Significance
Double fertilization of the female gametophyte is vital for angiosperm reproduction and requires delivery of sperm cells through a pollen tube. The sperm cells and pollen vegetative nucleus together are called the male germ unit. The pollen tube grows a long distance from the stigma to the ovary, and the mode of male germ unit movement and sperm cell delivery is largely unknown. This study reveals that WPP domain-interacting protein, a plant analog of animal proteins known to control nuclear movement, and WPP domain-interacting tail-anchored protein are responsible for movement of the vegetative nucleus within pollen tubes and contribute to sperm cell-to-ovule migration. This finding marks a leap forward in our understanding of double fertilization and the mechanism of nuclear movement in plants as a whole.
Keywords: nuclear envelope, Arabidopsis, male gametophyte, LINC complex
Abstract
Increasing evidence suggests that nuclear migration is important for eukaryotic development. Although nuclear migration is conserved in plants, its importance for plant development has not yet been established. The most extraordinary plant nuclear migration events involve plant fertilization, which is starkly different from that of animals. Instead of evolving self-propelled sperm cells (SCs), plants use pollen tubes to deliver SCs, in which the pollen vegetative nucleus (VN) and the SCs migrate as a unit toward the ovules, a fundamental but barely understood process. Here, we report that WPP domain-interacting proteins (WIPs) and their binding partners the WPP domain-interacting tail-anchored proteins (WITs) are essential for pollen nuclear migration. Loss-of-function mutations in WIT and/or WIP gene families resulted in impaired VN movement, inefficient SC delivery, and defects in pollen tube reception. WIPs are Klarsicht/ANC-1/Syne-1 Homology (KASH) analogs in plants. KASH proteins are key players in animal nuclear migration. Thus, this study not only reveals an important nuclear migration mechanism in plant fertilization but also, suggests that similar nuclear migration machinery is conserved between plants and animals.
Nuclear migration is essential for cell differentiation, polarization, and migration, which influence organism development (1–3). Examples range from Caenorhabditis elegans P-cell development to mammalian neural development (1–3). The key players in opisthokont nuclear migration are the inner nuclear membrane Sad1/UNC-84 (SUN) proteins and outer nuclear membrane Klarsicht/ANC-1/Syne-1 Homology (KASH) proteins. SUN and KASH proteins form the linkers of the nucleoskeleton and the cytoskeleton complexes at the nuclear envelope (NE) and transfer cytoplasmic forces to the nucleus (1–3). In plants, nuclear migration is associated with a number of developmental events and environmental responses, including fertilization, root and leaf hair formation, and plant–microbe interactions (4, 5). So far, little is known about the mechanism of plant nuclear migration. Although SUN proteins are conserved in plants (6, 7), absence of animal KASH homologs in plants suggests that plants may have evolved different molecular solutions to achieve nuclear migration. Recently, WPP domain-interacting proteins (WIPs) were identified as KASH proteins in plants (8), and their outer nuclear membrane binding partners WPP domain-interacting tail anchored proteins (WITs) were shown to interact with myosin XI-I (9). The WIT–myosin XI-I complexes regulate nuclear movement in root and mesophyll cells, but no developmental events have been linked to these nuclear movements (9).
Essential for plant fertility, pollen tube growth harbors the most dramatic nuclear movement in plants. Unlike animals, which have sperm cells (SCs) that travel through self-propelled flagellum, flowering plants use pollen tubes to deliver SCs to ovules (10–13). In Arabidopsis, pollen tube growth is guided by chemical cues in carpel tissues and attracted by small peptides secreted by synergid cells in the vicinity of ovules (14–18). Pollen tube reception is completed by pollen tube burst, SC release, and degeneration of synergid cells (12). If this process fails, a second pollen tube can be attracted to the same ovule for a second attempt, resulting in polytubey (19). The SCs [or their progenitor the generative cell (GC)] are enclosed by an endocytic membrane tethered to the pollen vegetative nucleus (VN) (20). During pollen tube elongation, the VN and the SCs/GC are usually closely associated and move as a male germ unit (MGU) (13, 21). For decades, the movement of the MGU has been analyzed using cytoskeleton-depolymerizing reagents or heterogeneous antimyosin antibodies (22–27). However, no genes have been implicated in MGU movement, and the function of the joint migration of VN and GC/SC remains hypothetical.
Here, we have identified the Arabidopsis WIT and WIP protein families as key players in VN movement. WIP1 and WIT1 are localized at the vegetative nuclear envelope (VNE). Loss of either WIT or WIP family proteins impaired VN movement, resulting in defective pollen tube reception and inefficient SC-to-ovule migration. This study has not only identified a molecular mechanism regulating the VN movement but also, revealed an important function of the VN in plant fertilization.
Results
WIT and WIP Are Required for Full Seed Set.
