Significance
Despite the importance of trichothecene mycotoxins in food safety and environmental exposure, molecular mechanism of their toxicity is not well-understood and there is a critical gap in our knowledge about cellular protection mechanisms against trichothecenes. To identify cellular functions that provide resistance to trichothecenes, we screened the yeast deletion library for increased sensitivity to trichothecin and identified a vital role for mitochondrial oxidative stress in trichothecene sensitivity and for mitophagy in protecting cells against trichothecenes. Our data show that enhancing degradation of trichothecene-damaged mitochondria by mitophagy reduces mitochondrial oxidative stress and increases cell survival. These results reveal a previously unidentified mechanism against trichothecenes.
Keywords: Fusarium head blight, deoxynivalenol, Fusarium graminearum
Abstract
Trichothecene mycotoxins are natural contaminants of small grain cereals and are encountered in the environment, posing a worldwide threat to human and animal health. Their mechanism of toxicity is poorly understood, and little is known about cellular protection mechanisms against trichothecenes. We previously identified inhibition of mitochondrial protein synthesis as a novel mechanism for trichothecene-induced cell death. To identify cellular functions involved in trichothecene resistance, we screened the Saccharomyces cerevisiae deletion library for increased sensitivity to nonlethal concentrations of trichothecin (Tcin) and identified 121 strains exhibiting higher sensitivity than the parental strain. The largest group of sensitive strains had significantly higher reactive oxygen species (ROS) levels relative to the parental strain. A dose-dependent increase in ROS levels was observed in the parental strain treated with different trichothecenes, but not in a petite version of the parental strain or in the presence of a mitochondrial membrane uncoupler, indicating that mitochondria are the main site of ROS production due to toxin exposure. Cytotoxicity of trichothecenes was alleviated after treatment of the parental strain and highly sensitive mutants with antioxidants, suggesting that oxidative stress contributes to trichothecene sensitivity. Cotreatment with rapamycin and trichothecenes reduced ROS levels and cytotoxicity in the parental strain relative to the trichothecene treatment alone, but not in mitophagy deficient mutants, suggesting that elimination of trichothecene-damaged mitochondria by mitophagy improves cell survival. These results reveal that increased mitophagy is a cellular protection mechanism against trichothecene-induced mitochondrial oxidative stress and a potential target for trichothecene resistance.
Trichothecene mycotoxins are highly toxic secondary metabolites produced by Trichothecium, Myrothecium, Trichoderma, and Fusarium. Fusarium graminearum and Fusarium culmorum cause Fusarium head blight (FHB), which is one of the most damaging diseases of small grain cereals. FHB adversely affects the food supply because trichothecene mycotoxins, such as deoxynivalenol (DON), accumulate in the infected grain, presenting a food safety risk and health hazard to humans and animals (1). Controlling their accumulation in small grains remains a huge challenge. Trichothecenes cause growth retardation, hemorrhagic lesions, immune dysfunction, and emesis (2, 3) and are neurotoxic (4–6). Trichothecene poisoning causes acute gastroenteritis and has been linked to alimentary toxic aleukia (ATA) and Kashin–Beck disease, an endemic and chronic degenerative osteoarthritis (3).
Trichothecenes inhibit protein synthesis by targeting ribosomal protein L3 in yeast (7–9). However, their toxicity is not entirely due to inhibition of cytosolic protein synthesis. In mammalian cells, DON induces activation of double-stranded RNA-associated protein kinase (PKR), promotes degradation of 28S rRNA, and up-regulates a number of microRNAs (3, 10). DON exposure stabilizes mRNAs encoding proinflammatory mRNAs (3, 10). In plants, T-2 toxin (T-2) and DON cause oxidative stress damage by increasing reactive oxygen species (ROS) levels (11).
Despite the importance of trichothecenes in food safety and chronic environmental exposure, the molecular mechanism of their toxicity is not well-understood and there is a critical gap in our knowledge about the mechanisms that can protect cells against trichothecenes. To understand the trichothecene mechanism of action, we previously carried out a genome-wide screen of Saccharomyces cerevisiae for resistance to trichothecin (Tcin) and showed that the largest group of resistant strains were affected in mitochondrial functions (12). We showed that trichothecenes inhibit mitochondrial translation, before depolarization and fragmentation of the mitochondrial membrane and independent of the cytosolic translation inhibition (12, 13). Previous studies showed a link between ROS generation and mitochondrial translation (14, 15). Yeast mutants with impaired mitochondrial translation exhibited faulty oxidative phosphorylation resulting in toxic levels of ROS, overwhelming the cell’s antioxidant capacity, and causing oxidative stress (14). These results suggested that mitochondrial dysfunction and the resulting oxidative stress might contribute to trichothecene sensitivity. To obtain a comprehensive view of the cellular functions needed for tolerance to trichothecenes, we screened the complete set of viable S. cerevisiae deletion strains for increased sensitivity to Tcin, a representative type B trichothecene that has a similar IC50 for Vero cells (0.5 µM) and yeast grown on nonfermentable media (0.75 µM) (12, 13). Analysis of the identified strains revealed a vital role for mitochondrial oxidative stress in trichothecene sensitivity and provided the first evidence to our knowledge for a prosurvival role for the autophagic degradation of damaged mitochondria or mitophagy in the reduction of trichothecene-mediated mitochondrial oxidative stress.
