Significance
UV-B is a key component of sunlight. In plants, UV-B is perceived by the photoreceptor protein UVR8. Along with regulating photoprotective responses, UV-B dramatically inhibits stem elongation. Here we show how UV-B regulates plant architecture, using the agriculturally important shade avoidance response. When grown in dense stands, plants use reflected far-red light signals from neighbors to detect the threat of shading. In many species, these signals drive rapid elongation responses to overtop competitors. UV-B perceived by UVR8 provides a potent sunlight signal that inhibits shade avoidance. UVR8 activation stimulates multiple pathways that converge to block biosynthesis of the plant growth hormone auxin. Understanding how UV-B regulates plant architecture is central to our understanding of plant growth and development in sunlight.
Abstract
Plants detect different facets of their radiation environment via specific photoreceptors to modulate growth and development. UV-B is perceived by the photoreceptor UV RESISTANCE LOCUS 8 (UVR8). The molecular mechanisms linking UVR8 activation to plant growth are not fully understood, however. When grown in close proximity to neighboring vegetation, shade-intolerant plants initiate dramatic stem elongation to overtop competitors. Here we show that UV-B, detected by UVR8, provides an unambiguous sunlight signal that inhibits shade avoidance responses in Arabidopsis thaliana by antagonizing the phytohormones auxin and gibberellin. UV-B triggers degradation of the transcription factors PHYTOCHROME INTERACTING FACTOR 4 and PHYTOCHROME INTERACTING FACTOR 5 and stabilizes growth-repressing DELLA proteins, inhibiting auxin biosynthesis via a dual mechanism. Our findings show that UVR8 signaling is closely integrated with other photoreceptor pathways to regulate auxin signaling and plant growth in sunlight.
In many plant species, the detection of neighboring vegetation provokes a suite of elongation responses, collectively termed the “shade avoidance syndrome.” Encroaching neighbors absorb red (R) light and reflect far-red (FR) light, lowering the ratio of R:FR sensed by phytochrome photoreceptors (1). This leads to the stabilization and activation of a subset of bHLH transcription factors, PHYTOCHROME INTERACTING FACTORS (PIFs), which drive auxin biosynthesis and elongation growth (2, 3). If the low R:FR signal persists, then flowering is accelerated to promote seed set (1).
UV-B, detected by the UVR8 photoreceptor, regulates plant morphogenesis (4, 5). UV-B is strongly absorbed by vegetation (6) and could signal to plants that they are in sunlight to counter shade avoidance responses. The potential role of UVR8 in regulating shade avoidance has not been explored, however.
To investigate the effects of UV-B in shade avoidance, we analyzed plant responses to simultaneous treatments of low R:FR and narrow-band UV-B. We show that low-dose UV-B perceived by UVR8 strongly inhibits hypocotyl and petiole elongation, even in the presence of a strong low R:FR signal. This occurs via a dual mechanism that simultaneously degrades and inactivates PIF4 and PIF5, thereby inhibiting auxin biosynthesis and cell elongation.
Results
UV-B Perceived by UVR8 Strongly Inhibits Shade Avoidance.
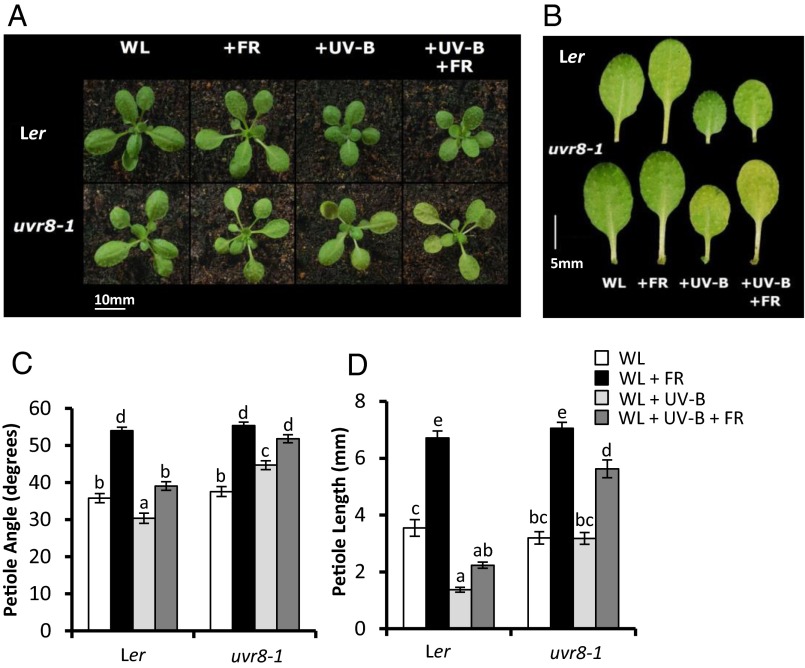
Arabidopsis plants respond to low R:FR (+FR) by elongating and elevating leaf stems (petioles). UV-B treatment has been shown to result in smaller, more compact plants (7). When low R:FR-treated plants were simultaneously exposed to low levels of UV-B (+UV-B) (Fig. S1), petiole elongation and leaf elevation were inhibited, despite the presence of a strong shade avoidance signal (Fig. 1). This inhibition was dependent on the UVR8 photoreceptor, because the UVR8-deficient mutant, uvr8-1 (8) showed very similar shade avoidance responses in the presence and absence of UV-B. In contrast to petiole elongation, leaf area was inhibited by individual low R:FR and UV-B treatments independent of UVR8 (Fig. S2A). Intriguingly, low R:FR promoted leaf expansion in the presence of UV-B in a UVR8-dependent manner (Fig. S2A). Along with the architectural responses, UV-B perceived by UVR8 also partially inhibited the low R:FR-mediated acceleration of flowering characteristic of shade avoidance (Fig. S2B).
