Abstract

The final step in lanthipeptide biosynthesis involves the proteolytic removal of an N-terminal leader peptide. In the class I lanthipeptide epilancin 15X, this step is performed by the subtilisin-like serine peptidase ElxP. Bioinformatic, kinetic, and mass spectrometric analysis revealed that ElxP recognizes the stretch of amino acids DLNPQS located near the proteolytic cleavage site of its substrate, ElxA. When the ElxP recognition motif was inserted into the noncognate lanthipeptide precursor NisA, ElxP was able to proteolytically remove the leader peptide from NisA. Proteolytic removal of the leader peptide by ElxP during the biosynthesis of epilancin 15X exposes an N-terminal dehydroalanine on the core peptide of ElxA that hydrolyzes to a pyruvyl group. The short-chain dehydrogenase ElxO reduces the pyruvyl group to a lactyl moiety in the final step of epilancin 15X maturation. Using synthetic peptides, we also investigated the substrate specificity of ElxO and determined the 1.85 Å resolution X-ray crystal structure of the enzyme.
Lanthipeptides are (methyl)lanthionine-containing peptides that belong to a growing class of natural products known as ribosomally synthesized and post-translationally modified peptides (RiPPs).1 As with most other RiPPs, lanthipeptides are synthesized as a precursor peptide (LanA) composed of an N-terminal leader peptide and a C-terminal core region harboring the different post-translational modification sites. When the posttranslational modifications result in a product with antimicrobial activity, the lanthipeptide is called a lantibiotic.2 The (methyl)lanthionine residues in lanthipeptides are installed in a two-step biosynthetic process. First, a lanthipeptide dehydratase catalyzes the elimination of water from Ser and Thr residues in the LanA core region to yield dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively. A lanthipeptide cyclase then catalyzes the Michael-type addition of cysteinyl thiols onto the dehydroamino acids. Following the modifications of the C-terminal core peptide, the modified precursor peptide is usually processed by a lanthipeptide peptidase that removes the N-terminal leader peptide (Figure 1a). Failure to remove the leader peptide usually results in a final product devoid of biological activity.3−5 In the case of epilancin 15X, a lanthipeptide produced by Staphylococcus epidermidis 15X154 that is remarkably potent against antibiotic-resistant strains of S. aureus and Enterococcus faecalis,6 leader peptide removal exposes an N-terminal Dha on the post-translationally modified core peptide. This Dha hydrolyzes to the corresponding pyruvyl group (Pyr), and the short chain dehydrogenase ElxO then reduces the ketone of the Pyr group to generate an N-terminal lactyl moiety (Lac) in the final step of maturation (Figure 1b).7 Here, we characterized the substrate specificity of the peptidase ElxP that removes the leader peptide during the biosynthesis of epilancin 15X.7 Our results reveal the importance of conserved residues in the precursor peptide ElxA for efficient cleavage by ElxP and provide insights into the substrate specificity of LanP enzymes. In addition, we also investigated the substrate specificity of the dehydrogenase ElxO and report its X-ray crystal structure.
Figure 1.
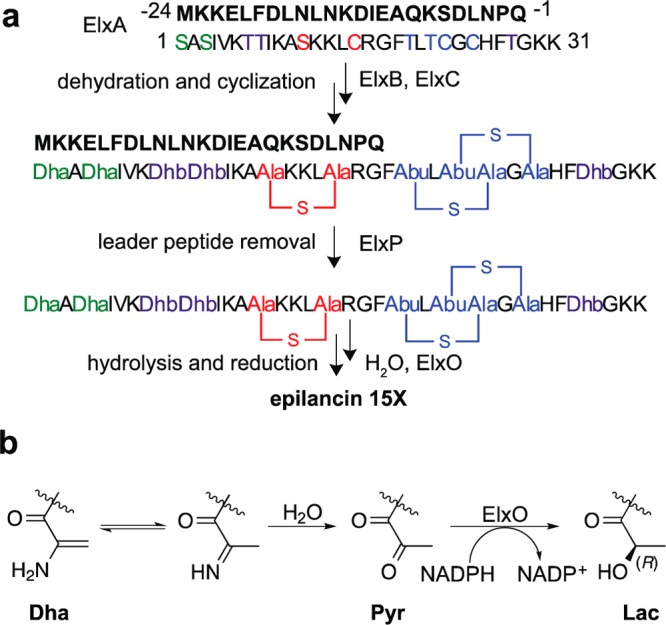
(a) Biosynthesis of epilancin 15X, involving dehydration of Ser and Thr residues by ElxB to yield dehydroalanine (Dha, green) and dehydrobutyrine (Dhb, purple), formation of lanthionine (red) or methyllanthionine (blue) rings catalyzed by ElxC, removal of the leader peptide by the peptidase ElxP, and (b) reduction of the N-terminal pyruvyl moiety catalyzed by ElxO. Abu (2-aminobutyric acid), Pyr (pyruvyl), Lac (lactyl). The leader peptide is shown in bold font.