Results
Genome-Wide Profiling of Yeast Knockout Mutants Reveals a Role for Oxidative Stress in Trichothecene Sensitivity.
To identify cellular genes that increase resistance to trichothecenes, we systematically screened the 4,720 nonessential S. cerevisiae deletion strains for increased sensitivity to 1 µM Tcin, which is nonlethal to the parental strain, BY4743 (12, 13). The screen was repeated four times, and 121 strains consistently exhibiting higher sensitivity than the parental strain were identified (Table S1). Classification of these strains using the Munich Information Center for Protein Sequences (MIPS) FunCat database revealed that they were disrupted in signaling and protein modification (SP; 20%), metabolism (19%), DNA repair and damage response (DNA; 16%), RNA degradation and stability (RNA; 15%), vacuolar protein sorting (VPS; 13%) pathway, cell cycle (CC;13%), and ribosome biogenesis and protein degradation (Protein; 12%) (Fig. 1A). At 1 µM Tcin, growth of BY4743 was only slightly inhibited (≤ 15%) (Fig. 1B). However, growth of the Tcin sensitive strains was severely inhibited, with the most sensitive 27 strains showing greater than 75% growth inhibition (Fig. 1B).
Fig. 1.
Analysis of yeast deletion mutants, which showed increased sensitivity to Tcin. (A) The mutants were classified based on MIPS FunCat Database terms such as signaling and protein modification (SP), ribosome biogenesis and protein degradation (Protein), RNA degradation and stability (RNA), DNA repair and damage response (DNA), VPS, mitochondria (Mito), unclassified (UC), cell cycle (CC), and the remaining characterized mutants were grouped as Other. (B) The top 27 mutants exhibiting the highest sensitivity to Tcin and BY4743 were grown with or without 1 µM Tcin and in the presence or absence of 4.5 mM vitamin E. At 18 h after treatment, growth at OD600 was measured and relative growth was calculated as the ratio of growth of treated cells to that of untreated cells, with a ratio of 1 indicating no effect on growth. (C) Following 1 h after treatment of the Tcin-sensitive strains and BY4743 with 1 µM Tcin, equal OD600 cells were stained with DCFH-DA. The ratio of ROS-positive cells for each Tcin-treated mutant was normalized to the ratio of ROS-positive cells for Tcin-treated BY4743, and fold increase was plotted. Error bars represent SE calculated from four independent experiments. Differences relative to BY4743 were assessed by using ANOVA followed by post hoc LSD tests (*P < 0.05).
A large group of the Tcin-sensitive mutants (42%) exhibited high sensitivity to oxidative stress based on the Saccharomyces genome database (SGD) (www.yeastgenome.org). ROS levels were measured by flow cytometry after 2′,7′-dichlorfluorescein-diacetate (DCFH-DA) staining in the parental strain, BY4743, and the 27 most sensitive mutants after treatment with 1 µM Tcin for 1 h. All 27 strains exhibited significantly higher ROS levels upon Tcin treatment with dep1Δ, snf6Δ, rei1Δ, sap30Δ, atg32Δ, and yor152CΔ showing a twofold or greater increase relative to the parental strain (Fig. 1C). We examined ROS levels in the previously identified 15 deletion strains that exhibited the highest resistance to Tcin (12). Treatment of BY4743 with 4 µM Tcin for 1 h led to a twofold increase in ROS levels (Fig. S1A). In contrast, ROS levels were significantly lower in the Tcin resistant mutants after treatment with 4 µM Tcin for 1 h (Fig. S1A).
A dose-dependent inhibition of yeast growth was observed upon treatment with type A trichothecenes, T-2 toxin (T-2) and diacetoxyscirpenol (DAS), and type B trichothecenes, Tcin and DON (Fig. 2A). Treated cells accumulated higher levels of ROS (two- to fourfold) relative to the untreated cells, and the response was dose-dependent (Fig. 2B). The increase in trichothecene-induced ROS levels was confirmed with the Amplex Red/peroxidase assay, which showed a significant increase in endogenous H2O2 levels in BY4743 after treatment with 300 µM T-2 or 8 µM Tcin relative to untreated cells (Fig. S1B).
Fig. 2.
