Significance
Liver resident activated hepatic stellate cells (aHSCs), and activated portal fibroblasts (aPFs) are the major source of the fibrous scar in the liver. aPFs have been implicated in liver fibrosis caused by cholestatic liver injury, whereas fibrosis in hepatotoxic liver injury is attributed to aHSCs. However, the contribution of aPFs to cholestatic fibrosis is not well characterized because of difficulties in cell purification and the lack of identified aPF-specific markers. We have developed a novel flow cytometry-based method of aPFs purification from the nonparenchymal cell fraction of collagen-α1(I)-GFP mice and have identified potential aPF-specific markers. The goal of this study is to determine whether aPFs contribute to cholestatic liver fibrosis and identify the mechanism(s) of their activation.
Keywords: ECM deposition, markers of fibrogenic myofibroblasts
Abstract
Hepatic myofibroblasts are activated in response to chronic liver injury of any etiology to produce a fibrous scar. Despite extensive studies, the origin of myofibroblasts in different types of fibrotic liver diseases is unresolved. To identify distinct populations of myofibroblasts and quantify their contribution to hepatic fibrosis of two different etiologies, collagen-α1(I)-GFP mice were subjected to hepatotoxic (carbon tetrachloride; CCl4) or cholestatic (bile duct ligation; BDL) liver injury. All myofibroblasts were purified by flow cytometry of GFP+ cells and then different subsets identified by phenotyping. Liver resident activated hepatic stellate cells (aHSCs) and activated portal fibroblasts (aPFs) are the major source (>95%) of fibrogenic myofibroblasts in these models of liver fibrosis in mice. As previously reported using other methodologies, hepatic stellate cells (HSCs) are the major source of myofibroblasts (>87%) in CCl4 liver injury. However, aPFs are a major source of myofibroblasts in cholestatic liver injury, contributing >70% of myofibroblasts at the onset of injury (5 d BDL). The relative contribution of aPFs decreases with progressive injury, as HSCs become activated and contribute to the myofibroblast population (14 and 20 d BDL). Unlike aHSCs, aPFs respond to stimulation with taurocholic acid and IL-25 by induction of collagen-α1(I) and IL-13, respectively. Furthermore, BDL-activated PFs express high levels of collagen type I and provide stimulatory signals to HSCs. Gene expression analysis identified several novel markers of aPFs, including a mesothelial-specific marker mesothelin. PFs may play a critical role in the pathogenesis of cholestatic liver fibrosis and, therefore, serve as an attractive target for antifibrotic therapy.
Chronic liver injury of many etiologies results in liver fibrosis. There are two general types of chronic liver diseases, hepatocellular (injury to hepatocytes, such as chronic viral hepatitis and nonalcoholic steatohepatitis) and cholestatic (obstruction to bile flow, such as primary biliary cirrhosis and primary sclerosing cholangitis) (1). Experimental rodent models of liver fibrosis mimic these two types of chronic liver injuries: Repeated carbon tetrachloride (CCl4) administration produces hepatocelluar injury, and common bile duct ligation (BDL) produces cholestatic injury (2). In all chronic liver diseases, myofibroblasts are embedded in the fibrous scar and are the source of this excessive extracellular matrix (ECM). Myofibroblasts, which are not present in normal liver, are characterized by distinct morphology, contractility with intracellular stress fibers [α-smooth muscle actin (α-SMA), nonmuscle myosin, and vimentin], and secretion of extracellular matrix (fibronectin and fibrillar collagens) (1, 2).
The cells of origin of hepatic myofibroblasts are unresolved, and perhaps the fibrosis induced by different types of liver injury results from different fibrogenic cells. Hepatic myofibroblasts may originate from bone marrow (BM)-derived mesenchymal cells and fibrocytes, but only a small contribution of BM-derived cells to the myofibroblast population has been detected in experimental liver fibrosis (3–5). Another potential source of myofibroblast is epithelial-to-mesenchymal transition (EMT), in which epithelial cells acquire a mesenchymal phenotype and may give rise to fully differentiated myofibroblasts. However, recent cell fate mapping studies have failed to detect any hepatic myofibroblasts originating from hepatocytes, cholangiocytes, or epithelial progenitor cells (3, 6–10). Thus, the major sources of myofibroblasts in liver fibrosis are the endogenous liver mesenchymal cells, which consist of portal fibroblasts and hepatic stellate cells.
Quiescent hepatic stellate cells (qHSCs) are located in the space of Disse, store retinoids in lipid droplets, and express neural markers, such as glial fibrillary acidic protein (GFAP), synaptophisin, and nerve growth factor receptor p75 (1). In response to injury, qHSCs down-regulate vitamin A-containing lipid droplets and neural markers, and differentiate into α-SMA–expressing myofibroblasts (1, 2). Portal fibroblasts normally comprise a small population of the fibroblastic cells that surround the portal vein to maintain integrity of portal tract. They were first described as “mesenchymal cells not related to sinusoids,” and since then have been called “periductular fibroblasts” or portal/periportal mesenchymal cells” (11) and implicated by association in the pathogenesis of cholestatic liver injury. In response to chronic injury, portal fibroblasts may proliferate, differentiate into α-SMA–expressing myofibroblasts, and synthesize extracellular matrix (11–14).
The contribution of portal fibroblasts (PFs) to liver fibrosis of different etiologies is not well understood, mainly because of difficulties in isolating PFs and myofibroblasts. The most widely used method of PF isolation from rats is based on liver perfusion with enzymatic digestion followed by size selection (15). Cell outgrowth from dissected bile segments is still used to isolate mouse PFs, and after 10–14 d in culture, PFs undergo progressive myofibroblastic activation (16). The disadvantage of this technique is that it requires multiple passaging and prolong culturing (11). A more physiological method of PF culturing in a precision-cut liver slice is designed to maintain cell–cell and cell–matrix interactions and mimic natural microenvironment of PFs, but it does not enable the study of purified PFs (17). Therefore, only a few markers of PFs are available to identify PFs in the myofibroblast population, including gremlin, Thy1, fibulin 2, interleukin 6 (IL-6), elastin, the ecto-AT-Pase nucleoside triphosphate diphosphohydrolase-2 (NTPD2), and coffilin 1. In addition, the lack of desmin, cytoglobin, α2-macroglobulin, neural proteins (GFAP, p75, synaptophysin), and lipid droplets distinguishes PFs from HSCs (1, 17–21).
Our study uses transgenic reporter mice and new flow cytometry protocols to identify the origin of myofibroblasts and quantify their numbers in two murine models of chronic liver injury (BDL and CCl4). Our study demonstrates that the origin of the myofibroblasts is determined by the type of liver injury. As previously reported using other methodologies, HSCs are the major source of myofibroblasts in CCl4 liver injury. In contrast, most of the myofibroblasts at the onset of BDL-induced liver injury originate from activated PFs (aPFs).
Results
BDL- and CCl4-Induced Liver Fibrosis Is Associated with Activation of Myofibroblasts in Mice.
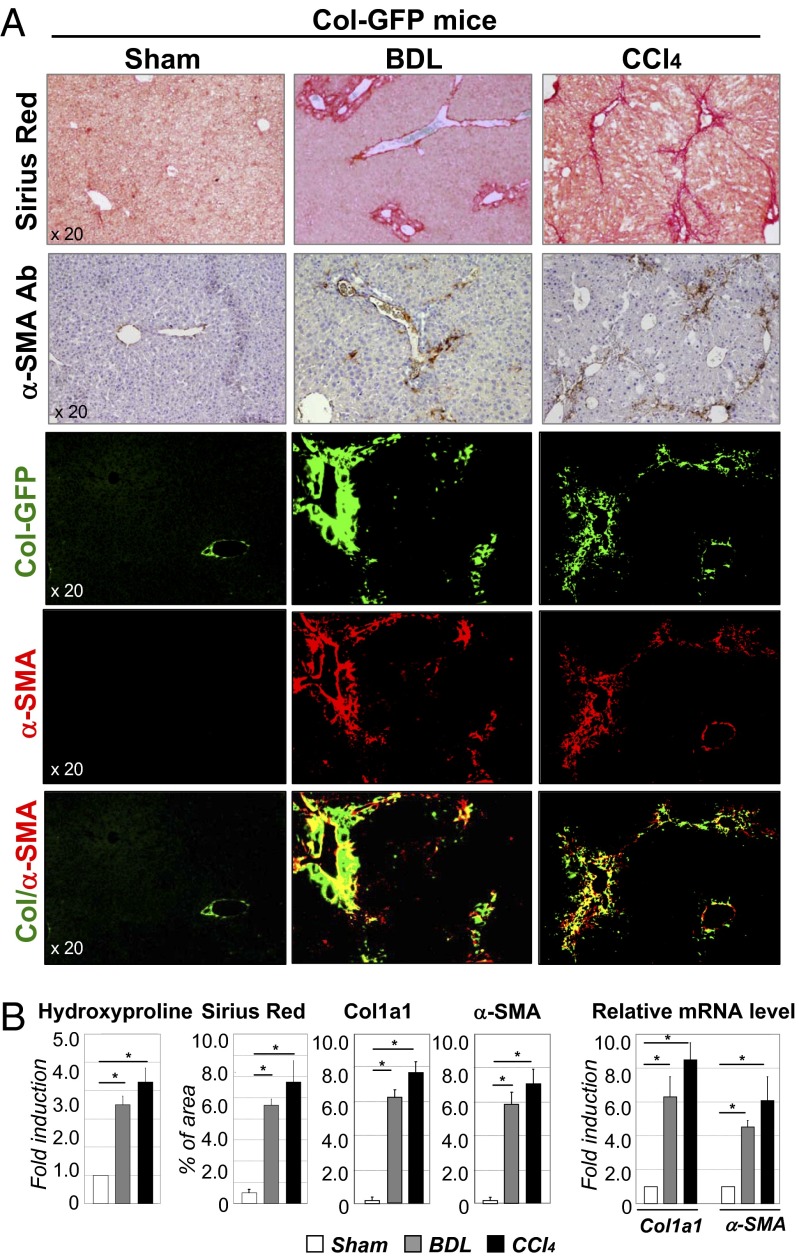
To study activation of hepatic myofibroblasts, Col-GFP mice expressing GFP under control of collagen α1(I) promoter/enhancer (22) were subjected to BDL (20 d) or CCl4 (1.5 mo) liver injury. Upon activation, hepatic myofibroblasts in these mice are visualized by GFP expression. Development of liver fibrosis was confirmed in Col-GFP mice by hydroxyproline content, Sirius Red staining (Fig. 1 A and B) and correlated with increased collagen-α1(I) (fold increase 6.1 ± 0.3 and 7.6 ± 0.4 in BDL- and CCl4-treated vs. control mice) and α-SMA mRNA expression (fold increase 4.2 ± 0.2 and 6.1 ± 0.7 vs. control mice, respectively; Fig. 1B). Development of liver fibrosis was also associated with activation of myofibroblasts, demonstrated by Col-GFP expression (6.5 ± 0.4% and 7.8 ± 0.5% of GFP+ area in BDL- and CCl4-treated vs. 0.3 ± 0.03% in control mice) and α-SMA expression (Fig. 1B). Thus, BDL and CCl4 induced comparable levels of fibrosis and activation of myofibroblasts in the liver, sufficient to isolate GFP+ myofibroblasts and determine their composition in response to two different injuries.