Lanthipeptides are classified into four classes (class I–IV) on the basis of differences in the biosynthetic machinery responsible for installing the (methyl)lanthionines.8,9 At present, three types of peptidases that are involved in the removal of the N-terminal leader peptide have been identified. In class II lanthipeptides, a bifunctional enzyme termed LanT removes the leader peptide. LanTs share a conserved N-terminal papain-like cysteine peptidase domain and are members of the ATP-binding cassette transporter, maturation, and secretion (AMS) proteins.10−12 Their main function is to transport the modified precursor peptide outside the cell with concomitant proteolysis of the N-terminal leader peptide at a highly conserved Gly–Gly motif.10,13−16 Recently, it was shown that the leader peptides in some class III lanthipeptides are removed by prolyl oligopeptidases.17 The peptidase involved in flavipeptin biosynthesis only processed the modified precursor substrate suggesting that the prolyl oligopeptidases recognize the modified core peptide.17
In class I lanthipeptides, the removal of the leader peptide is performed by a subtilisin-like serine peptidase termed LanP. In contrast to LanTs, little is known about the interactions that govern the substrate specificity of LanPs. A homology model of NisP, the peptidase involved in nisin biosynthesis, suggested that the substrate specificity of NisP relies on electrostatic and hydrophobic interactions between the S1/S4 NisP pockets and the residues in the −1 and −4 positions of nisin’s precursor peptide NisA (Figure 2a; negative numbers are used for the residues in the leader peptide counting back from the protease cleavage site).18 Mutating these two positions in the leader peptide of nisin precluded the removal of the leader peptide as demonstrated by the in vivo accumulation of post-translationally modified NisA with the leader peptide still attached.3 In addition, Kuipers and co-workers showed that NisP only removes the N-terminal leader peptide from the modified precursor peptide NisA and not from linear NisA.19 Whereas these studies provided the first insights into LanP enzymes, in general, our understanding of the substrate specificity of LanP enzymes is still limited in part because of the lack of detailed in vitro studies as a result of the intrinsic low expression and poor solubility associated with these enzymes.20−22 We report here in vitro studies on the substrate specificity of ElxP.
Figure 2.
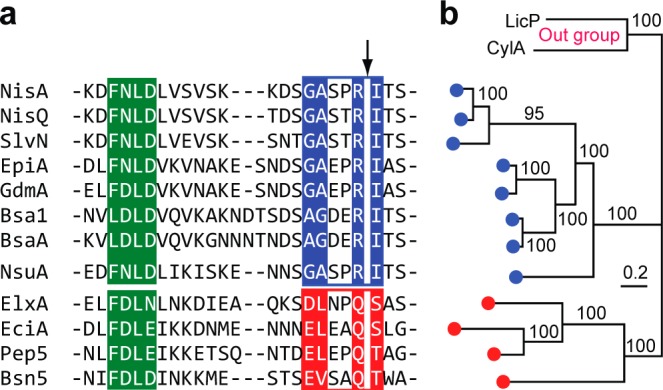
(a) Sequence alignment of selected LanA leader peptides for which the final products (class I lanthipeptides) have been structurally characterized. The FxLx motif is highlighted in green. The putative LanP recognition motifs are shown in blue and red boxes for the NisA-group and the ElxA-group, respectively. LanP cleavage sites are shown with an arrow. (b) MCMC phylogenetic tree of LanP enzymes corresponding to the LanA substrates shown in part a. Bayesian inferences of posterior probabilities are shown above or below the branches. Two LanPs involved in class II lanthipeptide biosynthesis (LicP for lichenicidin and CylA for cytolysin) served as the out group of the tree.
Results and Discussion
Determination of ElxP Substrate Specificity
To analyze the substrate specificity of ElxP, the sequences of several LanAs from class I lanthipeptides were aligned. The alignment clearly shows that the sequences near the proteolytic cleavage site group into two different types (Figure 2a). The first group, from here on named NisA-group, contains a conserved G/A-A/G-x-x-R motif at the C-terminus of the leader peptide and a Ile residue at the first position of the core peptide. The second group, from here on named ElxA-group, contains a conserved E/D-L/V-x-x-Q motif at the C-terminus of the leader peptide and a dehydratable residue (Ser/Thr) as the first amino acid of the core peptide. To investigate whether LanP enzymes are correlated with this grouping of LanAs, a Markov chain Monte Carlo (MCMC) phylogenetic tree of LanPs was constructed with high fidelity. Our analysis shows that LanPs involved in class I lanthipeptide biosynthesis (Supporting Information Table 1) fall into two major clades that correspond well with the grouping of LanAs (Figure 2b). This analysis suggests that the conserved motifs of LanAs near the proteolytic cleavage site likely are the recognition elements for LanP enzymes.