Fig. 1.
UV-B inhibits Arabidopsis shade avoidance in a UVR8-dependent manner. (A) WT (Ler) and uvr8-1 rosettes grown in WL in 16-h light/8-h dark cycles for 10 d before transfer to WL, +FR, +UVB, and +UV-B+FR for a further 9 d. (B) Dissected fourth leaf of plants grown as in A. (C and D) Petiole angle from the horizontal (C) and petiole length (D) of leaf 4 in plants grown as in A, at 19 d and 18 d, respectively. n ≥ 22 ± SE. Different letters indicate statistically significant (P < 0.05) differences between means.
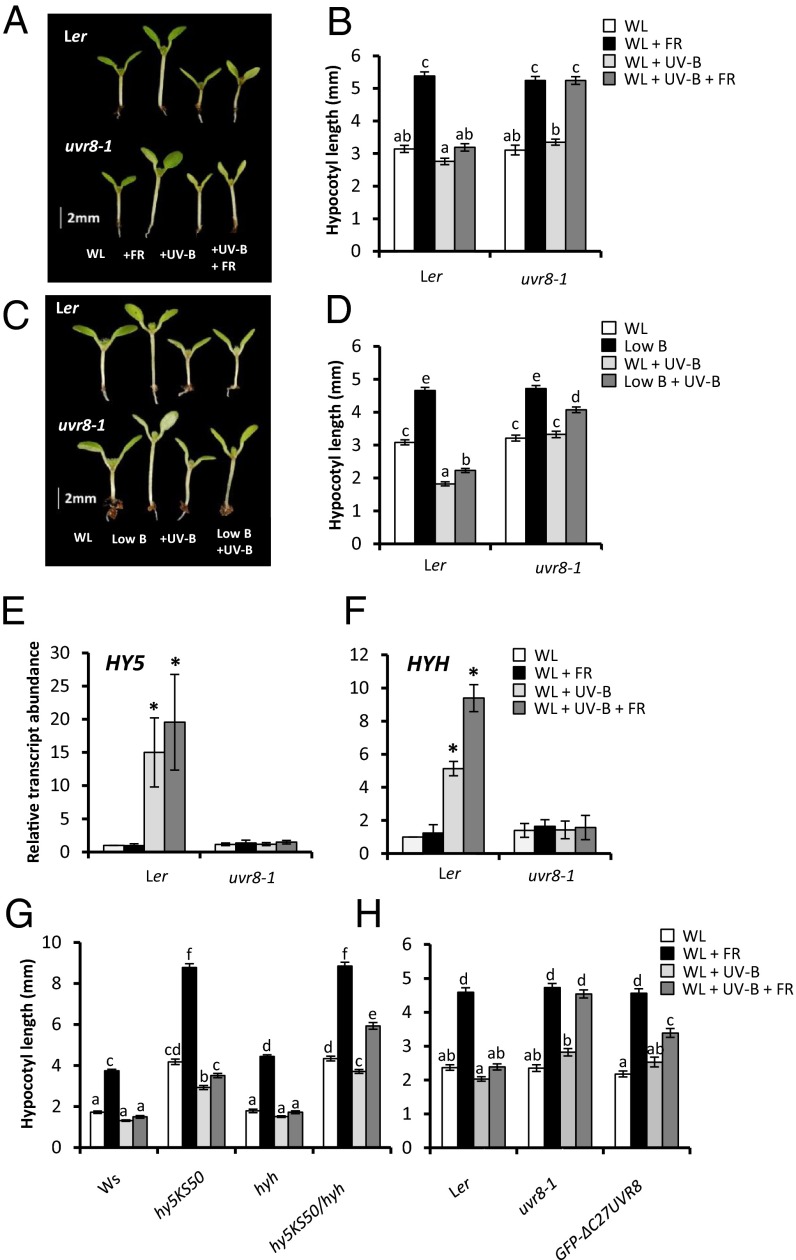
Low R:FR induces striking stem (hypocotyl) elongation in seedlings (Fig. 2 A and B). Thus, we investigated the effect of UV-B on this response. UV-B strongly inhibited low R:FR-mediated hypocotyl elongation in a UVR8-dependent manner (Fig. 2 A and B). Furthermore, low R:FR-mediated induction of the shade-avoidance “marker” genes ATHB2 (9) and PIF3-LIKE 1 (PIL1) (10) was strongly inhibited by UVR8 in the presence of UV-B (Fig. S2 C and D). In addition to low R:FR, vegetational shading reduces the quantity of blue (B) light, as sensed by the cryptochromes (11). Low B alone induces hypocotyl elongation in seedlings. This response also was limited by UV-B in a partially UVR8-dependent manner (Fig. 2 C and D and Fig. S3).
Fig. 2.