Fig. 1.
Development of liver fibrosis in Col-GFP mice in response to BDL and CCl4. (A) CCl4-treated and BDL-operated mice (but not sham mice, 8-wk-old, n = 10 per group) developed liver fibrosis, as shown by Sirius Red staining, fluorescent microscopy for collagen-GFP, and staining for α-SMA (20× objective). (B) Fibrosis was assessed by hydroxyproline and Sirius Red (positive area) content and by mRNA levels of fibrogenic genes (Col and α-SMA) in all groups of mice is shown, *P < 0.003; **P < 0.001.
Isolation of Myofibroblasts.
The reporter Col-GFP mice (22) have been extensively characterized and are widely used to visualize activated myofibroblasts in fibrotic liver, lungs, kidneys, and skin (3–5, 8, 23–36). Expression of GFP in these mice closely correlates with expression of collagen type I protein in hepatic myofibroblasts but is not expressed in endothelial, epithelial, or other cell types (37–39). Using Col-GFP mice we have demonstrated that activated hepatic stellate cells (aHSCs) (GFP+, vitamin A+, Desmin+ cells) comprise >92% of myofibroblasts in response to CCl4-induced or alcohol-induced fibrosis (1, 40).
Analysis of Activated Myofibroblasts by Flow Cytometry.
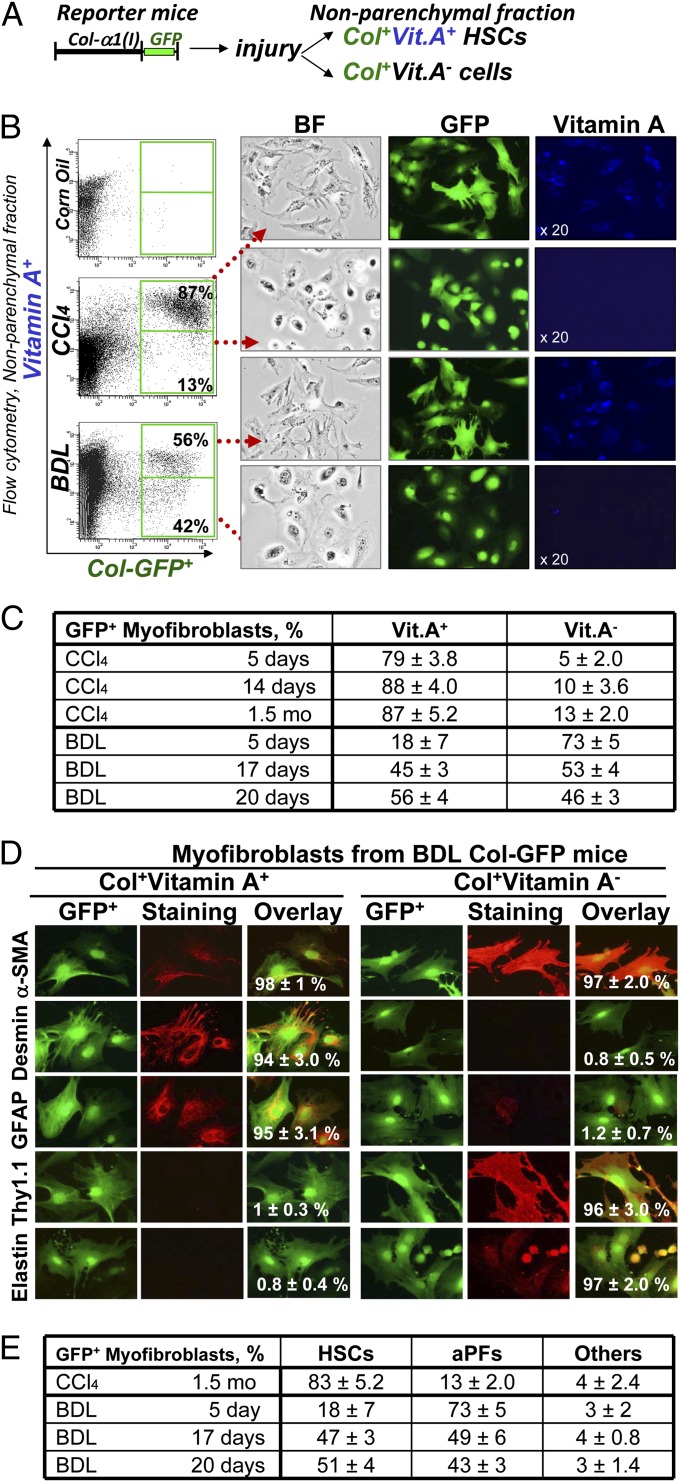
Our strategy to determine the composition of hepatic myofibroblasts is based on characterization of GFP+ cells in nonparenchymal liver fractions of BDL- and CCl4-treated Col-GFP mice (which contains all Col1a1+ and α-SMA+ myofibroblasts; for details, see Fig. S1A) (22). Although collagen-α1(I)-GFP is expressed in all activated myofibroblasts (40, 41), expression of vitamin A (Vit.A) droplets in the liver is solely attributed to HSCs (1) (Fig. 2A). The cell fate mapping of HSCs [using GFAPCre × Rosa26flox-TmRed-Stop-flox-GFP mice (40); Fig. S1 B and C] demonstrated that although HSCs down-regulate vitamin A upon activation (aHSCs), vitamin A is still detected in all aHSCs by flow cytometry (autofluorescent signal of vitamin A; Fig. S1D). We used flow cytometry to quantify the contribution of aHSCs (GFP+Vit.A+) and myofibroblasts of other origins (GFP+Vit.A−) in BDL and CCl4 injury (Fig. 2B). As expected, activation of hepatic myofibroblasts (GFP+ cells, 100%) was observed only in injured livers (Fig. 2B). CCl4-activated myofibroblasts contained 87 ± 6% GFP+Vit.A+ and 13 ± 3% GFP+Vit.A− cells. In contrast, the nonparenchymal fraction from BDL (20 d) mice consisted of 56 ± 4% GFP+Vit.A+ and 42 ± 5% GFP+Vit.A− myofibroblasts, suggesting that the composition of GFP+ myofibroblasts varies depending on the etiology of liver fibrosis. GFP+Vit.A+ and GFP+Vit.A− cells were sort purified and plated (Fig. 2B). Expression of GFP was confirmed in both fractions by fluorescent microscopy, whereas expression of Vit.A+ droplets was detected only in GFP+Vit.A+ cells.
Fig. 2.
Detection, quantification, and isolation of liver myofibroblasts. (A) Strategy to analyze myofibroblasts by flow cytometry: Collagen type I-expressing myofibroblasts were identified in nonparenchymal fraction by GFP expression and further fractionated to Vit.A+ and Vit.A− cells. (B) FACS analysis of nonparenchymal fraction from untreated and BDL-, and CCl4- treated Col-GFP mice: GFP+ cells were detected by argon laser at 488 nm wavelength, and Vit.A+ cells were detected by violet laser at 405 nm wavelength. Representative dot plots are shown, P < 0.03. GFP+Vit.A+ and GFP+Vit.A− cells were sort purified and analyzed by light and fluorescent microscopy for GFP and Vitamin A expression (UV laser, 20× objective). (C) Flow cytometry-based quantification of GFP+ myofibroblasts. Expression of vitamin A in GFP+ cells was analyzed in nonparenchymal fraction of Col-GFP mice at different time points (n = 6 per time point) of CCl4 and BDL, P < 0.01. (D) Immunophenotyping of GFP+ myofibroblasts isolated from BDL mice. GFP+Vit.A+ and GFP+Vit.A− fractions were sort purified from Col-GFP mice (n = 6) after BDL (20 d). Expression of myofibroblast marker (α-SMA), HSC markers (desmin, GFAP, CD146), and PF markers (elastin, Thy1) were analyzed by immunocytochemistry using specific antibodies or isotype matched controls (40× objective). GFP+Vit.A+ and GFP+Vit.A− cells were identified as aHSCs and aPFs, respectively. For each fraction, the percent of positively stained cells is calculated (compared with total cells, 100%, P < 0.05). (E) Quantification of GFP+Vit.A+ and GFP+Vit.A− fractions is based on expression of HSC- and PF-specific markers in GFP+ myofibroblasts (100%) as detected by immunocytochemistry, P < 0.05.
Activation of HSCs Differs in BDL- and CCl4-Induced Liver Injury.