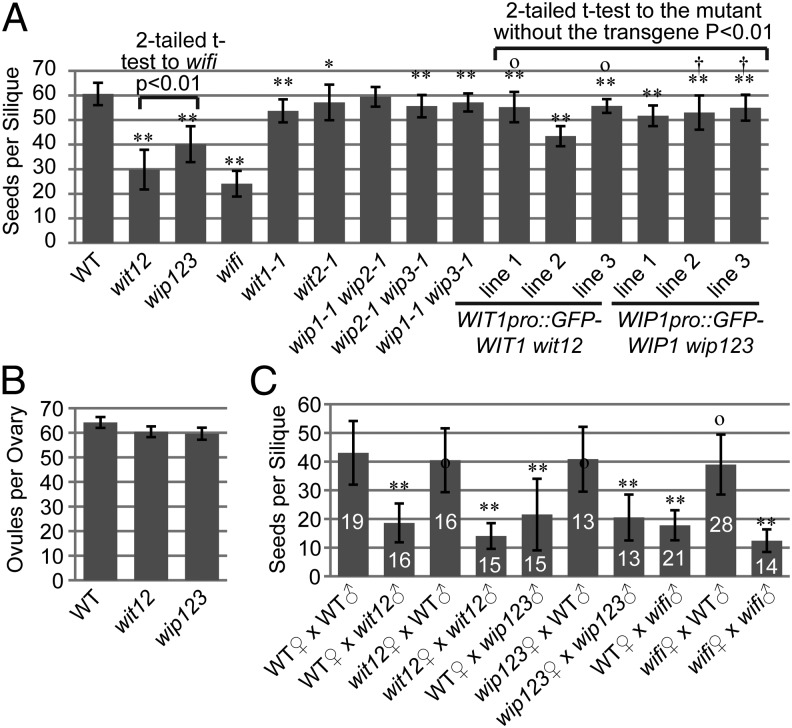
In Arabidopsis, WIT and WIP are two families of NE-associated proteins that synergistically anchor Ran GTPase-activating protein 1 (RanGAP1) to the NE (28, 29). Reduction in seeds per silique was observed in wit1-1 wit2-1 double null (wit12; ∼50% loss) (Fig. 1A) and wip1-1 wip2-1 wip3-1 triple null (wip123; ∼33% loss) (Fig. 1A). In contrast, the number of seeds per silique of wit1-1, wit2-1, wip1-1 wip2-1, wip2-1 wip3-1, and wip1-1 wip3-1 mutants was only slightly reduced compared with WT (Fig. 1A). Ovaries of wit12 and wip123 contain, on average, four ovules fewer than WT (Fig. 1B), which is unlikely to cause the significantly reduced seed production. The quintuple null mutant wip1-1 wip2-1 wip3-1 wit1-1 wit2-1 (wifi) has the most severe seed loss phenotype, suggesting that WIT and WIP synergistically affect seed production (Fig. 1A). As shown in Fig. 1A, WIT1 promoter-driven GFP-WIT1 (WIT1pro::GFP-WIT1) rescued (Fig. 1A, lines 1 and 3) or partially rescued (Fig. 1A, line 2) wit12, and WIP1 promoter-driven GFP-WIP1 (WIP1pro::GFP-WIP1) rescued (Fig. 1A, lines 2 and 3) or partially rescued (Fig. 1A, line 1) wip123. These data indicate that this phenotype is linked to the wit or wip mutations.
Fig. 1.
Reduction in seed production based on loss of WIT and/or WIP. (A) Number of seeds per silique compared between different mutants and lines. *0.01 < P < 0.05 compared with WT. **P < 0.01 compared with WT. OP > 0.05 compared with wit2-1. †P > 0.05 compared with wip2-1 wip3-1. n = 40 for all samples. (B) Number of ovules per ovary of WT, wit12, and wip123 (n = 10). (C) Number of seeds per silique after reciprocal crosses between WT and wit12, between WT and wip123, or between WT and wifi. **P < 0.01 compared with WT♀ × WT♂. OP > 0.05 compared with WT♀ × WT♂. The n for each dataset was indicated on the graph. In A–C, error bars in all histograms represent SD, and two-tailed t test was used for statistical analysis.
Loss of WIT or WIP Leads to Male Fertility Defects.
To analyze the parental origin of the seed loss, we performed reciprocal crosses between WT and wit12. As shown in Fig. 1C, when pollinated with WT pollen, both wit12 and WT developed similar numbers of seeds per silique. In contrast, when pollinated with wit12 pollen, both wit12 and WT developed only roughly one-half the amount of seeds per silique compared with hand-pollinated WT with WT pollen. Reciprocal crosses were also performed between WT and wip123 and between WT and wifi. Similar results were obtained (Fig. 1C).
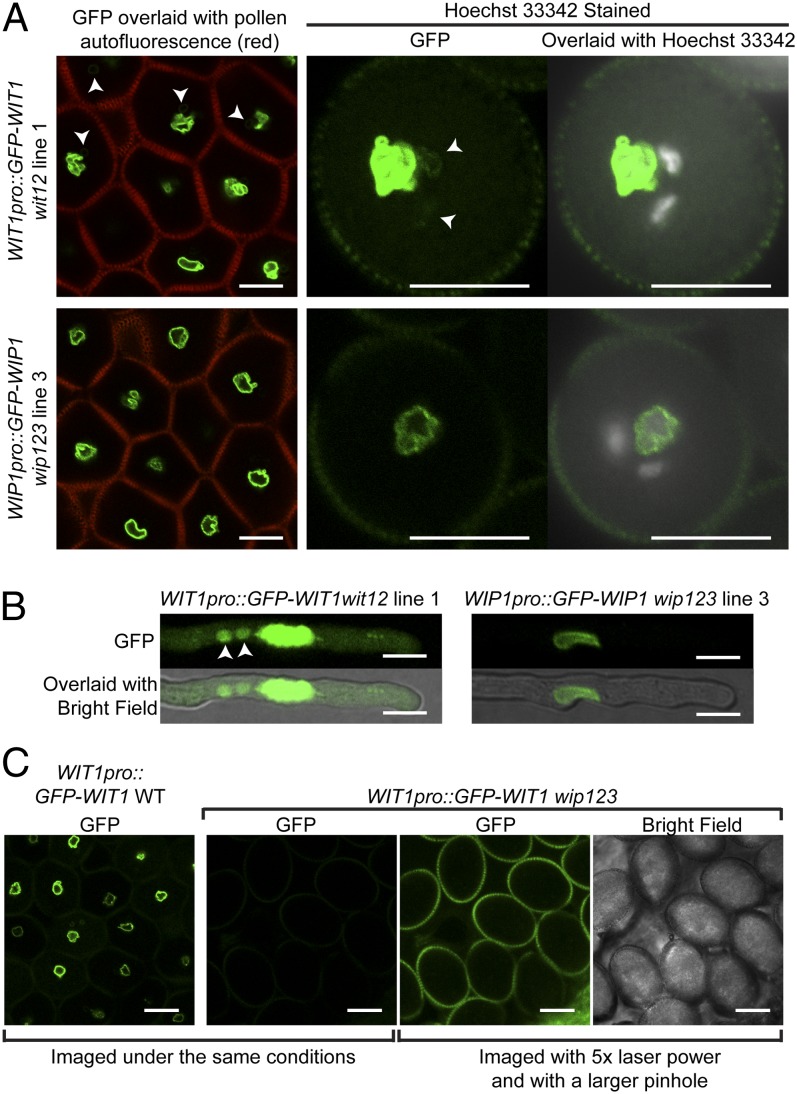
We then examined the expression and localization pattern of WIT1 and WIP1 in the pollen of WIT1pro::GFP-WIT1–rescued wit12 lines or the WIP1pro::GFP-WIP1–rescued wip123 lines. Both GFP-WIT1 and GFP-WIP1 signals were strongly associated with the VNE (Fig. 2A and Fig. S1 A and B). In addition, GFP-WIT1 weakly labeled the SC NE in some pollen grains (Fig. 2A and Fig. S1A). After 5 h of in vitro growth of pollen tubes, GFP-WIT1 and GFP-WIP1 were visible at the VNE, and WIT1 was also weakly visible in SCs, probably the SC NE (Fig. 2B). WT plants carrying the WIT1pro::GFP-WIT1 or WIP1pro::GFP-WIP1 transgene revealed similar results (Fig. S1B). It has