Significance
p53 is one of the most intensively studied tumor-suppressor genes. We identified a naturally occurring p53 isoform, generated by an alternative-splicing event, that, although lacking transcriptional activity and canonical tumor suppressor functions, is able to reprogram cells toward the acquisition of metastatic features via a cyclophilin D interaction in the mitochondria matrix. Interestingly, this isoform is expressed on tissue injury and in tumors characterized by increased metastatic spread. In some of these tumors, p53-like isoforms are generated by intron 6 mutations. This suggests a possible physiological origin of certain p53 mutations and indicates that mutations resulting in the generation of truncated p53Ψ-like proteins do more than create a 53-null state.
Keywords: reactive oxygen species, cancer
Abstract
Although much is known about the underlying mechanisms of p53 activity and regulation, the factors that influence the diversity and duration of p53 responses are not well understood. Here we describe a unique mode of p53 regulation involving alternative splicing of the TP53 gene. We found that the use of an alternative 3′ splice site in intron 6 generates a unique p53 isoform, dubbed p53Ψ. At the molecular level, p53Ψ is unable to bind to DNA and does not transactivate canonical p53 target genes. However, like certain p53 gain-of-function mutants, p53Ψ attenuates the expression of E-cadherin, induces expression of markers of the epithelial-mesenchymal transition, and enhances the motility and invasive capacity of cells through a unique mechanism involving the regulation of cyclophilin D activity, a component of the mitochondrial inner pore permeability. Hence, we propose that p53Ψ encodes a separation-of-function isoform that, although lacking canonical p53 tumor suppressor/transcriptional activities, is able to induce a prometastatic program in a transcriptionally independent manner.
An evolutionarily conserved transcription factor, p53 has origins that can be traced back to the early metazoans, ∼700 Mya (1). This transcription factor plays a critical role in regulating many fundamental aspects of reversible and irreversible cellular stress responses, genome surveillance, and suppression of oncogenic transformation (1). In response to strong cellular stresses such as DNA damage or oncogenic signals, p53 regulates the expression of a large cohort of genes that affect cell cycle arrest, senescence, and apoptosis (1). Recent work has uncovered additional roles for p53 under basal physiological conditions, such as regulation of development, reproduction, metabolism, and self-renewal capacity (2–5). However, the factors that influence the diversity and duration of p53 responses are not well understood. Here we describe a unique mode of p53 regulation that involves alternative splicing of the TP53 gene. We found that the use of an alternative 3′ splice site in intron 6 generates a previously uncharacterized p53 isoform that we named p53Ψ. Interestingly, this isoform is highly expressed in cells characterized by a CD44high/CD24low immune type. At the molecular level, p53Ψ lacks major portions of the DNA-binding domain, the nuclear localization sequence, and the tetramerization domain, features that are normally present in full-length p53 (p53FL). Consequently, this isoform proved incapable of sequence-specific DNA binding and transactivation of canonical p53 target genes. However, expression of the p53Ψ isoform attenuated the expression of E-cadherin, induced expression of markers associated with epithelial-mesenchymal transition (EMT), and enhanced the motility and invasive capacity of normal and malignant cells. Consistent with a role of these features in enhancing the prometastatic capabilities of cells, we observed that in patients with early-stage nonsmall cell lung carcinoma (NSCLC), expression of p53Ψ correlated with increased probability of relapse following surgical tumor resection. Such characteristics are similar to certain p53 gain-of-function missense mutants.
We also found that the reprogramming of cells toward acquisition of mesenchymal-like features—induced either by expressing p53Ψ or p53 with gain-of-function mutations—is contingent on increased production of reactive oxygen species (ROS) by virtue of interaction with cyclophilin D (CypD), a mitochondrial matrix peptidyl-prolyl isomerase known to modulate opening of the mitochondrial permeability transition pore (mPTP). Hence, we propose that p53Ψ encodes a “separation-of-function” isoform that lacks canonical p53 tumor suppressor/transcriptional activities but is capable of reprogramming cells toward acquisition of mesenchymal-like features in a transcriptionally independent manner. The remarkably similar activities of p53Ψ and certain p53 mutants also suggest that the latter “highjack” a regulated and reversible program (i.e., p53Ψ alternative splicing) that contributes to the biology of p53 mutations during tumorigenesis. Thus, in principle, this implies a possible physiological origin for certain p53 mutations.
Results
p53ψ Is a Unique p53 Isoform Generated from Use of an Alternative 3′ Splice Site.
In response to strong cellular stresses such as DNA damage or oncogenic signals, p53 regulates the expression of a large cohort of genes that affect cell cycle arrest, senescence, and apoptosis (1). However, recent analyses have uncovered essential roles for p53 also under basal physiological conditions such as regulation of development, reproduction, metabolism, and self-renewal capacity (2, 4–6). In particular, a recent publication by Godar et al. suggests that p53 specifically represses expression of CD44, a transmembrane glycoprotein that has been shown to regulate cell growth and cell motility in a variety of cells (6–10). Interestingly, increased expression of CD44 has been observed in CD24low, non–marrow-derived cells in tissues on injury. This observation prompted us to investigate whether p53 activity was deregulated in CD44high/CD24low cells. To investigate this, we used a murine lung injury model induced by i.p. naphthalene administration (Fig. 1A). Naphthalene treatment results in rapid necrotic changes within Clara cells of the terminal and respiratory bronchioles due to conversion of the drug into a toxic form by the cell-specific microsomal enzyme Cyp2F2 (11). As shown in Fig. 1B, on single i.p. injection of naphthalene, we observed an expansion of CD44high/CD24low non–marrow-derived (CD45−) and nonendothelial (CD31−) cells that increased in number in the lung in a time-dependent manner after injury from near undetectable levels in the vehicle-treated animals to almost 6% of total CD31−/CD45− cells by day 21. Interestingly, consistent with our initial hypothesis, gene expression analysis of FACS cells on injury revealed a reduction in the expression of known p53-regulated genes such as CDKN1A, Triap1, Sens1, Sens2, Pgam1, Pgam2, Sco1, Sco2, Tigar, and Gpx1 in CD44high/CD24low/CD31−/CD45− cells compared with CD44low/C24high/CD31−/CD45− cells (Fig. 1C and Fig. S1A).
Fig. 1.
Expression of p53ψ, a unique p53 isoform generated by the use of an alternative 3′ splice site, is enriched in CD44high/CD24low cells. (A) Schematic of naphthalene lung injury model. (B) Lung cell suspensions were sorted by FACS at different time points after injury with naphthalene. CD31- and CD45-negative cells were used to remove endothelial cells and bone marrow-derived cells, respectively. (Right) Accumulation of CD44high/CD24low cells over 21 d. Values in the right upper corners represent percentage of CD44high/CD24low cells relative to CD31−/CD45− cells. (C) The table reports expression of multiple p53 targets in CD44low/CD24high (CD44L) cells sorted from naphthalene-injured mice compared with levels in CD44high/CD24low (CD44H) cells. (D) RT-PCR analysis of lung tissue extracts obtained at the indicated time points after naphthalene treatment using oligonucleotide primers to exons 6 and 8 (ex6-ex8; Top) and primers specific for p53ψ (460-ex8; Middle). Actin was used for normalization. (E) Sequence analysis of the two PCR products amplified with p53 primers indicated the use of a unique splice junction between exon 6 and exon 8 in the shorter p53 transcript. (F) RT-PCR analysis of lung tissue obtained after naphthalene treatment at the indicated time points using primers specific for p53FL and p53ψ. (G) RT-PCR analysis of liver tissue after CCL4 treatment using oligonucleotide primers specific for p53FL and the p53ψ isoform at the indicated time points. Analyses of levels of TGF-β and IL-6 were used to confirm tissue injury. See Fig. S2C for further details on primer design. (H) RNA FISH confirmed expression of p53ψ in CCL4-injured livers. Tissue sections were hybridized with RNA FISH probes specific for p53ψ (red) and stained for smooth muscle actin (SMA, green) to highlight damaged area. DAPI (blue) was used as counterstain.