UV-B–mediated inhibition of seedling shade avoidance requires UVR8, but only partially involves HY5/HYH and UVR8-COP1 interaction. (A) WT (Ler) and uvr8-1 seedlings grown for 3 d in continuous WL before transfer to WL, +FR, +UV-B, and +FR+UV-B for a further 4 d. (B) Hypocotyl lengths of seedlings grown as in A. (C) Ler and uvr8-1 seedlings grown for 3 d in continuous WL before transfer to WL, low blue (low B), +UV-B, and low B+UV-B for a further 4 d. (D) Hypocotyl lengths of seedlings grown as in B. (E and F) HY5 (E) and HYH (F) transcript abundance of 7-d-old seedlings transferred to experimental conditions for 1 h. n = 3 ± SE. *Significant difference (P < 0.05) compared with WL. (G) Hypocotyl lengths of WT (Ws), hy5KS50, hyh, and hy5KS50/hyh double mutants grown as in A. (H) Hypocotyl lengths of Ler, uvr8-1, and GFP-ΔC27-UVR8 seedlings grown as in A. For hypocotyl assays, data represent mean length ± SE. n = 40. Different letters indicate statistically significant differences (P < 0.05) between means.
UV-B–Mediated Inhibition of Shade Avoidance Occurs Through HY5/HYH-Dependent and -Independent Mechanisms.
Shade avoidance responses can be inhibited by sun flecks through phytochrome-mediated induction of the bZIP transcription factor ELONGATED HYPOCOTYL 5 (HY5) and its close relative HY5 HOMOLOG (HYH) (12). HY5 and HYH are also components of UV-B signaling and control many UVR8-regulated genes (13, 14). UV-B increased transcript levels of HY5 and HYH in high and low R:FR backgrounds, in a UVR8-dependent manner (Fig. 2 E and F). To investigate the roles of HY5 and HYH in UV-B–mediated shade avoidance inhibition, we analyzed hypocotyl elongation in hy5 and hyh mutants. Single mutants displayed similar UV-B responses to WT controls (Fig. 2G). In contrast, the hy5KS50/hyh double mutant exhibited a significant low R:FR-mediated increase in hypocotyl length in the presence of UV-B. These data suggest that HY5 and HYH act redundantly to mediate the UV-B–induced inhibition of shade avoidance. The hy5KS50/hyh double mutant did retain some UV-B–mediated inhibition of hypocotyl elongation in low R:FR, however, suggesting the existence of HY5/HYH-independent mechanisms.
UV-B–mediated increases in HY5 expression require physical interaction between a 27-aa C-terminal region (C27) of UVR8 and the WD-40 domain of CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (4, 15, 16). GFP–ΔC27UVR8 plants express a deletion mutant of UVR8 that is unable to bind COP1 (16). To investigate the importance of UVR8–COP1 interaction in UV-B–mediated shade avoidance inhibition, we analyzed hypocotyl elongation in GFP–ΔC27UVR8 plants. These plants displayed less inhibition than WT plants, suggesting a requirement for UVR8–COP1 binding (Fig. 2H); however, the residual inhibition observed in GFP–ΔC27UVR8 plants compared with uvr8-1 suggests a unique UVR8 function, independent of COP1 interaction.
UVR8 Activation Suppresses Low R:FR-Mediated Auxin Biosynthesis Independent of HY5/HYH.
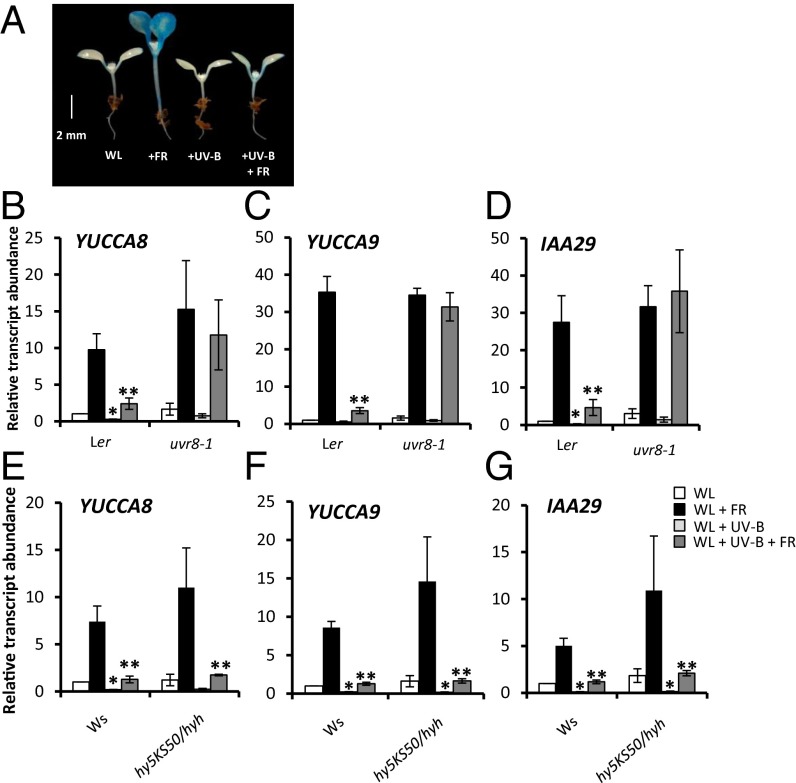
Auxin has been linked to leaf blade expansion and flavonoid levels in UV-B–treated Arabidopsis plants, although no molecular mechanism has been proposed (17). Elongation growth responses to low R:FR require increased auxin biosynthesis via TRYPTOPHAN TRANSAMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) (3). It was recently shown that members of the YUCCA family of flavin mono-oxygenases catalyze the rate-limiting step of this pathway (3, 18). YUCCA2, YUCCA5, YUCCA8, and YUCCA9 are rapidly up-regulated in response to low R:FR, forming a key regulatory mechanism controlling shade avoidance (3, 19). Thus, we investigated the role of auxin in UV-B–mediated shade avoidance inhibition. Analyses of transgenic plants expressing the auxin-responsive pDR5:GUS reporter (20) showed that low R:FR-mediated increases in auxin activity were inhibited in the presence of UV-B (Fig. 3A and Fig. S4A). Furthermore, UV-B strongly inhibited the low R:FR-mediated induction of YUCCA2, YUCCA5, YUCCA8, YUCCA9, and the auxin-responsive genes IAA29 and GH3.3 (Fig. 3 B–D and Fig. S4 B–D). The UV-B–mediated suppression of auxin biosynthesis/signaling genes observed in low R:FR was dependent on the presence of UVR8 (Fig. 3 B–D), but not on the presence of HY5/HYH (Fig. 3 E–G).