Analysis of all GFP+ myofibroblasts (100%) demonstrated that GFP+Vit.A+ aHSCs are the major source of activated myofibroblasts in response to CCl4 liver injury (Fig. 2B). Even at earlier time points of CCl4 treatment, 79 ± 3% (at 5 d) and 88 ± 4% (at 14 d) of the myofibroblasts were GFP+Vit.A+ HSCs (Fig. S2A). In contrast, BDL activated fewer HSCs (Fig. S2B). After 5 d of BDL, GFP+ myofibroblasts were mainly composed by GFP+Vit.A− cells (73 ± 5%), whereas GFP+Vit.A+ aHSCs represented only 18 ± 7% of GFP+ cells. After BDL (17 d), GFP+ myofibroblasts consisted of 53 ± 4% of GFP+Vit.A− cells and 45 ± 3% of GFP+Vit.A+ aHSCs, suggesting that activation of HSCs in BDL follows the induction of GFP+Vit.A− myofibroblasts. Flow cytomery-based statistical analysis of the number of Vit.A+ and Vit.A− myofibroblasts in response to BDL and CCl4 is summarized in Fig. 2C.
GFP+Vit.A+ Myofibroblast Originate from HSCs, Whereas GFP+Vit.A− Derive Predominantly from aPFs.
Sort-purified GFP+Vit.A− and GFP+Vit.A+ myofibroblasts were characterized by immunostaining for specific markers. As expected, all GFP+ cells expressed the myofibroblast marker α-SMA, demonstrating that only myofibroblasts express type I collagen in liver fibrosis. BDL-activated GFP+Vit.A+ myofibroblasts expressed the typical HSC markers GFAP (94 ± 2.6%), desmin (98 ± 2%), and mesenchymal marker CD146 (87 ± 3.0%), confirming that the GFP+Vit.A+ fraction consists solely of aHSCs (Fig. 2D). As expected, CCl4-induced GFP+Vit.A+ myofibroblasts were aHSCs (Fig. S3A). In contrast, GFP+Vit.A− myofibroblasts stained positive for the established portal fibroblast markers Thy1 (93 ± 4.0%) and elastin (86 ± 3.4%), but lacked markers of HSCs (GFAP, Desmin, CD146; Fig. 2D) and myeloid cells (CD11b, F4/80, CD68; Fig. S3B). Only a small number of GFP+Vit.A− cells expressed fibrocyte-like markers CD45 (3.1 ± 0.1%) and CD11b (2.4 ± 0.3%; Fig. S3B), suggesting that GFP+Vit.A− fraction predominantly (95 ± 4%) contains aPFs, and that less than 4 ± 1% of myofibroblasts originate from other sources (e.g., fibrocytes and BM derived mesenchymal progenitors). Immunocytochemistry-based analysis of myofibroblast composition in response to both BDL and CCl4 is summarized in Fig. 2E.
Gene Expression Profile Distinguishes BDL-Derived aPFs from CCl4-aHSCs and BDL-aHSCs.
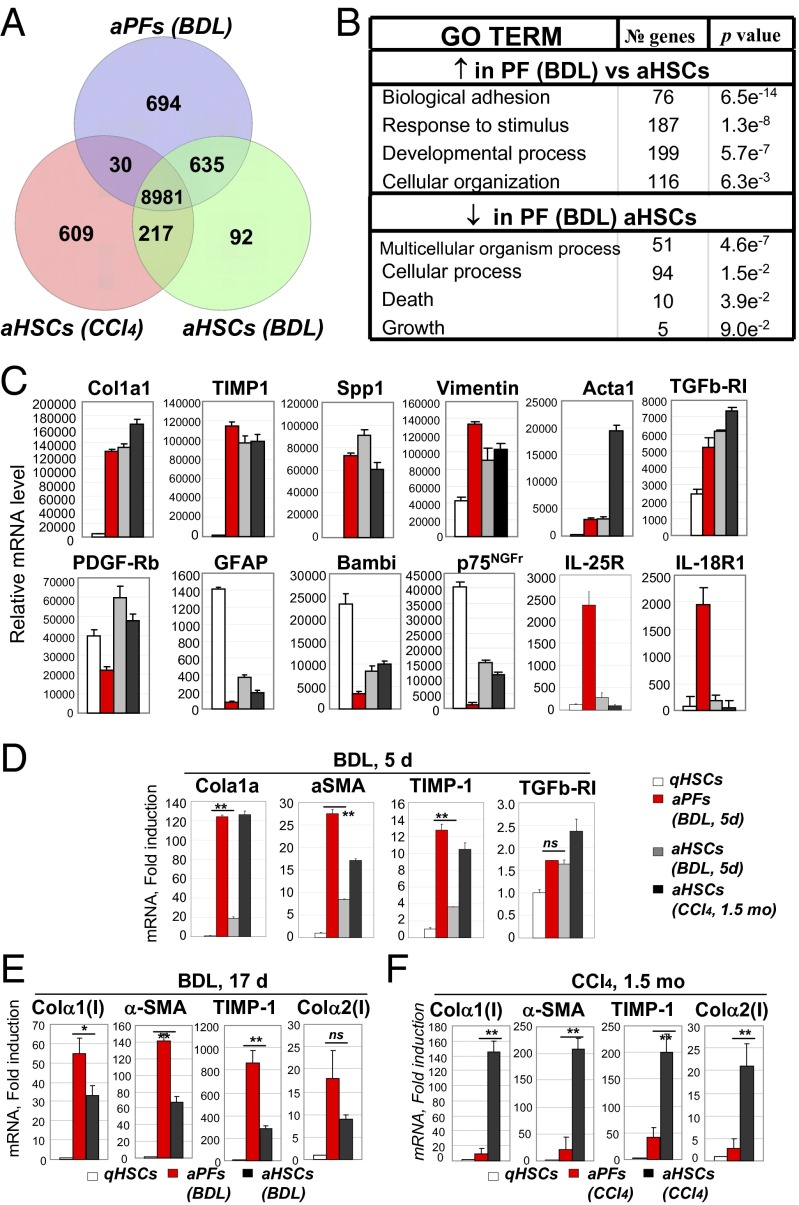
The gene expression profile of BDL-aPFs was compared with BDL-aHSCs and CCl4-aHSCs (Fig. 3A). Using a threshold defining confident detection of gene expression, we confirmed that aPFs exhibited a myofibroblast-like phenotype, sharing mRNA expression of 8,981 genes with aHSCs. These genes included Col1a1, Col1a2, Col2a1, TIMP-1, Spp1, TGFβ-RI, and Vimentin (Fig. 3C) and were induced in aPFs to a level comparable to BDL- and CCl4-aHSCs. As expected, GFAP and Bambi mRNAs were highly expressed in qHSCs, whereas PDGF-Rb was up-regulated in aHSCs. Meanwhile, the highest expression of Acta1 was detected in CCl4-aHSCs (Fig. 3C). aPFs up-regulated an additional 694 unique genes (Fig. 3A). This set of genes was enriched in Gene Ontology biological process annotations linked to biological adhesion, response to stimulus, developmental process and cellular organization (Fig. 3B), locomotion, focal adhesion, cell adhesion molecules, regulation of actin cytoskeleton, and were associated with the induction of the profibrogenic Wnt signaling pathway (Fig. S4). Furthermore, aPFs up-regulated expression of IL-18R, IL-25R (Fig. 3C), and other genes that distinguish them from aHSCs (Table 1, discussed below). Interestingly, BDL-aHSCs differentially expressed only 92 genes and shared more similarity with aPFs (635 genes) than with CCl4-aHSCs (217 genes; Fig. 3A), suggesting that in response to cholestatic liver injury, aHSCs may mimic the phenotype of aPFs (for comparison of BDL- and CCl4-aHSCs, see Fig. S5).
Fig. 3.
Characterization of aPFs and aHSCs. (A) BDL (20 d) GFP+Vit.A− aPFs and GFP+Vit.A+ aHSCs were analyzed by the whole mouse genome microarray, and their gene expression profile was compared with that in CCl4-activated GFP+Vit.A+ HSCs. Venn diagrams of the cell group-enriched genes that exhibited more than a twofold up-regulation compared with other groups. (B) GO TERM: demonstrates the signaling pathways that were up-regulated or down-regulated in BDL-aPFs versus BDL- or CCl4-aHSCs. (C) Expression of selected genes in qHSCs, BDL-aHSCs and BDL-aPFs, and CCl4-aHSCs. The results are relative mRNA level (average of normalized values/multiple probes/per gene) obtained by Agilant microarray, P < 0.001. (D) Expression of fibrogenic genes was analyzed by RT-PCR in BDL- (5 d) aPFs and BDL-aHSCs, isolated from the same mice (n = 6), and compared with that in qHSCs-aHSCs and CCl4 (1.5 mo)-aHSCs. The data are shown as fold induction compared with qHSCs, **P < 0.02 is shown for BDL-aPFs and BDL-aHSCs; ns is not significant. (E) Expression of fibrogenic genes was analyzed in BDL (17 d)-aPFs and BDL-aHSCs (isolated from the same mice, n = 6) by RT-PCR vs. qHSCs. The data are shown as fold induction compared with qHSCs, *P < 0.05; **P < 0.01; ns, nonsignificant. (F) Similarly, CCl4- (1.5 mo)aPFs and CCl4-aHSCs, isolated from the same mice (n = 4) were analyzed by RT-PCR. The data are shown as fold induction over qHSCs, *P < 0.05; **P < 0.01. The data in D–F represent at least three independent experiments.
Table 1.