Prompted by this observation, we evaluated expression of p53 in total lung tissue homogenates taken from naphthalene-treated and untreated animals. RT-PCR analysis using primers spanning exon 6 to exon 8 indicated a slower migrating p53 band in cell extracts obtained from injured lungs, which intensified in the days following naphthalene injection (Fig. 1D). Sequence analysis revealed no mutations in exons 6, 7, or 8 or in intron 6, but indicated that this band was a unique p53 mRNA variant generated by the use of an alternative 3′ splice acceptor site within intron 6 (Fig. 1E and Fig. S1B). We refer to this p53 variant as p53 psi (p53Ψ). Comparison across species revealed that the sequence surrounding the alternative 3′ splice acceptor site in intron 6 is highly conserved (Fig. S1C). This observation is of particular interest given that intronic sequences are usually highly divergent (12). Using p53Ψ-specific primers, we verified p53Ψ was enriched in CD44high/CD24low/CD31−/CD45− cells compared with CD44low/C24high/CD31−/CD45− cells on lung injury (Fig. S1 D and E).
To determine whether p53Ψ was unique to the naphthalene lung injury model, we extended our analysis to other organs and to an additional tissue injury model. Specifically we determined the expression of p53Ψ in the thymus, salivary gland, small intestine, brain, heart, kidney, skeletal muscle, spleen, stomach, liver, and lung, as well as in the liver by RT-PCR analysis, using primers designed to amplify p53Ψ and the full-length p53 (p53FL) mRNAs (Fig. S1D). We were not able to detect expression of p53Ψ in any of the organs at steady state (Fig. S1F). However, similarly to what we observed in the case of lungs of naphthalene-treated mice (Fig. 1 D and F), p53Ψ was observed in CCL4-injured livers (Fig. 1G). Using RNA FISH, we confirmed expression of p53Ψ in CCL4-treated livers and observed that its expression was localized in the proximity of tissue lesions (i.e., the SMA-α–positive areas; Fig. 1H and Fig. S1G).
In sum, these observations indicate the existence of a unique p53 isoform generated through an alternative splicing event that is conserved across species and whose expression appears particularly enriched on tissue injury.
p53Ψ Is Expressed in Human Tumors and Tumor-Derived Cell Lines.
Human tumor analyses have shown that TP53 is mutated in approximately half of all cancers (13). Somatic p53 mutations occur in almost every type of tumor, at rates ranging from 30% to 50% depending on tumor type (13). Mutations are more frequent in advanced-stage cancers or in cancer subtypes that are highly metastatic (13). Hence, the generation of p53Ψ could, in principle, represent an alternative mode of p53 regulation in tumorigenesis. To investigate this, we assessed expression of p53Ψ and p53FL levels using RNA-FISH on a human tissue microarray (TMA) comprised of NSCLC tissues from 233 patients mainly with early-stage adenocarcinomas (Fig. S2 A and B). Lung tumor samples were stained with p53Ψ-specific probes. Approximately 22% of tumors clearly expressed p53Ψ (Fig. 2A and Fig. S2 C–F). Interestingly, the majority of tumor cores that were positive for p53Ψ were constituted primarily of CD44high/CD24low cells (Fig. 2B and Fig. S2). Univariate Kaplan–Meier survival analysis indicated that patients with tumors expressing p53Ψ displayed a decrease in overall survival compared with the p53Ψ-low group and to p53-null tumors (Fig. 2C and Fig. S2G). In principle this suggests that the generation of p53Ψ does more than create a p53-null state.
Fig. 2.
p53ψ is expressed in tumors and tumor-derived cell lines. (A) Expression of p53Ψ in two representative lung adenocarcinoma tumor cores characterized by high percentages of CD44highCD24low cells (Upper) or CD44lowCD24high cells (Lower). CD44 staining was pseudocolored in green and CD24 is in red. Staining with p53Ψ sense (SE) and antisense (AS) probes are shown in orange. DAPI (blue) as counterstain. See Fig. S2 for details on probe design. (B) The chart represents the distribution of p53Ψ mRNA expression in CD44highCD24low NSCLC tumors. (C) Kaplan–Meyer distribution of p53Ψ-positive and -negative NSCL tumors. See Fig. S2 for further details. (D) Semiquantitative RT-PCR analysis of p53FL and p53ψ in CD44high/CD24low cells sorted from multiple human cancer-derived cell lines. (E) The pie chart represents the distribution of mutations at position c.673–2A in 28,581 tumors as reported in the IARC p53 database. (F) A collection of 172 upper urinary tract transitional carcinoma cases was analyzed for mutations in the TP53 gene. The number of missense mutations (Upper) and mutations predicted to affect the TP53 splicing pattern (Lower) is shown. (G) Schematic of the minigene used in this study. (H) RT-PCR analysis of transcripts from the minigene using primers to the CMV promoter and exon 8 indicated that the presence of a G in position -2 relative to the first nucleotide in exon 7 resulted in the generation of an alternative transcript of the expected size of a p53Ψ-like transcript (PSI). Sequence analysis confirmed that this transcript was the result of the use of the same cryptic acceptor site in intron 6 used for the generation of p53Ψ. (I) RT-PCR analyses of cells expressing p53FL (A549) and a TP53 c.673–2A/G mutation (HOP62) indicate that the latter induces the generation of a p53Ψ-like transcript. Primers to exons 4 and 7 were used for PCR amplification of transcripts. (L) Western blot analysis with an N-terminal p53 antibody (DO1) of A549 and HOP62 cells extracts indicates that the HOP62 cells inherently express a p53Ψ-like protein of the expected size.
It has recently been shown that tumor cells characterized by a CD44high/CD24low immune type can be generated through epigenetic/stochastic events in virtually all tumor-derived cell lines and primary tumors (14). Given the high abundance of p53Ψ in cells of this immune type in NSCLC and in injured normal tissues, we investigated whether stochastically generated CD44high/CD24low cells also expressed p53Ψ. RT-PCR analysis of FACS-sorted cells from multiple tumor-derived human cell lines using primers specific for p53FL and p53Ψ revealed that p53Ψ was expressed predominantly in the CD44highCD24low cell fraction (Fig. 2D). Sequence analysis confirmed the identity of this presumed p53 isoform as an ortholog of the murine p53Ψ gene (Fig. S2H). In addition, Western blot analysis revealed the presence of a band of the expected p53Ψ size in CD44high/CD24low cell extracts (Fig. S2I).
