of a library of nearly all human transcription factors fused to the GAL4 DNA binding domain (DBD) (SI Appendix, Fig. S1) (19). Surprisingly, the C1/M2 coactivator fusion selectively activated several proteins within the MYC:MAX network including MYCN, MXI (a.k.a. MAD2), and MAX (Fig. 1A). In contrast, neither parental CRTC1 nor MAML2 coactivator proteins displayed this activity. The catalytic subunit of PKA served as a control for the assay and displayed, as expected (20, 21), highly specific activation of GAL4–ATF1 and repression of GAL4–ATF4 reporter activity. Furthermore, as expected, expression of CRTC1 activated GAL4–ATF1 and repressed GAL4–ATF4 reporter activity, whereas MAML2 activated GAL4–MEF2, which are known binding partners for these coactivators (22–24). Finally, transfection experiments with a 5×GAL4::UAS-luciferase reporter and GAL4–DBD fusions of MYC, MYCN, or MAX selected from the transcription factor library confirmed these screen results (Fig. 1B).
Fig. 1.
The C1/M2 chimeric oncoprotein selectively coactivates the MYC network. (A) Heat map of screen data displaying luciferase reporter activation (red) and repression (green). GAL4–DBD library proteins are along the y axis, with coexpressed C1/M2 and controls along the x axis. (B) HEK293T cells were transiently cotransfected with CRTC1, MAML2, C1/M2, or empty expression vectors, and GAL4–DBD library proteins and the 5×GAL4::UAS-luciferase reporter and luciferase assays were performed 24 h posttransfection (n = 4; mean ± SEM). (C) Odc1 or Odc1ΔE-Box luciferase reporters were cotransfected in NIH 3T3 cells with CRTC1, MAML2, C1/M2, or empty expression vectors, and luciferase assays were performed 24 h posttransfection (n = 3; mean ± SEM). (D) Transient cotransfections of HEK293T cells were performed with MYC (MYCsi_A) or nonspecific siRNAs and the indicated E-Box–containing luciferase reporters, and luciferase assays were performed 72 h posttransfection (n = 4; mean ± SEM). (Inset) Western blot analysis demonstrated efficient knockdown of MYC with MYCsi_A. (E) Co-IP with FLAG-tagged C1/M2 and GAL4–MAX or GAL4–MYC was performed in transiently transfected HEK293T cells. Whole-cell lysates were immunoprecipitated using anti-FLAG–M2 magnetic beads followed by Western blot analysis with anti-FLAG or anti-GAL4–DBD antibodies. As a negative control, empty GAL4–DBD vector was cotransfected with FLAG–C1/M2. (F) C1/M2 interacts with MYC via its bHLH-Zip domain. GST pull-down assay using FLAG–C1/M2 lysates incubated with immobilized GST or GST–MYC Exon3 encompassing the bHLH-Zip domain (Upper Left). Binding of C1/M2 was analyzed by Western blotting with a FLAG antibody. HEK293T cells were transiently cotransfected with C1/M2, GAL4–DBD vector, or GAL4–DBD MYC proteins, and the 5×GAL4::UAS-luciferase reporter and luciferase assays were performed 24 h posttransfection (Upper Right). (G) Co-IP of endogenous C1/M2 with endogenous c-MYC from human H3118 MEC tumor cells that harbor the t (11, 19) translocation. Immunoprecipitation of C1/M2 was performed using a MAML2 antibody followed by Western blotting with anti-MAML2 or anti–c-MYC antibodies. (H) GST pull-down assay with full-length GST–MYC incubated with 35S-labeled, in vitro translated CREB, CRTC1, MAML2, C1/M2, C1/MΔ8–27, or C1/MΔ28–41. Shown are 10% input labeled polypeptides.
MYC oncoproteins are overexpressed in over 50% of all cancers, leading to aberrant expression of MYC target genes (25, 26). To assess the effect of the C1/M2 chimera on a native bona fide MYC target, transient transfections were conducted using the well-characterized, MYC-responsive Ornithine decarboxylase (Odc1) gene promoter driving luciferase (27). This promoter–reporter or one bearing mutations that disrupt Myc:Max binding to the Odc E-Box sequences CACGTG was cotransfected with vector or test expression constructs. The C1/M2 chimera potently activated the Odc promoter, and this was dependent on Myc:Max binding sites (Fig. 1C). In contrast, neither CRTC1 nor MAML2 proteins display this activity. Furthermore, C1/M2 induces the promoter activity of Mct1 and TRPM1, which harbor E-Box promoters and are bound by MYC (28, 29), and this C1/M2-mediated activation of Mct1 and TRPM1 promoters is effectively blocked by siRNA-directed knockdown of MYC (Fig. 1D and SI Appendix, Fig. S2 A–E). In contrast, MYC knockdown does not affect C1/M2 activation of either the EVX1 or 3×CRE luciferase reporters, indicating that the observed effects are selective for Myc-responsive promoters (SI Appendix, Fig. S2 F and G). Thus, C1/M2 activates MYC transcription targets in a MYC-dependent fashion.
To test if C1/M2 forms direct complexes with MYC and/or MAX, we performed coimmunoprecipitation (co-IP) studies. Lysates of HEK293 cells expressing GAL4–MYC or GAL4–MAX and FLAG-tagged C1/M2 were immunoprecipitated with anti-FLAG antibody, and complex formation was assessed by immunoblotting with GAL4 antibody. Both MYC and MAX coimmunoprecipitated along with C1/M2, whereas no interaction was seen with the GAL4–DBD alone (Fig. 1E). Because MYC and MAX heterodimerize via a common basic helix–loop–helix leucine zipper (bHLH-Zip) domain but MAX lacks the N-terminal transactivation domain present in MYC, we next tested if C1/M2 binds via the conserved bHLH-Zip domain. We expressed the C-terminal (exon 3) region of MYC (amino acids 266–453) encompassing the bHLH-Zip domain as a GST fusion protein and performed GST pull-down experiments with lysates from HEK293 cells expressing FLAG-tagged C1/M2. C1/M2 protein bound to GST–MYC Ex3 but not to GST alone (Fig. 1F, Upper) (30). Moreover, deletion of the MYC bHLH-Zip domain attenuates functional activation by C1/M2 (Fig. 1F, Lower). To determine if endogenous C1/M2 and MYC proteins interact, we performed immunoprecipitation of endogenous C1/M2 using anti-MAML2 antibodies and performed immunoblot analyses. Notably these studies established that C1/M2 coimmunoprecipitates with endogenous c-MYC in human H3118 mucoepidermoid tumor cells that harbor the t (11, 19) translocation and that express the C1/M2 fusion oncoprotein (Fig. 1G). To confirm that the interaction of C1/M2 with MYC is direct and to test if MYC binding is unique to the C1/M2 fusion, we performed GST pull-down assays using purified recombinant full-length MYC. Incubation with 35S-labeled in vitro translated full-length C1/M2 resulted in strong binding to GST–MYC, whereas full-length CREB, CRTC1, and MAML2 failed to bind to MYC (Fig. 1H). Importantly, microdeletions within the N-terminal region of CRTC1 disrupted MYC binding, confirming that MYC binds directly to the region of C1/M2 encompassing the fusion domain. Thus, the C1/M2 oncoprotein can complex with MYC and MAX and activate MYC transcription targets.
C1/M2 Functions as a MYC Coactivator.