Expression of signature genes distinguishes BDL-aPFs from BDL- and CCl4-aHSCs
| Maximum induction (up-regulation) in aPF (BDL, 20 d) | Fold |
| Calcitonin α (Calca) | 66 |
| Glycoprotein m6a (Gpm6a) | 35 |
| Uroplakin 1β | 28 |
| Basonuclin 1 (Bnc1) | 24 |
| Mesothelin (msln) | 24 |
| Frizzled-related protein 4 (Sfrp4) | 21 |
| Cyp2s1 | 20 |
| Proteoglycan 4 (Prg4) | 18 |
| Asporin (aspn) | 18 |
| Mucin 16 (Muc16) | 16 |
| IL-18R1 | 14 |
| Myosin light peptide7 (Myl7) | 14 |
| Vitrin (Vit) | 12 |
| Glipican 3 (Gpc3) | 12 |
| CD200 | 11 |
| Apolipoprotein D (ApoD) | 10 |
| IL-25R | 9.7 |
| Dermokin (Dmkn) | 9.3 |
| Vanin (Vnn1) | 8.5 |
| Thrombospondin 4 (Thbs4) | 7.0 |
| Integrin β4 (Itgb4) | 6.5 |
| CD55 | 5.6 |
| Gremlin 1 (Grem1) | 4.8 |
| NTPD2 | 4.6 |
| PDGFc | 4.6 |
| Fibulin 2 (Fbln2) | 4.4 |
| CD9 | 3.1 |
| Elastin (Eln) | 2.3 |
| Thy1 (CD90) | 1.8 |
| Cytoglobin | 0.6 |
Using the whole mouse genome microarray, expression of signature genes was determined for BDL-aPFs. Expression of genes previously identified as PF-specific (underlined) was confirmed. Fold induction (compared with the highest value observed in BDL- or CCl4-aHSCs) is shown for each gene. Full list of genes is shown in Fig. S7.
PFs Are Activated in Early BDL-Induced Liver Injury.
Our data indicate that aPFs and aHSCs exhibit similar level of activation in response to BDL (20 d; Fig. 3C). To further characterize the fibrogenic properties of aPF and aHSC, earlier time points of BDL were examined. After 5 d of BDL (Fig. 3D), expression levels of Col1a1, aSMA, and TIMP1 mRNA were much higher in aPFs than in aHSCs, suggesting that the activation of PF precedes the activation of HSCs in BDL injury. For example, Co1la1 was 120-fold induced in aPFs over the level in qHSCs, compared with 20-fold induction in aHSCs. After 17 d of BDL (Fig. 3E), activation of HSCs became more prominent (i.e., Col1a1 mRNA: 33-fold induction in aHSCs, vs. 55 in aPFs). Meanwhile, CCl4-aPFs exhibited a much lower level of Col1a1 mRNA than CCl4-aHSCs (fold induction 20 and 160, respectively; Fig. 3F), demonstrating that PFs are only minor contributors to toxic CCl4-induced liver injury. These data are in concordance with our previous results obtained by flow cytometry (Fig. 2) and demonstrate that there is a correlation between increased number of BDL-aPFs and the level of their activation.
Functional Properties of BDL-Derived aPFs Differ from aHSCs.
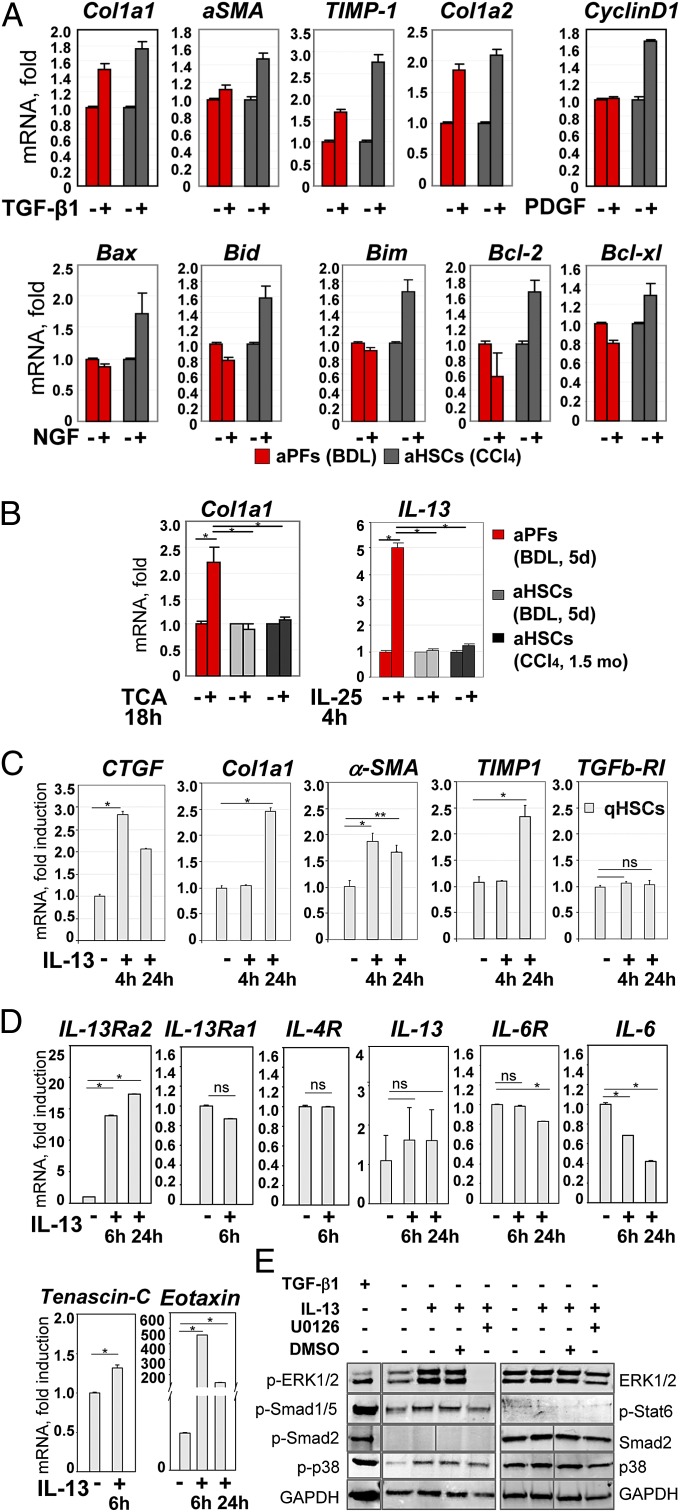
Previous studies have proposed differences in aPFs and aHSCs that underlie fibrogenesis of different etiologies (42). Therefore, we assessed how aPFs and aHSCs responded to fibrogenic stimuli in vitro. As expected, the fibrogenic cytokine TGF-β1 had similar effects on aPF and aHSC (Fig. 4A). However, aPFs were unresponsive to the known HSC agonists PDGF and NGF (demonstrated by mRNA expression of target genes CyclinD1; Bax, Bid, Bim, Bcl-2, and Bcl-xl, respectively). Despite high expression of IL-18R, treatment of aPFs with IL-18 (100 ng/mL; 8 h) did not induce expression of tested IL-18 target genes (MMP3, MMP8, and MMP13, Cox-2, iNOS, IL-6). Meanwhile, only PFs responded to the bile acid TCA, with increased Col1a1 mRNA expression (>2.2-fold induction over control aPFs), suggesting that TCA may directly mediate PF activation (Fig. 4B). Furthermore, aPFs responded to IL-25 stimulation by induction of IL-13 [similar to IL-13 induction by IL-25–treated macrophages (43) and fibroblasts (44)]. Although IL-13 is implicated in HSC activation, and IL-13 levels are up-regulated in patients with liver cirrhosis (3, 4, 27), the role of IL-13 in cholestatic liver injury has not been well defined. We hypothesize that IL-25–mediated IL-13 production by BDL-aPFs may stimulate activation of HSCs. To assess the effect of aPF-produced IL-13 on HSCs, qHSCs were incubated in the presence of IL-13. As we predicted (45), IL-13 increased CTCF (after 4 h) mRNA expression, and also induced up-regulation of Co1la1, aSMA, TIMP1, and mRNA (after 24 h) in HSCs (Fig. 4C), suggesting that aPFs may locally facilitate HSC activation via production of IL-13. A more detailed analysis (Fig. 4D) demonstrated that stimulation of HSCs with IL-13 causes up-regulation of IL-13Ra2 expression (but not IL-13Ra1 or IL-4) and transcription of IL-13 target genes Tenascin-C and Eotaxin (46, 47). Because IL-13–treated HSCs did not express IL-13 or IL-6, we concluded that IL-13 directly mediated HSC activation, and this effect was associated with phosphorylation of ERK1/2 (which is completely blocked by ERK inhibitor U0126; Fig. 4E) and activation of the p38 and Smad1/5 signaling pathways. Similar results were obtained in human primary HSCs. hIL-13 induced a dose-dependent secretion of CCL11/eotaxin (Fig. S6A) in hHSCs. In a separate experiment, hIL-13 alone (or in combination with TGF-β1) mediated an increase in IL-13Ra2, Tenascin C, Col1a1, Col3a1, fibronectin, and LoxL2 genes (Fig. S6B). In turn, TGF-β1 and serum stimulation did not result in IL-13 secretion by hHSCs (Fig. S6C), suggesting that aPFs may serve as a source of IL-13 in liver fibrosis.
Fig. 4.
Functional properties of aPFs and aHSCs. (A) Response to cytokines was compared in BDL-aPFs and CCl4-aHSCs. Both aHSCs and aPFs responded to TGF-β1 (10 ng/mL). aHSCs, but not aPFs, responded to PDGF (100 pg/mL) and NGF (100 ng/mL). The data are fold induction compared with untreated aPFs (or aHSCs), P < 0.01. (B) BDL-aPFs (but not BDL-aHSCs or CCl4-aHSCs) responded to bile acid taurocholic acid (TCA; 1,200 nmol/mL) by up-regulation of Col1a1, and to IL-25 (100 ng/mL) by IL-13 secretion, P < 0.05. Stimulation of aPFs with Tauro-ursodeoxycholate (TUDCA; 25 nmol/mL), deoxycholic acid (DCA; 0.1 nmol/mL), taurochenodeoxycholate (TCDCA; 60 nmol/mL), Tauro b-muricholate (TbMCA; 2,000 nmol/mL), and cholic acid (CA; 20 nmol/mL) did not result in Col1a1 induction. The data are fold induction compared with untreated aPFs (or aHSCs), *P < 0.05. (C) The effect of IL-13 on HSC activation was evaluated. qHSCs were incubated with IL-13 (100 ng/mL) for 4 h and 24 h. Gene expression was evaluated by RT-PCR, *P < 0.01; **P < 0.02; ns, nonsignificant. The data (A–C) represent three independent experiments. For each experiment, the cells were isolated from three mice. (D) IL-13 signaling in mouse HSCs: IL-13–stimulated HSCs (100 ng/mL, 6 h) up-regulate IL-13Rα2, tenascin C, and eotaxin, but do not express IL-13 or IL-6, as shown by RT-PCR. (E) IL-13 signaling in HSCs (2 h) causes phosphorylation of ERK1/2 (which is blocked by ERK inhibitor U0126, 10 μM), p38, and Smad1/5, as shown by Western blot. TGF-β1–stimulated HSCs served as a control.