Splicing is carried out by the spliceosome, a massive structure comprised of five small nuclear ribonucleoprotein particles (snRNPs) and a large number of auxiliary proteins that accurately recognizes the splice sites and catalyzes the two steps of the splicing reaction (15). The decisions as to which splice sites are used and which exons are included involve intronic and exonic RNA sequence elements (cis-regulatory elements) and their cognate protein regulators (trans-regulatory factors) (15). Our data indicate that a natural, regulated switch in p53FL/p53Ψ splice site selection occurs in stochastically generated CD44high /CD24low cells and in injured normal tissues. This observation suggests that in physiological conditions, controlled changes in abundance or activity of trans-acting factors modulate the p53Ψ alternative splicing events. However, in tumors, in addition to the deregulation of the proper splicing factor balance, the occurrence of genetic aberrations affecting cis-regulatory elements could also result in aberrant p53Ψ expression. The splicing reaction requires the presence of a highly conserved AG intronic dinucleotide at the intron/exon boundary at the 3′ acceptor site. Hence, mutations in the normal acceptor site at the intron 6/exon 7 boundary (−1G to A/T/C or −2A to G/T/C with respect to the splice junction) could favor the use of the cryptic acceptor site in intron 6 generating the p53Ψ isoform. To test this hypothesis, we probed the International Agency for Research on Cancer (IARC) TP53 database for mutations at the intron 6/exon 7 boundary. Our analysis indicated that the G/T/C mutations at position c.673-2A indeed occur in multiple tumors including NSCLC (Fig. 2E and Fig. S2L) and that these are the most frequent intronic mutations observed in the TP53 gene (Fig. S2M). Among different tumor types, these mutations are particularly enriched in upper urinary tract transitional cell carcinoma (UUTCC) (Fig. S2L). Mutation analysis of 172 UUTCC samples indicated these are the most frequent TP53 mutations in this type of cancer (Fig. 2F). Given the conservation of AG residues at all splicing sites, the high frequency of mutations at the intron 6/exon 7 boundary acceptor site clearly suggests a functional selection of this particular mutation.
Several studies have demonstrated a significant correlation between the spectrum of p53 mutation and exposure to certain types of carcinogens. For example, transversions in codon 157, although uncommon in other types of cancer, are mutation hotspots in lung, breast, and head and neck cancers and are associated with smoking in lung cancer patients. In the case of UUTCC, the occurrence of carcinoma in this highly unusual location has been associated with dietary exposure to aristolochic acid (16, 17). Following metabolic activation, aristolochic acid reacts with genomic DNA to form aristolactam-DNA adducts that generate A to T transversions with high frequency in the p53 gene (17). Because all patients with tissues part of the cohort analyzed in our study had been exposed to aristolochic acid, this could explain the prevalence of c.673-2A mutations observed in this tumor compared with other.
To verify that the presence of mutation at position c.673-2A alters p53 splicing, we generated a minigene containing the genomic fragment from exons 5–8 under cytomegalovirus (CMV) promoter control (Fig. 2G). Detection and analysis of minigene-derived transcripts were achieved by RT-PCR. Consistent with the observation that normal cells preferentially express p53FL, the minigene expressed the expected transcript with the expected sequence at the exon/intron junction (Fig. 2H). However, mutation of the invariant A residue to G at the 3′ acceptor site of intron 6 resulted in the generation of a p53Ψ isoform (Fig. 2H).
To provide further evidence that the presence of intron 6 mutations could result in the generation of a p53Ψ-like transcript, we extended our analysis to a NSCLC-derived cell line (HOP62) that was reported to harbor a c.673-2A to G mutation. We verified the presence of the mutation by sequence analysis (Fig. S2O) and confirmed expression of a p53Ψ-like transcript by RT-PCR analysis using primers complementary to sequences in exon 4 and exon 7 (Fig. 2I). Western blot analysis of HOP62 cell extracts with a p53 N-terminal antibody also indicated the presence of a protein of 27 kDa, the size of the p53Ψ protein (Fig. 2L).
Multiple alternative p53 isoforms generated by virtue of alternative splicing mechanisms have been previously described (Fig. S2P) (18). To determine whether p53Ψ co-occurs with any of these p53 splicing isoforms, we performed RT-PCR analysis using oligonucleotides spanning the entire p53 coding sequence in cells harboring a WT p53 allele compare with cells harboring a homozygous c.673-2A mutation (Fig. S2P). Although we cannot exclude that in other cell types or under different experimental conditions p53Ψ may occur with other known p53 isoforms, the TA p53Ψ-α isoform was the main isoform expressed in HOP62 cells.
p53ψ Is Devoid of Transcriptional Activity.
Due to an early stop codon, the p53Ψ isoform encodes a p53 protein that is devoid of critical residues required for DNA binding, oligomerization, and localization to the nucleus (Fig. 3 A and B). Indeed, when we examined the cellular distribution of p53Ψ by immunofluorescence in HOP62 cells and in p53-null cells (H1299) ectopically expressing p53Ψ and p53FL, we detected remarkable differences (Fig. 3C and Fig. S3A). Whereas p53FL was mainly localized in the nucleus, p53Ψ was excluded from the nucleus and predominantly localized in the cytoplasm in a partly punctate pattern. We confirmed these observations by biochemical fractionation in (wtp53) A549 cells ectopically expressing p53Ψ and p53FL (Fig. 3D). As expected, p53FL was found in nuclear fractions and p53Ψ in cytoplasmic fractions.
Fig. 3.
p53ψ is devoid of transcriptional activity. (A) Schematic representation of p53ψ and p53FL. (B) Western blot analysis indicated that in A549 cells ectopic expression of the unique p53 isoform generated a protein of an apparent size of 27 kDa. (C) Immunostaining analysis revealed a predominantly cytoplasmic, partly punctate localization of p53Ψ (green). Phalloidin (red) and DAPI (blue) were used as counterstains to highlight actin fibers and the nucleus, respectively. (D) Subcellular fractionation of A549 cells expressing vector or p53ψ indicates a cytoplasmic distribution of the p53ψ protein isoform. Equal amounts of whole cell lysate (T) and cytoplasmic (C) and nuclear (N) protein fractions were analyzed by Western blotting using an antibody directed against the N-terminal domain of p53 (DO1). Tubulin and uncleaved PARP were used as controls for cytoplasmic and nuclear fractions, respectively. (E) The chart represents expression of known p53 targets (p21, Puma, Tigar) in H1299 cells ectopically expressing vector, p53ψ, and p53FL. mRNA levels were quantified by SYBR green-based real-time RT-PCR in tetracycline inducible p53-null cells (H1299) ectopically expressing p53FL or p53Ψ on induction with doxycycline (0.5 μg/mL) for 5 d. Columns represent relative expression values (P < 0.0001, Student t test). Levels of expression of p53FL and p53Ψ are provided in Fig. 3A. (F) A dual luciferase reporter assay in H1299 cells indicated that ectopic expression of p53ψ fails to activate the synthetic p53-responsive promoter p21Cip1-luc. Luciferase activity was normalized to Renilla activity. Data shown are representative of three independent experiments (P = 0.03241). Cells were treated with doxycycline (0.5 μg/mL) for 3 d before the assay. (G) p53ψ is unable to modify the transcriptional activity of p53FL. Ectopic expression of p53Ψ in cells expressing endogenous p53FL (A549 cells) did not induce expression of known p53 targets. To increase p53 activity cells were treated with the DNA damaging agent doxorubicin for 24 h. The chart represents relative mRNA levels of the indicated p53 targets on treatment with doxorubicin (1 μM). Data shown represent relative (compared with actin) expression levels (mean ± SD, n = 6; P < 0.0001, Student t test) as measured by SYBR green-based real-time PCR. Similar results were observed at steady state (Fig. 3C). Expression levels of p53FL and p53Ψ are provided in Fig. 3C.