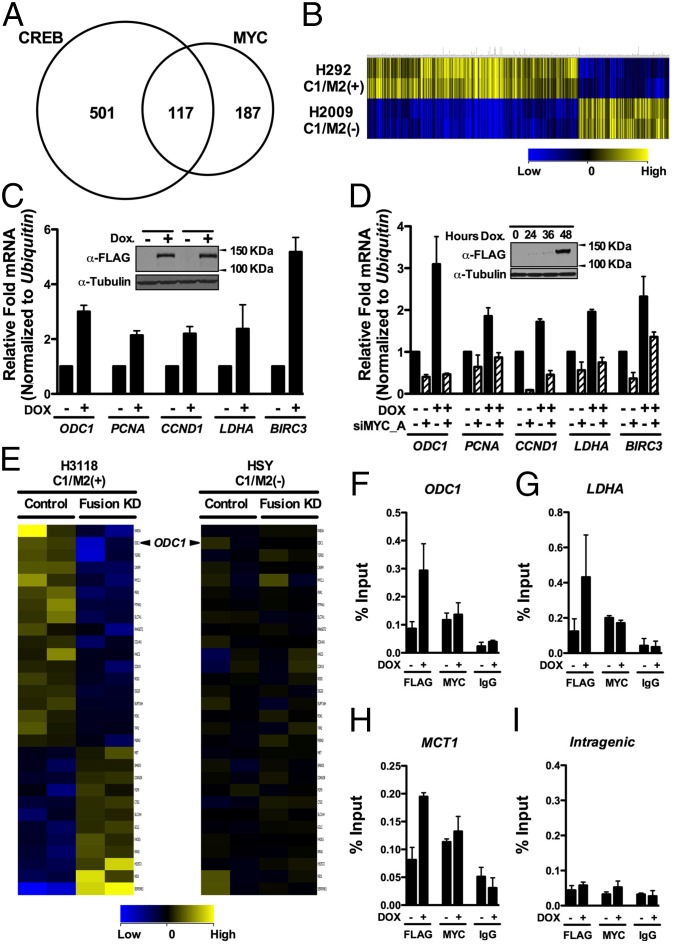
To determine the effects of C1/M2 on the transcriptome, we performed high-throughput Illumina RNA sequencing (RNA-seq). Given the epithelial nature of most tumors bearing the t (11, 19) translocation, we generated an isogenic doxycycline (Dox)-inducible stable cell line by flippase recombinase-directed integration of the C1/M2 oncogene at one genomic location in a human epithelial cell background (HEK293-CMVTetRTetOC1/M2). Samples of total RNA isolated from mock (no dox) versus dox-treated cells were analyzed and generated an average of 13.9 million reads using Illumina GA IIx 76-bp single reads that mapped to 23,455 annotated human Ref Seq genes. The identified gene list was filtered for targets previously characterized by chromatin immunoprecipitation microarray (ChIP-Chip) analysis to be directly bound by CREB or MYC transcription factors (31, 32). Of the 4,741 genes differentially regulated up or down more than 1.5-fold by C1/M2, a total of 805 genes were identified as direct targets of CREB or MYC (Fig. 2A and SI Appendix, Table S1). Specifically, 501 versus 187 of the expressed genes are direct CREB or MYC targets, respectively, whereas 117 of these genes are bound by both transcription factors. In contrast, of the 4,741 genes differentially regulated by C1/M2, a total of 79 genes were identified as direct NOTCH targets (33), despite the absence of a NOTCH binding domain within C1/M2, and 20 of these genes are shared with the C1/M2-regulated CREB–MYC gene signature (SI Appendix, Fig. S3 A and B). Importantly, application of Ingenuity Pathway Analysis to the 4,741 differentially regulated genes confirmed that CREB (P = 1.75−6) and both MYC (P = 3.04−10) and MYCN (P = 5.01−13) are top-ranked pathways that are significantly regulated by the C1/M2 oncoprotein.
Fig. 2.
MYC target genes are regulated by the C1/M2 oncoprotein. (A) RNA-seq analysis of genes regulated by a stably integrated, Dox-inducible C1/M2 transgene in FLP-In T-Rex HEK293 cells (HEK293-CMVTetRTetOC1/M2) reveals that C1/M2 regulates genes of the CRTC:CREB and MYC:MAX networks. A Venn diagram is shown, with values representing the number of differentially regulated genes that are direct CREB and/or direct MYC targets. In total, 187 genes induced by C1/M2 were scored as direct MYC targets. (B) Hierarchical clustering of differentially expressed C1/M2-regulated CREB–MYC signature genes in H292 MEC (C1/M2+) versus H2009 non-MEC tumor cells. guanine cytosine robust multi-array analysis (GCRMA) quantile normalization was applied to the raw CEL files for two biological replicates from H2009 and H292 MEC cell line samples with baseline transformation set to the median of all samples. (C) Real-time qPCR analysis of endogenous MYC target genes in HEK293-CMVTetRTetOC1/M2 cells +/− Dox. Fold induction is shown; expression was normalized to Ubiquitin mRNA levels (n = 4). (Inset) Western blot analysis of C1/M2 levels following Dox treatment in independently derived stable cell clones. (D) Knockdown of MYC (siMYC_A) blocks the induction of Myc target genes by C1/M2 in double stable TRE-Tight-C1/M2 Tet-On Advanced A549 NSCLC lung cancer cells (A549-CMVrtTA2-M2TRE-TightC1/M2 cells). These cells were transfected with siMYC_A or nonspecific siRNAs (NSsi; −siMYC lanes) and then were treated +/− Dox for 48 h, and expression of MYC targets was assessed by real-time qPCR. The fold induction relative to nonspecific silencing RNA (NSsi) no Dox treatment is shown, and data were normalized to Ubiquitin mRNA levels (n = 4). (Inset) Western blot analysis of C1/M2 levels following Dox treatment. (E) Gene expression profiling of the C1/M2-regulated MYC signature genes in H3118 MEC tumor cells that harbor the t (11, 19) translocation and express C1/M2 compared with HSY tumor cells that lack the translocation and do not express C1/M2 following lentiviral-mediated delivery of C1/M2 (Fusion KD) or control shRNAs (n = 2 biological replicates). (F–I) C1/M2 is recruited to endogenous MYC-responsive promoters. Chromatin from HEK293-CMVTetRTetOC1/M2 cells +/− Dox (48 h) was immunoprecipitated with α-FLAG or α-MYC antisera, or with isotype-matched normal IgG, and analyzed for occupancy of the promoter-regulatory regions of the (F) ODC1, (G) LDHA, and (H) MCT1 genes relative to occupancy of a nonspecific (I) intragenic region of CCNB1 (84). Real-time qPCR quantification of C1/M2 and MYC occupancy is expressed as percent chromatin precipitated relative to input (n = 4). Data in C and D as well as F–I represent mean ± SEM.
Annotation of the C1/M2-regulated CREB–MYC target genes by their Gene Ontology (GO) classification indicates that the C1/M2 oncoprotein regulates genes involved in several biological, cellular, and molecular processes common to either CREB- or MYC-regulated targets, including genes involved in cell cycle, signal transduction, energy metabolism, and biosynthesis of DNA, RNA, and protein (SI Appendix, Fig. S4). These results suggest that coactivation of the MYC:MAX pathway is an important component of the oncogenic process provoked by C1/M2.
To validate the relevance of our identified C1/M2-regulated CREB–MYC gene targets (805 direct CREB and/or MYC target genes), we used a bioinformatics approach to test if this gene list could identify MEC tumor cell lines with known C1/M2 involvement. Analysis of H292 lung MEC cells that harbor the t (11, 19) translocation and express C1/M2, and of H2009 lung tumor cells that lack C1/M2, demonstrated that this signature accurately discriminates these distinct MEC tumors based solely on regulation of our identified C1/M2-regulated CREB–MYC gene targets (Fig. 2B and SI Appendix, Table S2).
Collectively, these interaction and expression analyses suggested that the C1/M2 oncoprotein would activate MYC transcription targets. To test this, real-time qPCR was performed for several well-characterized MYC targets in HEK293-CMVTetRTetOC1/M2 cells +/− Dox. A set of MYC target genes were selected based on their functional relevance to the “hallmarks of cancer” as they relate to those biological capabilities acquired by tumor cells to promote tumorigenesis (1, 2). These capabilities include genes that regulate DNA replication, cellular proliferation, growth, survival, metabolism, and migration. For example, Dox induction of C1/M2 activated expression of the MYC metabolic target genes ODC1 and LDHA, the proliferative genes PCNA and CCND1, and the cell survival gene BIRC3 (Fig. 2C). C1/M2 expression also activated several CREB targets including the master transcriptional regulator NR4A2, proliferative genes FOS and JUNB, the transcriptional coactivator and metabolic regulator PGC-1α, and the glucose transporter SLC2A3 (SI Appendix, Fig. S5A). With respect to gain-of-function MYC coactivation, synergy is observed between C1/M2 and MYC in inducing Odc1 transcripts in NIH 3T3 fibroblasts transduced with Dox-inducible C1/M2 retrovirus and that were stably engineered to express the tamoxifen-regulated Myc–ER transgene (34) along with the reverse Tet transactivator rtTA2-M2 (SI Appendix, Fig. S5B). Cooperation between C1/M2 and MYC for regulation of direct MYC target genes was further confirmed by MYC siRNA knockdown in a Dox-inducible C1/M2 human lung cell line stably expressing rtTA2-M2 (Fig. 2D). Importantly, induction of C1/M2 expression does not affect endogenous levels of CRTC1 or MAML2, or the levels of MYC or MAX (SI Appendix, Fig. S5C). Thus, C1/M2 activates endogenous MYC target genes involved in several key aspects of tumor development, and this activation is dependent on the presence of MYC.