been reported that the WIT1 protein level is significantly reduced in wip123 vegetative tissues, likely because the protein is destabilized/degraded in the absence of the WIP–WIT complex (29). The GFP-WIT1 signal is also barely detectable in wip123 pollen (three lines were examined, and a representative image is shown in Fig. 2C), which agrees with the previous report and suggests that the reduced fertility phenotype of wip123 is, to a large extent, caused by the loss of WIT proteins.
Fig. 2.
WIT1 and WIP1 subcellular localization in pollen grains and pollen tubes. (A) The localization of WIT1 and WIP1 in pollen grains was examined using WIT1pro::GFP-WIT1 wit12 line 1 and WIP1pro::GFP-WIP1 wip123 line 3, respectively. GFP signal was imaged by confocal microscopy, and the Hoechst 33342 signal was imaged by fluorescence microscopy. The autofluorescence from pollen walls was collected through the RFP channel and merged with the GFP signal in the images in Left. GFP-WIT1 and GFP-WIP1 were both localized at the VNE. In addition, GFP-WIT1 also weakly labeled the SC NE (arrowheads). (B) WIT1pro::GFP-WIT1 wit12 line 1 and WIP1pro::GFP-WIP1 wip123 line 3 pollen tubes after 5 h of pollen germination. GFP-WIT1 was strongly localized at the VNE, and it was also visible in the SCs, probably the SC NE (arrowheads), whereas GFP-WIP1 was only detectable at the VNE. Imaging conditions were tuned to view the signal in the SCs, and therefore, the GFP signals at the VNE are saturated and appear nuclear. (C) Compared with its protein level in WT, GFP-WIT1 was barely detectable in wip123 pollen grains. (Scale bars: 10 µm.)
To assess the influence of the wit or wip mutation on male fertility, pollen competition assays were performed between WT pollen and (i) wit12 pollen, (ii) WIT1pro::GFP-WIT1 wit12 line 1 pollen, (iii) wip123 pollen, or (iv) WIP1pro::GFP-WIP1 wip123 line 3 pollen. As shown in pollen competition experiments (Fig. S1C), wit12 and wip123 have a much reduced transmission efficiency (3.9% and 13.3%, respectively), whereas WIT1pro::GFP-WIT1 and WIP1:GFP-WIP1 rescued the transmission efficiency to the WT level (50%).
These data indicate that WIT and WIP are essential for male fertility. This defect is clearly different from the female gametophyte lethality caused by mutating RanGAP1 and RanGAP2 (30), suggesting that maintaining pollen fertility is a function of WIT and WIP independent of their reported role as RanGAP NE anchors.
WIT and WIP Are Required for Proper Nuclear Movement in Pollen Tubes.
We then examined wit12, wip123, and wifi pollen in detail. The morphology of mutant anthers and pollen grains is normal, and no significant amount of dead pollen was observed using Alexander staining (Fig. S1D). The three nuclei—one VN and two SC nuclei (SN)—are also normal (Fig. S1E). Because WIPs are plant KASH proteins and WITs are involved in root and leaf nuclear movement (8, 9), we monitored nuclear movement in pollen tubes germinated in vitro. mCherry tagged with a nuclear localization signal and driven by the ubiquitin 10 promoter was used as a VN marker (VN-RFP) (31). MGH3/HRT10, a male gamete-specific histone H3 gene fused with GFP and driven by the MGH3 promoter, was used as an SN marker (SN-GFP) (32).