Because the nuclear localization sequence and oligomerization and DNA binding domains of p53 are critical for sequence-specific DNA binding and transactivation of target genes (19), we reasoned that p53Ψ would be devoid of a canonical p53 transcription activity. As a first step to provide experimental evidence for this hypothesis, we ectopically expressed p53Ψ and p53FL in p53-null cells (H1299) and measured expression of known p53 targets. Although overexpression of p53FL was sufficient to augment PUMA, TIGAR, and p21 mRNA (Fig. 3E) and p21 protein levels (Fig. S3B), overexpression of p53Ψ failed to elicit such a response. These observations prompted us to directly compare p53Ψ and p53FL transcriptional activity. To this end we measured the activation of a p53-responsive promoter in cells ectopically expressing either p53FL or p53Ψ. As shown in Fig. 3F, luciferase reporter assays using a synthetic p21CIP1 promoter (e.g., p21CIP1-luc) confirmed that p53Ψ was transcriptionally inactive.
It seemed possible, however, that p53Ψ could affect p53 target gene transcription by acting in a dominant negative fashion (e.g., by titrating p53 interacting proteins). Hence we sought to determine whether ectopic expression of p53Ψ in cells expressing p53FL (A549 cells) would modify the expression of p53 target genes. Because in principle a putative dominant negative effect of p53Ψ could be promoter specific, in addition to PUMA, BAX, and p21, we extended our analysis to a broader array of p53 targets (i.e., tiger, sod2, sgo2, cycg2, sharp1, gpx1, sens1, and sens2). However, no differences in the expression of any of these p53 targets were observed in cells harboring WT p53FL on overexpression of p53Ψ, either in basal conditions (Fig. S3D) or on stimulation with doxorubicin (Fig. 3G).
In sum, based on this experimental evidence, we concluded that p53Ψ is devoid of transcriptional activity. Thereby, in principle, promoting the generation of p53Ψ at the expense of p53FL could represent a unique physiologically relevant mode to limit p53 tumor suppression function. Indeed, in cells expressing p53Ψ, we consistently observed decreased expression of p53 target genes and increased expression of genes repressed by p53 (Fig. 1C).
p53ψ, Like Certain p53 Gain-of-Function Mutations, Is Sufficient to Reprogram Epithelial Cells Toward the Acquisition of Prometastatic Features.
Somatic alterations of the p53 tumor suppressor gene located on chromosome 17p often occur in cancers and are associated with poor prognosis (20). In these tumors, p53 mutations disable p53-mediated inhibition of proliferation and promotion of apoptosis in response to stress (20). In addition, missense p53 mutations may have gain-of-function activities that lead to an increased metastatic spread (21). The observed decrease in average disease-free and overall survival times in patients with tumors expressing p53Ψ (Fig. S2G) and the high frequency of the c.673–2A to G mutation in cancer patients and in patients with UUTCC (Fig. 2F and Fig. S2M) suggests that cancer-associated p53Ψ does more than create a p53-null state. To test this hypothesis, we silenced p53Ψ expression in HOP62 cells. These mesenchymal cells are homozygous for the c.673-2A to G mutation and inherently express exclusively a p53Ψ-like isoform (Fig. 2I and Fig. S2O). On knockdown of p53Ψ in these cells, we observed phenotypic and molecular changes distinctive of cells undergoing mesenchymal to epithelial transition. Cells in which p53Ψ expression was silenced lost the elongated appearance typical of mesenchymal-like cells and instead acquired a cobblestone morphology characteristic of epithelial cells (Fig. 4A). At the molecular level, these changes were associated with increased expression of E-cadherin and diminished expression of vimentin and the master regulators of the EMT program zeb1, twist, and slug (Fig. 4B and Fig. S4A).
Fig. 4.
p53Ψ, like certain p53 gain-of-functions mutations, is sufficient to reprogram cells toward the acquisition of prometastatic features. (A) Silencing of p53Ψ in cells inherently and exclusively expressing p53Ψ (HOP62) resulted in loss of mesenchymal-like features and the acquisition of an epithelial morphology. Representative pictures of cells 4 d after transfection with a mixture of two independent siRNA oligonucleotides targeting p53 are shown. Knockdown efficiency is shown as part of the RT-PCR analysis in B. (B) The chart represents qRT-PCR analysis of the canonical EMT markers E-cadherin (ECAD) and vimentin (VIM), as well as the EMT master regulators Slug, Twist, and Zeb1 in HOP62 cells on inhibition of p53 with two different siRNAs. No difference in Snail expression was observed. Data shown represent relative (compared with actin) expression levels (mean ± SD, n = 6; P < 0.0001, Student t test) as measured by SYBR green-based real-time RT-PCR. Note these cells do not express p53FL. (C) Ectopic expression of p53Ψ resulted in the acquisition of morphological features characteristic of cells undergoing EMT. Scanning electron micrographs of representative MCF7 and A549 cells are shown. Level of expression of p53FL and p53Ψ in MCF7are provided in Figs. S4B and S3E, respectively. (D) The chart represents qRT-PCR analysis of the canonical EMT markers E-cadherin (ECAD) and vimentin (VIM), as well as the EMT master regulators Snail, Slug, Twist, and Zeb1 in H1299 cells ectopically expressing p53Ψ or p53FL. Data shown represent relative (compared with actin) expression levels (mean ± SD, n = 6; P < 0.0001, Student t test) as measured by SYBR green-based real-time RT-PCR. (E and F) Expression of p53Ψ in A549 cells leads to increased cell motility and invasion. In E, the percentage of closure of a wound at the indicated time points in a 2D cell monolayer is depicted. Each bar is the average of four individual wounds. The histogram shows the mean value ± SD (P ≤ 0.0001 by Student t test). The invasion potential of cells in F was determined in a standard Matrigel invasion assay. Filter chambers were coated with 40 µL Matrigel and invasion was assessed after 30 h. TGF-β–treated cells were used as a positive control. Motility and invasion were determined in A549 cells after induction for 5 d with doxycycline (0.5 μg/mL).
Conversely, ectopic expression of p53Ψ was sufficient to induce morphological (Fig. 4C) and molecular (Fig. 4D and Fig. S4) changes typical of cells undergoing EMT. These changes were independent of their preexisting p53 status, as cells that expressed p53FL (MCF7) and p53-null cells (H1299) both showed decreased expression of E-cadherin but enhanced expression of vimentin, snail, zeb1, twist, and slug on p53Ψ expression (Fig. 4D and Fig. S4C).
TP53 is a tumor suppressor whose FL ectopic expression transcriptionally activates stress responses. Although p53Ψ is devoid of transcriptional activity, to further exclude the possibility that the phenotypes we observed were due a selection mechanism in response to cellular stress, we tested the effect of p53Ψ expression in a tetracycline inducible A549-based cell line. Like cells that constitutively express p53Ψ, transient expression of p53Ψ in these cells was also able to reduce the expression of E-cadherin (Fig. S4 C and D). Of note, changes in E-cadherin were also apparent when analyzing its distribution. Whereas in A549 cells at confluence, E-cadherin was mostly localized at cell-cell junctions, in A549 cells ectopically expressing p53Ψ, E-cadherin was mainly localized in the cytoplasm (Fig. S4E).