Effect of trichothecenes on growth and ROS generation. (A) BY4743 cells were treated with increasing concentrations of trichothecenes for 18 h, and the ratio of OD600 for treated cells relative to untreated cells (UT) was plotted as relative growth. (B) For ROS measurement, equal OD600 cells were stained with DCFH-DA at 1 h after treatment with trichothecenes. (C) BY4743 cells were cotreated with either vitamin E or vitamin C and trichothecenes overnight, and relative growth was calculated as the ratio of OD600 for treated cells relative to untreated cells. Error bars represent SE calculated from three independent experiments.
To determine whether scavenging trichothecene-induced ROS would alleviate cytotoxicity, yeast cells were cotreated with trichothecenes and either ascorbic acid (vitamin C) or vitamin E overnight, and relative growth was examined (Fig. 2C). Treatment with the antioxidants alone did not substantially affect growth. Survival of T-2–treated cells increased from 10 to 36% upon cotreatment with vitamin E and from 10 to 40% upon cotreatment with vitamin C (Fig. 2C). A greater increase in cell growth was observed when Tcin-exposed cells were treated with antioxidants (Fig. 2C). Cell survival increased from 12 to 97% with vitamin E and from 12 to 62% with vitamin C (Fig. 2C). We found a twofold or higher increase in cell survival in 19 of the 27 most sensitive strains after treatment with 1 µM Tcin in the presence of vitamin E (Fig. 1B). These results strongly implicate ROS production in the trichothecene sensitivity.
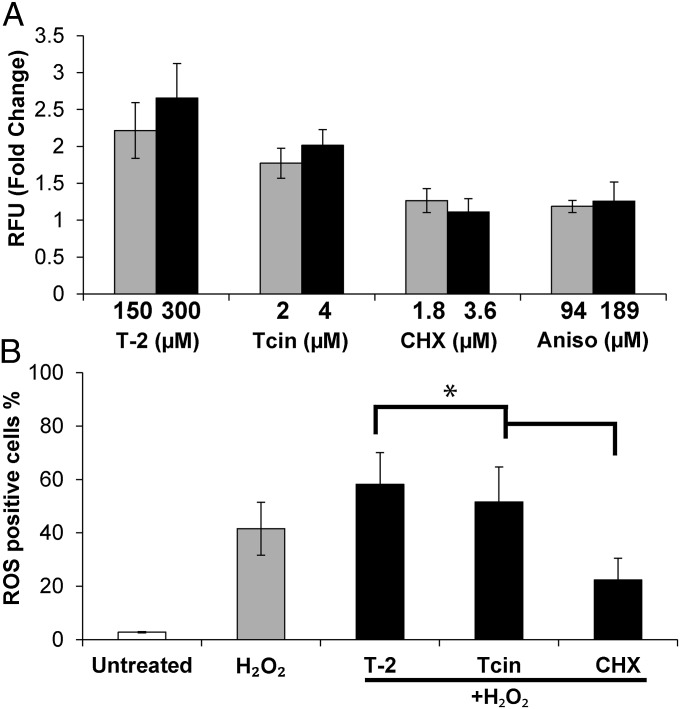
To determine whether ROS generation is a secondary effect of translation inhibition by trichothecenes, we examined ROS levels in cells treated with increasing concentrations of cycloheximide or anisomycin for 1 h (Fig. 3A). Trichothecene treatment increased ROS levels 2.2–2.7-fold with T-2 and 1.7–2.0-fold with Tcin. ROS levels did not increase when total translation was inhibited with cycloheximide or anisomycin (Fig. 3A), suggesting that trichothecene-triggered ROS generation was not solely due to the inhibitory effects of trichothecenes on cytosolic protein synthesis. We further examined ROS levels in cells cotreated with 2.5 mM H2O2 and either trichothecenes or cycloheximide (Fig. 3B). A 42% increase in ROS levels was seen in H2O2-treated cells at 1 h after treatment relative to untreated cells (3%). The ROS levels in cells cotreated with H2O2 and trichothecenes were significantly higher than in cells cotreated with H2O2 and cycloheximide (CHX) (Fig. 3B). These results demonstrate that trichothecene treatment increases the sensitivity to oxidative stress in a manner that does not solely depend on the inhibition of cytosolic translation.
Fig. 3.
ROS generation in response to translation inhibition. (A) BY4743 cells were treated with increasing concentrations of T-2, Tcin, cycloheximide (CHX), or anisomycin (Aniso) for 1 h, and equal OD600 cells were stained with DCFH-DA. The ratio of mean fluorescence of treated cells to that of untreated control were plotted as relative fluorescence units (RFU). (B) BY4743 cells were cotreated with 2.5 mM H2O2 and 300 µM T-2, 4 µM Tcin, or 3.6 µM cycloheximide (CHX) for 1 h and stained with DCFH-DA. Error bars represent SE calculated from three independent experiments. Significant difference between cotreatment with H2O2/T-2 and H2O2/Tcin relative to H2O2/CHX is indicated (ANOVA with post hoc LSD tests, *P < 0.05).