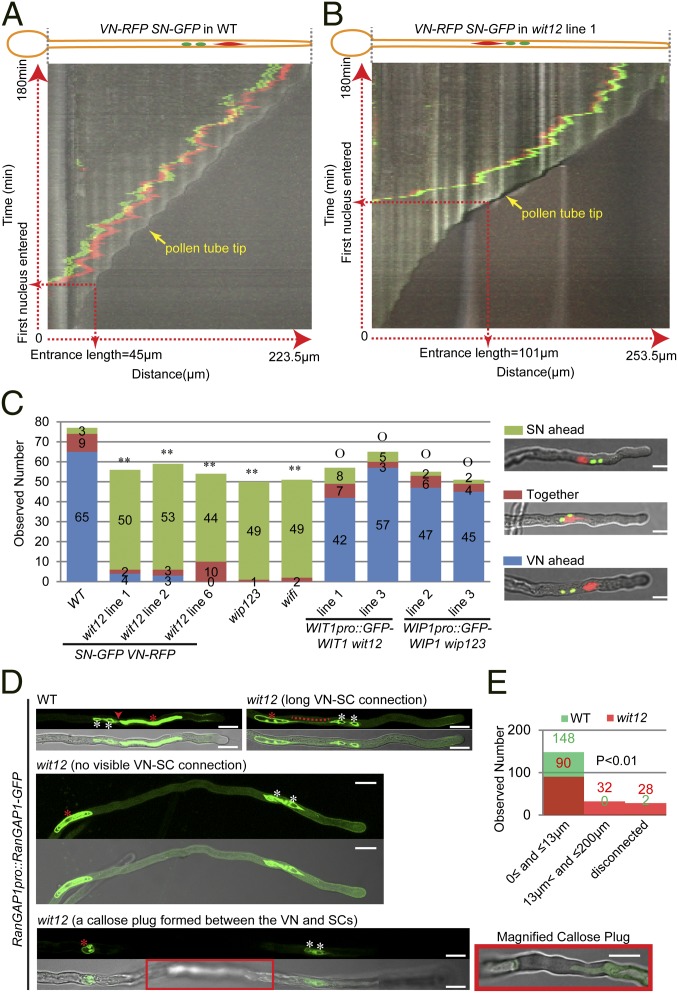
As shown in Fig. 3A and Movie S1, during WT pollen germination, typically, the VN entered the pollen tube first when the pollen tube reached a certain length (we define the tube length when the first nucleus permanently enters the pollen tube as the entrance length, which is quantified in Fig. S2A). As observed in many plant species (13), the VN precede the SCs as they move along the growing pollen tube. The VN also remained at a certain distance from the growing tip followed by the SN. In contrast to WT, as shown in Fig. 3B and Movie S2, when wit12 pollen tubes had reached their much greater entrance length (Fig. S2A), the two SN typically entered the pollen tube first (Fig. S2B), and the two SN then led the VN in the growing pollen tubes (Fig. 3B and Movie S2). Measuring the leading SN-to-tip distance every 1 min for 60 min after the two SN had entered the pollen tube, a mean value of 56.2 µm (SD = 31.2 µm, 10 pollen tubes were measured) was obtained, much larger than that of WT (Fig. S2C).
Fig. 3.
Nuclear movement in pollen tubes is affected in wit12, wip123, and wifi. (A) Kymograph of nuclear movement in a VN-RFP SN-GFP WT pollen tube. Vertical axis represents time (total = 180 min), and horizontal axis represents the distance along the pollen tube (total pollen tube length = 223.5 µm). (B) Kymograph of nuclear movement in a VN-RFP SN-GFP wit12 pollen tube. Vertical axis represents time (total = 180 min), and horizontal axis represents the distance along the pollen tube (total pollen tube length = 253.5 µm). (C) Nuclear order in pollen tubes after 5 h of germination. An example of each category is shown in Right. For wip123, wifi, and WIP1pro::GFP-WIP1 wip123 pollen tubes, Hoechst 33342 was used to stain DNA, and confocal microscopy was used for the others. Two-tailed Fisher exact test was used for statistical analysis, and numbers for each category are indicated on the graph. **P < 0.01 compared WT. OP > 0.05 compared with WT. (Scale bars: 10 µm.) (D) Position of the VN (red asterisks) relative to the SCs (white asterisks) after 16 h of semi-in vivo germination. In WT pollen tubes, the VN was closely associated with the SCs, and the connection (red arrowhead) was visible. However, in the wit12 pollen tubes, the connections were longer (red dotted line), undetectable, or interrupted by a callose plug (red box that is magnified at a focal plane clearly showing the plug). For each example, GFP signal is shown in the upper part, and the overlay with a bright-field image is shown in the lower part. The two examples in Middle and Bottom are maximum projections of z-stack images. Bottom was assembled from three continuous images by image stitching. (Scale bars: 10 µm.) (E) Quantification of the VN–SC distance after 16 h of semi-in vivo pollen germination. The shortest distance between the VNE and SC NE was measured. For either WT or wit12, one VN-RFP SN-GFP line and two RanGAP1pro::RanGAP1-GFP lines were used. Fifty pollen tubes were measured for each line. The VN, which were blocked by a callose plug or undetectable in over 300-µm pollen tubes, were considered disconnected. The numbers are indicated (green, WT; red, wit12). P < 0.01 represents the statistically significant difference between WT and wit12 (P < 0.01, n = 150, two-tailed Fisher exact test).
We then examined the nuclear position in in vitro-germinated pollen tubes at 5 h, a time point when in vivo pollen tubes would start entering the micropyle (33). In nearly all wit12 pollen tubes, the two SN were still ahead of the VN, significantly different from WT (Fig. 3C). A similar phenotype was also observed in Hoechst 33342-stained wip123 and wifi pollen tubes after 5 h in vitro germination (Fig. 3C). Consistently, the transgenic mutant lines with seed loss phenotype that was rescued showed normal nuclear positioning after 5 h of in vitro germination (Fig. 3C). Recently, myosin XI-I was reported to be recruited to the NE by WIT1 and WIT2 and regulate nuclear movement in roots and mesophyll cells (9). However, kaku1-4, a reported myosin xi-i null mutant (9), exhibits normal seeds per silique and normal nuclear order after 5 h of in vitro pollen germination (Fig. S2 D and E), consistent with the report that no nuclear migration defects were observed in kaku1-4 (9).