Fig. 3.
UV-B inhibits auxin biosynthesis in a UVR8-dependent and HY5/HYH-independent manner. (A) pDR5:GUS seedlings grown for 3 d in continuous WL before transfer to WL, +FR, +UV-B, and +FR+UV-B for a further 4 d, stained for 24 h. (B–D) Relative transcript abundance of YUCCA8, YUCCA9, and IAA29 in 7-d-old WT (Ler) and uvr8-1 seedlings transferred to experimental conditions for 4 h. (E–G) Relative transcript abundance of YUCCA8, YUCCA9, and IAA29 in 7-d-old WT (Ws) and hy5ks50/hyh seedlings transferred to experimental conditions for 4 h. n = 3 ± SE. *Significant difference (P < 0.05) in transcript abundance compared with WL. **Significant difference (P < 0.05) in transcript abundance compared with +FR.
Taken together, our data suggest that a major HY5/HYH-independent role of UV-B in shade avoidance is to suppress low R:FR-mediated increases in auxin biosynthesis. Consistent with hypocotyl elongation data (Fig. 2H), ΔC27UVR8 plants displayed less UV-B–mediated inhibition of low R:FR-induced YUCCA8 and YUCCA9 transcript abundance compared with WT plants. A significant residual inhibition response was observed, suggesting that UVR8 can suppress auxin biosynthesis independent of COP1 binding (Fig. S5).
UV-B–Mediated DELLA Stabilization Contributes to Shade Avoidance Inhibition.
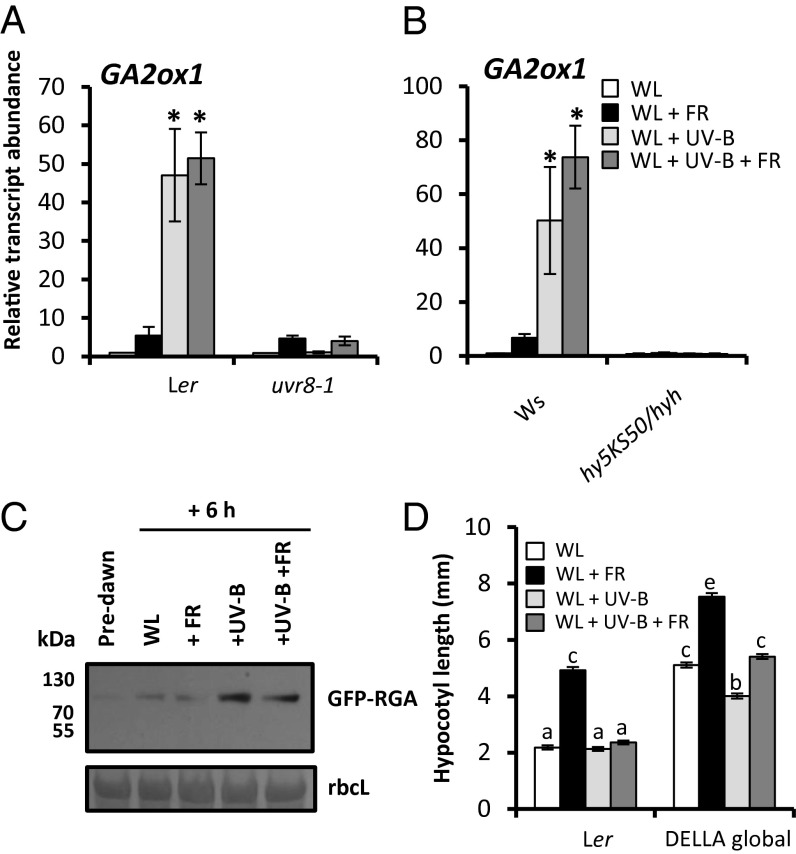
Along with increased auxin activity, shade avoidance also involves gibberellic acid (GA) signaling (1). In low R:FR environments, GA promotes elongation growth by triggering the degradation of growth-repressing DELLA proteins. Indeed, DELLA degradation is essential for low R:FR-induced hypocotyl elongation (21). DELLAs function in part by physically interacting with growth-promoting PIF proteins, preventing them from binding to DNA targets (22, 23). A key mechanism controlling bioactive GA levels in Arabidopsis involves 2β-hydroxylation of GA to an inactive form by GA2-oxidases (GA2ox) (24). UV-B induced a rapid increase in GA2ox1 transcript abundance in a response requiring both UVR8 (Fig. 4A) and HY5/HYH (Fig. 4B) (25). Increased GA2ox1 levels were accompanied by enhanced stability of the DELLA protein, RGA (Fig. 4C). Furthermore, UV-B inhibited transcript accumulation of the GA biosynthesis gene GA20ox2 in a UVR8-dependent manner. Inhibition of GA3ox1 transcript abundance was observed only in low R:FR + UV-B, however, suggesting that inhibition effects may be gene-specific (Fig. S6).