RNAi-mediated knockdown of endogenous C1/M2 in mucoepidermoid (MEC) tumor cell lines harboring t (11, 19) blocks tumor cell growth (35, 36). We therefore assessed effects of shRNA-directed C1/M2 knockdown on C1/M2-regulated MYC signature genes in H3118 lung MEC cells that harbor the t (11, 19) translocation and express C1/M2 (Fig. 2E, Left and SI Appendix, Table S3). Specifically, we used a previously characterized retrovirus that expresses shRNA targeting C1/M2 (Fusion KD) or a control shRNA (36) to transduce H3118 MEC cells that express C1/M2+ and HSY MEC cells that lack the fusion (C1/M2−). C1/M2 knockdown in H3118 MEC reduced the levels of transcripts for several of the identified C1/M2-regulated MYC signature genes, including ODC1. In contrast, C1/M2 knockdown in HSY tumor cells that lack the t (11, 19) had no effect on these targets relative to those of the control shRNA (Fig. 2E, Right and SI Appendix, Table S3). Moreover, C1/M2 knockdown or overexpression in fusion-positive or -negative MEC tumor cell lines had no affect on MYC levels (SI Appendix, Fig. S6 A and B). Importantly, although two recent reports have claimed that MYC is a global amplifier of transcription of active genes (37, 38), Myc does not affect the expression of C1/M2 (SI Appendix, Fig. S6C).
Collectively, these data suggest that C1/M2–MYC:MAX complexes bind to endogenous MYC-responsive promoters. To test this, ChIP analyses were performed using HEK293-CMVTetRTetOC1/M2 cells to assess if C1/M2 inducibly bound to several promoters shown in the USCS Genome Browser are occupied by MYC (SI Appendix, Fig. S7A) (39, 40). Real-time quantitative PCR (qPCR) analysis confirmed MYC occupancy at the promoter regulatory regions of ODC1, LDHA, and MCT1 and showed that with Dox-induced FLAG–C1/M2 it was recruited to these same promoters, relative to a nonspecific intragenic region, which lacked this enrichment (Fig. 2 F–I). Direct comparison of C1/M2 occupancy at E-Box motifs in the LDHA promoter relative to its occupancy at cAMP response element (CRE) motifs in the NR4A2 promoter established C1/M2 recruitment to both MYC- and CREB-responsive promoters in Dox-treated HEK293-CMVTetRTetOC1/M2 cells, respectively (SI Appendix, Fig. S7 B and C). Thus, C1/M2 binds to MYC and directs transcription of MYC-responsive target genes by binding to E-Box–containing promoters.
The N-Terminal Gain-of-Function Domain of C1/M2 Directs MYC Activation and Is Required for Epithelial Cell Transformation.
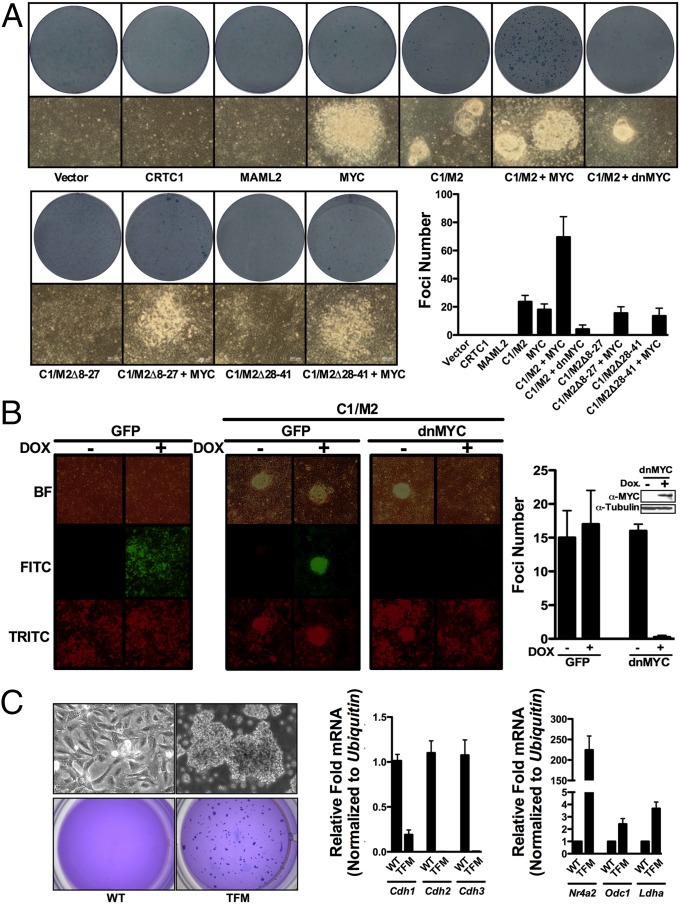
Forced expression of the C1/M2 oncoprotein fusion transforms rat kidney epithelial cells (RK3E), whereas cooverexpression of CRTC1 or MAML2 does not (6, 17) (SI Appendix, Fig. S8). To test if MYC is necessary for C1/M2 transformation, we performed focus formation assays with RK3E cells (Fig. 3A). As previously observed, neither CRTC1 nor MAML2 overexpression provoked foci formation, whereas C1/M2 promoted the formation of nearly symmetrical, punctate foci with clearly defined edges (6, 17). These foci were morphologically distinct from those observed following MYC overexpression alone, which were irregularly shaped. Notably, cotransfection with a dominant-negative (In373; dnMYC) form of MYC (41–43) or a dominant-negative (A-CREB; dnCREB) form of CREB (44) impaired C1/M2-induced focus formation (Fig. 3A and SI Appendix, Fig. S8).
Fig. 3.
MYC is necessary for CTRC1/MAML2-directed transformation of epithelial cells. (A) Rat kidney epithelial cells (RK3E) were transfected with the indicated expression plasmids, plated, grown for 3 wk, and then fixed with methanol and stained with methylene blue to quantify foci formation. Representative images of foci morphology before staining (bottom photographs) along with wells after staining (Lower photographs) and quantitation of observed foci numbers are shown (n = 3). (B) TRE-Tight-C1/M2 Tet-On Advanced RK3E cells expressing Dox-inducible GFP (RK3E-CMVrtTA2-M2TRE-TightGFP-PGKtdTomato) or In373-dnMYC (RK3E-CMVrtTA2-M2TRE-TightdnMYC-PGKtdTomato) were assessed for foci formation following the induction of C1/M2 by Dox treatment. RK3E cells engineered to inducibly express GFP were allowed to form monolayers and then treated with Dox or vehicle for 72 h and imaged (Left). RK3E cells engineered to inducibly express GFP or In373-dnMYC were transfected with C1/M2, allowed to form monolayers, and then were treated with Dox or vehicle, and foci formation imaged and quantitated at 3 wk posttransfection (Right) (n = 2). (C) Myc augments the transforming potential of C1/M2 in primary RK3E cells. (Upper) Phase contrast of parental RK3E cells (WT) or RK3E transformed by C1/M2 plus MYC (TFM). (Lower) Colony formation in soft agar was assessed for RK3E WT or TFM cells. A total of 5 × 103 cells were plated in 0.7% agar medium over agar underlayers. At 3 wk plates were stained with crystal violet to quantify colony numbers and were photographed. (Right) Real-time qPCR analysis of Cdh1-3, Nr4a2, Odc1, or Ldha levels in parental RK3E cells (WT) or in cells transformed with C1/M2 plus Myc (TFM). Fold changes are expressed relative to WT, and data are normalized to Ubiquitin mRNA levels (n = 4). Data in A–C represent mean ± SEM.