Investigating semi-in vivo germinated pollen tubes 8 h after germination—the time when double fertilization starts in vivo (33)—we discovered that the nuclear order was still reversed in wit12 pollen tubes (Fig. S3A). More interestingly, unlike the closely associated VN and SN in WT, isolated VN (Fig. S3A, dotted line) and solely migrating SN (Fig. S3A, arrowheads) were observed in wit12 pollen tubes (Fig. S3A). We examined this phenotype in detail 16 h after semi-in vivo pollen germination. RanGAP1pro::RanGAP1-GFP was used as an additional marker to visualize the physical connection between the VN and the SCs, because RanGAP1 labels both the VNE and the SC cytoplasm. As shown in Fig. 3D, a close VN–SC connection was visible in WT pollen tubes. The VN–SN distance (the closest distance between the VNE and the SC NE) was quantified (Fig. 3E), and 99% of WT pollen tubes had a VN–SN distance ≤13 µm. In contrast, for wit12 pollen tubes, only 60% fell in this range, and 21% had a much longer VN–SN distance (>13 but ≤200 µm), and in 19% of pollen tubes, the VN was either blocked by a callose plug or undetectable (disconnected) (Fig. 3 D and E). However, the VN movement defect of wit12 does not impair pollen germination or pollen tube length (details are in Fig. S3 B–D).
Loss of WIT or WIP Impaired the Pollen Tube Reception and SC-to-Ovule Migration.
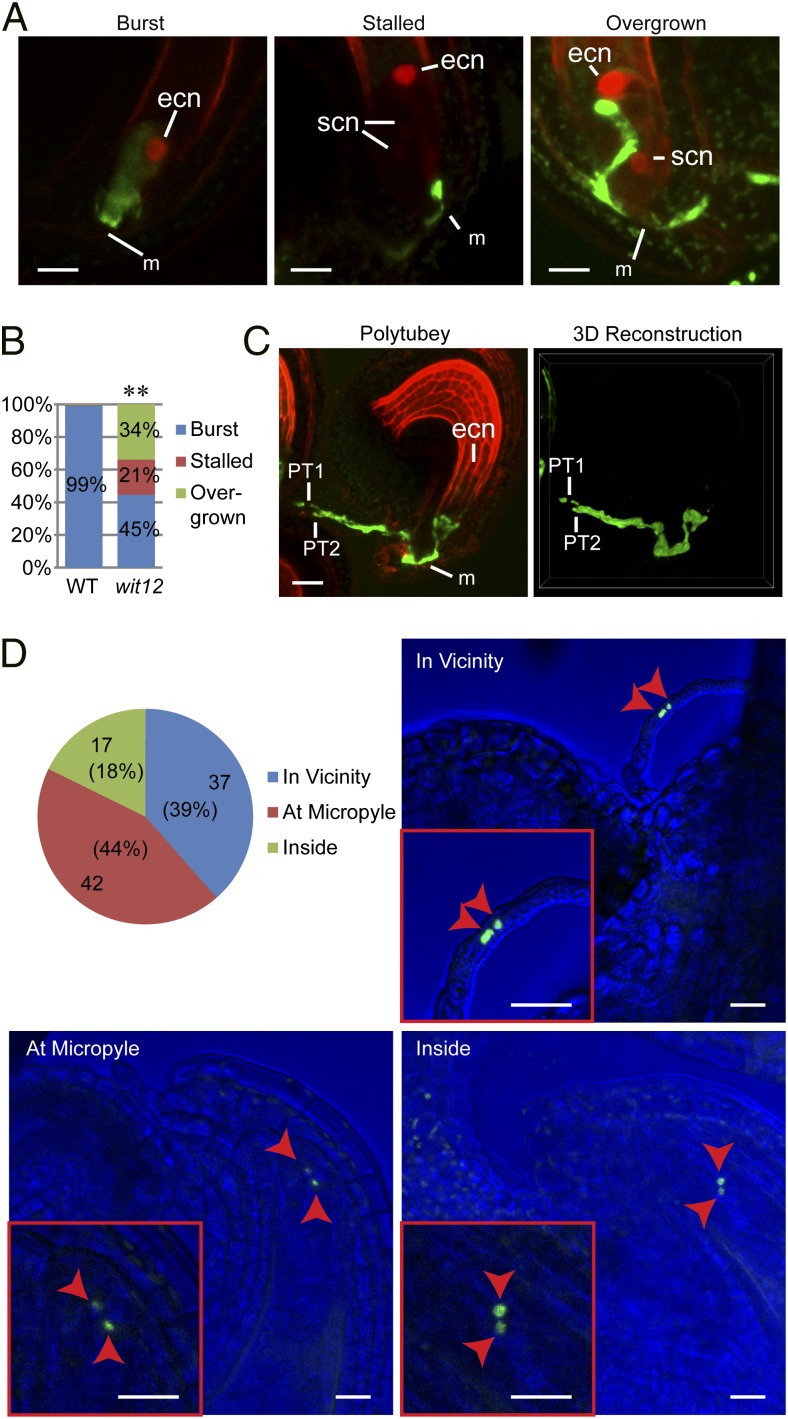
To determine the cause of male fertility defects in wit12, WT pistils expressing NLS-2xRFP (nuclear localization signal fused to double red fluorescent protein) under the Egg Cell 1 promoter (EC1pro::NLS-2xRFP) were pollinated with wit12 pollen expressing GFP under the Lat52 promoter (Lat52pro::GFP). The ovules were examined after 24 h. As shown in Fig. 4 A and B, only 45% of pollen tubes burst, whereas 21% stalled at the micropyle, and 34% overgrew into the ovules. Ovules that had attracted two pollen tubes were also observed occasionally (Fig. 4C). In contrast, ∼99% of Lat52pro::GFP WT pollen tubes burst.
Fig. 4.