Expression of Novel Markers Distinguishes BDL-Derived aPFs from BDL-aHSCs and CCl4-aHSCs.
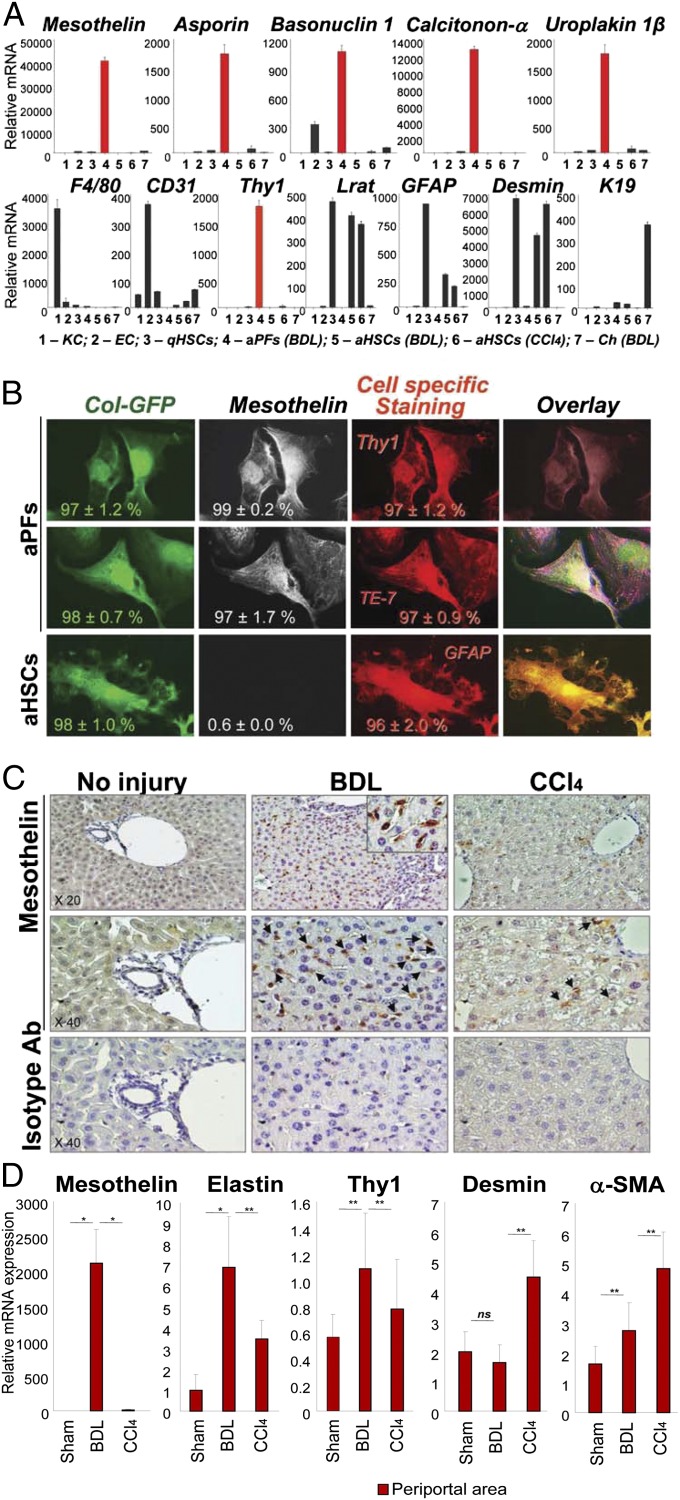
To further distinguish aPFs from aHSCs and other myofibroblasts, we interrogated the whole mouse genome microarray to determine “signature genes” for aPFs (Table 1). In concordance with previous studies, we confirmed that aPFs lack expression of cytoglobin (an HSC marker), but express Thy1, elastin, Gremlin 1, Fibulin 2, and NTPD2 mRNAs (the markers that have been reported to discriminate between aPFs and aHSCs) (2, 11, 17–21). However, expression of cofilin-1 (21) distinguished aPFs from CCl4-aHSCs, but not from BDL-aHSCs, which limits the usefulness of this marker. Furthermore, aPFs uniquely expressed calcitonin α (fold induction >48 over the highest value in BDL-aHSCs or CCl4-aHSCs), mesothelin (>28), uroplakin 1β (>22), basonuclin 1 (>18), asporin (>14), proteoglycan 4 (>14), glipican 3 (>12), and CD200 (>11) mRNA (Fig. S7). Up-regulation of these genes specifically in aPFs [but not in quiescent or aHSCs, endothelial cells, Kupffer cells, and hepatocytes (Fig. 5A and Fig. S8A) or BDL-activated cholangiocytes (Fig. 5A and Fig. S8C)] was confirmed by RT-PCR and immunohistochemistry, suggesting that these genes may serve as potential novel markers of aPFs. Some of these genes (including basonuclin 1, glycoprotein m6a, uroplakin 3b and 1b, mesothelin, IL-18R, calcitonin-related peptides, and vitrin) were reported as signature genes of murine hepatic mesothelial (48) and epicardial cells (49) (Fig. S7), supporting the theory that PFs originate from mesothelial cells (50, 51).
Fig. 5.
Expression of mesothelin in aPFs is associated with cholestatic liver fibrosis in mice. (A) Expression of selected signature genes was compared by RT-PCR in aPFs and other cells in the liver. Mesothelin, asporin, basonuclin 1, calcitonin-α, and uroplakin 1β mRNA were up-regulated in BDL- (17 d) aPFs, but not in KC, endothelial cells (EC), BDL- and CCl4-aHSCs and qHSCs, or BDL-induced cholangiocytes (Ch). The purity of each fraction was estimated by expression of F4/80 in KC, CD31 in EC, Lrat, GFAP, and Desmin in HSCs, Thy1 in aPFs, and K19 in cholangiocytes. The data (from three independent experiments) are shown as relative mRNA expression, P < 0.01. (B) aPFs and aHSCs were isolated from BDL (17 d)-injured Col-GFP mice and stained with anti-mesothelin Ab. Expression of Mesothelin was detected only in aPFs (but not in GFAP+ aHSCs) and colocalized with Elastin (TE-7) and Thy1 staining. The percent of immunostained cells is calculated, P < 0.05 (four independent experiments; Fig. S7B). (C) Paraffin sections of liver tissue from BDL- (17 d) or CCl4- (1.5 mo)treated mice (n = 4 per group) were immunostained with anti-mesothelin antibody or isotype-matched control. Expression of mesothelin was detected in BDL mice but not in sham-operated mice. Only a few mesothelin positive cells were detected in CCl4-treated mice. Representative images are shown using 20× and 40× objective, (Fig. S7C). (D) Up-regulation of mesothelin is detected by laser capture microdissection in BDL-induced (but not CCl4-induced) liver fibrosis. Laser capture microdissection was used to isolate periportal myofibroblasts from BDL (20 d) mice and CCl4 (1.5 mo)-treated mice (n = 3 per group), cells were analyzed by RT-PCR for expression of aPF- and aHSC-specific markers. Mesothelin, elastin, and Thy1 were highly expressed in myofibroblasts obtained from periportal area of BDL liver. Desmin was expressed at high levels in CCl4-treated liver. Unlike desmin, mesothelin was not expressed in CCl4-treated periportal area. The data (from three independent experiments) are mRNA fold induction compared with periportal area of sham mice, *P < 0.01; **P < 0.05; ns, nonsignificant.
The role of most of these genes in liver fibrosis has not been evaluated, with the exception of calcitonin α and mesothelin. Calcitonin α, a calcium metabolism regulating hormone, was implicated in pathogenesis of cholestatic injury, and mice devoid of calcitonin α are more resistant to BDL-induced liver fibrosis (52). In turn, mesothelin, a glycosylphosphatidylinositol-linked glycoprotein, is expressed in hepatic mesothelial cells and malignant mesotheliomas (53) and mediates intracellular adhesion and metastatic spread (54). Mesothelin knockout mice are viable and exhibit no obvious abnormalities (55). Expression of mesothelin was detected only in isolated aPFs but not in other cellular fractions (Fig. 5A).
Expression of Mesothelin Is Up-Regulated in aPFs in Response to Injury.
We examined the expression of mesothelin in isolated aPFs and aHSCs. Unlike GFP+GFAP+ aHSCs, GFP+ aPFs expressed mesothelin (97 ± 1.7%). Mesothelin+ aPFs coexpressed elastin (detected with TE-7 Ab) and Thy1, and immunostaining with mesothelin colocalized with Elastin+Thy1+ aPFs (Fig. 5B and Fig. S8B). Next, expression of mesothelin was evaluated in livers of BDL- and CCl4-injured mice (Fig. 5C and Fig. S8B). In concordance with our previous findings, very few mesothelin+ cells were detected in CCl4-injured livers. In contrast, mesothelin was highly expressed in livers from BDL-injured mice, with an expression pattern similar to the other PF markers Thy1 and elastin (Fig. S8 B and C). In support of our findings, expression of mesothelin mRNA was also detected in laser capture microdissected portal areas from BDL (20 d)-treated mice but not from CCl4-treated mice (Fig. 5D). In addition, mesothelin was not expressed in sham-operated mice, suggesting that mesothelin identifies the aPF phenotype.