Because RK3E cells are traditionally transfected with the desired expression plasmids to perform focus formation assays, the possibility exists that foci that develop in the dnMYC condition represent those cells that did not receive dnMYC and therefore escape the block imposed by dnMYC. To test whether dnMYC is capable of completely blocking C1/M2-induced transformation, we generated RK3Es that stably express Dox-inducible dnMYC. Universal expression of dnMYC revealed that these cells were totally refractory to C1/M2-induced focus formation in the presence of Dox compared with control, vector-only RK3E cells (Fig. 3B). Finally, combined overexpression of both C1/M2 and MYC significantly increased the size and number of irregular-shaped foci (Fig. 3A). Moreover, C1/M2 and MYC also induced anchorage-independent growth of RK3E cells and led to transformed colonies (TFMs) that expressed markedly reduced levels of cadherin receptor genes (Cdh1, Cdh2, Cdh3) and increased levels of the Creb-responsive gene Nr4a2 and the Myc-responsive genes Odc1 and Ldha (Fig. 3C).
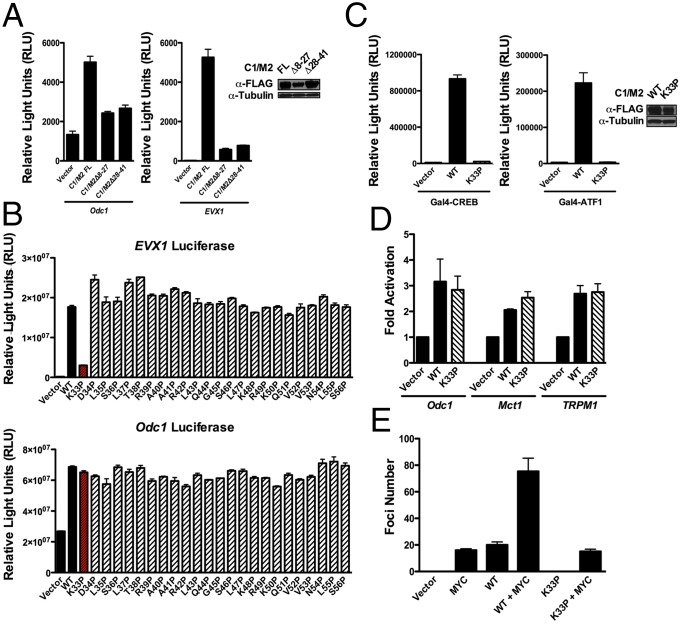
The N-terminal 42 amino acids of the C1/M2 fusion encompassing the CREB binding domain (CBD) of CRTC1 are required for RK3E transformation and focus formation (16). Indeed, forced expression of N-terminal in-frame deletion mutants of C1/M2 (Δ8–27 and Δ28–41) failed to synergize with MYC in RK3E cell transformation and did not activate the Odc1 reporter (Figs. 3A and 4A). However, because these mutants also block coactivation of the CREB-responsive EVX1 promoter (Fig. 4A, Right), we used proline-scanning mutagenesis to create a library of point mutants spanning the C1/M2 fusion domain, to screen for mutations that discriminate between CREB and MYC coactivation (SI Appendix, Fig. S9). This library was screened for loss-of-function activity on the EVX1 and Odc1 luciferase reporters. These analyses identified K33 as a site sensitive to proline substitution, where the K33P C1/M2 mutant failed to activate the CREB target EVX1 yet maintains the ability to activate Odc1 (Fig. 4B). Importantly, K33 is immediately adjacent to two residues conserved across all three CRTC family members that were recently shown to be important for interactions with the CREB bZip domain (45). The disabling effects of the K33P mutant on the native EVX1 promoter were confirmed using a completely defined 5×GAL4::UAS promoter, which revealed that the C1/M2 K33P mutant could not activate either CREB or its paralog ATF1 (Fig. 4C). In contrast, the C1/M2 K33P mutant is comparable to wild-type C1/M2 in coactivating MYC-responsive promoters (Fig. 4D). Notably, the C1/M2 K33P mutant was totally defective in inducing RK3E cell transformation (Fig. 4E). Collectively, these results indicate that coactivation of both MYC and CREB is necessary for C1/M2-induced epithelial cell transformation.
Fig. 4.
Coactivation of CREB and MYC by C1/M2 is functionally separable. (A) NIH 3T3 cells were transiently transfected with Odc1 or EVX1 luciferase reporters along with C1/M2, C1/M2Δ8–27, C1/M2Δ28–41, or empty expression vector, and luciferase assays were performed 24 h posttransfection (n = 4). (B) Functional screen for site-directed mutants that can discriminate between CREB and MYC transcription factor targets. Transient assays of EVX1-luciferase or Odc1-luciferase reporters in HEK293T cells cotransfected with wild-type (WT) C1/M2 versus the library of C1/M2 proline point mutants (n = 4). (C) Effect of wild-type (WT) C1/M2 versus proline point mutant (C1/M2-K33P) in a transient assay of 5×GAL4::UAS-luciferase reporter in HEK293T cells cotransfected with GAL4–CREB or GAL4–ATF1 (n = 3). (Inset) Western blot analysis of C1/M2 WT and K33P mutant expression levels. (D) HEK293T cells were cotransfected with WT C1/M2 or the C1/M2-K33P mutant and the indicated MYC-responsive luciferase reporters (n = 4). (E) Quantitation of observed foci numbers in RK3E focus assays comparing C1/M2 WT and C1/M2-K33P (n = 2). Data in A–E represent mean ± SEM.
The C1/M2 Gene Signature Predicts Tumors with MYC and CREB Involvement.
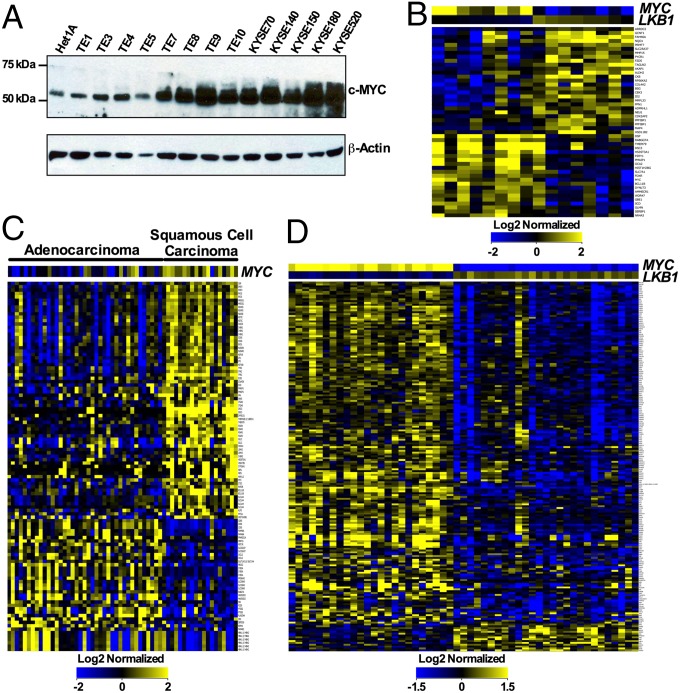
Activation of CREB by CRTCs is harnessed by CRTC phosphorylation, which retains these coactivators in the cytoplasm (46). Therefore, constitutive CREB activity is manifest following inactivation of upstream regulatory kinases such as LKB1 that phosphorylate CTRCs (47–49). Indeed, several human esophageal tumor cell lines express reduced levels of LKB1 and CRTC1 phosphorylation relative to Het1 immortalized, normal esophageal epithelial cells (48). Notably, Western blot analysis revealed increased MYC levels in well over half (9/13) of such tumors, suggesting that activation of both MYC and CREB pathways is indeed manifest in some tumor types (Fig. 5A).