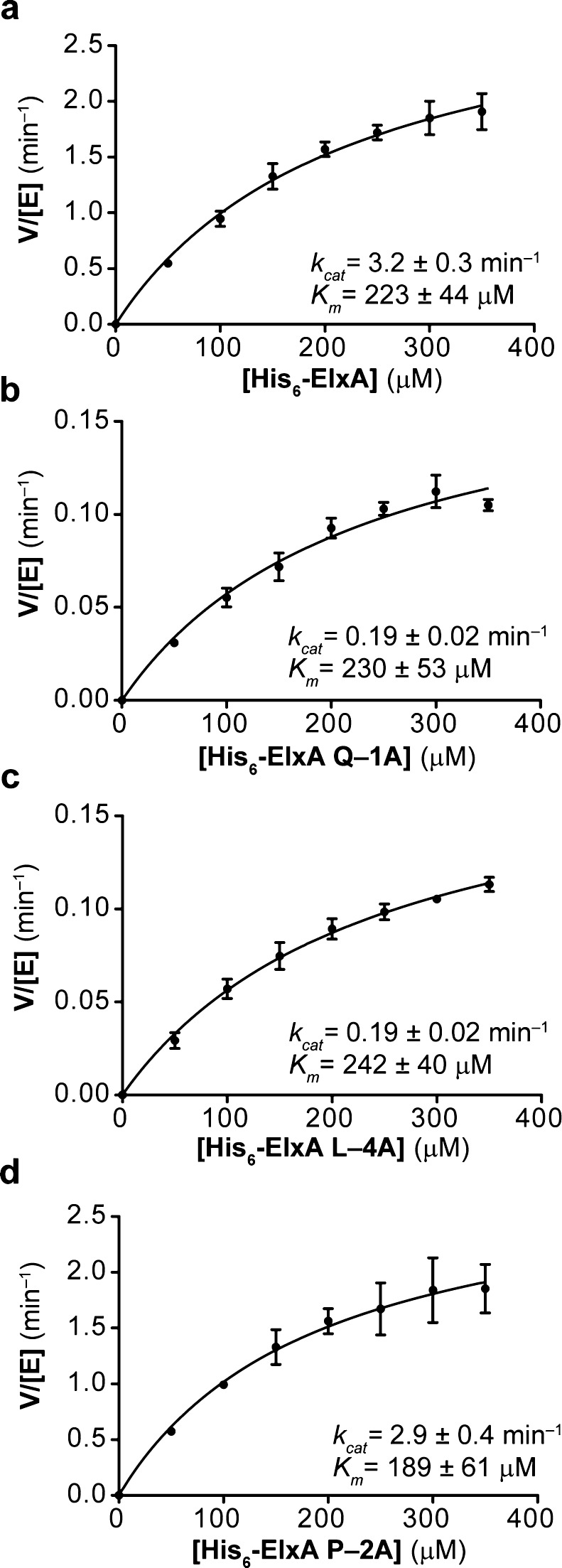
To test this hypothesis, ElxA was expressed in Escherichia coli as an N-terminally hexahistidine tagged peptide and purified by metal affinity chromatography, as previously described.7 ElxP was expressed in E. coli fused to the C-terminus of maltose binding protein (MBP-ElxP; Supporting Information Figure 1). We then performed alanine scanning mutagenesis on the E/D-L/V-x-x-Q motif present in the ElxA leader peptide. Indeed, single alanine mutations at the Gln−1, Leu−4, and Asp−5 positions in the leader peptide of ElxA significantly reduced the cleavage efficiency of MBP-ElxP as observed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Supporting Information Table 2). To quantify and distinguish the contribution of each amino acid to the substrate specificity of ElxP, we next determined the kinetic parameters of MBP-ElxP by using the wild type peptide and three ElxA mutants Q−1A, P−2A and L−4A as substrates. Reversed phase high performance liquid chromatography (RP-HPLC) was used to monitor N-terminal leader peptide formation at different substrate concentrations (Supporting Information Figures 2 and 3). MBP-ElxP cleaved wild type His6-ElxA with a catalytic efficiency of ∼240 M–1 s–1 (Figure 3a), whereas the mutants Q−1A or L−4A exhibited ∼14-fold reduced catalytic efficiency (Figure 3b and c).
Figure 3.
Kinetic characterization of ElxP peptidase activity for His6-ElxA and mutant peptides. Purified (a) ElxA, (b) ElxA Q−1A, (c) ElxA L−4A, and (d) ElxA P−2A were digested with MBP-ElxP and leader peptide formation was monitored at different time points by HPLC. The rate of MBP-ElxP catalysis was plotted as a function of different substrate concentrations. The data was fit to the Michaelis–Menten equation to give the kinetic parameters shown, presented as the average ± standard error of two independent experiments.
In contrast, single alanine substitutions at the Pro−2 and Asn−3 positions in ElxA did not obviously decrease the efficiency of MBP-ElxP to remove the N-terminal leader peptide from ElxA (Supporting Information Table 2 and Figure 3d), supporting the hypothesis based on sequence conservation that the amino acids in the E/D-L/V-x-x-Q motif of ElxA are the most important for substrate recognition by ElxP. Although it is possible that proteolysis by wild type ElxP of the ElxA peptide possessing the thioether rings would be more efficient, the catalytic efficiency of MBP-ElxP observed in this study with linear peptides is sufficient for application of the enzyme as a sequence specific protease.