Discussion
Our study was designed to determine the origin of hepatic myofibroblasts activated in response to chronic injury of two different etiologies. We demonstrate that hepatotoxic (CCl4) and cholestatic (BDL) liver injuries activate distinct subsets of fibrogenic myofibroblasts. Thus, CCl4 activates preferentially aHSCs, whereas BDL initially preferentially aPFs. We developed a reliable method of isolation and quantification of hepatic myofibroblast fractions by using flow cytometry. Based on the distinctive expression of Vitamin A and GFAP in HSCs and Thy1 and elastin in PFs, this study establishes cell sorting as a robust method to purify distinct populations of myofibroblasts in mice, providing a nonbiased approach to purify and characterize all myofibroblasts. By demonstrating that HSCs are the major source of myofibroblasts in hepatotoxic liver injury (CCl4), we confirmed the previous cell fate mapping studies that used GFAP-Cre (56, 57), PDGFRb-Cre (58), and Lrat-Cre (59).
In contrast to CCl4-induced injury, our study demonstrates that PFs rapidly activate at the onset of cholestatic injury and up-regulate fibrogenic genes. Furthermore, early activation of PFs during BDL injury may affect HSCs, and BDL-aHSCs exhibit more similarity to aPFs than to CCl4-aHSCs. Gene expression profiling demonstrated novel signature genes for aPFs. According to cell fate mapping, PFs originate from the mesothelium (51, 60), and our data suggest that aPFs share similarity in signature gene expression with other cells of mesothelial origin. One of these genes, mesothelin, is highly induced specifically in aPFs in response to BDL injury, suggesting that mesothelin may become a new target for antifibrotic therapy.
aHSCs and aPFs Are the Major Source of Myofibroblasts in Fibrotic Liver.
Although vitamin A-rich lipid droplets are a distinctive characteristic of HSCs, activation results in a decrease in these droplets (1). However, in vivo aHSCs do not lose their vitamin A droplets completely, and vitamin A-induced buoyancy has become a standard way to purify quiescent and aHSCs in vivo, as confirmed by gene expression profiling (25, 41). Our current study provides additional proof that vitamin A is a reliable marker for identification, quantification, and purification of aHSCs, making flow cytometry using vitamin A autofluorescence as the method of choice to purify aHSCs from myofibroblasts of other origins. Flow cytometry enables identification of hepatic myofibroblasts and isolation of distinct subsets of myofibroblasts (HSCs and PFs) with high purity from the same mouse liver.
Using collagen-GFP reporter mice, we demonstrate that the total population of GFP+ myofibroblasts isolated in the nonparenchymal fraction consists of two major populations: Vit.A+ aHSCs and Vit.A− aPFs. These results were confirmed by immunostaining for cell-specific markers, RT-PCR, and gene expression microarrays. Specifically, aHSCs were identified as Vit.A+, GFAP+, Desmin+, and CD146+ cells that exhibit specific morphology. In turn, Vit.A− aPFs lacked GFAP or Desmin expression, but were characterized by expression of Thy1 and Elastin, and a more round-shaped morphology. Collectively, HSCs and PFs contribute to more than 94% of GFP+ myofibroblasts. This type of analysis should now be extended to other experimental models of liver fibrosis, such as alcohol-induced liver disease and nonalcoholic steatohepatitis.
aHSCs and aPFs Contribute Differently to Liver Fibrosis of Different Etiologies.
Although the role of aPFs in the development of portal fibrosis has been discussed (42, 61), our study is the first to our knowledge to quantify the myofibroblast populations over a time course. Consistent with previous studies (62, 63), we demonstrate that aPFs play an important role at early stages of BDL-induced liver fibrosis (13) by contributing >70% of myofibroblasts. Moreover, even at later stages (BDL, 17–20 d), aPFs contribute ∼50% of myofibroblasts and exhibit a more activated phenotype than aHSCs. Thus, the composition of myofibroblasts varies depending on the etiology and time course of liver injury and fibrosis.
Cholestatic Injury Induces Predominant Activation of aPFs.
The mechanism of fibrogenesis differs in CCl4 and BDL models of liver injury. Treatment with CCl4 is hepatotoxic, causing necrosis of hepatocytes and inflammation in the pericentrolobular area. However, BDL induces obstruction of bile flow with increased biliary pressure, moderate inflammation, and cytokine secretion by biliary epithelial cells (64). Diffusion (accumulation) of free bile acids may trigger ductular reaction (hyperplastic response of bile duct epithelial cells), resulting in activation of cholangiocytes and portal fibroblasts. The mechanism of PF activation is poorly understood. Here, we propose that TCA bile acid can directly activate PFs (but not HSCs) into myofibroblasts, and this effect may rely on TCA-induced induced cytotoxicity, because PFs have been reported to lack the bile acid receptors FXR (farnesoid X receptor) and TGR5 (the membrane G protein-coupled receptor) (65, 66). TCA-induced activation of PFs appears to be specific, and stimulation with other bile acids (TUDCA, DCA, TCDCA, TbMCA, CA) did not induce fibrogenic gene expression in PFs. However, unresponsiveness of PFs to tested bile acids may result from already high activation of isolated PFs (5 d after BDL), the lack of corresponding receptors (65), or poor experimental conditions (67). In addition, individual bile acids may produce other effects on PFs, such as cellular proliferation and cytokine secretion (17), which were not evaluated in this study. Furthermore, our in vitro conditions may not mimic the complex liver microenvironment required for bile acid stimulation of PFs (17). Alternatively, bile acids may indirectly induce PF activation by affecting cholangiocytes (68) or hepatocytes (65) that, in turn, may facilitate selective aPF activation via cell-cell signaling or cytokine secretion (64). In addition, specific factors produced by activated cholangiocytes may presensitize PFs for bile acid stimulation (69).
aPFS May Facilitate Activation of HSCs in BDL Model of Liver Injury.
Another characteristic feature of aPFs is expression of IL-25R. Up-regulation of proinflammatory IL-17A, IL-25, IL-22, and IL-6 in the serum and in the liver accompany development of BDL-induced liver fibrosis (28). Therefore, it is not surprising that IL-25 may stimulate aPFs. Similar to other cell types, IL-25 induced secretion of IL-13 by aPFs, but did not further their activation. IL-13 has been implicated in pathogenesis of Schistosoma mansoni infection-induced liver fibrosis (70), and recently IL-13 was shown to directly stimulate HSCs to produce CTGF and subsequently upegulate fibrogenic genes in response to nonparasite liver injury (71). Therefore, we hypothesized that following BDL, IL-25–stimulated aPFs secrete IL-13, which facilitates HSC activation (via induction of IL-13Ra2, Col1a1, Eotaxin, Tenascin-C, fibronection, and phosphorylation of ERK1/2). Supporting this notion, bone marrow transplantation in Abcb4−/− mice lessened hepatic fibrosis via Th1 responses, but did not alter the level of IL-13 production (72), suggesting there must be an endogenous source of IL-13 in these mice. Further studies are required to determine the mechanism of HSC activation in response to cholestatic liver injury.
Proposed Novel Markers of Portal Fibroblasts.
Robust markers of aHSCs and aPFs are needed. Our data confirmed that expression of Thy1 and Elastin distinguishes Vit.A−GFAP−Desmin−CD146− aPFs from Vit.A+GFAP+Desmin+CD146+Thy1−Elastin− aHSCs. Using gene expression profiling of in vivo aHSCs and aPFs, we have identified that mesothelin, calcitonin α, uroplakin 1β, basonclin 1, asporin, IL-18R1, and IL-25R may serve as additional useful markers to distinguish aPFs from aHSCs and myofibroblasts of other origins. We determined that these genes are highly expressed in portal fibroblasts but not in other cell types in fibrotic liver.
Interestingly, aPFs express mesothelin, calcitonin α, uroplakin 1β, basonclin 1, asporin, and IL-18R1 genes. The hepatic mesothelium is the source of HSCs and PFs during development (51, 60). Previous studies have demonstrated that the genes mentioned above and other genes [e.g., glycoprotein m6a, mesothelin, Uroplakin 1β and 3 β, Cyp2s1, mucin 16, crystalline, Prss12, Slipi, Caveolin, Dermokin, Calcitonin-related peptide, vanin, cytokeratin 7, Slc9a3r1, and Slc39a8 (metal ion transporter)], Igfbp6, see Fig. S6) are expressed in hepatic mesothelium (48). Furthermore, the gene expression profiles of epicardium isolated from adult mouse infarction-injured hearts identified the same genes among epicardium-specific signature genes, and for the first time, to our knowledge, implicated these genes (alone or in combination) in wound healing (49). Morphological studies have suggested that septum transversum mesenchyme (STM) is the source of hepatic mesenchymal cells (HSCs and perivascular mesenchymal cells) (73) and cardiac mesoderm [that gives rise to epicardium (74)]. Therefore, a common origin of hepatic mesothelium and epicardium may explain the similarity of gene expression profile of these tissues. During development, hepatic mesothelium undergoes an epithelial-to-mesenchymal (EMT) transition to produce PFs and HSCs. Furthermore, the expression of WT1, a mesothelial-specific factor (60), is expressed in aPFs (vs. aHSCs; Fig. S6). Because both hepatic mesothelium and epicardium can contribute to myofibroblasts in their respective organs, the contribution of the aforementioned genes to repair and fibrosis should be addressed.
Mesothelin is a glycosyl phosphatidylinositol (GPI)-anchored membrane glycoprotein that is expressed in normal mesothelial cells. It is also highly expressed in several species of malignant tumors, such as mesothelioma as well as ovarian and pancreatic cancers (75–77). We determined that mesothelin (Msln)-deficient mice are less susceptible to liver fibrosis compared with the wild-type mice. Previous studies have implicated mesothelin in mediation of cellular interaction and metastatic dissemination. Because of a strong induction in different types of cancer, mesothelin is considered as a tumor-associated antigen, which serves as a prognostic marker of disease progression, and became a therapeutic target for anti-cancer therapy. Here we demonstrate that mesothelin is highly expressed in aPFs in response to BDL, so that mesothelin may serve as a novel marker of aPFs and a potential target for antifibrotic therapy.