Fig. 5.
The C1/M2 gene signature predicts human malignancies with dual activation of MYC and CREB transcription networks. (A) Western blot analysis reveals increased MYC levels in several human esophageal cancer cell lines relative to immortal normal esophageal Het1A epithelial cells. (B) Hierarchical clustering of differentially expressed C1/M2-regulated CREB–MYC signature genes from multiple esophageal (n = 16) tumor samples based on an inverse correlation between MYC and LKB1 expression levels. The esophageal samples (75 total samples) were log2 normalized, the baseline transformation was set to the median of all samples, and then samples were divided into two groups relative to median MYC and LKB1 signals: HighMYC–LowLKB1 (eight samples) and LowMYC–HighLKB1 (eight samples). (C) Hierarchical clustering based solely on the presence of differentially expressed C1/M2-regulated CREB–MYC signature genes in lung adenocarcinoma and squamous cell carcinoma tumor samples. GCRMA quantile normalization was applied to the raw CEL files (58 total) for 40 lung adenocarcinoma and 18 squamous cell carcinoma samples, with baseline transformation set to the median of all samples. (D) Hierarchical clustering of differentially expressed C1/M2-regulated CREB–MYC signature genes from multiple lung adenocarcinoma samples (n = 51) based on an inverse correlation between MYC and LKB1 expression levels. GCRMA quantile normalization was applied to the raw CEL files (462 total), with baseline transformation set to the median of all samples. A total of 51 samples (either HighMYC–LowLKB1 or LowMYC–HighLKB1) were selected for further analysis based on identification of top (high) and bottom (low) quartile samples. Data in B–D represent fold change greater than 1.5 (P < 0.05).
To more broadly define the importance of coactivating both MYC and CREB transcription targets for transformation and tumorigenesis, we first validated scenarios leading to activation of direct CREB or MYC target genes. Gene lists of targets directly bound by CREB or MYC transcription factors were generated by analyzing previously characterized ChIP microarray (ChIP-Chip) datasets (31, 32). LKB1-dependent regulation of CREB “On/Off” scenarios were then validated by mining previously published microarray datasets generated from a total of 35 pathologically scored primary or metastatic tumors from transgenic mice intercrossed with a conditionally activated Kras allele (Lox–Stop–Lox–KrasG12D) and either Lkb1 wild-type, germ-line null, or conditionally null alleles (50). Gene expression profiling analyses established that a CREB “On” scenario is clearly manifest in primary and metastatic lung tumors from conditional LKB1-null mice when queried for our CREB target gene signature, where these tumors have marked induction in CREB target genes (SI Appendix, Fig. S10A and Table S4) (50). Moreover, mining previously published microarray datasets generated from LKB1-null human H2126 lung carcinoma cells where LKB1 expression was restored (50) blocks CREB target gene activation (a CREB “Off” scenario) (SI Appendix, Fig. S10B and Table S5). MYC activity is manifest following amplification or overexpression, and thus, MYC On/Off scenarios were validated by mining previously published microarray datasets generated from primary mouse B cells following lipopolysaccharide stimulation of MYC expression (37). Gene expression profiling analyses established that a MYC On scenario is clearly manifest upon MYC overexpression when we queried our MYC target gene signature (SI Appendix, Fig. S10C and Table S6).
Next, we investigated the utility of our identified C1/M2-regulated CREB–MYC gene signature (Fig. 2A and SI Appendix, Table S1) in predicting human cancers that possess simultaneous activation of both CREB and MYC through independent mechanisms (e.g., MYC amplification or overexpression coupled with aberrant CRTC activation). The rationale for pursuing this analysis is supported by data indicating that LKB1 expression (CREB Off) maintains epithelial cell integrity and blocks tumor cell proliferation by inducing degradation of MYC protein (51, 52). We used the C1/M2-derived CREB–MYC gene signature to query several tumor microarray datasets from the Gene Expression Omnibus, National Cancer Institute caArray, or The Cancer Genome Atlas repositories (53). Application of this signature to primary esophageal tumor microarray datasets having an inverse correlation of MYC (high, a MYC On scenario) and LKB1 (low or absent, a CREB On scenario) levels revealed that a subset of these tumors share significant overlap with the C1/M2 CREB–MYC signature (Fig. 5B and SI Appendix, Table S7). Over 25% of all lung tumors display mutations affecting MYC expression (54). Analysis independent of high MYC and low LKB1 levels and based solely on the C1/M2-regulated CREB–MYC signature confirmed combined activation of the CREB and MYC networks in a large cohort of lung tumor samples, and this analysis discriminated tumor subtypes, where lung squamous cell carcinomas that are known to have amplifications of MYC expression (55) display the CREB–MYC signature (Fig. 5C and SI Appendix, Table S8). However, this analysis revealed that several lung adenocarcinoma tumor samples also display the CREB–MYC signature. Indeed, application of this signature to primary lung adenocarcinoma microarray datasets having an inverse correlation of MYC (high) and LKB1 (low or absent) levels confirmed that a subset of lung adenocarcinomas display significant overlap with the C1/M2 CREB–MYC signature, and this connotes reduced overall survival in these same lung adenocarcinoma patients (Fig. 5D and SI Appendix, Table S9 and Fig. S11A). Moreover, analysis of overall survival for The Cancer Genome Atlas lung adenocarcinoma datasets where LKB1 mutation status and MYC expression status were available confirms that LKB1 loss-of-function coupled with high MYC levels connotes reduced overall survival (SI Appendix, Fig. S11B). Thus, dual activation of the MYC and CREB transcription factor networks is a common feature of some human tumor types, validating the relevance of the identified C1/M2-regulated CREB–MYC target genes. Furthermore, these data support the functional relevance of de novo interactions between C1/M2 and MYC in activating MYC target genes that are important for transformation and tumorigenesis.
Discussion
Oncoprotein fusions are conventionally thought to augment the activity of one partner of the chimera, for example as seen in the activation of the ABL1 tyrosine kinase in BCR–ABL1, or to activate both components of the fusion, for example the transcription factors E2A and PBX1 in the E2A–PBX1 oncoprotein found in pre–B-cell acute lymphocytic leukemia (56, 57). The latter is true for the C1/M2 translocation where both CREB and NOTCH targets are activated in these tumors (16, 17, 35). However, here we demonstrate a gain-of-function de novo interaction between the C1/M2 coactivator fusion and MYC family oncoproteins that drives cellular transformation via functional complementation of the MYC and CREB transcription networks. These findings suggest that targeting C1/M2–MYC interactions represents an attractive strategy for developing effective and safe anticancer therapeutics in tumors harboring the t (11, 19) translocation.
A widely held notion is that proteins can be divided into distinct modular domains that enable or carry out specific functions (58, 59). Given our findings, it is tempting to speculate that the functional nature of some chimeric oncoproteins is not simply the additive sum of the modular parts recruited by each fusion partner, but rather includes the generation of de novo functions. In fact, the trained predictive algorithm Globplot (60) revealed an extensive, unique region of folded globular structure corresponding to residues 1–84 of C1/M2 compared with CRTC1 and MAML2 (SI Appendix, Fig. S12), suggesting this domain might confer the identified de novo interactions with MYC. Future studies directed at structural analyses will be important for defining the mechanism of this gain-of-function interaction.
Activation of MYC by C1/M2 presents an attractive mechanism for coopting proliferative signals and for coordinating metabolic changes that accompany the transformed state. The C1/M2 oncoprotein may induce the transcription of MYC target genes by stabilizing interactions of the MYC:MAX complex with transcriptional coactivators such as CBP/ p300. Specifically, because the N-terminal transactivation domain of MYC also interacts with histone acetyl transferases (61), the C1/M2 oncoprotein fusion may coordinate interactions with both the MYC bHLH-Zip domain and CBP/p300 to augment MYC activity at its target gene promoters by promoting histone acetylation. The clinical relevance of this gain-of-function interaction with MYC is supported by CGH+SNP microarray analyses of C1/M2 fusion-positive patients with unfavorable prognoses that have CNV gains and losses, including deletions of the CDKN2A locus that harbors both p16INK4a and p14ARF (18). MYC-driven tumorigenesis activates the Arf–p53 tumor suppressor pathway (62), and loss of ARF is a hallmark of MYC-driven tumors. Therefore, the unfavorable prognoses for C1/M2 fusion-positive MECs are likely due to selection of deletions within CDKN2A that are driven by, and cooperate with, C1/M2-mediated activation of MYC.