Many leader peptides of class I lanthipeptides share an F-D/N-L-N/D sequence motif (Figure 2a). Previous studies have shown that this conserved motif is important for substrate recognition by the enzymes involved in (methyl)lanthionine incorporation.21,23−26 To probe whether this region is also important for recognition and efficient cleavage by ElxP, we performed alanine scanning mutagenesis on the F-D-L-N sequence in ElxA and analyzed the cleavage efficiency by MALDI-TOF MS. Our results show that the F-D-L-N motif is not essential for ElxP recognition (Supporting Information Table 2), suggesting that LanP enzymes do not recognize the same amino acid motif needed by the enzymes responsible for installing the lanthionine rings (e.g., LanBs and LanCs for the biosynthesis of class I lanthipeptides). This finding is in line with other studies that have found that different parts of the leader peptide are recognized by different post-translational modification enzymes during RiPP biosynthesis.27,28
Insertion of the ElxP Recognition Motif into NisA
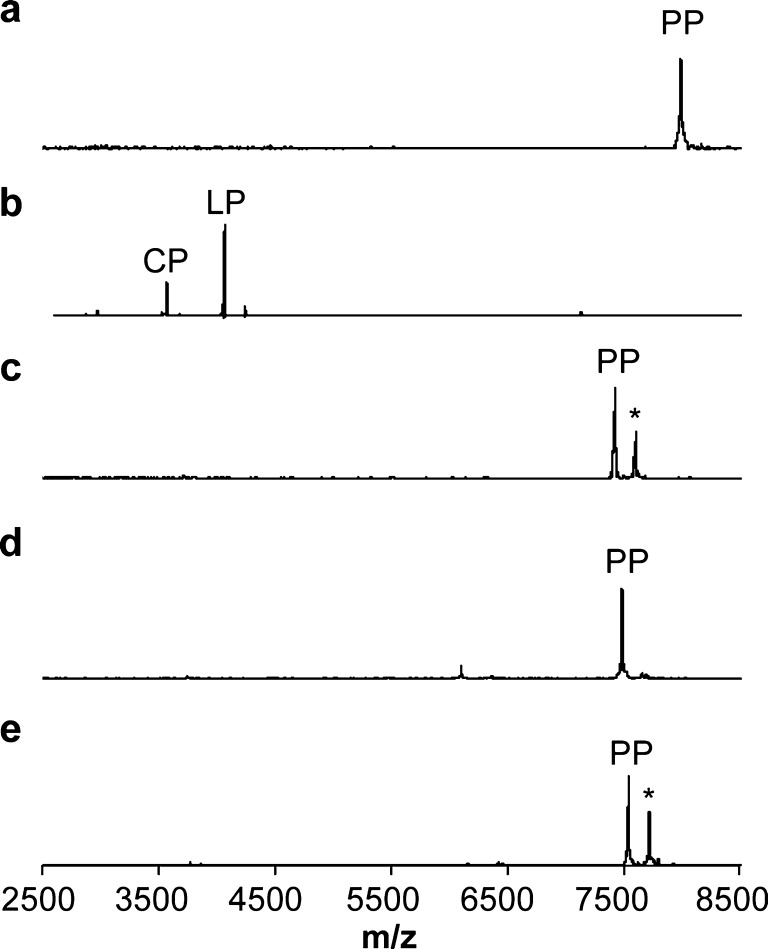
Based on our data, the conserved E/D-L/V-x-x-Q-T1/S1 motif present in the ElxA-group of LanAs likely serves as the main recognition element for their LanPs. This sequence could possibly be used as a tool to selectively remove tags from fusion proteins or leader peptides from other RiPPs. To determine this potential, we analyzed the ElxP activity on the noncognate lanthipeptide precursor peptide NisA. Upon incubation of wild type NisA with ElxP, no new peaks corresponding to the N-terminal leader or core peptide masses were observed by MALDI-TOF MS analysis, demonstrating that ElxP does not cleave wild type NisA (Figure 4a). However, when we replaced the G-A-S-P-R-I sequence of NisA with the corresponding sequence present in ElxA (D-L-N-P-Q-A, in which an Ala residue is used to mimic the dehydrated Ser1 residue of ElxA), the resultant mutant peptide (NisA-G−5D/A−4L/S−3N/R−1Q/Q−1_I1insA) was completely cleaved by ElxP (Figure 4b; for the sequences of the NisA mutants, see Supporting Information Figure 4). Other NisA mutants with partial permutations in the G-A-S-P-R-I sequence, including NisA-R−1Q, NisA-G−5D/A−4L/S−3N/R−1Q, and NisA-R−1Q/Q−1_I1insA, were not cleaved by ElxP (Figure 4c–e). These results support the model that the ElxP specificity relies on the complete D-L-x-x-Q-T1/S1 sequence motif.
Figure 4.
MALDI-TOF MS data on cleavage of NisA and NisA mutants by ElxP. (a) NisA (m/z 7992) treated with ElxP, (b) NisA-G−5D/A−4L/S−3N/R−1Q/Q−1_I1insA treated with ElxP, (c) NisA-R−1Q (m/z 7412) treated with ElxP, (d) NisA-R−1Q/Q−1_I1insA (m/z 7485) treated with ElxP, and (e) NisA-G−5D/A−4L/S−3N/R−1Q (m/z 7542) treated with ElxP. PP-precursor peptide, CP-core peptide, and LP-leader peptide. His6-NisA-G−5D/A−4L/S−3N/R−1Q/Q−1_I1insA unmodified core peptide, m/z 3568; leader peptide, m/z 4064. *Ion corresponding to peptide with gluconoylation of the His6-tag of NisA.