Defects of pollen tube reception and SC-to-ovule migration in wit12. (A) EC1pro::NLS-2xRFP WT pistils were pollinated with Lat52pro::GFP WT pollen or Lat52pro::GFP wit12 pollen. The pistils were dissected and imaged 24 h later. The wit12 pollen tubes showed three types of fate: burst (WT like), stalled [stopped at the micropyle (m) but did not burst], and overgrown inside ovules. The stalled and overgrown images are maximum projections of z-stack images, which were processed with a 5-pixel median filter to reduce background noise. NLS-2xRFP strongly labeled the egg cell nuclei (ecn) and also weakly labeled the synergid cell nuclei (scn). (Scale bars: 10 µm.) (B) Quantification of the three types of wit12 pollen tube fate observed in A. The percentage of each category is indicated on the graph. For WT, n = 270, and for wit12, n = 170. **P < 0.01 compared with WT (two-tailed Fisher exact test). (C) Two Lat52pro::GFP wit12 pollen tubes were attracted to one EC1pro::NLS-2xRFP WT ovule 24 h after pollination. Left is a maximum projection of a z-stack image processed with 5-pixel median filter to reduce background noise. Right is a 3D reconstruction of its GFP channel using α-blending. PT1, pollen tube 1; PT2, pollen tube 2. (Scale bar: 20 µm.) (D) The SC fate in the pollen tubes that targeted small unfertilized ovules of self-pollinated VN-RFP SN-GFP wit12 was examined 72 h after flower opening. The SN were found in the vicinity of an ovule, at the micropyle of an ovule, or inside an ovule. These three categories are quantified in the pie chart, and representative examples are shown. The bright field was tuned to dark blue for better exhibition of the SN. Arrowheads indicate the SN. (Scale bars: 20 µm.)
We then dissected and examined wit12 ovaries ∼72 h after flower opening. Two types of ovules were found in wit12 ovaries—large fertilized ovules with visible developing embryos and small unfertilized ovules with central cell nuclei that were clearly visible (Fig. S4 A and B). A similar phenotype was also observed in the ovaries of wip123 and wifi (Fig. S4A). The number of large fertilized ovules per ovary of WT and wit12 was slightly lower than their average seeds per silique (compare Fig. 1A with Fig. S4C), suggesting that a small fraction of the unfertilized ovules scored would still become fertilized. Aniline blue staining of the ovules of wit12 or imaging of the ovules of the Lat52pro::GFP wit12 at this stage revealed both overgrowth and polytubey abnormalities (Fig. S5 A and B), similar to the pollen tube defects of wit12 24 h after pollination (Fig. 4A). These two abnormalities are more common in unfertilized ovules— among 302 examined ovules, 64 (21%) ovules displayed polytubey phenotype, and 187 (62%) ovules exhibited overgrown pollen tubes (Table 1). Similar abnormalities were also observed in the ovaries of wip123 (Table 1 and Fig. S5A).
Table 1.
Quantification of pollen tube reception defects of wit12 and wip123
| Pollen tube morphology | WT | wit12 | wip123 | |||
| Big fertilized ovules | Small unfertilized ovules | Big fertilized ovules | Small unfertilized ovules | Big fertilized ovules | Small unfertilized ovules | |
| With pollen tubes; not overgrown | 265 | 2 | 138 | 77 | 118 | 39 |
| With overgrown pollen tubes | 0 | 1 | 3 | 187 | 12 | 48 |
| Without pollen tubes | 0 | 4 | 0 | 38 | 0 | 10 |
| Total | 265 | 7 | 141 | 302 | 130 | 97 |
| Among total: ovules with polytubey | 10 | 1 | 14 | 64 | 16 | 19 |
To determine the localization of SN and VN in the unfertilized ovules, confocal microscopy was used to image the small unfertilized ovules of VN-RFP SN-GFP wit12. As shown in Fig. 4D and Fig. S5C, the two SN were arrested in the vicinity of the ovules (39%), at the micropyle (44%), or inside the ovules (18%), reflecting a severe SC-to-ovule migration defect. In none of the analyzed ovules was a VN visible. Because the semi-in vivo assay shows that the VN can be disconnected from the SCs during migration (Fig. 3 D and E and Fig. S3A), we hypothesize that the VN might never reach the vicinity of these ovules. To assess this possibility under more native conditions, pollen grains were germinated 24 h on pistils with ovaries that were cut to a length of ∼600 µm, and the pollen tubes that grew through the pistils were examined. Pollen tubes with both the VN and the SCs detectable within 250 µm from the tips were considered as intact MGU emerged; otherwise, they were considered as no intact MGU emerged. As shown in Fig. S5D, 40% of wit12 pollen tubes had no intact MGU emerged, which supports our hypothesis. Together, these data suggest that wit12 exhibits severe impairment in pollen tube reception and SC-to-ovule migration.
Discussion
The MGU has been observed in many plant species (13), and our analysis of in vitro-geminated Arabidopsis pollen shows that the VN and SCs do form a linked unit that moves together along growing pollen tubes (Movie S1). Even in the wit12 mutant, with a VN that seemingly loses its driving force, the VN was still connected to and moved in conjunction with the SCs (Movie S2), reflecting a functional linkage between the VN and SCs. During WT pollen tube growth, the VN entered the pollen tube first and led the movement of SCs with occasionally backward movement when the MGU was extremely close to the tip of the growing tube (Fig. 3A and Movie S1). SCs coordinated with this movement exhibited transient position shifts with the VN (Fig. 3A and Movie S1). Although the mobility of the VN is impaired in wit12 and the entrance length is longer than that of WT, the distance-keeping ability of the MGU is not totally lost: the SCs led the movement of the MGU and made back-and-forth adjustment according to the distance to the growing tube apex (Fig. 3B and Movie S2). Even when the VN was disconnected from the SCs, the SCs still moved forward (Fig. 3D and Fig. S3A). These observations indicate that there are (i) a main driving force transferred through WITs that moves the VN, (ii) a secondary driving force facilitating the movement of the SCs, and (iii) a signaling mechanism between the VN–SC and the growing tip regulating the two driving forces to maintain proper nucleus–tip distance. Because the VN and the two SCs are connected, these two forces coordinate with each other to move the MGU forward. This double-driving force hypothesis is consistent with a reported observation that the SC projection that links the VN exhibited somewhat independent movement to the overall momentum of SCs (34). In addition, the bidirectional movement of the MGU was proposed to be a result of the dynamic balance between acropetal and basipetal forces (35). Although the nuclear order defects and the increased leading nucleus-to-tip distance of wit12 mimics the nuclear movement in the Galanthus nivalis pollen tubes after microtubule depolymerization (24), the question of whether the MGU movement is regulated by the F-actin network or the microtubule network still needs additional investigation, because myosin has also been proposed to move the MGU in pollen tubes of Lilium longiflorum and Nicotiana alata (26).