Our studies conclusively demonstrate that both MYC and CREB activation are necessary for C1/M2-driven transformation. Ectopic interactions between C1/M2 and MYC coupled with CREB activation in epithelial cells may contribute to tumor progression and/or to maintenance of the malignant state, especially given that CRTC1–CREB activity has been implicated in the proliferation of epithelial stem cells and MYC is important for cell “stem-ness” and pluripotency (63–68). An ordered N-terminal coiled-coil domain of CRTC1 directs its binding to the CREB bZip domain at promoter-proximal CRE motifs and coordinates interactions with the histone acetyl transferases CBP/p300 (24, 46, 69, 70). Notably, this 42-amino-acid N-terminal CBD is the only region of CRTC1 present in the C1/M2 fusion oncoprotein (6, 24) and contributes to C1/M2-induced cell transformation (16).
In addition to identifying ectopic interactions between C1/M2 and MYC, our characterization of a C1/M2 gene signature composed of direct CREB and MYC target genes successfully predicted human cancers, with dual CREB–MYC involvement arising by mechanisms that are independent of the C1/M2 oncogene. Common methods of MYC activation include amplification, overexpression, or translocation (25), whereas CREB activation may occur via loss of negative feedback loops and/or gain-of-positive feedback loops that directly affect CREB or indirectly affect upstream signaling components (71, 72). For example, CRTC1 is a potent activator of CREB-dependent transcription complexes that are assembled in response to hormone and stress signaling (19, 24, 71, 73). Specifically, we show that increased MYC expression coupled with alterations in CRTC function arising from loss-of-function or silencing of the upstream signaling kinase LKB1 leads to the coordinate activation of the MYC and CREB transcription networks, respectively. Aberrant CREB activity coupled with activation of an oncogene such as MYC may provide a selective advantage, as CREB directly induces several prosurvival factors including BCL2 and the NR4A family members, thereby counteracting MYC-induced apoptosis (16, 35, 74, 75). This is supported by studies showing a role for CREB activation in several cancers including melanoma, acute myeloid leukemia, and nonsmall cell lung cancer (76–78).
In summary, de novo interactions between the C1/M2 chimeric fusion and Myc family oncoproteins drive cellular transformation via functional complementation of the CREB and MYC transcription networks. Our investigation into the mechanisms of C1/M2 function provides insight into the function of other chimeric gene fusions and more broadly highlights the convergence of CREB and MYC pathways in other cancers.
Materials and Methods
Cell Lines.
HEK293T, FLP-In T-Rex HEK293, EcoPack 2–293, NIH 3T3, RK3E, and A549 cells were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (Gemini), GlutaMAX (Invitrogen), and penicillin, streptomycin, and l-glutamine (PSG) (Invitrogen). NIH 3T3–MycER was maintained in DMEM without Phenol Red (Cellgro) supplemented with 10% FBS, GlutaMAX, and PSG. NCI-H292 and NCI-H3118 cells were maintained in RPMI-1640 medium supplemented with 10% FBS, nonessential amino acids, sodium pyruvate, sodium bicarbonate (all from Invitrogen), GlutaMAX, and PSG. All cells were maintained at 37 °C with 5% CO2 in a humidified incubator.
High-Throughput Transcription Factor Interaction Screen.
Conditions for the high-throughput mammalian Coactivator Trap assay were as described, with minor modifications (19). Briefly, reverse transfections of HEK293T cells were performed with CRTC1, MAML2, C1/M2, or empty expression vectors; the 5×GAL4::UAS-luciferase reporter; and GAL4–DBD library proteins arrayed into 384-well plates. Luminescence was measured 24 h posttransfection on an Analyst GT plate reader (Molecular Devices) following addition of BriteLite Plus (PerkinElmer).
Transfection and Luciferase Reporter Assays.
Transfection of HEK293T cells for luciferase assays was carried out using Lipofectamine 2000 as described, and where indicated, RNAi was performed using control siRNAs (NSsi; Qiagen 1027280 or GFP-22; Qiagen 1022176) or target siRNAs (MYCsi_A, Qiagen SI02662611 or MYCsi_B, Qiagen SI00300902) (19, 24). Luminescence was measured 24 h posttransfection for standard assays or 72 h posttransfection for RNAi assays on an Envision plate reader (Perkin–Elmer) following addition of BriteLite Plus (PerkinElmer). Transfections of RK3E, A549, NCI-H292, and NCI-H3118 cells were performed with Fugene HD (Promega), whereas transfection of NIH 3T3 cells used Lipofectamine LTX with PLUS reagent (Invitrogen). The GAL4–DBD constructs, 5×GAL4::UAS-luciferase, EVX1-luciferase, and Odc1-luciferase reporters were as described (19, 27, 79).
Plasmid Construction and Generation of Stable Cell Lines.
The N-terminal FLAG-tagged C1/M2 cDNA was directionally cloned with 5′ KpnI and 3′ XhoI restriction enzyme sites into the pcDNA5/FRT/TO© vector. This construct was then used with the FLP-In T-Rex Core kit (Invitrogen) to generate the HEK293-CMVTetRTetOC1/M2 stable cells according to the manufacturer’s instructions. Stable rtTA2-M2–expressing A549, NIH 3T3–MycER, and RK3E cells were generated using pTet-On Advanced (Clontech), and clonal stable expressing lines were selected in G418R medium using sterile cloning discs (Spectrum). An N-terminal FLAG-tagged C1/M2 cDNA was directionally cloned with 5′ NotI and 3′ MluI restriction enzyme sites into pRetroX-Tight-Pur (Clontech), and ecotropic retrovirus was packaged in EcoPack 2–293 cells. A549 cells that stably express rtTA2-M2 were then transduced with RetroX-Tight-FLAG_C1/M2 virus, and clonal A549-CMVrtTA2-M2TRE-TightC1/M2 stable expressing lines were selected in PuromycinR medium using sterile cloning discs. For analysis of the effects of inducible expression of C1/M2 in cells that also express the Myc–ER transgene, an N-terminal FLAG-tagged C1/M2 cDNA was directionally cloned with 5′ NotI and 3′ MluI restriction enzyme sites into pRetroX-Tight-tdTomato, and ecotropic retrovirus was packaged and was then used to transduce NIH 3T3–MycER–CMVrtTA2-M2 cells. The dominant-negative MYC mutant (dnMYC, In373) affecting the basic region of the DBD was as described (41). For focus formation assays, GFP or dnMYC cDNAs were cloned into pRetroX-Tight-tdTomato, and ecotropic retrovirus was packaged and used to transduce RK3E-CMVrtTA2-M2 cells.
Western Blots.
Analysis of protein expression was performed using conventional methodologies. Antibodies included FLAG–M2 HRP conjugate (Sigma; A8592), MAML2 (Cell Signaling; 4618), MAML2 (Bethyl Labs; A300-681A), MYC N-262 (Santa Cruz; sc-764), MYC 9E10 (Santa Cruz; sc-40), and Tubulin (Sigma; T9026).
Co-IP and GST Pull-Down Assays.
Assays were performed as previously described with minor modifications (19, 24). Briefly, co-IP experiments were performed using HEK293T cells grown in 10-cm dishes transfected with Opti-MEM, Lipofectamine 2000, and 5 μg each of the indicated plasmids. The cells were lysed 24 h posttransfection in IP lysis buffer (Pierce) containing complete EDTA-free protease inhibitor mixture (Roche), and the cell lysates were immunoprecipitated using FLAG–M2 magnetic beads (Sigma) and a DynaMag2 magnetic particle concentrator (Invitrogen) followed by immunoblotting using GAL4–DBD (Abcam) or FLAG–M2 HRP conjugate (Sigma) antisera.