Mitochondria Are the Main Site of ROS Production in Trichothecene-Treated Cells.
Because mitochondria are a major source of ROS (16, 17), we examined ROS levels in trichothecene-treated BY4743 (grande, rho+) or a petite strain (rho0) derived from BY4743 (12). Grande (rho+) cells exhibited a significant increase in the number of ROS-positive cells 1 h after treatment with T-2 or Tcin (Fig. 4A). In contrast, trichothecenes failed to induce similar levels of ROS in the petite strain (rho0) (Fig. 3A). When grande cells were treated with carbonyl-cyanide p-trifluoromethoxyphenylhydrazone (FCCP), a mitochondrial membrane uncoupler that permeabilizes the inner mitochondrial membrane, ROS levels did not increase in T-2 or Tcin-treated cells (Fig. 4A). FCCP pretreatment increased survival of yeast treated with 150 µM and 300 µM T-2 by 1.8- and 1.7-fold, respectively (Fig. 4B). Similarly, survival of FCCP pretreated cells increased by 2- and 2.6-fold after treatment with 2 µM and 4 µM Tcin, respectively (Fig. 4B). These results are consistent with mitochondria as the main site of ROS production in trichothecene-treated cells.
Fig. 4.
The role of mitochondria in trichothecene-induced ROS generation and growth inhibition. (A) BY4743 cells that are either grande (rho+), petite (rho0), or grande (rho+) pretreated with 0.4 µM FCCP for 1 h were treated with trichothecenes for 1 h and stained with DCFH-DA. Differences relative to the no toxin treatment were assessed by using ANOVA followed by the Fisher Test (LSD) (*P < 0.05). (B) BY4743 cells were pretreated with 1 µM FCCP before treating with increasing concentrations of trichothecenes overnight. Relative growth is calculated as the ratio of OD600 of treated cells to the untreated control. Error bars represent SE calculated from three independent experiments.
We showed that Tcin treatment led to fragmentation of the tubular mitochondrial network (12). The ROS generated by mitochondria upon mycotoxin exposure may be responsible for the disruption of mitochondrial membrane morphology. To address this hypothesis, we examined mitochondrial membrane morphology of yeast cells constitutively expressing a green fluorescent protein (GFP)-tagged mitochondrial matrix protein (18) after treatment with 300 µM T-2 or 4 µM Tcin in the presence of either vitamin C or vitamin E. Significant fragmentation of the tubular network of mitochondria was observed in trichothecene-treated cells in the absence of the antioxidants (Fig. 5). However, this fragmentation was only moderately visible when cells were cotreated with vitamin C (Fig. 5). The tubular network remained largely intact with little to no fragmentation when cells were cotreated with vitamin E (Fig. 5). Treatment with antioxidants protected mitochondrial membranes, suggesting that trichothecene-induced mitochondrial ROS leads to mitochondrial membrane damage.
Fig. 5.
Effect of antioxidants on mitochondrial membrane morphology in trichothecene-treated cells. BY4743 cells constitutively expressing mitochondrial targeted GFP were cotreated with either vitamin C or vitamin E and 300 µM T-2 or 4 µM Tcin for 6 h. Cells were photographed by using an epifluorescence microscope. (Magnification: 100×.) (Scale bar: 5 µm.)
Rapamycin Treatment Reduces Mitochondrial ROS Levels and Rescues Trichothecene-Treated Cells.
A number of highly Tcin-sensitive strains (vps25Δ, vps20Δ, vps8Δ, snf7Δ, vps36Δ, pep12Δ, bro1Δ, vps4Δ, vac7Δ, vps3Δ, did4Δ, vps34Δ, vps16Δ) (Table S1) encode components of the VPS pathway, which is critical for selective autophagic processes, including mitophagy (19, 20). The VPS pathway is required for broad resistance to oxidative stress (21). We identified components of the ubiquitin-mediated protein degradation pathway (doa1Δ, ubp2Δ, ubp6Δ, shp1Δ, rad6Δ, bre5Δ, ubp3Δ), including the Ubp3–Bre5 complex (22) and Doa1/Ufd3 (23), which are involved in the selective autophagic degradation of mature ribosomes in yeast, also called ribophagy. Mutants defective in the ubiquitin-proteasome system and ribophagy are hypersensitive to trichothecenes, possibly because damaged ribosomes produce defective translation products that cause stress to the cell’s degradation machinery. Mutants defective in mitophagy (atg32Δ, slt2Δ, bck1Δ) (Table S1) exhibited higher sensitivity to Tcin (Fig. 1B) and accumulated significantly higher levels of ROS than the parental strain (Fig. 1C), suggesting that impaired mitophagy causes an increase in trichothecene-mediated oxidative damage.