The fact that the VN and GC/SCs migrate as a unit has been proposed to be important for efficient SC-to-ovule migration (13, 21, 36). This model is supported by the inefficient migration of SCs in wit12 with VN movement that is impaired (Figs. 3 and 4D and Fig. S5D). However, positioning the VN close to the growing pollen tube tip probably plays additional important roles in pollen tube reception, which was implied by the overgrown and stalled pollen tubes (Fig. 4 A–C and Fig. S5 A–C). Pollen tube reception involves players from both male and female gametophytes. In Arabidopsis, synergid-expressed FERONIA, NORTIA, and LORELEI are essential for pollen tube reception, and their mutants display overgrown pollen tubes (37–41). On the pollen side, ANXUR1 and -2 may be involved in pollen reception (42, 43). Three pollen tube-expressed MYB transcription factors (MYB97, MYB101, and MYB120) were identified as regulating pollen tube gene expression and SC release (44, 45). These findings suggest that, on pollen tube arrival, signaling cascades are triggered in both pollen tubes and synergids. Positioning the VN close to the pollen tube tip might ensure that (i) transcripts from the VN required for sensing synergid contact have to be translated at the pollen tube tip and/or (ii) synergid feedback signals can only reach a tip-positioned VN for an adequate gene expression response. In support of the latter scenario, in neuroepithelium progenitor cells, the nuclei were shown to oscillate between apical and basal positions to sense the Notch gradient (46).
In summary, WIT and WIP family proteins are localized at the VNE and mediate VN migration during pollen tube growth. Our data support that a mechanism that regulated VN movement is important for SC-to-ovule migration and pollen tube reception. To our knowledge, WITs and WIPs are the first genes identified in pollen nuclear migration and can now serve as a base to reveal the full molecular picture of this important process.
Methods
For in vitro pollen germination, pollen grains from the stamens of fully opened flowers were germinated on a pollen germination medium. For semi-in vitro pollen germination, stage 14 male sterility 1 stigmas were saturated with pollen and incubated on pollen germination medium. For ovules imaged at 72 h after flower opening, a magnifier was used to identify early-stage 13 flower buds (47), which were tagged and collected 72 h later. All procedures are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. Xianfeng Xu, Qiao Zhao, and Jelena Brkljacic for help in constructing the wifi mutant; Ms. Anna Griffis for wording the significance statement; Drs. R. Keith Slotkin and Anna Dobritsa for numerous helpful discussions and donation of the pollen nuclear markers; and Dr. Ravishankar Palnivelu for help with the pollen in vitro germination assay. Financial support was through National Science Foundation Grant NSF-MCB 1243844.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. U.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323104111/-/DCSupplemental.
References
- 1.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152(6):1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothballer A, Schwartz TU, Kutay U. LINCing complex functions at the nuclear envelope: What the molecular architecture of the LINC complex can reveal about its function. Nucleus. 2013;4(1):29–36. doi: 10.4161/nucl.23387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takagi S, Islam MS, Iwabuchi K. Dynamic behavior of double-membrane-bounded organelles in plant cells. In: Kwang WJ, editor. International Review of Cell and Molecular Biology. Vol 286. London: Academic; 2011. pp. 181–222. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Meier I. How plants LINC the SUN to KASH. Nucleus. 2013;4(3):206–215. doi: 10.4161/nucl.24088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oda Y, Fukuda H. Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 2011;66(4):629–641. doi: 10.1111/j.1365-313X.2011.04523.x. [DOI] [PubMed] [Google Scholar]
- 7.Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2010;61(1):134–144. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol. 2012;196(2):203–211. doi: 10.1083/jcb.201108098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura K, et al. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol. 2013;23(18):1776–1781. doi: 10.1016/j.cub.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 10.Beale KM, Johnson MA. Speed dating, rejection, and finding the perfect mate: Advice from flowering plants. Curr Opin Plant Biol. 2013;16(5):590–597. doi: 10.1016/j.pbi.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Lausser A, Kliwer I, Srilunchang KO, Dresselhaus T. Sporophytic control of pollen tube growth and guidance in maize. J Exp Bot. 2010;61(3):673–682. doi: 10.1093/jxb/erp330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessler SA, Grossniklaus U. She’s the boss: Signaling in pollen tube reception. Curr Opin Plant Biol. 2011;14(5):622–627. doi: 10.1016/j.pbi.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 13.McCue AD, Cresti M, Feijó JA, Slotkin RK. Cytoplasmic connection of sperm cells to the pollen vegetative cell nucleus: Potential roles of the male germ unit revisited. J Exp Bot. 2011;62(5):1621–1631. doi: 10.1093/jxb/err032. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu KK, Okada K. Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development. 2000;127(20):4511–4518. doi: 10.1242/dev.127.20.4511. [DOI] [PubMed] [Google Scholar]
- 15.Johnson MA, Preuss D. Plotting a course: Multiple signals guide pollen tubes to their targets. Dev Cell. 2002;2(3):273–281. doi: 10.1016/s1534-5807(02)00130-2. [DOI] [PubMed] [Google Scholar]