Substrate Specificity of ElxO
Previous attempts to use the dehydratase ElxB, the cyclase ElxC, and the peptidase ElxP to generate dehydroepilancin 15X, the substrate of the dehydrogenase ElxO, were unsuccessful.7 However, we showed that His6-ElxO catalyzes the reduction of the synthetic peptide Pyr-AAIVK, resembling the N-terminal region of dehydroepilancin 15X (Figure 1a), to D-Lac-AAIVK, demonstrating that the (methyl)lanthionine residues and full length ElxA peptide are not strictly required for substrate recognition by ElxO.7 Thus, ElxO could potentially be used to introduce N-terminal alcohols to other peptides or proteins that contain N-terminal Pyr or 2-oxobutyryl (Obu) groups, thus enhancing their chemical stability and resistance against degradation by aminopeptidases. To explore the substrate specificity of ElxO, a series of small potential substrates were synthesized by Fmoc-based solid phase peptide synthesis (SPPS) followed by coupling of pyruvic acid using hydroxybenzotriazole (HOBt) and diisopropylcarbodiimide (DIC) as activating reagents to produce the Pyr-containing substrates. Single residues of the originally tested substrate, Pyr-AAIVK, were replaced systematically with Ala, and Ala2 was changed to a wide variety of amino acids, including polar, nonpolar, acidic, and basic residues to obtain a set of alternative substrates (Table 1). In addition, the Pyr group was replaced with an N-terminal Obu group, which is generated upon hydrolysis of an N-terminal Dhb residue in the lanthipeptides Pep5,29,30 lacticin 3147 A2,31 lichenicidin,32−35 and prochlorosin 1.7.36 To release the peptides from the solid support, the resin-linked peptides were treated with TFA cleavage cocktails that did not contain triisopropylsilane since the presence of this reagent resulted in the chemical reduction of the ketone-containing peptides, as was also observed previously in other work.37 The peptides were purified by RP-HPLC and the identities of the compounds were confirmed by electrospray ionization mass spectrometry (ESI-MS).
Table 1. Substrates Tested for Reduction by His6-ElxO.

| entry | substrate | kcat/Km (M–1 s–1) | relative (kcat/Km) |
|---|---|---|---|
| 1 | Pyr-AAIVK | 2.43 ± 0.06 | 1.00 |
| 2 | Pyr-AAIV | 1.13 ± 0.00 | 0.47 |
| 3 | Pyr-AAI | 0.06 ± 0.00 | 0.02 |
| 4 | Pyr-AA | <0.03 | <0.01 |
| 5 | Pyr-A | <0.03 | <0.01 |
| 6 | Pyr-AAIVKBBIKAAKK | 14.2 ± 0.4 | 5.83 |
| 7 | Pyr-AAIVA | 1.33 ± 0.01 | 0.55 |
| 8 | Pyr-AAIAK | 1.59 ± 0.02 | 0.65 |
| 9 | Pyr-AAAVK | 0.29 ± 0.01 | 0.12 |
| 10 | Pyr-RAIVK | 5.50 ± 0.05 | 2.26 |
| 11 | Pyr-KAIVK | 4.22 ± 0.04 | 1.74 |
| 12 | Pyr-DAIVK | 0.29 ± 0.02 | 0.12 |
| 13 | Pyr-NAIVK | 7.60 ± 0.04 | 3.13 |
| 14 | Pyr-PAIVK | 0.13 ± 0.01 | 0.05 |
| 15 | Pyr-MAIVK | 15.5 ± 0.1 | 6.40 |
| 16 | Obu-AAIVK | 0.92 ± 0.03 | 0.38 |
| 17 | Obu-RAIVK | 1.51 ± 0.02 | 0.62 |
| 18 | Glx-AAIVK | <0.03 | <0.01 |
The purified peptides were then incubated with His6-ElxO in the presence of NADPH and the change in the absorbance at 340 nm over time was monitored by UV spectrophotometry. Attempts to determine the steady state kinetic parameters kcat and Km using a subset of peptides were not successful, since it was not possible to saturate the enzyme with the substrates before reaching the peptide solubility limits (Supporting Information Figure 5). Therefore, the kinetic constant kcat/Km was determined by measuring the reaction rates at various peptide concentrations (Table 1). For all tested substrates, the values of kcat/Km were relatively small, presumably because the peptides are lacking structural features compared to the expected physiological substrate, such as the thioether rings or additional amino acids. The smaller peptides Pyr-AAIV and Pyr-AAI (entries 2 and 3), but not Pyr-AA and Pyr-A (entries 4 and 5), were reduced by His6-ElxO in the presence of NADPH based on LC-MS analysis, although with considerably lower reaction rates compared with Pyr-AAIVK. In contrast, the peptide Pyr-AAIVKBBIKAAKK, where B stands for l-2-aminobutyric acid to mimic the dehydrobutyrines in dehydroepilancin 15X (Figure 1a), was converted at a higher rate (entry 6), suggesting that a longer peptide is beneficial for substrate recognition. The Ala scanning analysis performed along the sequence Pyr-AAIVK (Table 1, entries 7–9) indicated that the enzyme was able to reduce all the peptides tested albeit with a lower kcat/Km for Pyr-AAAVK (Table 1, entry 9). Next, we evaluated peptides containing amino acids with nonpolar, polar, acidic, or basic residues at position 2, and found that they were all transformed to the reduced products (Table 1 and Supporting Information Table 3).