Rapamycin has been shown to enhance autophagy and specifically mitophagy after preculturing yeast in nonfermentable medium, which induces proliferation of mitochondria (24). We treated yeast cells with 5 nM rapamycin and either 300 µM T-2 or 4 µM Tcin for 1 h and stained for ROS (Fig. 6A). The number of ROS-positive cells significantly increased from 3% (untreated) to 15% after T-2 treatment, to 9% after Tcin treatment, and to 42% after H2O2 treatment (Fig. 6A). Cotreatment with rapamycin reduced trichothecene-induced ROS levels from 15 to 9% after treatment with T-2 and from 9 to 6% after treatment with Tcin (Fig. 6A). Rapamycin treatment also reduced H2O2-induced ROS levels from 42 to 19% (Fig. 6A).
Fig. 6.
Effect of rapamycin on trichothecene sensitivity and ROS generation. (A) BY4743 cells were cotreated with 5 nM rapamycin and 300 µM T-2, 4 µM Tcin, or 2.5 mM H2O2 for 1 h and stained with DCFH-DA. Significant difference among T-2–, Tcin-, and H2O2-treated samples relative to untreated is indicated (ANOVA with post hoc LSD tests, *P < 0.05, **P < 0.01). BY4743 and mutants were cotreated with 5 nM rapamycin and either 300 µM T-2 (B) or 4 µM Tcin (C) overnight. Relative growth is calculated as the ratio of OD600 of treated cells to the untreated control. Error bars represent SE calculated from three independent experiments.
Cotreatment with rapamycin increased survival of T-2–treated cells from 17 to 41% (Fig. 6B) and Tcin-treated cells from 15 to 23% (Fig. 6C). Rapamycin treatment also increased survival of H2O2 treated cells from 57 to 74%. Several strains (slt2Δ, whi2Δ, uth1Δ, atg32Δ, and hog1Δ) deficient in mitophagy (25) were treated with trichothecenes in the presence or absence of rapamycin. Treatment with 5 nM rapamycin alone did not affect the growth of these mutants. Survival of the mitophagy-defective strains did not increase when they were cotreated with 5 nM rapamycin and T-2 (Fig. 6B) or 5 nM rapamycin and Tcin (Fig. 6C). We cotreated the mutants, which showed greater than 75% growth inhibition upon 1 µM Tcin treatment, with rapamycin (Fig. S2). Although the growth of these mutants was not affected by treatment with 5 nM rapamycin alone, rapamycin alleviated toxicity of Tcin moderately in some mutants (mbf1Δ, pho23Δ, ubp3Δ, vac7Δ) but not in the others (Fig. S2). These results suggest that enhancing the efficiency with which cells clear damaged mitochondria through mitophagy decreases ROS levels, allowing cells to escape oxidative damage due to trichothecene exposure.
Discussion
Mitochondrial Oxidative Stress Contributes to Trichothecene Sensitivity.
Our genome-wide screen of the yeast deletion library identified 121 genes that encode functions critical for Tcin tolerance. A substantial number of gene deletions (42%) affected resistance to various oxidants including H2O2, menadione, diamide, and cumene hydroperoxide (21), indicating a potential overlap between cellular response to oxidative stress and trichothecene tolerance. Consistent with our results in yeast, F. graminearum inoculation of barley induced expression of proteins associated with oxidative stress (26). DON treatment of wheat elicited hydrogen peroxide production and induced defense responses (27) and T-2–induced hydrogen peroxide production in Arabidopsis leaves (28). Tcin induced significantly higher levels of ROS in the most highly sensitive strains relative to the parental strain (Fig. 1C). In contrast, Tcin failed to generate ROS in previously identified Tcin-resistant strains (Fig. S1A). Other trichothecenes (T-2, DAS, and DON) increased ROS levels in the parental strain in a dose-dependent manner (Fig. 2 A and B). These results provided evidence for a critical role for oxidative stress in trichothecene sensitivity.
Exogenous addition of antioxidants alleviated growth inhibition by trichothecenes (Fig. 2C), indicating that toxicity of trichothecenes is a consequence of increased ROS production. Addition of ascorbic acid, which scavenges ROS (29) or vitamin E, which stabilizes and maintains membrane integrity against lipid peroxidation induced by ROS (30), significantly increased survival of the parental strain (Fig. 2C). Vitamin E also increased growth of the Tcin-sensitive mutants (Fig. 1B). Both antioxidants protected against trichothecene-mediated disruption of the tubular mitochondrial membrane network (12) with vitamin E providing the greatest protection (Fig. 5). The protection of mitochondrial membranes from trichothecene damage correlated with the reduction in trichothecene-induced growth inhibition after antioxidant treatment (Fig. 2C), suggesting that mitochondrial membrane is a critical target for oxidative damage.