Fig. 4.
DELLA proteins are stabilized by UV-B and contribute to shade avoidance inhibition. (A and B) Relative transcript abundance of GA2ox1 in 7-d-old Ler and uvr8-1 (A) and Ws and hy5ks50/hyh (B) seedlings transferred to experimental conditions for 4 h. n = 3 ± SE. *Significant increase in transcript abundance by +UV-B treatment. (C) Western blot showing RGA-GFP abundance in pRGA:GFP-RGA seedlings grown for 10 d in WL in 16-h light/8-h dark cycles. Protein extracts from tissue samples harvested predawn and after 6 h of WL, +FR, +UVB, and +UV-B +FR. A Ponceau stain of rbcL is shown below as a loading control. (D) Hypocotyl lengths of Ler and DELLA global null seedlings grown as in A. n = 40 ± SE. Different letters indicate statistically significant differences (P < 0.05) between means.
We assessed the contribution of DELLA proteins to UV-B–mediated shade avoidance inhibition through analysis of a DELLA global null mutant (26). DELLA-deficient plants displayed long hypocotyls that were still elongated in low R:FR (Fig. 4D). This response was partially attenuated by UV-B, suggesting the existence of both DELLA-dependent and -independent mechanisms (Fig. 4D).
PIF4 and PIF5 Are Degraded in the Presence of UV-B.
The promotion of auxin biosynthesis in shade avoidance is regulated by PIF proteins. In low R:FR, PIF4 and PIF5 levels are stabilized, whereas PIF7 is activated via dephosphorylation (2, 19). These PIFs display enhanced binding to G-boxes in the promoter regions of YUCCA8 and YUCCA9, driving auxin biosynthesis (19, 27). Other, minor roles have been reported for PIF1 and PIF3 in parallel (28). Consistent with previous studies (2), we found that pif4/5 double mutants displayed significantly impaired hypocotyl elongation responses to low R:FR, confirming a role for these transcription factors in shade avoidance in our experimental conditions (Fig. S7A). UV-B suppressed the elongated phenotypes of phytochrome-deficient (phyABCDE) mutants (Fig. S7B), PIF4-overexpressing plants (Fig. S7C) and PIF5-overexpressing plants (Fig. S7D), suggesting that UV-B may inhibit PIF activity independent of phytochromes. Thus, we investigated whether UVR8 could physically interact with PIFs. We used a yeast two-hybrid (Y2H) approach to investigate UVR8 interactions with PIF4, PIF5, and PIF7. Control experiments confirmed UV-B–dependent UVR8–COP1 interaction and PIF heterodimerization (4, 16, 29) (Fig. S8), but after growth on selective media, no interaction was observed between UVR8 and PIF4, PIF5, or PIF7 (Fig. S8).
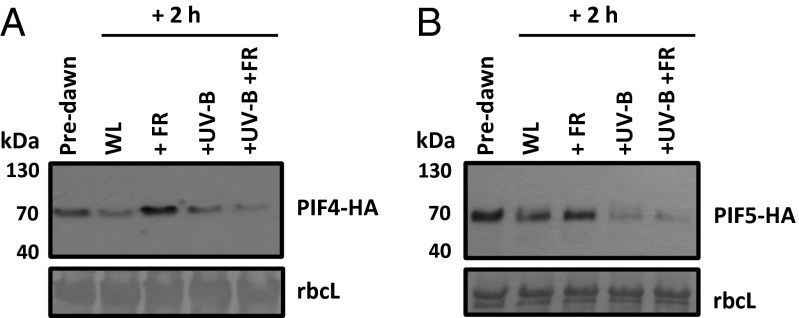
We next investigated whether UV-B affects PIF protein abundance. Consistent with previous observations, PIF4-HA and PIF5-HA accumulated in the dark, were degraded in white light (WL), and were stabilized by low R:FR (2). Surprisingly, UV-B significantly decreased the stability of PIF4 (Fig. 5A) and PIF5 (Fig. 5B) in both high and low R:FR backgrounds.
Fig. 5.
UV-B decreases PIF4 and PIF5 protein abundance. Western blots showing PIF4-HA (A) and PIF5-HA (B) abundance in 35S:PIF-HA seedlings grown for 10 d in WL in 16-h light/8-h dark cycles. Proteins were extracted from tissue samples harvested predawn and after 2 h of WL, +FR, +UVB, and +UV-B+FR. A Ponceau stain of rbcL is shown below each blot as a loading control.
When plants were subject to continuous light treatments for 4 d, a reduction in PIF5 protein abundance was maintained in +FR +UV-B samples compared with +FR alone (Fig. S9A). UV-B–mediated inhibition of low R:FR-induced IAA29 transcript abundance was also maintained in a UVR8-dependent manner (Fig. S9B). These data are consistent with the reduced pDR5:GUS activity observed at this time point (Fig. 3A). In contrast, UV-B–mediated increases in GA2ox1 transcript abundance were not maintained after 4 d of continuous UV-B treatment, suggesting that the DELLA-mediated inhibition component of the inhibition response may be more transient (Fig. S9C).
Decreased PIF4 and PIF5 abundance in UV-B–treated plants would contribute to the reduced auxin activity (Fig. 3 A–G) (19, 27, 30), decreased stem elongation (Figs. 1 and 2 A and B) (19, 27), and delayed flowering (Fig. S2B) (2) observed in both high and low R:FR. The lack of physical interaction between UVR8 and PIFs in yeast is intriguing and suggests the existence of a unique UV-B–mediated mechanism regulating PIF4 and PIF5 abundance.