In general, cells acquiring mesenchymal-like features tend to be more motile and more invasive. Indeed, when we scored cell migration in vitro in a standard wound-healing assay, A549 cells expressing p53Ψ closed the cell monolayer opening more rapidly than cells expressing p53FL (Fig. 4E). Similarly, A549 cells expressing p53Ψ had enhanced capability to migrate through a complex extracellular matrix compared with cells expressing p53FL (Fig. 4F). The EMT transition, increased motility, and cell invasion are important hallmarks of metastatic cells (22, 23). Hence, in principle, the observed decrease in average disease-free and overall survival times in patients with tumors expressing p53Ψ (Fig. 2C) supports a general relevance of p53Ψ in reprogramming cells toward the acquisition of prometastatic features.
Mitochondrial Localization of p53Ψ Is Required for Induction of the Epithelial to Mesenchymal Transition.
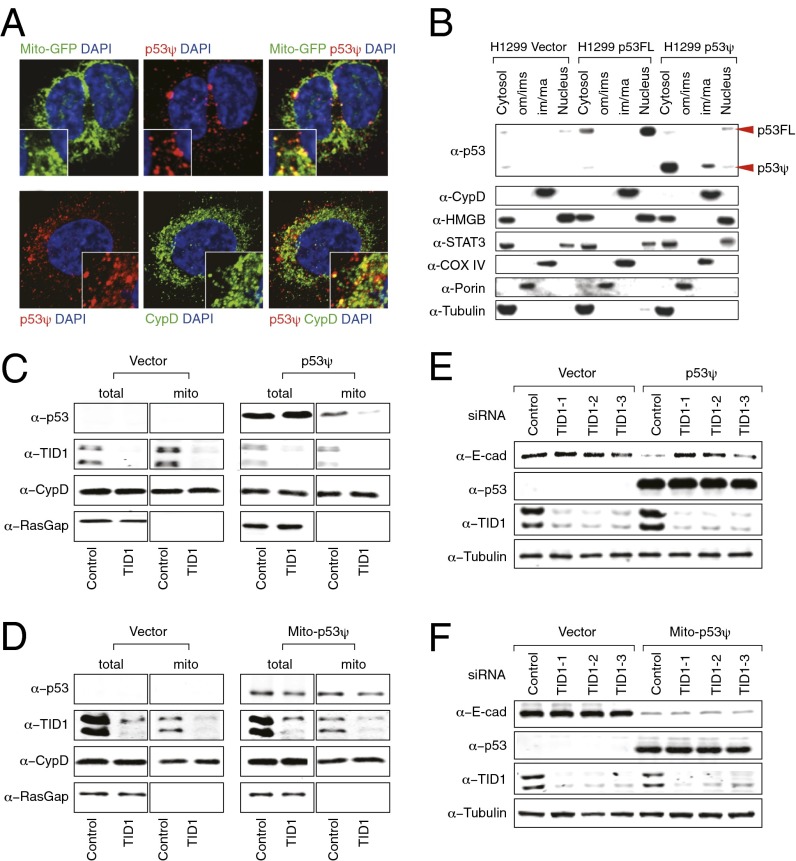
In the case of endogenous p53FL, p53 mitochondrial localization has been observed under stress conditions and on MDM2-induced p53 ubiquitination (24, 25). In particular, a growing body of evidence has highlighted the importance of p53 localization to the mitochondria in mediating certain transcription-independent activities of p53 (24, 26, 27). Because p53Ψ was entirely excluded from the nucleus and devoid of transcriptional activity, we undertook studies to determine whether p53Ψ was localized to the mitochondria and, if so, whether this was required for the p53Ψ-induced epithelial to mesenchymal transition. Of note, when we examined the subcellular distribution of p53Ψ by immunofluorescence and biochemical fractionation, we observed that p53Ψ was partially localized into the mitochondrial matrix under basal growing conditions (Fig. 5 A and B).
Fig. 5.
Mitochondrial localization of p53Ψ is required for p53ψ-induced epithelial to mesenchymal transition. (A) Immuno-staining analysis of H1299 cells revealed a partial mitochondrial localization of p53Ψ (red). Mitochondrial GFP (pseudocolored in green in Upper) and cyclophilin D (CypD), a mitochondrial matrix protein (pseudocolored in green in Lower) were used as counterstains to highlight the mitochondria. The cell nuclei were stained with DAPI (blue). (B) Western blot analysis of H1299 cells was used to characterize the different submitochondrial fractions, indicating localization of p53Ψ within the inner membrane/matrix fraction (im/ma). CypD and COX IV staining were used to control for purity of the inner membrane/matrix fraction, the high-mobility group box 1 (HMGB) antibody for the nuclear fraction, PORIN antibody for the outer membrane fraction, and TUBULIN antibody for the cytosolic fraction. (C and D) TID-1 is required for localization of p53Ψ in the mitochondria. Biochemical fractionation followed by Western blot analysis was used to analyze the distribution of (C) p53Ψ and (D) mito-p53Ψ on TID-1 knockdown in p53Ψ-expressing A549 cells. At 72 h after transfection with Tid-1–specific siRNA, p53Ψ mitochondiral localization was determined. Analyses of CypD (a mitochondrial matrix protein) and p120 RasGAP (a cytoplasmic protein) localizations were used as controls for purity of the mitochondrial fractions. (E and F) Western blot analysis of protein extracts from A549 cells ectopically expressing (E) p53Ψ and (F) mito-p53Ψ on inhibition of Tid1 expression indicate that Tid-1 is required for p53Ψ-induced reduction of E-cadherin levels.
In eukaryotic organisms, about 10–15% of nuclear genes encode mitochondrial proteins (28). These proteins are synthesized in the cytosol and are then translocated to the mitochondrial inner or outer membranes or to the mitochondrial intermembrane space or matrix (28). Although many proteins that translocate to the mitochondrial matrix possess an N-terminal targeting sequence called matrix-targeting sequence (MTS), many mitochondrial precursors do not contain an MTS. For precursors without an MTS, chaperone proteins may stabilize and assist in their transport to mitochondria (28, 29). In the case of p53, the chaperone protein Tid1 has been shown to interact with the N-terminal domain of p53 and to mediate its translocation into the mitochondria matrix (30, 31). Tid1, also known as mitochondrial Hsp40 (mtHsp40), is the mammalian homolog of the Drosophila tumor suppressor Tid56 (32). Tid1 contains a conserved DnaJ domain through which it interacts with the cytosolic Hsp70 family of chaperone proteins that are also engaged in mitochondrial transport of MTS-deficient proteins (32). DnaJ-like proteins function as cochaperones with DnaK-like ATPases to promote the (un)folding and translocation of polypeptides.
To determine whether Tid1 is involved in the translocation of p53Ψ to mitochondria, we silenced Tid1 and determined its effect on the subcellular localization of p53Ψ. Biochemical fractionation indicated that decreased expression of Tid1 was sufficient to reduce the amount of p53Ψ transported into the mitochondria without affecting the localization of CypD, a protein localized in the mitochondrial matrix via an MTS (Fig. 5C). In further support of Tid1-mediated p53Ψ mitochondrial localization, we generated a p53Ψ construct that was constitutively localized to the mitochondria independent of Tid1 by virtue of the presence of an N-terminal MTS tag (mito-p53Ψ). As predicted, the presence of an MTS sequence was sufficient to overcome the effect of Tid1 silencing (Fig. 5D).