Evidence for the mitochondrial origin of trichothecene-induced ROS generation was provided by the lack of increase in ROS levels after trichothecene treatment of a petite version of the parental strain and after cotreatment of the parental strain with FCCP and trichothecenes (Fig. 4A). The increase in ROS levels could not be attributed to inhibition of cytosolic translation by the trichothecenes (Fig. 3). We propose that trichothecene-mediated inhibition of mitochondrial translation (13) may be responsible for the increase in ROS levels. Genetic perturbations in yeast, which cause imbalanced and reduced mitochondrial translation, lead to increased sensitivity to H2O2-mediated oxidative stress, high levels of intracellular ROS, and, subsequently, to complete inactivation of respiration (14, 15). Mitochondrial translation inhibition would cause formation of ROS by affecting translation of mitochondrial DNA-encoded oxidative phosphorylation complexes, leading to an increase in oxygen concentration, favoring formation of ROS (14, 15). ROS produced above a certain threshold level can overwhelm cell’s antioxidant capacity, resulting in damage to mitochondrial DNA and respiratory chain components (14, 31). The initial increase in ROS levels at 1 h after treatment with trichothecenes (Fig. 2B) dropped at 6 h, possibly due to depolarization of the mitochondrial membrane (13). The decrease in mitochondrial membrane potential (13) may ultimately lead to further damage to mitochondria and other cellular functions and to eventual cell death.
Trichothecene mycotoxins function as virulence factors allowing the spread of F. graminearum within infected wheat heads (32, 33). An oxidative burst of hydrogen peroxide has been shown to enhance DON production by F. graminearum via up-regulation of various Tri genes involved in trichothecene biosynthesis (34). Our results provide evidence that inhibiting trichothecene-induced oxidative stress may protect against FHB by scavenging excess ROS, which may decrease mycotoxin production by F. graminearum, leading to a reduction in the spread of the disease.
Elimination of Trichothecene Damaged Mitochondria Protects Cells Against Trichothecenes.
Autophagy and particularly mitophagy have been reported to play an essential role in reducing mitochondrial production of ROS, allowing mitochondria to escape oxidative damage in yeast (35). Deletion of components of the endosomal sorting complex required for transport (ESCRT) machinery (vps20Δ, vps25Δ, snf7Δ, vps36Δ, bro1Δ, vps4Δ, did4Δ), class C core vacuole/endosome tethering (CORVET) complex (vps8Δ, vps3Δ, vps16Δ), and t-SNARE protein, Pep12, which binds to the CORVET complex, led to increased sensitivity to Tcin (Table S1). These complexes are needed for tethering and fusion of the autophagasome with the vacuole (36). They may control trafficking of proteins that respond to trichothecene-mediated oxidative damage and trafficking of trichothecene-damaged proteins to the vacuole for degradation. Mutants defective in mitophagy (atg32Δ, slt2Δ, bck1Δ) (Table S1) exhibited higher sensitivity to Tcin (Fig. 1B) and accumulated higher levels of ROS than the parental strain (Fig. 1C). Atg32 is a mitophagy-specific receptor necessary for the recognition of mitochondria in yeast (37). During mitophagy, the mitogen-activated protein kinases (MAPKs) Slt2 and Hog1 activate Atg32, which facilitates the removal of damaged mitochondria (38). Bck1 is a MAPK kinase kinase identified in a screen for mitophagy defective strains (19). Deletion of genes involved in mitophagy may increase trichothecene-mediated oxidative damage by increasing the accumulation of damaged mitochondria.
Rapamycin, which inhibits the target of rapamycin (TOR) kinases (38, 39), reduced trichothecene and H2O2-induced ROS levels (Fig. 6A) and alleviated growth inhibition of the parental strain by trichothecenes (Fig. 6 B and C). In contrast, rapamycin did not improve growth of the mitophagy-defective strains after treatment with trichothecenes, suggesting that rapamycin reduces intracellular ROS most likely by up-regulating mitophagy. Inhibition of TOR signaling has been shown to enhance life span in yeast by increasing respiration via enhanced mitochondrial translation (15). It is possible that increased mitochondrial translation due to rapamycin treatment also contributes to improved growth of the trichothecene-treated parental strain.
We propose that Tcin inhibits mitochondrial translation, leading to the formation of ROS. If trichothecene-damaged mitochondria are not efficiently cleared by mitophagy, then they accumulate, leading to further production of ROS and oxidative stress, resulting in a dramatic loss of viability. Although increasing the antioxidant capacity of the cell is one way to protect against trichothecenes, our results provide the first evidence, to our knowledge, that mitophagy is a cellular protection mechanism to reduce trichothecene-mediated mitochondrial oxidative damage and to increase tolerance to trichothecenes. Our findings linking mitophagy to trichothecene tolerance in yeast provide novel insights into trichothecene resistance in higher organisms. Mitochondrial dysfunction and defective mitophagy have been implicated in several neurodegenerative diseases, including Parkinson, Alzheimer’s, and Huntington disease (40). It will be important to investigate whether enhancing selective removal of dysfunctional mitochondria through mitophagy would reduce trichothecene sensitivity in mammalian cells.