Materials and Methods
Mice and Liver Injury.
Collagen α1(I)-GFP mice (22) and wild-type littermates were used at 8 wk of age, in C57BL/6 background. Liver injury was induced in mice by CCl4 (1:4 dilution in corn oil, 60 µL × 14 injections; ref. 41) or ligation of the common bile duct (20 d) (41). Mice were maintained under specific pathogen free conditions at the animal facilities of University of California, San Diego (protocol S07088 approved by Institutional Animal Care and Use Committee).
Isolation of Nonparenchymal Fraction.
Livers were perfused and digested by using the pronase/collagenase method (41), and cells were centrifuged to pellet the hepatocytes. The remaining nonparenchymal cell fraction [containing hepatic myofibroblasts (HSCs, portal fibroblasts, and others), Kupffer cells, BM cells, and endothelial cells] (41). aPFs and aHSCs were isolated by using cell sorting for Col-GFP+Vit.A− and Col-GFP+Viat.A+ cells. Kupffer cells (KC) and endothelial cells were isolated by gradient centrifugation (15% Nycodenz) following by magnetic sorting with anti-CD11b and anti-CD31 antibodies, respectively (Miltenyi Biotec). Cholangiocytes were a gift of Gianfranco Alpini (Texas A&M Health Science Center, Central Texas Veterans Health Care System, Temple, TX) and were isolated from BDL mice (78).
Flow cytometry.
Flow cytometry was based on simultaneous detection of collagen-α1(I)-GFP (488 nm) and vitamin A (autofluorescent signal detected by violet laser at 405 nm; Fig. 2B) in Col-GFP mice (40). Phenotyping of the nonparenchymal fraction isolated from Col-GFP mouse livers (n = 6 time point) was performed on Canto (BD). Cell sorting was performed on a MoFlo (Beckman Coulter).
Immunofluorescence and immunohistochemistry.
Formalin-fixed frozen livers were stained with Sirius Red and anti–α-SMA Ab (Abcam). Immunohistochemistry was performed by using DAB staining (Vector) and counterstaining with Hematoxilin. Immunocytochemistry is described in SI Materials and Methods.
Whole Mouse Genome Gene Expression Microarray.
The gene expression profile of qHSCs, CCl4- (1.5 mo) aHSCs, BDL- (20 d) aHSCs, and PFs was studied by using Whole Mouse Genome Microarray (Agilent) (40). See SI Materials and Methods for details.
Characterization of IL-13 Signaling in Human HSCs.
Human stellate cells (ScienCell) were plated overnight, then serum-starved for 6 h and stimulated with IL-13, TGFβ1 (R&D Systems), or a combination of both. CCL11/eotaxin was measured in cell-free supernatants 48 h after stimulation with IL-13 by sandwich ELISA (RnD Systems). Gene expression was assessed at 24 h by quantitative RT-PCR.
Quantitative RT-PCR.
Total RNA was isolated from the nonparenchymal fraction, hepatocyte fraction, or purified Col+Vit.A+ and Col+Vit.A+ cells or hepatic stellate cells by using RNeasy columns (Qiagen). Gene expression levels were calculated after normalization to the standard housekeeping gene 18S by using the ∆∆ CT method as described by the manufacturer (Invitrogen) and expressed as relative mRNA levels compared with control. The results are represented as mean ± SEM, P < 0.0001.
Laser Capture Microdissection and RNA Extraction.
Livers from sham-, CCl4- and BDL-injured mice were snap-frozen in FSC 22 Frozen Section Media (Leica Microsystems) and stored at −80 °C. Transverse sections (10 µm) were cut with a cryostat at −20 °C. Cryosections were mounted on membrane-coated slides. A Leica LMD7000 system (Leica Microsystems) was used to cut periportal or centrilobular area on sections. Microdissected sections were collected in the lid of a 0.5-mL microtube containing RLT buffer from the RNeasy (Qiagen). Total RNA was extracted by using the same kit and following the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Dr. Sato and Dr. Uehiro for initial technical assistance with laser capture microdissection and Tom Kisby and Rebecca Dunmore for technical assistance with the in vitro studies using human HSCs. This work was supported by National Institutes of Health Grants DK088837, GM41804, AA15055, DK72237, AI0777802, and P50 AA011999; the Japanese Ministry of Health, Labour, and Welfare; and the American Liver Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400062111/-/DCSupplemental.
References
- 1.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115(2):209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forbes SJ, Parola M. Liver fibrogenic cells. Best Pract Res Clin Gastroenterol. 2011;25(2):207–217. doi: 10.1016/j.bpg.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Kisseleva T, et al. Bone marrow-derived fibrocytes participate in pathogenesis of liver fibrosis. J Hepatol. 2006;45(3):429–438. doi: 10.1016/j.jhep.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Kisseleva T, et al. Fibrocyte-like cells recruited to the spleen support innate and adaptive immune responses to acute injury or infection. J Mol Med (Berl) 2011;89(10):997–1013. doi: 10.1007/s00109-011-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholten D, et al. Migration of fibrocytes in fibrogenic liver injury. Am J Pathol. 2011;179(1):189–198. doi: 10.1016/j.ajpath.2011.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalluri R. EMT: When epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119(6):1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology. 2009;50(6):2007–2013. doi: 10.1002/hep.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taura K, et al. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology. 2010;51(3):1027–1036. doi: 10.1002/hep.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholten D, et al. Genetic labeling does not detect epithelial-to-mesenchymal transition of cholangiocytes in liver fibrosis in mice. Gastroenterology. 2010;139(3):987–998. doi: 10.1053/j.gastro.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu AS, et al. Lineage tracing demonstrates no evidence of cholangiocyte epithelial-to-mesenchymal transition in murine models of hepatic fibrosis. Hepatology. 2011;53(5):1685–1695. doi: 10.1002/hep.24206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dranoff JA, Wells RG. Portal fibroblasts: Underappreciated mediators of biliary fibrosis. Hepatology. 2010;51(4):1438–1444. doi: 10.1002/hep.23405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhunchet E, Wake K. Role of mesenchymal cell populations in porcine serum-induced rat liver fibrosis. Hepatology. 1992;16(6):1452–1473. doi: 10.1002/hep.1840160623. [DOI] [PubMed] [Google Scholar]
- 13.Desmoulière A, et al. Extracellular matrix deposition, lysyl oxidase expression, and myofibroblastic differentiation during the initial stages of cholestatic fibrosis in the rat. Lab Invest. 1997;76(6):765–778. [PubMed] [Google Scholar]
- 14.Uchio K, et al. Cellular retinol-binding protein-1 expression and modulation during in vivo and in vitro myofibroblastic differentiation of rat hepatic stellate cells and portal fibroblasts. Lab Invest. 2002;82(5):619–628. doi: 10.1038/labinvest.3780456. [DOI] [PubMed] [Google Scholar]
- 15.Wen JW, Olsen AL, Perepelyuk M, Wells RG. Isolation of rat portal fibroblasts by in situ liver perfusion. J Vis Exp. 2012;(64):3669. doi: 10.3791/3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruglov EA, Jain D, Dranoff JA. Isolation of primary rat liver fibroblasts. J Investig Med. 2002;50(3):179–184. doi: 10.2310/6650.2002.33431. [DOI] [PubMed] [Google Scholar]
- 17.Clouzeau-Girard H, et al. Effects of bile acids on biliary epithelial cell proliferation and portal fibroblast activation using rat liver slices. Lab Invest. 2006;86(3):275–285. doi: 10.1038/labinvest.3700386. [DOI] [PubMed] [Google Scholar]
- 18.Knittel T, et al. Rat liver myofibroblasts and hepatic stellate cells: Different cell populations of the fibroblast lineage with fibrogenic potential. Gastroenterology. 1999;117(5):1205–1221. doi: 10.1016/s0016-5085(99)70407-5. [DOI] [PubMed] [Google Scholar]
- 19.Goodpaster T, et al. An immunohistochemical method for identifying fibroblasts in formalin-fixed, paraffin-embedded tissue. J Histochem Cytochem. 2008;56(4):347–358. doi: 10.1369/jhc.7A7287.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dranoff JA, et al. The ecto-nucleoside triphosphate diphosphohydrolase NTPDase2/CD39L1 is expressed in a novel functional compartment within the liver. Hepatology. 2002;36(5):1135–1144. doi: 10.1053/jhep.2002.36823. [DOI] [PubMed] [Google Scholar]
- 21.Bosselut N, et al. Distinct proteomic features of two fibrogenic liver cell populations: Hepatic stellate cells and portal myofibroblasts. Proteomics. 2010;10(5):1017–1028. doi: 10.1002/pmic.200900257. [DOI] [PubMed] [Google Scholar]
- 22.Yata Y, et al. DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology. 2003;37(2):267–276. doi: 10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- 23.Brenner DA, Rippe RA, Veloz L. Analysis of the collagen alpha 1(I) promoter. Nucleic Acids Res. 1989;17(15):6055–6064. doi: 10.1093/nar/17.15.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Österreicher CH, et al. Fibroblast specific protein 1 identifies a subpopulation of liver inflammatory macrophages. Proc Natl Acad Sci USA. 2010;108(1):308–13. doi: 10.1073/pnas.1017547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Minicis S, et al. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132(5):1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 26.Inokuchi S, et al. Toll-like receptor 4 mediates alcohol-induced steatohepatitis through bone marrow-derived and endogenous liver cells in mice. Alcohol Clin Exp Res. 2011;35(8):1509–1518. doi: 10.1111/j.1530-0277.2011.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin SL, Kisseleva T, Brenner DA, Duffield JS. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol. 2008;173(6):1617–1627. doi: 10.2353/ajpath.2008.080433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng F, et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology. 2012;143(3):765–776. doi: 10.1053/j.gastro.2012.05.049. e761-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meurer SK, et al. Overexpression of endoglin modulates TGF-β1-signalling pathways in a novel immortalized mouse hepatic stellate cell line. PLoS ONE. 2013;8(2):e56116. doi: 10.1371/journal.pone.0056116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons CJ, et al. Mutation of the 5′-untranslated region stem-loop structure inhibits α1(I) collagen expression in vivo. J Biol Chem. 2011;286(10):8609–8619. doi: 10.1074/jbc.M110.189118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefanovic B, Brenner DA. 5′ stem-loop of collagen alpha 1(I) mRNA inhibits translation in vitro but is required for triple helical collagen synthesis in vivo. J Biol Chem. 2003;278(2):927–933. doi: 10.1074/jbc.M209175200. [DOI] [PubMed] [Google Scholar]