Discussion
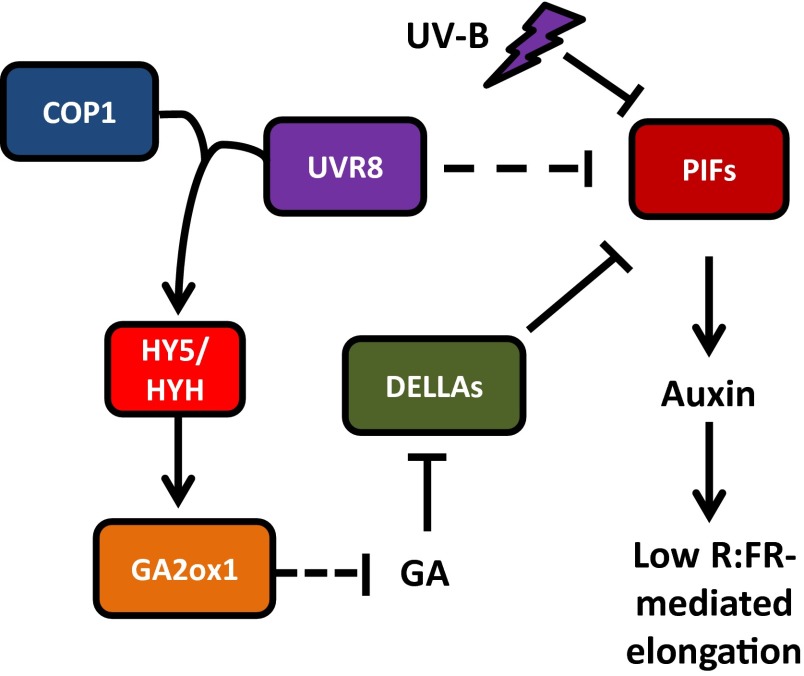
In this work, we show that UV-B, perceived by the UVR8 photoreceptor, acts as a potent brake on plant shade avoidance (Figs. 1 and 2 A–D). This action occurs through multiple pathways (Fig. 6). UVR8 interaction with COP1 leads to elevated levels of HY5 and HYH (15, 16, 25) (Fig. 2 E and F), which in turn leads to increased GA2ox1 transcript abundance (Fig. 4 A and B). Increased GA catabolism in UV-B without a concomitant increase in GA biosynthesis likely will contribute to increased DELLA protein stability (Fig. 4C), which suppresses PIFs (22, 23). In a parallel HY5/HYH-independent pathway, perception of UV-B by UVR8 inhibits low R:FR-mediated induction of YUCCA2, YUCCA5, YUCCA8, and YUCCA9 (Fig. 3 B, C, E, and F and Fig. S4 B and C). This likely occurs in part through UV-B–mediated turnover of PIF4 and PIF5 (Fig. 5). UVR8 does not physically interact with PIF4 or PIF5 in Y2H (Fig. S8), suggesting that an as-yet unidentified pathway may link UVR8 activation to PIF degradation. The reduced abundance and activity of PIF4 and PIF5 in UV-B decreases auxin activity (Fig. 3A), which limits elongation growth, suppressing shade avoidance.
Fig. 6.
Hypothesized role of UV-B in shade avoidance. UV-B perceived by the photoreceptor UVR8 interacts with COP1 and up-regulates transcription of HY5 and HYH. GA2ox1 levels increase, reducing GA, and increasing DELLA stability, which inhibits PIF function. In a separate pathway, UV-B signaling blocks low R:FR-mediated up-regulation of auxin biosynthesis, in part through enhanced degradation of PIF4 and PIF5. Together, UV-B inhibits auxin biosynthesis and elongation growth, inhibiting shade avoidance.
The residual shade avoidance observed in pif4/5 double mutants was inhibited by UV-B (Fig. S7A), suggesting that a PIF4/5-independent mechanism must exist. This mechanism may include UV-B–mediated dephosphorylation of PIF7 (19). In this way, UV-B perception by UVR8 provides plants with an unambiguous signal of sunlight, triggering a dual growth restraint mechanism to prevent the unnecessary allocation of resources toward neighbor competition. Furthermore, the inhibition of auxin signaling by UVR8 activation provides a molecular explanation for the compact architecture of UV-B–treated plants; however, shade avoidance responses to reflected FR signals have been observed in multiple species in the field, where UV-B is clearly present (31, 32). It is possible that the magnitude of UV-B–mediated shade avoidance inhibition varies with both species and background levels of photosynthtically active radiation (PAR). The impact of UV-B:PAR ratio on shade avoidance will be an important area of future study.
Taken together, our data show how plants integrate UV-B signals with other photoreceptor pathways to interpret sunlight. PIF proteins function as signaling “hubs” (29), regulating auxin and GA activity to control plant development. Understanding how plants integrate the full sunlight spectrum is essential to provide a holistic understanding of growth and development in fluctuating natural light environments.
Materials and Methods
Plant Material.
All mutants and transgenic lines used in this study have been described previously. The uvr8-1 mutant (8), ΔC27UVR8 line (16), phyABCDE mutant (33), and DELLA global mutant (26) are in the Landsberg erecta (Ler) background. The hy5KS50, hyh, and hy5KS50/hyh mutants are in the Wassilewskija (Ws) background (34). The pif4/pif5 mutant (2), PIF4- and PIF5-overexpressing lines (2), and DR5::GUS reporter line are in the Columbia-0 (Col-0) background (20).