Materials and Methods
Yeast Strains and Reagents.
Homozygous diploid yeast strain BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ) was used in all experiments. Ascorbic acid and vitamin E were purchased from Sigma-Aldrich. FCCP was purchased from Enzo Life Sciences.
Trichothecene Isolation.
Tcin was isolated from Trichothecium roseum and prepared as described (12). DAS, T-2, and DON were prepared as described (41).
Yeast Knockout Library Screen.
The homozygous diploid (BY4743) knockout collection was obtained from Thermo Scientific and screen was carried out as described (12) except using 1 µM Tcin as described in SI Materials and Methods.
ROS Measurement.
The ROS levels were quantified by staining with DCFH-DA (Sigma-Aldrich). An equal number of cells were collected after treatment with trichothecenes or antioxidants, cells were washed and resuspended in 1× PBS with 2% glucose (wt/vol) and stained with DCFH-DA for 30 min at 30 °C, and were washed with water and analyzed by using the Accuri C6 Flow Cytometer (Accuri Cytometers) as described in SI Materials and Methods. H2O2 levels were quantified by using the Amplex Red/peroxidase assay (42) as described in SI Materials and Methods.
Microscopy.
Mitochondrial morphology was examined with an Olympus BX41 epifluorescence microscope as described (12). Cells were transformed with pVT100U-mtGFP, which encodes GFP targeted to the mitochondria (18).
Statistics.
The data are shown as means ± SEM, graphed by using Microsoft Excel, and analyzed by ANOVA using the Fisher Test (least significant difference; LSD) to test differences between individual treatments with Origin 9.1 (OriginLab) software.
Supplementary Material
Acknowledgments
We thank Dr. Benedikt Westermann for pVT100U-mtGFP, Dr. Jennifer Nielsen Kahn for determining the sensitivity of Vero cells to Tcin and for helpful comments, and Dr. Mike Pierce at the School of Environmental and Biological Sciences Core Facility for help with flow cytometry. This study is Cooperative Project 59-0206-1-121 supported by the US Department of Agriculture (USDA) in cooperation with the US Wheat and Barley Scab Initiative and by the New Jersey Agricultural Experiment Station and the USDA–National Institute for Food and Agriculture, Hatch Project Number NJ12117.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403145111/-/DCSupplemental.
References
- 1.McCormick SP, Alexander NJ, Proctor RH. Phytochemicals, Plant Growth, and the Environment. New York: Springer; 2013. Trichothecene triangle: Toxins, genes, and plant disease; pp. 1–17. [Google Scholar]
- 2.Sudakin DL. Trichothecenes in the environment: Relevance to human health. Toxicol Lett. 2003;143(2):97–107. doi: 10.1016/s0378-4274(03)00116-4. [DOI] [PubMed] [Google Scholar]
- 3.Wu F, Groopman JD, Pestka JJ. Public health impacts of foodborne mycotoxins. Annu Rev Food Sci Technol. 2014;5:351–372. doi: 10.1146/annurev-food-030713-092431. [DOI] [PubMed] [Google Scholar]
- 4.Weidner M, et al. Influence of T-2 and HT-2 toxin on the blood-brain barrier in vitro: New experimental hints for neurotoxic effects. PLoS ONE. 2013;8(3):e60484. doi: 10.1371/journal.pone.0060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corps KN, Islam Z, Pestka JJ, Harkema JR. Neurotoxic, inflammatory, and mucosecretory responses in the nasal airways of mice repeatedly exposed to the macrocyclic trichothecene mycotoxin roridin A: Dose-response and persistence of injury. Toxicol Pathol. 2010;38(3):429–451. doi: 10.1177/0192623310364026. [DOI] [PubMed] [Google Scholar]
- 6.Karunasena E, Larrañaga MD, Simoni JS, Douglas DR, Straus DC. Building-associated neurological damage modeled in human cells: A mechanism of neurotoxic effects by exposure to mycotoxins in the indoor environment. Mycopathologia. 2010;170(6):377–390. doi: 10.1007/s11046-010-9330-5. [DOI] [PubMed] [Google Scholar]
- 7.Fried HM, Warner JR. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci USA. 1981;78(1):238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickner RB, Ridley SP, Fried HM, Ball SG. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1982;79(15):4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cundliffe E, Cannon M, Davies J. Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins. Proc Natl Acad Sci USA. 1974;71(1):30–34. doi: 10.1073/pnas.71.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestka JJ. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch Toxicol. 2010;84(9):663–679. doi: 10.1007/s00204-010-0579-8. [DOI] [PubMed] [Google Scholar]