- 32.Stefanovic B, et al. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol Cell Biol. 1997;17(9):5201–5209. doi: 10.1128/mcb.17.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefanovic B, Lindquist J, Brenner DA. The 5′ stem-loop regulates expression of collagen alpha1(I) mRNA in mouse fibroblasts cultured in a three-dimensional matrix. Nucleic Acids Res. 2000;28(2):641–647. doi: 10.1093/nar/28.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefanovic B, Schnabl B, Brenner DA. Inhibition of collagen alpha 1(I) expression by the 5′ stem-loop as a molecular decoy. J Biol Chem. 2002;277(20):18229–18237. doi: 10.1074/jbc.M108065200. [DOI] [PubMed] [Google Scholar]
- 35.Stefanovic L, Brenner DA, Stefanovic B. Direct hepatotoxic effect of KC chemokine in the liver without infiltration of neutrophils. Exp Biol Med (Maywood) 2005;230(8):573–586. doi: 10.1177/153537020523000809. [DOI] [PubMed] [Google Scholar]
- 36.Taura K, et al. Hepatic stellate cells secrete angiopoietin 1 that induces angiogenesis in liver fibrosis. Gastroenterology. 2008;135(5):1729–1738. doi: 10.1053/j.gastro.2008.07.065. [DOI] [PubMed] [Google Scholar]
- 37.Magness ST, Bataller R, Yang L, Brenner DA. A dual reporter gene transgenic mouse demonstrates heterogeneity in hepatic fibrogenic cell populations. Hepatology. 2004;40(5):1151–1159. doi: 10.1002/hep.20427. [DOI] [PubMed] [Google Scholar]
- 38.Magness ST, Brenner DA. Targeted disruption of the mouse ferrochelatase gene producing an exon 10 deletion. Biochim Biophys Acta. 1999;1453(1):161–174. doi: 10.1016/s0925-4439(98)00096-9. [DOI] [PubMed] [Google Scholar]
- 39.Bataller R, et al. NADPH oxidase signal transduces angiotensin II in hepatic stellate cells and is critical in hepatic fibrosis. J Clin Invest. 2003;112(9):1383–1394. doi: 10.1172/JCI18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kisseleva T, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci USA. 2012;109(24):9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seki E, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13(11):1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 42.Perepelyuk M, et al. Hepatic stellate cells and portal fibroblasts are the major cellular sources of collagens and lysyl oxidases in normal liver and early after injury. Am J Physiol Gastrointest Liver Physiol. 2013;304(6):G605–G614. doi: 10.1152/ajpgi.00222.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fort MM, et al. IL-25 induces IL-4, IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity. 2001;15(6):985–995. doi: 10.1016/s1074-7613(01)00243-6. [DOI] [PubMed] [Google Scholar]
- 44.Gregory LG, et al. 25 drives remodelling in allergic airways disease induced by house dust mite. Thorax. 2012;68(1):82–90. doi: 10.1136/thoraxjnl-2012-202003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, et al. IL-13 induces connective tissue growth factor in rat hepatic stellate cells via TGF-β-independent Smad signaling. J Immunol. 2011;187(5):2814–2823. doi: 10.4049/jimmunol.1003260. [DOI] [PubMed] [Google Scholar]
- 46.Pope SM, et al. Identification of a cooperative mechanism involving interleukin-13 and eotaxin-2 in experimental allergic lung inflammation. J Biol Chem. 2005;280(14):13952–13961. doi: 10.1074/jbc.M406037200. [DOI] [PubMed] [Google Scholar]
- 47.Jinnin M, et al. Upregulation of tenascin-C expression by IL-13 in human dermal fibroblasts via the phosphoinositide 3-kinase/Akt and the protein kinase C signaling pathways. J Invest Dermatol. 2006;126(3):551–560. doi: 10.1038/sj.jid.5700090. [DOI] [PubMed] [Google Scholar]
- 48.Onitsuka I, Tanaka M, Miyajima A. Characterization and functional analyses of hepatic mesothelial cells in mouse liver development. Gastroenterology. 2010;138(4):1525–1535. doi: 10.1053/j.gastro.2009.12.059. e1521–1526. [DOI] [PubMed] [Google Scholar]
- 49.Bochmann L, et al. Revealing new mouse epicardial cell markers through transcriptomics. PLoS ONE. 2010;5(6):e11429. doi: 10.1371/journal.pone.0011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asahina K. Hepatic stellate cell progenitor cells. J Gastroenterol Hepatol. 2012;27(Suppl 2):80–84. doi: 10.1111/j.1440-1746.2011.07001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asahina K, et al. Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development. Hepatology. 2009;49(3):998–1011. doi: 10.1002/hep.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glaser SS, et al. Knockout of alpha-calcitonin gene-related peptide reduces cholangiocyte proliferation in bile duct ligated mice. Lab Invest. 2007;87(9):914–926. doi: 10.1038/labinvest.3700602. [DOI] [PubMed] [Google Scholar]
- 53.Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci USA. 1996;93(1):136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grigoriu BD, Grigoriu C, Chahine B, Gey T, Scherpereel A. Clinical utility of diagnostic markers for malignant pleural mesothelioma. Monaldi Arch Chest Dis. 2009;71(1):31–38. doi: 10.4081/monaldi.2009.374. [DOI] [PubMed] [Google Scholar]
- 55.Bera TK, Pastan I. Mesothelin is not required for normal mouse development or reproduction. Mol Cell Biol. 2000;20(8):2902–2906. doi: 10.1128/mcb.20.8.2902-2906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lujambio A, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153(2):449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henderson NC, et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med. 2013;19(12):1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mederacke I, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asahina K, Zhou B, Pu WT, Tsukamoto H. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology. 2011;53(3):983–995. doi: 10.1002/hep.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Desmoulière A. Hepatic stellate cells: The only cells involved in liver fibrogenesis? A dogma challenged. Gastroenterology. 2007;132(5):2059–2062. doi: 10.1053/j.gastro.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 62.Tang L, Tanaka Y, Marumo F, Sato C. Phenotypic change in portal fibroblasts in biliary fibrosis. Liver. 1994;14(2):76–82. doi: 10.1111/j.1600-0676.1994.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 63.Tuchweber B, Desmoulière A, Bochaton-Piallat ML, Rubbia-Brandt L, Gabbiani G. Proliferation and phenotypic modulation of portal fibroblasts in the early stages of cholestatic fibrosis in the rat. Lab Invest. 1996;74(1):265–278. [PubMed] [Google Scholar]
- 64.Syal G, Fausther M, Dranoff JA. Advances in cholangiocyte immunobiology. Am J Physiol Gastrointest Liver Physiol. 2012;303(10):G1077–G1086. doi: 10.1152/ajpgi.00227.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fickert P, et al. Farnesoid X receptor critically determines the fibrotic response in mice but is expressed to a low extent in human hepatic stellate cells and periductal myofibroblasts. Am J Pathol. 2009;175(6):2392–2405. doi: 10.2353/ajpath.2009.090114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fausther M, Dranoff JA. New insights on the pathogenesis of biliary cirrhosis provided by studies in FXR knockout mice. J Hepatol. 2011;55(4):939–940. doi: 10.1016/j.jhep.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guyot C, et al. Fibrogenic cell phenotype modifications during remodelling of normal and pathological human liver in cultured slices. Liver Int. 2010;30(10):1529–1540. doi: 10.1111/j.1478-3231.2010.02342.x. [DOI] [PubMed] [Google Scholar]
- 68.Keitel V, et al. The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology. 2009;50(3):861–870. doi: 10.1002/hep.23032. [DOI] [PubMed] [Google Scholar]
- 69.Fabris L, Strazzabosco M. Epithelial-mesenchymal interactions in biliary diseases. Semin Liver Dis. 2011;31(1):11–32. doi: 10.1055/s-0031-1272832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Munker S, Müllenbach R, Weng HL. IL-13 signaling in liver fibrogenesis. Front Immunol. 2012;3:116. doi: 10.3389/fimmu.2012.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roderfeld M, et al. Bone marrow transplantation improves hepatic fibrosis in Abcb4-/- mice via Th1 response and matrix metalloproteinase activity. Gut. 2012;61(6):907–916. doi: 10.1136/gutjnl-2011-300608. [DOI] [PubMed] [Google Scholar]
- 73.Enzan H, et al. Development of hepatic sinusoidal structure with special reference to the Ito cells. Microsc Res Tech. 1997;39(4):336–349. doi: 10.1002/(SICI)1097-0029(19971115)39:4<336::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 74.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454(7200):109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hassan R, Bera T, Pastan I. Mesothelin: A new target for immunotherapy. Clin Cancer Res. 2004;10(12 Pt 1):3937–3942. doi: 10.1158/1078-0432.CCR-03-0801. [DOI] [PubMed] [Google Scholar]
- 76.Scholler N, et al. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci USA. 1999;96(20):11531–11536. doi: 10.1073/pnas.96.20.11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sapede C, et al. Aberrant splicing and protease involvement in mesothelin release from epithelioid mesothelioma cells. Cancer Sci. 2008;99(3):590–594. doi: 10.1111/j.1349-7006.2007.00715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo K, et al. Adenosine triphosphate release and purinergic (P2) receptor-mediated secretion in small and large mouse cholangiocytes. Hepatology. 2010;52(5):1819–1828. doi: 10.1002/hep.23883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.