Endogenous co-IP experiments were performed using 40 mg whole-cell lysate harvested from NCI-H3118 MEC cells and immunoprecipitated with 25 μL MAML2 antibody (CST 4618) and 50 μL Protein A/G agarose. After extensive washing, the agarose beads were boiled with 30 μL SDS/PAGE loading buffer, and the immunoprecipitates were analyzed by SDS/PAGE followed by silver staining and Western blot with indicated antibodies.
GST–MYC Ex3 (30) and GST proteins were expressed in Escherichia coli using standard techniques. GST pull-down assays were performed as described previously (30) followed by immunoblot using FLAG–M2 HRP conjugate (Sigma) or MYC 9E10 (Santa Cruz) antisera. We incubated 1 μg of purified, full-length GST–MYC protein (Abnova) with products of TnT-coupled reticulate lysate in vitro transcription/translation assays (Promega) as previously described (80).
Transcriptome Analysis.
HEK293-CMVTetRTetOC1/M2 cells were cultured for 72 h +/− Dox (Sigma), and total RNA was extracted using RNeasy RNA extraction kit (Qiagen) followed by on-column DNase digestion (Qiagen). The mRNA pool was purified from 1 μg of total RNA using poly T beads, and Illumina Next-Generation Sequencing (NGS) was performed following Illumina protocols. Sequence data were exported as qseq files, uploaded into both Geospiza and CLC Bio software, and mapped using Burrows–Wheeler algorithm against the human reference genome (Human Build 36) (81, 82). Data for each gene are shown as scaled gene counts within each lane divided by the total lane count, the standard reads per kilobase of exon model per million mapped reads (RPKM). The RPKM values were then used to directly compare each sample. Genes with less than 10 total reads were considered absent and eliminated from analysis.
Bioinformatics.
GlobPlot (http://globplot.embl.de/html/abstract.html) was used to identify putative ordered domains for CRTC1, MAML2, and C1/M2, and the PSIPRED protein structure prediction server (http://bioinf.cs.ucl.ac.uk/psipred) was used to predict secondary structure (60, 83). The default settings were used for each web service to investigate the amino acid sequences.
Custom java software using the HtmlUnit library (http://htmlunit.sourceforge.net) was used to query the complement of direct CREB and/or MYC target genes identified by Illumina NGS. A list of gene symbols corresponding to target genes that were regulated less than or greater than 1.5-fold was searched against ChIP-Chip databases, the CREB Target Gene Database (http://natural.salk.edu/CREB/, “Ave. HEK CREB Binding Ratio” > 2), and the MYC Cancer Gene Database (www.myc-cancer-gene.org/site/mycTargetDB.asp) (31, 32). The proportions of genes within the CREB or MYC networks were then plotted as a Venn diagram (http://omics.pnl.gov/software/VennDiagramPlotter.php). For each identified gene symbol, a local copy of the GO database was queried to obtain a list of GO Slim terms associated with that symbol. The frequency of the GO Slim terms was plotted as pie charts.
All heatmaps were generated using GeneSpring GX11 (GS GX11, Agilent) hierarchical clustering algorithm. The similarity measure of genes was set to Pearson centered, and the Linkage rule was set to average. Yellow indicates higher than median and blue indicates lower than median signal for each probe set. Additional details of bioinformatics data processing, procedures, and analysis are provided in SI Appendix, SI Materials and Methods.
Real-Time qPCR.
Total RNA was extracted from the indicated cell lines using RNeasy RNA extraction kit followed by on-column DNase digestion and reverse transcribed to cDNA using SuperScript III (Invitrogen) according to the manufacturer’s instructions. Real-time qPCR was performed using SYBR Green I (Roche) on a Roche LightCycler 480 instrument. PCR amplicons for each target were designed using Roche Applied Science Universal Probe Library Assay Design Center software (SI Appendix, Table S9). Relative quantification was as described (19, 73).
ChIP.
Conditions for the ChIP assay were as described with minor modifications (19). Briefly, HEK293–CMVTetRTetOC1/M2 cells were cultured +/− Dox and processed for ChIP. The chromatin bound by C1/M2 or MYC was immunoprecipitated using FLAG–M2 or MYC antisera, respectively, and the purified DNA was PCR amplified with promoter-specific primers (SI Appendix, Table S10).
Focus Formation and Soft Agar Assays.
Transformation-induced foci formation was performed as described with minor modifications (6). Specifically, 2 × 105 RK3E cells seeded in six-well plates were transfected with 2 μg each (4 μg total) of the indicated plasmids using Fugene HD (Promega; E2311) with duplicate wells per condition and incubated at 37 °C with 5% CO2 in a humidified incubator. Fresh medium was provided three times a week over the course of 3–4 wk, and macroscopic foci (≥50 μm) were stained with methylene blue, counted, and photographed.
To assess anchorage-independent colony formation, six-well plates were prepared with 0.7% agar medium underlays using Gene Pure Low melt agar (ISC BioExpress) and complete DMEM. Single-cell suspensions of parental RK3E or RK3E cells transformed by C1/M2 and/or MYC isolated from the foci formation assays were plated in 0.7% agar medium at 1 × 104 cells per well with triplicate wells per condition. Additional medium was kept on the surface and changed three times a week over the course of a 3–4-wk incubation at 37 °C with 5% CO2 in a humidified incubator, and colonies were stained with crystal violet and photographed.
Proline-Scanning Mutagenesis and Functional Screen.
A small library of point mutants spanning the t (11, 19) fusion junction in the FLAG–C1/M2 plasmid was created according to QuikChange site-directed mutagenesis protocols using primers designed with QuikChange Primer Design Software (Agilent Technologies). This library was spotted into 96-well plates and screened for functional activity by reverse transfection of HEK293T cells with EVX1-luciferase or Odc1-luciferase reporters and wild-type C1/M2 or empty expression vectors as controls. Luminescence was measured 24 h posttransfection on an Envision plate reader (Perkin–Elmer) following the addition of BriteLite Plus (PerkinElmer).