- 16.Jones-Rhoades MW, Borevitz JO, Preuss D. Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 2007;3(10):1848–1861. doi: 10.1371/journal.pgen.0030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda S, et al. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458(7236):357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi H, Higashiyama T. A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 2012;10(12):e1001449. doi: 10.1371/journal.pbio.1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruyama D, et al. Independent control by each female gamete prevents the attraction of multiple pollen tubes. Dev Cell. 2013;25(3):317–323. doi: 10.1016/j.devcel.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Hamamura Y, et al. Live-cell imaging reveals the dynamics of two sperm cells during double fertilization in Arabidopsis thaliana. Curr Biol. 2011;21(6):497–502. doi: 10.1016/j.cub.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Dumas C, Knox RB, Gaude T. The spatial association of the sperm cells and vegetative nucleus in the pollen grain of Brassica. Protoplasma. 1985;124(3):168–174. [Google Scholar]
- 22.Åström H, Sorri O, Raudaskoski M. Role of microtubules in the movement of the vegetative nucleus and generative cell in tobacco pollen tubes. Sex Plant Reprod. 1995;8(2):61–69. [Google Scholar]
- 23.Kaul V, Theunis CH, Palser BF, Knox RB, Williams EG. Association of the generative cell and vegetative nucleus in pollen tubes of Rhododendron. Ann Bot. 1987;59(2):227–235. [Google Scholar]
- 24.Heslop-Harrison J, Heslop-Harrison Y, Cresti M, Tiezzi A, Moscatelli A. Cytoskeletal elements, cell shaping and movement in the angiosperm pollen tube. J Cell Sci. 1988;91(1):49–60. [Google Scholar]
- 25.Tirlapur UK, et al. Confocal imaging and immunogold electron microscopy of changes in distribution of myosin during pollen hydration, germination and pollen tube growth in Nicotiana tabacum L. Eur J Cell Biol. 1995;67(3):209–217. [PubMed] [Google Scholar]
- 26.Miller DD, Scordilis SP, Hepler PK. Identification and localization of three classes of myosins in pollen tubes of Lilium longiflorum and Nicotiana alata. J Cell Sci. 1995;108(Pt 7):2549–2563. doi: 10.1242/jcs.108.7.2549. [DOI] [PubMed] [Google Scholar]
- 27.Heslop-Harrison J, Heslop-Harrison Y. Myosin associated with the surfaces of organelles, vegetative nuclei and generative cells in angiosperm pollen grains and tubes. J Cell Sci. 1989;94(2):319–325. [Google Scholar]
- 28.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17(13):1157–1163. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell. 2008;20(6):1639–1651. doi: 10.1105/tpc.108.059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigo-Peiris T, Xu XM, Zhao Q, Wang H-J, Meier I. RanGAP is required for post-meiotic mitosis in female gametophyte development in Arabidopsis thaliana. J Exp Bot. 2011;62(8):2705–2714. doi: 10.1093/jxb/erq448. [DOI] [PubMed] [Google Scholar]
- 31.Schönberger J, Hammes UZ, Dresselhaus T. In vivo visualization of RNA in plants cells using the λN₂₂ system and a GATEWAY-compatible vector series for candidate RNAs. Plant J. 2012;71(1):173–181. doi: 10.1111/j.1365-313X.2012.04923.x. [DOI] [PubMed] [Google Scholar]
- 32.Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol. 2007;17(12):1032–1037. doi: 10.1016/j.cub.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 33.Faure JE, Rotman N, Fortuné P, Dumas C. Fertilization in Arabidopsis thaliana wild type: Developmental stages and time course. Plant J. 2002;30(4):481–488. doi: 10.1046/j.1365-313x.2002.01305.x. [DOI] [PubMed] [Google Scholar]
- 34.Ge L, et al. Migration of sperm cells during pollen tube elongation in Arabidopsis thaliana: Behavior during transport, maturation and upon dissociation of male germ unit associations. Planta. 2011;233(2):325–332. doi: 10.1007/s00425-010-1305-8. [DOI] [PubMed] [Google Scholar]
- 35.Heslop-Harrison J, Heslop-Harrison Y. Actomyosin and movement in the angiosperm pollen tube: An interpretation of some recent results. Sex Plant Reprod. 1989;2(4):199–207. [Google Scholar]
- 36.Russell SD, Cass DD. Ultrastructure of the sperms of Plumbago zeylanica 1. Cytology and association with the vegetative nucleus. Protoplasma. 1981;107(1-2):85–107. [Google Scholar]
- 37.Huck N, Moore JM, Federer M, Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003;130(10):2149–2159. doi: 10.1242/dev.00458. [DOI] [PubMed] [Google Scholar]
- 38.Escobar-Restrepo JM, et al. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317(5838):656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- 39.Kessler SA, et al. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330(6006):968–971. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- 40.Capron A, et al. Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell. 2008;20(11):3038–3049. doi: 10.1105/tpc.108.061713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukamoto T, Qin Y, Huang Y, Dunatunga D, Palanivelu R. A role for LORELEI, a putative glycosylphosphatidylinositol-anchored protein, in Arabidopsis thaliana double fertilization and early seed development. Plant J. 2010;62(4):571–588. doi: 10.1111/j.1365-313X.2010.04177.x. [DOI] [PubMed] [Google Scholar]
- 42.Miyazaki S, et al. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol. 2009;19(15):1327–1331. doi: 10.1016/j.cub.2009.06.064. [DOI] [PubMed] [Google Scholar]
- 43.Boisson-Dernier A, et al. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development. 2009;136(19):3279–3288. doi: 10.1242/dev.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leydon AR, et al. Three MYB transcription factors control pollen tube differentiation required for sperm release. Curr Biol. 2013;23(13):1209–1214. doi: 10.1016/j.cub.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang Y, et al. MYB97, MYB101 and MYB120 function as male factors that control pollen tube-synergid interaction in Arabidopsis thaliana fertilization. PLoS Genet. 2013;9(11):e1003933. doi: 10.1371/journal.pgen.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134(6):1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2(8):755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.