The observation that Tid1 was required for p53Ψ mitochondrial localization prompted us to determine whether the translocation of p53Ψ into the mitochondrial matrix was in fact required for p53Ψ-induced EMT. Indeed, silencing of Tid1 in cells expressing p53Ψ resulted in a failure of p53Ψ to decrease the expression of E-cadherin and in a decreased p53Ψ-mediated increase in cell motility (Fig. 5E and Fig. S5A). Notably, no difference in E-cadherin expression was observed in cells in which Tid1 was silenced but p53Ψ was not expressed (i.e., vector-transfected cells). Because the effect of Tid1 silencing on the regulation of EMT could be independent of p53Ψ, we forced the translocation of p53Ψ into the mitochondrial matrix in a Tid1-independent manner (i.e., using mito-p53Ψ). Even in the absence of Tid1, mito-p53Ψ–expressing cells were characterized by decreased E-cadherin levels (Fig. 5F). On the basis of this evidence, we concluded that under basal conditions, p53Ψ is partially localized to the mitochondrial matrix in a Tid1-dependent fashion and that localization to the mitochondria is both necessary and sufficient for induction of an EMT phenotype by p53Ψ.
p53ψ Interaction with Cyclophilin D Is Sufficient to Increase Mitochondrial Pore Permeability and Reactive Oxygen Production.
When localizing to the mitochondria under apoptotic conditions, p53 modulates the activities of antiapoptotic (Bcl-xL and Bcl-2) and proapoptotic (BAK/BAX) members of the Bcl-2 family to regulate the integrity of the outer mitochondrial membrane (27). p53FL interactions with Bcl-xL/Bcl-2 on the one hand and BAK on the other hand result in BAK oligomerization with subsequent outer membrane permeabilization (MOMP) and release of cytochrome C and other proapoptotic factors into the cytoplasm, mediating apoptosis (27). Within the matrix, p53 has been shown to interact with MnSOD, the primary antioxidant enzyme in mitochondria, and with CypD, an obligatory activator of the mPTP that is closed in healthy cells. To determine whether p53Ψ retained interacts with any of these proteins, we conducted immunoprecipitation experiments. Because CypD is localized in the mitochondrial matrix, we used as input materials fractions containing outer mitochondrial membrane/intermembranous space and inner mitochondrial membrane/matrix. We found that mitochondrial p53Ψ was unable to bind to BAX, BAK, and MnSOD (Fig. S5A) but reproducibly interacted with CypD (Fig. 6A). Consistent with a failure to interact with BAX and BAK, cells expressing p53Ψ were viable and did not release cytochrome C from their mitochondria (Fig. S5E).
Fig. 6.
p53ψ interaction with cyclophylin D is sufficient to increase the mPTP pore permeability and reactive oxygen production. (A) Immunoprecipitation analysis indicates an interaction between p53Ψ and CypD in the mitochondrial fraction. The mitochondrial fraction of A549 ectopically expressing p53Ψ was immunoprecipitated with a CypD-specific antibody and probed with a p53 N-terminal antibody or as controls with CypD and Smurf1 antibodies. (B) Schematic of calcein AM assay. (C) The pictures depict representative fluorescence microscopy images. A549 cells were loaded with calcein at 10 nM, and fluorescence was detected by laser confocal microscopy after 15 min. Nonmitochondrial calcein fluorescence was quenched by cotreatment with CoCl2. Treatment with the ionophore ionomycin (50 nM) is shown as control. Cyclosporin A (CsA) was used at 2 mM. (D) The bars represent the percentage of drop of calcein fluorescence on quenching of cytosolic calcein with CoCl2 in three independent experiments. Median calcein fluorescence was assessed by FACS. Calcein was loaded at 10 nM and detected at 515 nM after 15 min on excitation with Red HeNe at 495 nM. Treatment with ionomycin was used as control to estimate basal fluorescence. FACS plots are provided in Fig. S6. (E) Representative fluorescence microscopy images of A549 cells loaded with MitoSOX (1 μM). Images show the fluorescence on excitation at 390 nm, mainly from the hydroxyethidium derivative (red). DAPI (blue) was used as a counterstain. (F) The histogram represents the quantification of the MitoSox-positive fractions in three independent FACS experiments from E.
Vaseva et al. previously showed that the interaction with CypD is mediated by a part of the DNA binding domain that is fully retained in p53Ψ (24). To determine whether its residual DNA binding domain (amino acids 102–243) mediated the p53Ψ interaction with CypD and whether the interaction is direct, we performed pull-down experiments. As shown in Fig. S5C, glutatione S-transferase (GST)-tagged CypD selectively precipitated recombinant p53Ψ; hence, the interaction appears to be direct.
Having shown that p53Ψ was able to bind directly to CypD, we next explored a possible functional role for the p53Ψ/CypD interaction in cells. Because the only known activity of CypD is regulation of the mPTP opening, we measured the effect of p53Ψ on the mitochondrial permeability transition using calcein release, a highly selective indicator of sustained PTP opening in situ (33, 34). Of note, MOMP and the Bax/Bak lipid pore are completely incompetent for calcein release (35). In this assay, cells are loaded with calcein (acetomethylester) together with its quencher cobalt ions. Calcein freely diffuses throughout the cell including into the mitochondrial matrix. However, the cobalt quencher cannot diffuse across the inner mitochondrial membrane, therefore quenching Calcein fluorescence everywhere except in the mitochondrial matrix. Thus, calcein fluorescence comes only from the matrix. Only on PTP opening will cobalt ions gain access to the matrix and quench calcein, resulting in a sharp drop of mitochondrial fluorescence, which can be measured by FACS or under the microscope (Fig. 6B). Notably, cells expressing p53Ψ exhibited increased mPTP permeability relative to cells that did not express this p53 isoform (Fig. 6 C and D). This effect was CypD dependent, because treatment with cyclosporine A (CsA), a highly selective pharmacological inhibitor of CypD, reduced CoCl2-mediated quenching of calcein fluorescence (Fig. 6 C and D). Although CsA also inhibits other cytosolic cyclophilins in addition to mitochondrial matrix-specific CypD, none play any role in the regulation of the mPTP pore opening. As a control, cells that had been loaded with calcein AM were treated with the ionophore ionomycin. In the presence of ionomycin, we observed a rapid loss of mitochondria calcein fluorescence in all experimental settings (Fig. 6 C and D).
An increase in time that mPTP is in the open state will result in an increased outflow of electrons with increased accumulation of ROS inside the mitochondria (36). To assess levels of superoxide in the mitochondria of live cells, we used the fluorogenic dye MitoSOX (37). When added to cells, the MitoSOX reagent is rapidly and selectively targeted to mitochondria, where MitoSOX is oxidized by superoxide. When exited with a light at 390-nm frequencies, it emits red fluorescence. On loading with MitoSOX, cells expressing p53Ψ exhibited increased MitoSOX staining compared with p53FL-expressing cells and/or vector control (Fig. 6 E and F). Consistent with increased mPTP opening mediated by the interaction of p53Ψ with CypD, the increased MitoSOX fluorescence was again decreased on inhibition of CypD with CsA (Fig. 6 E and F).