Collectively, these results suggest that the flanking residue is not critical for activity and that the minimal length of the peptide to be accepted as substrate is four residues. Although all mutants at position 2 proved substrates for ElxO, Pyr-RAIVK, Pyr-KAIVK, Pyr-NAIVK, and Pyr-MAIVK were better substrates than Pyr-AAIVK (entries 10, 11, 13, 15), whereas Pyr-DAIVK and Pyr-PAIVK were converted considerably less efficiently (entries 12 and 14), suggesting that negatively charged residues and Pro in position 2 are not as well tolerated.
Obu-AAIVK and Obu-RAIVK were also accepted as substrates leading to the formation of an N-terminal 2-hydroxybutyryl group (Hob), although at lower rates (Table 1, entries 16 and 17). Similarly, the peptides Obu-AAAVK and Obu-AAIAK were substrates for the enzyme (Supporting Information Table 3). However, when Pyr was substituted by a glyoxyl (Glx) group, such as in the peptide Glx-AAIVK (Table 1, entry 18), no significant formation of the reduced peptide was observed.
Use of ElxO to Reduce Other Lanthipeptides
In addition to epilancin 15X, two other lantibiotics, epilancin K7, and epicidin 280, contain an N-terminal Lac moiety. To explore the potential of using His6-ElxO for the synthesis of other lantibiotics, peptides mimicking the N-terminal portion of dehydroepilancin K7 (Pyr-AAVLK), and dehydroepicidin 280 (Pyr-LGPAIK) were synthesized and tested as substrates (Supporting Information Table 3). His6-ElxO reduced both peptides, even though their sequences are quite different from the N-terminus of epilancin 15X. Similarly, peptides resembling the N-terminus of lactocin S (Pyr-APVLA and Pyr-BPVLAAVAVAKKK) and Pep5 (Obu-AGPAIR) were reduced by His6-ElxO in the presence of NADPH as confirmed by LC-MS analysis (Supporting Information Table 3).
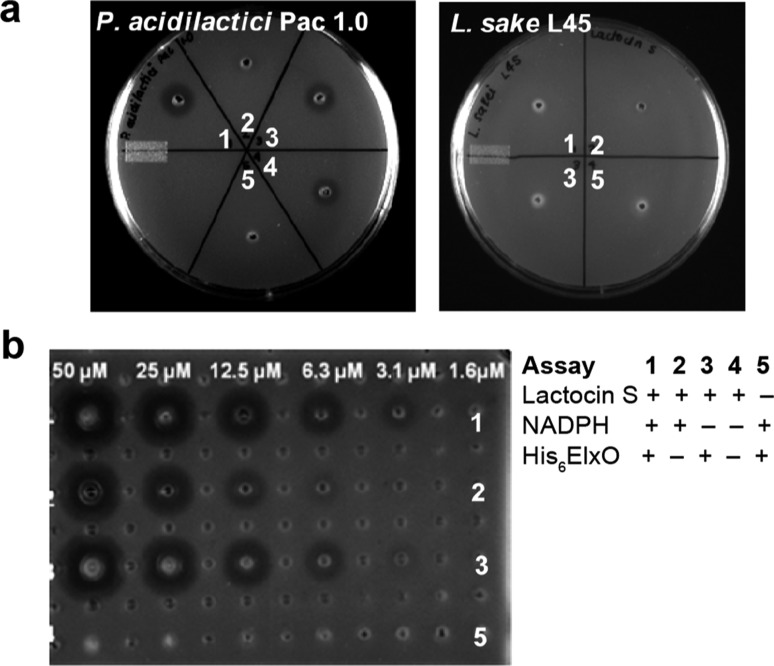
Encouraged by these results with short peptides we next turned to lactocin S, a 37-residue lantibiotic (Figure 5a) produced by Lactobacillus sake L45 that contains an N-terminal Pyr.38 To evaluate if lactocin S would be a substrate for ElxO, a synthetic sample of the lantibiotic39 was incubated with NADPH in the presence of His6-ElxO and the reduction of the peptide was monitored by high-resolution LC-MS (Figure 5b), confirming the formation of dihydrolactocin S. Samples containing the reduced peptide and control samples containing lactocin S were tested by agar diffusion bioactivity assays using either Pediococcus acidilactici Pac1.0 as indicator strain (Figure 6a left and 6b) or the lactocin S producer strain (Figure 6a right). All the peptides were active against P. acidilactici Pac1.0 but not against L. sake L45 suggesting that the N-terminal Pyr is not involved in self-immunity. From the serial dilution assay (Figure 6b), the sizes of the inhibition zones were determined and the critical concentrations at which no inhibition zones were observed were calculated (Supporting Information Figure 6).40 The sample containing dihydrolactocin S produced slightly larger inhibition zones and smaller critical concentrations than control samples containing the native peptide, illustrating that the N-terminal Pyr of lactocin S is not essential for bioactivity. Similar results were obtained upon determination of apparent minimal inhibitory concentrations (MIC) by a serial dilution bioactivity assay in liquid media (Supporting Information Figure 6).