- 11.Arunachalam C, Doohan FM. Trichothecene toxicity in eukaryotes: Cellular and molecular mechanisms in plants and animals. Toxicol Lett. 2013;217(2):149–158. doi: 10.1016/j.toxlet.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 12.McLaughlin JE, et al. A genome-wide screen in Saccharomyces cerevisiae reveals a critical role for the mitochondria in the toxicity of a trichothecene mycotoxin. Proc Natl Acad Sci USA. 2009;106(51):21883–21888. doi: 10.1073/pnas.0909777106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bin-Umer MA, McLaughlin JE, Basu D, McCormick S, Tumer NE. Trichothecene mycotoxins inhibit mitochondrial translation—implication for the mechanism of toxicity. Toxins(Basel) 2011;3(12):1484–1501. doi: 10.3390/toxins3121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol Cell Biol. 2006;26(13):4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5(4):265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: Central role of complex III. J Biol Chem. 2003;278(38):36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 17.Bhatia-Kiššová I, Camougrand N. Mitophagy in yeast: Actors and physiological roles. FEMS Yeast Res. 2010;10(8):1023–1034. doi: 10.1111/j.1567-1364.2010.00659.x. [DOI] [PubMed] [Google Scholar]
- 18.Westermann B, Neupert W. Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16(15):1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Kanki T, Wang K, Klionsky DJ. A genomic screen for yeast mutants defective in mitophagy. Autophagy. 2010;6(2):278–280. doi: 10.4161/auto.6.2.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reggiori F, Klionsky DJ. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics. 2013;194(2):341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: Oxidative-stress-response genes. Proc Natl Acad Sci USA. 2004;101(17):6564–6569. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 23.Ossareh-Nazari B, et al. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010;11(7):548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: The Om45-GFP processing assay. Autophagy. 2009;5(8):1186–1189. doi: 10.4161/auto.5.8.9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhatia-Kiššová I, Camougrand N. Mitophagy: A process that adapts to the cell physiology. Int J Biochem Cell Biol. 2013;45(1):30–33. doi: 10.1016/j.biocel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Geddes J, Eudes F, Laroche A, Selinger LB. Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host, Hordeum vulgare. Proteomics. 2008;8(3):545–554. doi: 10.1002/pmic.200700115. [DOI] [PubMed] [Google Scholar]
- 27.Desmond OJ, et al. The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol Plant Pathol. 2008;9(4):435–445. doi: 10.1111/j.1364-3703.2008.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiuchi T, et al. Fusarium phytotoxin trichothecenes have an elicitor-like activity in Arabidopsis thaliana, but the activity differed significantly among their molecular species. Mol Plant Microbe Interact. 2006;19(5):512–520. doi: 10.1094/MPMI-19-0512. [DOI] [PubMed] [Google Scholar]
- 29.Asada K. Ascorbate peroxidase—a hydrogen peroxide—scavenging enzyme in plants. Physiol Plant. 1992;85(2):235–241. [Google Scholar]
- 30.Fryer M. The antioxidant effects of thylakoid Vitamin E (α-tocopherol) Plant Cell Environ. 1992;15(4):381–392. [Google Scholar]
- 31.Davidson JF, Schiestl RH. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(24):8483–8489. doi: 10.1128/MCB.21.24.8483-8489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proctor RH, Hohn TM, McCormick SP. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact. 1995;8(4):593–601. doi: 10.1094/mpmi-8-0593. [DOI] [PubMed] [Google Scholar]
- 33.Jansen C, et al. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc Natl Acad Sci USA. 2005;102(46):16892–16897. doi: 10.1073/pnas.0508467102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponts N, Pinson-Gadais L, Barreau C, Richard-Forget F, Ouellet T. Exogenous H2O2 and catalase treatments interfere with Tri genes expression in liquid cultures of Fusarium graminearum. FEBS Lett. 2007;581(3):443–447. doi: 10.1016/j.febslet.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Kurihara Y, et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem. 2012;287(5):3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solinger JA, Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013;280(12):2743–2757. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- 37.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendl N, et al. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci. 2011;124(Pt 8):1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- 39.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13(24):3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Chan DC. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desjardins AE, McCormick SP, Appell M. Structure-activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. J Agric Food Chem. 2007;55(16):6487–6492. doi: 10.1021/jf0709193. [DOI] [PubMed] [Google Scholar]
- 42.Ojovan SM, et al. Accumulation of dodecyltriphenylphosphonium in mitochondria induces their swelling and ROS-dependent growth inhibition in yeast. J Bioenerg Biomembr. 2011;43(2):175–180. doi: 10.1007/s10863-011-9345-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.