Recently, Vaseva et al. reported that on oxidative stress, p53FL translocates to the mitochondrial matrix and triggers sustained mPTP opening by engaging in a physical interaction with CypD, thereby inducing necrotic cell death (26). Confirming Vaseva et al., when we forced p53FL into the mitochondrial matrix by generating an N-terminal MTS fusion protein, we observed massive cell death. However, this was not the case when we expressed either mitochondria-targeted p53 gain-of-function mutants or p53Ψ either in p53-null cells or in cells expressing p53FL (Fig. S5D). We did not observe major differences in CypD binding or in mitochondrial permeability when p53FL, p53Ψ, and p53 gain-of-function mutants were compared, suggesting that interaction with CypD and/or an increased mitochondrial permeability, although required for mitochondrial p53FL-mediated cell death, is not sufficient but requires oxidative damage.
CypD and ROS Are Required for EMT Induction by p53ψ and p53 Gain-of-Function Mutants.
Although initially considered to be detrimental to cells, recent studies suggest that ROS are necessary for the activation of multiple cellular pathways and for induction of different cell states (38, 39). Specifically, increased levels of ROS induce EMT and increase the metastatic competence of cells (40). To determine whether CypD and ROS have a causative role in EMT mediated by p53Ψ, we modulated CypD and ROS levels in cells ectopically expressing p53Ψ (A549, MCF7, and H1299) and determined their effects on expression of EMT markers and cell invasion. Silencing CypD with either of two siRNAs (Fig. 7A) or pharmacologically inhibiting CypD by treating cells with CsA (Fig. 7B and Fig. S6A) was sufficient to prevent EMT and to diminish cell motility in cells expressing p53Ψ (Fig. 7C). As in the case of CypD, inhibition of ROS production with N-acetyl cysteine (NAC) or Tempol, two well-known ROS scavengers, augmented expression of E-cadherin (Fig. 7E and Fig. S6C) and decreased invasive capabilities of p53Ψ-expressing cells compared with control cells (Fig. S6D). In contrast, low doses of H2O2 decreased the expression of E-cadherin without compromising cell viability (Fig. 7D).
Fig. 7.
Cyclophylin D and ROS are required for EMT induction by p53ψ. (A) Western blot analysis of A549 cells ectopically expressing p53Ψ on transfection with two independent siRNA targeting cyclophilin D (CypD). (B) Treatment with CsA, a highly specific and potent pharmacological inhibitor of CypD, is sufficient to restore expression of E-cadherin to a level similar to that observed in control cells and to reduce expression of EMT markers in cells ectopically expressing p53Ψ. The chart represents qRT-PCR analysis of the canonical EMT markers E-cadherin (ECAD) and vimentin (VIM), as well as the EMT master regulators Snail, Slug, Twist, and Zeb1 in H1299 cells ectopically expressing p53Ψ on treatment for 5 d with 2 mM CsA. Data shown represent relative (compared with actin) expression levels (mean ± SD, n = 6; P < 0.0001, Student t test) as measured by SYBR green-based real-time RT-PCR. Similar results were observed in MCF7 cells. (C) The chart represents the motility of A549 cells ectopically expressing p53FL or p53Ψ after treatment for 5 d with CsA. Cell motility was measured in a standard wound healing experiment as previously described. The charts indicate the percentage of closure at 48 h in the presence or absence of 2 mM CsA. Each bar is the average of four individual wounds. The histogram shows the mean value ± SD (P ≤ 0.0001 by Student t test). (D) Treatment with low but increasing concentrations of H2O2 is sufficient to decrease expression of E-cadherin to levels similar to those observed in cell lines ectopically expressing p53Ψ. mRNA levels of E-cadherin were assessed by SYBR green-based real-time RT-PCR on treatment for 5 d with H2O2. Data shown represent relative expression compared with vector (mean ± SD, n = 6; P < 0.0001, Student t test). (E) Reduction of ROS levels is sufficient to enhance expression of E-cadherin levels in cells expressing p53Ψ. Cells were treated for 5 d with 10 mM NAC. Similar results were obtained in cells treated with Tempol, another ROS scavenger (Fig. S6C). (F) Schematic of proposed mechanism to explain p53Ψ-induced EMT. On acute oxidative stress, p53FL was previously shown to interact with CypD and trigger necrotic cell death by opening mPTP pore (Vaseva Cell 2012, Left).
To exclude the possibility that ROS-induced EMT was independent of CypD in p53Ψ-expressing cells, we performed epistasis experiments. Specifically, we measured E-cadherin expression on manipulation of ROS levels (e.g., by NAC treatment) in cells ectopically expressing p53Ψ in the presence and absence of CypD (Fig. S6B). We reasoned that if CypD and p53Ψ act on parallel pathways, we would observe an additive effect of CypD silencing and ROS inhibition. In contrast, if p53Ψ acts solely through CypD, then CypD silencing would be sufficient to reduce ROS levels and no additive effect would be observed. In agreement with our original hypothesis, silencing of CypD blocked p53Ψ-induced EMT, and treatment of cells with NAC did not increase E-cadherin expression in cells in which CypD was silenced (Fig. S6B). Together these data strongly support a model in which the p53Ψ interaction with CypD increases mPTP permeability and increases ROS production that in turn is necessary and sufficient for p53Ψ-induced EMT (model in Fig. 7F, Right).
Discussion
In summary, we provide evidence describing a unique, evolutionarily conserved mode of p53 regulation involving alternative splicing of the TP53 mRNA. The use of an alternative 3′ splice site in intron 6 generates a previously uncharacterized p53 isoform. The protein product of this alternatively spliced mRNA, p53Ψ, is incapable of sequence-specific DNA binding and transactivation of canonical p53 target genes but does induce the acquisition of mesenchymal-like characteristics. Because p53Ψ is still capable of binding to CpD and to regulate the mitochondria pore permeability, we propose that p53Ψ encodes a separation-of-function isoform. Importantly, our data also indicate that the cytoplasmic p53 activity is sufficient to reprogram cells toward acquisition of features associated with prometastatic phenotypes.
At the molecular level, we provide evidence suggesting that p53Ψ-induced EMT does not depend on p53 transcriptional activity but instead relies on its mitochondrial matrix localization and its physical interaction with mPTP regulator CypD. In particular we found that ROS play a pivotal role as second messengers in mediating p53Ψ-induced EMT.
Regulation of ROS by p53 is not a novel concept. In fact, p53 has been shown to regulate the expression of many genes (e.g., Pig3, Tigar, Sens1, Sens2, Gpx1, Sco2, Bax, and Puma) that modify the production of ROS in cells. Although p53Ψ ectopic expression failed to modify the expression of any of these genes, the fact that p53Ψ is generated at the expense of p53FL indicates that in cells in which p53Ψ is generated as a result of an alternative splicing events, ROS production can in principle be attributed to both a CypD-dependent mechanism and to changes in the expression of these p53FL target genes. Indeed, in cells that inherently express p53Ψ (CD44high/CD24low cells sorted from injured lung), we observed decreased expression of these p53 targets.