Growth Conditions.
Seeds were sown directly onto a 3:1 mixture of compost: horticultural silver sand. After 4 d of stratification in darkness at 4 °C, seedlings were germinated in controlled growth cabinets (Microclima 1600E; Snijder Scientific) in continuous WL (R:FR >10) at 20 °C and 70% humidity. For adult plant experiments and protein stability assays, plants were grown in the same cabinets under a 16-h light/8-h dark cycle. WL was provided by cool-white fluorescent tubes (400–700 nm) at a photon irradiance of 90 μmol m−2s−1 for low R:FR experiments, decreased to 65 μmol m−2s−1 with neutral density filters (Lee Filters) for low B experiment controls. Low R:FR experiments were performed with supplementary arrays of far-red LEDs positioned overhead (λmax, 735 nm). For these experiments, plants received the same photon irradiance of photosynthetically active radiation, but with an R:FR of 0.05. For low B experiments, white fluorescent tubes were filtered through two layers of yellow filter (010 Medium Yellow; Lee Filters) to reduce blue light (400–500 nm) from ∼12 μmol m−2s−1 to ∼2 μmol m−2s−1 (total PAR, 65 μmol m−2s−1).
All light measurements were performed using an EPP2000 fiberoptic spectrometer with a planar sensor (Stellarnet). Supplementary narrow-band UV-B was provided at a photon irradiance of 400 mWm (2) (∼1 µmol m−2s−1) by Philips TL100W/01 tubes filtered through a 3-mm-thick Perspex clear acrylic sheet. The biologically effective UV-B dose (BE-UV-B) was calculated as 3.6 µWm−2nm−1, following Flint and Caldwell (35). Control experiments were performed with UV-B tubes filtered through 3-mm-thick extruded acrylic tubes, which selectively removed all UV-B. UV-B measurements were performed using a SpectroSense 2 fitted with a Skye Instruments SKU430 UV-B sensor. The spectra of all eight light conditions are shown in Figs. S1 and S3.
Plant Measurements.
Measurements of hypocotyl length, petiole length, leaf angle, and leaf area were recorded using ImageJ software (rsb.info.nih.gov/ij/). For hypocotyl measurements, a minimum of 34 seedlings were measured for each genotype in each condition. For leaf area and petiole length measurements, the largest fully expanded rosette leaf (leaf 4) was excised from each plant at day 19. Flowering times were recorded by counting rosette leaves when plants displayed a 10-mm bolt. Leaf angles were measured from the horizontal soil surface at day 18. Measurements were recorded from a minimum of 22 plants per treatment. All experiments were repeated multiple times, with similar results.
RNA Extraction and Quantitative PCR Analysis.
Seedlings were initially grown for 7 d in WL before being transferred to different light conditions for the indicated times. RNA extraction, cDNA synthesis, and quantitative PCR were performed as described previously (10). Transcript abundance values were normalized to ACTIN2 using the primers ActinF and ActinR (Table S1). Selected results were confirmed with normalization to UBIQUITIN CONJUGATING ENZYME 21 (UBC21) using the primers UBC21F and UBC21R (Table S1). These experiments produced similar results. The primers used for analysis of HY5, HYH, YUCCA2, YUCCA5, YUCCA8, YUCCA9, IAA29, GH3.3, GA2ox1, GA20ox2, GA3ox1, GUS, PIL1, and ATHB2 are listed in Table S1. Three biological replicates were performed for each experiment.
GUS Assay.
GUS activity was assayed in 7 d-old DR5::GUS seedlings (3 d in WL, followed by 4 d in light treatments). Samples were immersed in 1 mL of assay buffer containing 0.1 M NaPO4 (pH 7.0), 10 mM EDTA, 0.1% Triton X-100, 1 mM K3Fe(CN)6, and 2 mM 5-bromo-4-chloro-3-indolyl glucuronide salt (X-Gluc; Melford Laboratories) at 37 °C and incubated for 24 h in the dark. Samples were then washed three times in 50% ethanol and photographed.
Western Blot Analysis.
Seedlings were grown in 16-h light/8-h dark cycles of high R:FR (90 μmol m−2s−1, 20 °C, 70% humidity). At 10 d, samples were harvested predawn and following the indicated time in controlled experimental conditions. 35S:PIF4-HA and 35S:PIF5-HA lines were extracted in 100 mM Tris⋅HCl pH 8, 4 M urea, 5% (wt/vol) SDS, 15% (vol/vol) glycerol, 10 mM β-ME, and 30 µL/mL protease inhibitor cocktail (Sigma-Aldrich; P9599), before boiling at 95 °C for 4 min and centrifugation at top speed for 15 min. pRGA:GFP-RGA lines were extracted in 1.1 M glycerol, 5 mM EGTA, 1.5% (wt/vol) PVP, 1% (wt/vol) ascorbic acid, 400 mM BTP, and 10 µL/mL protease inhibitor mixture (Sigma-Aldrich; P9599), pH 7.6, before centrifugation at top speed for 10 min. Protein levels were quantified either by the RC DC Lowry assay (Bio-Rad) for PIF4-HA and PIF5-HA or by the Bradford assay (Bio-Rad) for GFP-RGA. SDS/PAGE sample buffer [4×; 8% (wt/vol) SDS, 0.4% (wt/vol) bromophenol blue, 40% (vol/vol) glycerol, 200 mM Tris⋅HCl pH 6.8, and 400 mM β-ME] was added to supernatants to a final dilution of 1×. Samples were then heated for 10 min at 95 °C. Aliquots containing 30 µg (PIF4-HA and PIF5-HA) or 40 µg (GFP-RGA) of total protein were then loaded onto 10% SDS/PAGE gels and blotted onto nitrocellulose membrane (Bio-Rad).