Figure 5.
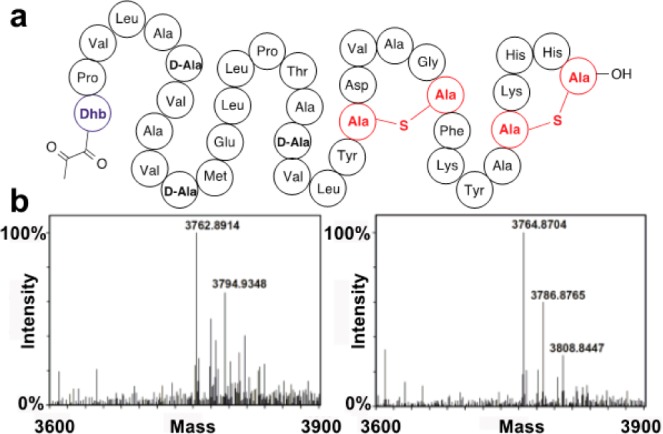
Schematic structure of lactocin S and formation of dihydrolactocin S. (a) The lantibiotic lactocin S contains an N-terminal Pyr group. (b) Lactocin S was converted to dihydrolactocin S. Left panel: MS analysis of lactocin S (calculated m/z = 3762.8851) incubated with NADPH in the absence of His6-ElxO. The peak at m/z = 3794.9348 corresponds to oxidized lactocin S (M+O). Right panel: MS analysis of dihydrolactocin S (calculated m/z = 3764.8851) generated after incubation of lactocin S with both NADPH and His6-ElxO. The peaks at m/z = 3786.8765 and 3808.8447 correspond to the sodium and disodium adducts of dihydrolactocin S.
Figure 6.
(a) Single concentration and (b) serial dilution agar diffusion bioactivity assays. The samples spotted were enzymatically synthesized dihydrolactocin S (sample 1) and control samples lacking enzyme (sample 2), cofactor (sample 3), or both (sample 4) and incubated under the same reaction conditions. Sample 5 was a control assay lacking lactocin S. See also Supporting Information Figure 6.
X-ray Crystal Structure of ElxO
At present, very little is known about the structural biology of lantibiotic biosynthetic enzymes, as structures are available only for LanC4 and LanD41,42 enzymes. Here, we determined the X-ray cocrystal structure of ElxO with NADPH bound at its active site to 1.9 Å resolution and the structure of a Y152F inactive variant to 1.85 Å resolution (Supporting Information Table 4). As expected from primary sequence analysis, ElxO is a member of the short-chain dehydrogenase/reductase (SDR) superfamily of NAD(P)(H)-dependent oxidoreductases. The overall structure is composed around a core consisting of a “Rossmann-fold” dinucleotide-binding motif (Figure 7a). Clear, continuous density, corresponding to a bound NADP(H) was observed in the vicinity of the active site. The specificity for the cofactor is established by Arg35, which engages the phosphate group of NADP(H). The cofactor is bound in an extended conformation to allow transfer of the pro-S hydrogen on C4 of the nicotinamide to the ketone of the peptide substrate. A surface representation of the ElxO structure demonstrates the presence of an extended groove along the active site that likely harbors the binding site for the substrate peptide (Figure 7c). Unfortunately, attempts to obtain a structure in complex with peptides (Pyr-AAIVK, Pyr-DAIVK, Pyr-VAIVK, Obu-AAIVK, or Obu-RAIVK) were unsuccessful, possibly because of relatively high Km values for these substrates. Co-crystallization and soaking efforts with both the wild-type enzyme and an inactive Y152F variant yielded only the unbound structures.
Figure 7.
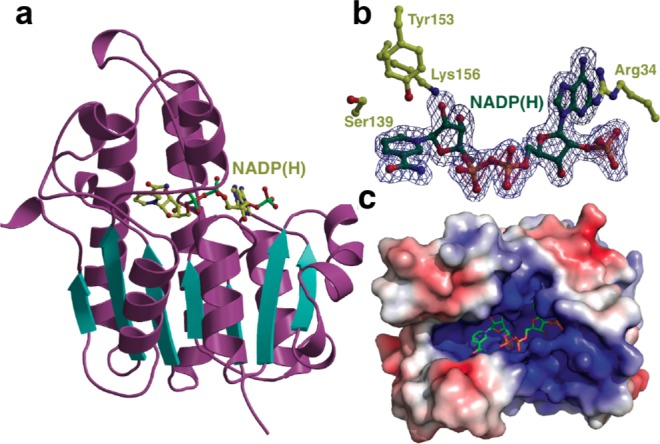
(a) X-ray crystal structure of ElxO bound to NADPH. (b) Side chain residues important for binding of NADPH and catalysis in ElxO. c) ElxO surface representation depicting groove, which might serve as the putative peptide-binding site.