Supplementary Material
Acknowledgments
We thank Min Guo and Mark Hall for providing the TRPM1-Luc and Mct1-Luc reporters, respectively; Joanne Doherty for the Mct1 ChIP primers; Min Guo, Kendall Nettles, and Matjaz Rokavec for technical assistance; and members of the M.D.C. and J.L.C. laboratories for helpful comments, suggestions, and scientific review of this manuscript. This work was supported by a Howard Temin Pathway to Independence Award in Cancer Research from the National Cancer Institute (NCI) (K99-CA157954) (to A.L.A.), National Institutes of Health/NCI R01 Grant CA100603 (to J.L.C.), a PGA National WCAD Cancer Research Fellowship and Ruth L. Kirschstein National Research Service Award from the National Cancer Institute (F32-CA134121) (to A.L.A.), the Margaret Q. Landerberger Research Foundation (to M.D.C.), a Swiss National Foundation Postdoctoral Fellowship (to F.X.S.), and monies from the State of Florida to Scripps Florida.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319176111/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 4.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 5.Rabbitts TH. Commonality but diversity in cancer gene fusions. Cell. 2009;137(3):391–395. doi: 10.1016/j.cell.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 6.Tonon G, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33(2):208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 7.Martins C, et al. A study of MECT1-MAML2 in mucoepidermoid carcinoma and Warthin’s tumor of salivary glands. J Mol Diagn. 2004;6(3):205–210. doi: 10.1016/S1525-1578(10)60511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behboudi A, et al. Clear cell hidradenoma of the skin-a third tumor type with a t(11;19)-associated TORC1-MAML2 gene fusion. Genes Chromosomes Cancer. 2005;43(2):202–205. doi: 10.1002/gcc.20168. [DOI] [PubMed] [Google Scholar]
- 9.Winnes M, et al. Frequent fusion of the CRTC1 and MAML2 genes in clear cell variants of cutaneous hidradenomas. Genes Chromosomes Cancer. 2007;46(6):559–563. doi: 10.1002/gcc.20440. [DOI] [PubMed] [Google Scholar]
- 10.Kazakov DV, et al. Skin-type hidradenoma of the breast parenchyma with t(11;19) translocation: Hidradenoma of the breast. Am J Dermatopathol. 2007;29(5):457–461. doi: 10.1097/DAD.0b013e318156d76f. [DOI] [PubMed] [Google Scholar]
- 11.Lennerz JKM, Perry A, Mills JC, Huettner PC, Pfeifer JD. Mucoepidermoid carcinoma of the cervix: Another tumor with the t(11;19)-associated CRTC1-MAML2 gene fusion. Am J Surg Pathol. 2009;33(6):835–843. doi: 10.1097/PAS.0b013e318190cf5b. [DOI] [PubMed] [Google Scholar]
- 12.El-Naggar AK, Lovell M, Killary AM, Clayman GL, Batsakis JG. A mucoepidermoid carcinoma of minor salivary gland with t(11;19)(q21;p13.1) as the only karyotypic abnormality. Cancer Genet Cytogenet. 1996;87(1):29–33. doi: 10.1016/0165-4608(95)00266-9. [DOI] [PubMed] [Google Scholar]
- 13.Tirado Y, et al. CRTC1/MAML2 fusion transcript in high grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin’s tumors: Implications for histogenesis and biologic behavior. Genes Chromosomes Cancer. 2007;46(7):708–715. doi: 10.1002/gcc.20458. [DOI] [PubMed] [Google Scholar]
- 14.Kaye FJ. Emerging biology of malignant salivary gland tumors offers new insights into the classification and treatment of mucoepidermoid cancer. Clin Cancer Res. 2006;12(13):3878–3881. doi: 10.1158/1078-0432.CCR-06-0791. [DOI] [PubMed] [Google Scholar]
- 15.O’Neill ID. t(11;19) translocation and CRTC1-MAML2 fusion oncogene in mucoepidermoid carcinoma. Oral Oncol. 2009;45(1):2–9. doi: 10.1016/j.oraloncology.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Coxon A, et al. Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res. 2005;65(16):7137–7144. doi: 10.1158/0008-5472.CAN-05-1125. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, et al. Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J. 2005;24(13):2391–2402. doi: 10.1038/sj.emboj.7600719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anzick SL, et al. Unfavorable prognosis of CRTC1-MAML2 positive mucoepidermoid tumors with CDKN2A deletions. Genes Chromosomes Cancer. 2010;49(1):59–69. doi: 10.1002/gcc.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amelio AL, et al. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci USA. 2007;104(51):20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: Activating transcription factor proteins and homeostasis. Gene. 2001;273(1):1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 21.Rehfuss RP, Walton KM, Loriaux MM, Goodman RH. The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J Biol Chem. 1991;266(28):18431–18434. [PubMed] [Google Scholar]
- 22.Shen H, et al. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20(6):675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemaigre FP, Ace CI, Green MR. The cAMP response element binding protein, CREB, is a potent inhibitor of diverse transcriptional activators. Nucleic Acids Res. 1993;21(12):2907–2911. doi: 10.1093/nar/21.12.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conkright MD, et al. TORCs: Transducers of regulated CREB activity. Mol Cell. 2003;12(2):413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22(56):9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 27.Packham G, Cleveland JL. Induction of ornithine decarboxylase by IL-3 is mediated by sequential c-Myc-independent and c-Myc-dependent pathways. Oncogene. 1997;15(10):1219–1232. doi: 10.1038/sj.onc.1201273. [DOI] [PubMed] [Google Scholar]
- 28.Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103(47):17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doherty JR, et al. Blocking lactate export by inhibiting the Myc target MCT1 disables glycolysis and glutathione synthesis. Cancer Res. 2014;74(3):908–920. doi: 10.1158/0008-5472.CAN-13-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrova N, et al. The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol Cell Biol. 1995;15(9):5188–5195. doi: 10.1128/mcb.15.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eilers M, Picard D, Yamamoto KR, Bishop JM. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340(6228):66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 35.Komiya T, et al. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene. 2006;25(45):6128–6132. doi: 10.1038/sj.onc.1209627. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, et al. Aberrantly activated AREG-EGFR signaling is required for the growth and survival of CRTC1-MAML2 fusion-positive mucoepidermoid carcinoma cells. Oncogene. 2013 doi: 10.1038/onc.2013.348. [DOI] [PubMed] [Google Scholar]
- 37.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita PA, et al. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2011;39(Database issue):D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ, et al. The human genome browser at UCSC. Genome Research. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stone J, et al. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang CV, McGuire M, Buckmire M, Lee WM. Involvement of the ‘leucine zipper’ region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- 43.Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70(6):901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 44.Ahn S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18(2):967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Q, et al. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc Natl Acad Sci USA. 2012;109(51):20865–20870. doi: 10.1073/pnas.1219028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119(1):61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Shaw RJ, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310(5754):1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu Y, et al. Altered LKB1/CREB-regulated transcription co-activator (CRTC) signaling axis promotes esophageal cancer cell migration and invasion. Oncogene. 2012;31(4):469–479. doi: 10.1038/onc.2011.247. [DOI] [PubMed] [Google Scholar]
- 49.Komiya T, et al. Enhanced activity of the CREB co-activator Crtc1 in LKB1 null lung cancer. Oncogene. 2010;29(11):1672–1680. doi: 10.1038/onc.2009.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji H, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 51.Partanen JI, Nieminen AI, Mäkelä TP, Klefstrom J. Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci USA. 2007;104(37):14694–14699. doi: 10.1073/pnas.0704677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang X, Nan K-J, Li Z-L, Xu Q-Z. Overexpression of the LKB1 gene inhibits lung carcinoma cell proliferation partly through degradation of c-myc protein. Oncol Rep. 2009;21(4):925–931. doi: 10.3892/or_00000305. [DOI] [PubMed] [Google Scholar]
- 53.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Cespedes M. The role of LKB1 in lung cancer. Fam Cancer. 2011;10(3):447–453. doi: 10.1007/s10689-011-9443-0. [DOI] [PubMed] [Google Scholar]
- 55.Network CGAR, et al. Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 57.Van Dijk MA, Voorhoeve PM, Murre C. Pbx1 is converted into a transcriptional activator upon acquiring the N-terminal region of E2A in pre-B-cell acute lymphoblastoid leukemia. Proc Natl Acad Sci USA. 1993;90(13):6061–6065. doi: 10.1073/pnas.90.13.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee D, Grant A, Buchan D, Orengo C. A structural perspective on genome evolution. Curr Opin Struct Biol. 2003;13(3):359–369. doi: 10.1016/s0959-440x(03)00079-4. [DOI] [PubMed] [Google Scholar]
- 59.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14(2):208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 60.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31(13):3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20(2):556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jaskoll T, Htet K, Abichaker G, Kaye FJ, Melnick M. CRTC1 expression during normal and abnormal salivary gland development supports a precursor cell origin for mucoepidermoid cancer. Gene Expr Patterns. 2011;11(1-2):57–63. doi: 10.1016/j.gep.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2(1):18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 66.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 67.Wilson A, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18(22):2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16(20):2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iourgenko V, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci USA. 2003;100(21):12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bittinger MA, et al. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14(23):2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 71.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 73.Amelio AL, Caputi M, Conkright MD. Bipartite functions of the CREB co-activators selectively direct alternative splicing or transcriptional activation. EMBO J. 2009;28(18):2733–2747. doi: 10.1038/emboj.2009.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16(10):5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ke N, et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004;64(22):8208–8212. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- 76.Jean D, Harbison M, McConkey DJ, Ronai Z, Bar-Eli M. CREB and its associated proteins act as survival factors for human melanoma cells. J Biol Chem. 1998;273(38):24884–24890. doi: 10.1074/jbc.273.38.24884. [DOI] [PubMed] [Google Scholar]
- 77.Shankar DB, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005;7(4):351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 78.Aggarwal S, Kim S-W, Ryu S-H, Chung W-C, Koo JS. Growth suppression of lung cancer cells by targeting cyclic AMP response element-binding protein. Cancer Res. 2008;68(4):981–988. doi: 10.1158/0008-5472.CAN-06-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Conkright MD, et al. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003;11(4):1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 80.Parker D, et al. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol Cell. 1998;2(3):353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- 81.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 84.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99(9):6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.