For PIF4-HA and PIF5-HA detection, blots were incubated overnight in 1:1,000 anti-HA antibody conjugated to peroxidase (Roche). For RGA-GFP detection, blots were incubated overnight in 1:1,000 mouse anti-GFP antibody (Roche) and for 1 h in 1:2,000 anti-mouse antibody conjugated to peroxidase (Dako). Signals were detected using Pierce ECL2 Western blotting substrate (Thermo Scientific). Two or three biological repeats of each Western blot were performed, with identical results.
Statistical Analyses.
Statistical analyses were carried out using SPSS version 21.0 (IBM). Morphological and flowering time assays were analyzed using one-way ANOVA, treating genotype and light condition together as a single factor. Tukey’s post hoc tests were used to deduce statistically significant means (P < 0.05), as indicated by letters in the figures. For gene expression analyses, relative expression values were first log2-transformed. Student t tests were then performed to investigate differences between the means indicated in the figure legends (P < 0.05).
Supplementary Material
Acknowledgments
We acknowledge the G.I.J. and Christie laboratories at the University of Glasgow for technical advice on cloning, Christian Fankhauser (University of Lausanne) for PIF5ox lines, Clark Lagarias (University of California, Davis) for phyABCDE mutants, and Nicholas Harberd (University of Oxford) for DELLA global mutants. We thank the G.I.J. and Christie laboratories, Severine Lorrain (University of Lausanne), and Patrick Achard (Centre National de la Recherche Scientifique) for advice on Western blot analysis. We also thank James Foster (University of Bristol) for help with BE-UV-B calculations and Paul Parsons (University of Bristol) for help with statistics. S.H. is supported by a studentship from the UK Natural Environment Research Council. C.N.V. is supported by the State Scholarship Foundation of Greece. K.A.F. is a Royal Society Research Fellow. Work in G.I.J.’s laboratory is supported by the Biotechnology and Biological Sciences Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403052111/-/DCSupplemental.
References
- 1.Franklin KA. Shade avoidance. New Phytol. 2008;179(4):930–944. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- 2.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53(2):312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 3.Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133(1):164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 5.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335(6075):1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell MM, Robberecht R, Flint SD. Internal filters: Prospects for UV-acclimation in higher plants. Physiol Plant. 1983;58:445–450. [Google Scholar]
- 7.Jansen M. Ultraviolet-B radiation effects on plants: Induction of morphogenic responses. Physiol Plant. 2002;116:423–429. [Google Scholar]
- 8.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130(1):234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steindler C, et al. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999;126(19):4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- 10.Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426(6967):680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- 11.Keuskamp DH, et al. Blue-light–mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 2011;67(2):208–217. doi: 10.1111/j.1365-313X.2011.04597.x. [DOI] [PubMed] [Google Scholar]
- 12.Sellaro R, Yanovsky MJ, Casal JJ. Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J. 2011;68(5):919–928. doi: 10.1111/j.1365-313X.2011.04745.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown BA, Jenkins GI. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008;146(2):576–588. doi: 10.1104/pp.107.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown BA, et al. A UV-B–specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA. 2005;102(50):18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B–induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28(5):591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloix C, et al. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA. 2012;109(40):16366–16370. doi: 10.1073/pnas.1210898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hectors K, van Oevelen S, Guisez Y, Prinsen E, Jansen MA. The phytohormone auxin is a component of the regulatory system that controls UV-mediated accumulation of flavonoids and UV-induced morphogenesis. Physiol Plant. 2012;145(4):594–603. doi: 10.1111/j.1399-3054.2012.01590.x. [DOI] [PubMed] [Google Scholar]
- 18.Won C, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(45):18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26(8):785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djakovic-Petrovic T, de Wit M, Voesenek LACJ, Pierik R. DELLA protein function in growth responses to canopy signals. Plant J. 2007;51(1):117–126. doi: 10.1111/j.1365-313X.2007.03122.x. [DOI] [PubMed] [Google Scholar]
- 22.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451(7177):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 23.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451(7177):475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rieu I, et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 2008;20(9):2420–2436. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18(8):1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19(5):408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 27.Hornitschek P, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71(5):699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 28.Leivar P, et al. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 2012;24(4):1398–1419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leivar P, Quail PH. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16(1):19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011;156(1):357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballaré CL, et al. Early detection of neighbour plants by phytochrome perception of spectral changes in reflected sunlight. Plant Cell Environ. 1987;10:551–557. [Google Scholar]
- 32.Ballaré CL, Scopel AL, Sánchez RA. Far-red radiation reflected from adjacent leaves: An early signal of competition in plant canopies. Science. 1990;247(4940):329–332. doi: 10.1126/science.247.4940.329. [DOI] [PubMed] [Google Scholar]
- 33.Hu W, et al. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci USA. 2013;110(4):1542–1547. doi: 10.1073/pnas.1221738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001;20(1-2):118–127. doi: 10.1093/emboj/20.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flint SD, Caldwell MM. A biological spectral weighting function for ozone depletion research with higher plants. Physiol Plant. 2003;117:137–144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.