The observation that p53Ψ is inherently expressed in tumors and during tissue injury is particularly intriguing. Generation of cells bearing characteristics of those observed in wound healing has been previously described in tumors and has been suggested to reflect a maintenance in physiologic response of tumors to regulatory pathways governing the tissue from which they are derived (41, 42). In the 1860s, Virchow famously referred to tumors as “wounds that never heal” (43). Hence, the remarkably similar activities of p53Ψ and certain p53 mutants, which also reduce E-cadherin expression and facilitate invasion in a CypD-dependent manner, suggest that these p53 mutations highjack a highly regulated program that during tissue injury leads to the generation of p53Ψ. In principle, this implies a possible physiological origin for certain p53 gain-of-function mutants.
Materials and Methods
Cell Culture.
A549, H1299 (NCI-H1299), MCF7, PC9, H460 (NCI-H460), H4006 (NCI-H4006), and Phoenix-AMPHO cells were obtained from the American Type Culture Collection (ATCC) repository. The Hop62 cells were obtained from the National Cancer Institute (NCI) repository. All of the cell lines except for Phoenix-AMPHO, HEK293T, and MCF7 were cultured in RPMI supplemented with 5% (vol/vol) FBS, glutamine, penicillin, and streptomycin. Phoenix-AMPHO, HEK293, and MCF7 cells were cultured in DMEM containing 10% (vol/vol) FBS, penicillin, streptomycin, and sodium pyruvate.
Antibodies and Reagents.
The following antibodies were used in this study: mouse anti-E-cadherin antibody (BD Transduction Laboratories), anti-p53 antibody (DO-1; Calbiochem), mouse anti–β-tubulin antibody (2-28-33; Santa Cruz Biotechnology), rabbit anti-PARP antibody (46D11; Cell Signaling Technology), rabbit anti-BAX antibody (D2E11; Cell Signaling Technology), rabbit anti-PUMA antibody (#4976; Cell Signaling Technology), rabbit anti-p21waf1/cip1 antibody (12D1; Cell Signaling Technology), anti-CpD (ab110324; Abcam), anti-CD31 (clone 390; EBioscience), anti-CD45 (Clone 30-F11; EBioscience), anti-CD24 (Clone MI-69; EBioscience), anti-CD44 (Clone IM-7; Biolegend), and Tid-1 (Neomarkers MS-1564-P0). The chemical reagents used for cell treatment were cyclosporin A from Sigma-Aldrich (30024), calcein AM and MitoSox from Molecular Probes/Life Technologies, H2O2 and doxorubicin hydrochloride from Sigma-Aldrich (#D1515), NAC from Sigma-Aldrich (A9165), and tempol from Tocris (#3082). p53 siRNA (GGGTTAGTTTACAATCAGC; GGTGAACCTTAGTACCTAA), Tid1 siRNA (CTACATCCACATCAAGATA;GAAAGCCTATTATCAGCTT; AGCGAGTGATGATCCCTGT), and CypD siRNA (AGGCAGATGTCGTCCCAAA; CGACTTCACCAACCACAAT) were purchased from Invitrogen.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11576.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321640111/-/DCSupplemental.
References
- 1.Junttila MR, Evan GI. p53—A Jack of all trades but master of none. Nat Rev Cancer. 2009;9(11):821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 2.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450(7170):721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 3.Aldaz CM, et al. Serial analysis of gene expression in normal p53 null mammary epithelium. Oncogene. 2002;21(41):6366–6376. doi: 10.1038/sj.onc.1205816. [DOI] [PubMed] [Google Scholar]
- 4.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 5.Meletis K, et al. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133(2):363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 6.Godar S, et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134(1):62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61(7):1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 8.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 9.Weber GF, et al. Absence of the CD44 gene prevents sarcoma metastasis. Cancer Res. 2002;62(8):2281–2286. [PubMed] [Google Scholar]
- 10.Ponta H, Sherman L, Herrlich PA. CD44: From adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 11.Buckpitt A, et al. Naphthalene-induced respiratory tract toxicity: Metabolic mechanisms of toxicity. Drug Metab Rev. 2002;34(4):791–820. doi: 10.1081/dmr-120015694. [DOI] [PubMed] [Google Scholar]
- 12.Kloek AP, McCarter JP, Setterquist RA, Schedl T, Goldberg DE. Caenorhabditis globin genes: Rapid intronic divergence contrasts with conservation of silent exonic sites. J Mol Evol. 1996;43(2):101–108. doi: 10.1007/BF02337354. [DOI] [PubMed] [Google Scholar]
- 13.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2(4):466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 16.Chen CH, et al. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci USA. 2012;109(21):8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grollman AP, et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci USA. 2007;104(29):12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourdon JC, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19(18):2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batinac T, et al. Protein p53—structure, function, and possible therapeutic implications. Acta Dermatovenerol Croat. 2003;11(4):225–230. [PubMed] [Google Scholar]
- 20.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 21.Hanel W, et al. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20(7):898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Zhou BP. New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 2008;40(7):643–650. doi: 10.1111/j.1745-7270.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 24.Vaseva AV, et al. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149(7):1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26(4):923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mihara M, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11(3):577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 27.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787(5):414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 29.Yogev O, Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta. 2011;1808(3):1012–1020. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Trinh DL, Elwi AN, Kim SW. Direct interaction between p53 and Tid1 proteins affects p53 mitochondrial localization and apoptosis. Oncotarget. 2010;1(6):396–404. doi: 10.18632/oncotarget.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn BY, et al. Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene. 2010;29(8):1155–1166. doi: 10.1038/onc.2009.413. [DOI] [PubMed] [Google Scholar]
- 32.Lu B, Garrido N, Spelbrink JN, Suzuki CK. Tid1 isoforms are mitochondrial DnaJ-like chaperones with unique carboxyl termini that determine cytosolic fate. J Biol Chem. 2006;281(19):13150–13158. doi: 10.1074/jbc.M509179200. [DOI] [PubMed] [Google Scholar]
- 33.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87(1):99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 34.Gillessen T, Grasshoff C, Szinicz L. Mitochondrial permeability transition can be directly monitored in living neurons. Biomed Pharmacother. 2002;56(4):186–193. doi: 10.1016/s0753-3322(02)00184-1. [DOI] [PubMed] [Google Scholar]
- 35.Petronilli V, et al. Imaging the mitochondrial permeability transition pore in intact cells. Biofactors. 1998;8(3-4):263–272. doi: 10.1002/biof.5520080314. [DOI] [PubMed] [Google Scholar]
- 36.Batandier C, Leverve X, Fontaine E. Opening of the mitochondrial permeability transition pore induces reactive oxygen species production at the level of the respiratory chain complex I. J Biol Chem. 2004;279(17):17197–17204. doi: 10.1074/jbc.M310329200. [DOI] [PubMed] [Google Scholar]
- 37.Robinson KM, et al. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci USA. 2006;103(41):15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Belle JE, et al. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8(1):59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurd TR, DeGennaro M, Lehmann R. Redox regulation of cell migration and adhesion. Trends Cell Biol. 2012;22(2):107–115. doi: 10.1016/j.tcb.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes LR, Terra LF, Sogayar MC, Labriola L. Epithelial-mesenchymal transition: implications in cancer progression and metastasis. Curr Pharm Biotechnol. 2011;12(11):1881–1890. doi: 10.2174/138920111798377102. [DOI] [PubMed] [Google Scholar]
- 42.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18(6):884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virchow R. Die Cellularpathologie in Ihrer Begründung auf Physiologische und Pathologische Gewebelehre. Berlin: A. Hirschwald; 1858. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.