Prior studies of SDR family enzymes suggest a tyrosinate-oxygen (and its conjugate acid) as acting as both the active site acid and base.43 An adjacent Lys lowers the pKa of the Tyr, and is also involved in binding to the 2′-nicotinamide ribose hydroxyl, and a conserved Ser polarizes the carbonyl of the substrate.44 The structure of ElxO reveals that Ser139, Tyr152, and Lys156 are located in the active site and are poised to function in catalysis (Figure 7b). In order to determine the importance of these residues for catalysis, the single mutants Y152F, S139A, K156A, and K156 M were prepared and purified. Enzymatic assays of wild type and mutant proteins were performed using Pyr-AAIVK as substrate and the consumption of NADPH over time was determined by UV spectrophotometry (Supporting Information Figure 7). The single mutations S139A, Y152F, K156A, or K156 M resulted in a strong reduction of the reaction rate (kcat/Km < 0.03 M–1 s–1), indicating important roles of these residues in catalysis.
Based on these findings and on previous studies on SDR enzymes,45 a catalytic mechanism is proposed for ElxO. Initially, the hydroxyl group of protonated Tyr152 and possibly the hydroxyl group of Ser139 are hydrogen bonded to the N-terminal carbonyl oxygen of dehydroepilancin 15X. Hydride transfer from C-4 of the nicotinamide ring of NADPH to the Si-face of the carbonyl with concomitant protonation of the resulting hydroxyl group by the catalytic acid Tyr152 then generates the (R)-Lac moiety7 of epilancin 15X and NADP+.
Conclusion
Our current work provides evidence that the LanA and LanP proteins likely coevolved and that LanP sequence specificity is mainly determined by the amino acids near the proteolytic site. Specifically, the conserved E/D-L/V-x-x-Q-T1/S1 sequence motif present in the ElxA-group of LanAs provides the full recognition elements for ElxP. This recognition sequence may find use in applications of ElxP for removing expression tags or removing leader peptides from RiPPs.
This study also demonstrates that ElxO is highly tolerant with respect to the structure of its substrates. This tolerance allowed the use of ElxO to modify lactocin S, a lantibiotic that is structurally unrelated to epilancin 15X but that contains an N-terminal Pyr. ElxO was also able to reduce Obu to the corresponding alcohol, providing a means for generating potentially useful lantibiotic analogues. In addition to these studies on substrate scope, the X-ray crystal structure of ElxO combined with the mutagenesis studies provide insights into the reduction process in which Ser139, Tyr152, and Lys156 play essential roles.
Methods
Organisms, Media, and Growth Conditions
All oligonucleotides used in this study were purchased from Integrated DNA Technologies (Supporting Information Table 5). For microorganisms used, see Supporting Information Table 6. Reagents used for molecular biology experiments were purchased from New England BioLabs, Thermo Fisher Scientific, or Gold Biotechnology. Plasmid sequencing was performed by ACGT Inc. unless otherwise noted. Escherichia coli DH5α and BL21 (DE3) were used for plasmid maintenance and protein or peptide overexpression, respectively. The strains L. sake L45 and P. acidilactici Pac 1.0 were grown in de Man–Rogosa–Sharpe (MRS) solid agar or broth. MALDI-TOF measurements were performed using a Bruker UltrafleXtreme MALDI-TOF-TOF instrument using a positive reflective mode and sinapic acid as a matrix unless otherwise noted. Detailed experimental procedures for the MCMC phylogenic tree analysis are provided in the Supporting Information.
Purification of MBP-ElxP and ElxA Peptides
Detailed experimental procedures are described in the Supporting Information. Primers used for the construction of mutant substrates are listed in Supporting Information Table 5.
MBP-ElxP Activity Assays by MS
A typical activity assay consisted of 50 mM Tris-HCl pH 8.0, 50 μM peptide, 5 μM MBP-ElxP in a final volume of 100 μL. The sample was incubated for 2 h at RT. To monitor cleavage activity, samples were desalted using a zip tip concentrator (Millipore), mixed in a 1:1 ratio with sinapic acid, and spotted on a MALDI-TOF Bruker plate. Ion intensities for the resulting precursor peptide, leader peptide and core peptide were normalized and the proteolytic efficiency was measured as the amount of substrate left after cleavage reaction (Supporting Information Table 2). Reactions were performed with tagged enzyme and substrates unless otherwise noted.
Determination of ElxP Kinetic Parameters
Detailed experimental conditions and procedures are described in the Supporting Information associated with this article.
Synthesis of ElxO Substrate Analogues
Peptides were synthesized by standard Fmoc-based solid phase peptide synthesis. Detailed experimental procedures are described in the Supporting Information associated with this article.
Purification of wt His6-ElxO and Mutant Variants
To generate pHis6-ElxO(S139A), pHis6-ElxO(Y152F), pHis6-ElxO(K156A), and pHis6-ElxO(K156M), the entire pHis6-ElxO reported previously7 was amplified by PCR using PfuTurbo hotstart DNA polymerase (Stratagene) or iProof high-fidelity polymerase (BioRad) and the appropriate mutagenesis primers: ElxO.S139A.FP and ElxO.S139.RP, ElxO.Y152F.FP and ElxO.Y152F.RP, ElxO.K156A.FP and ElxO.K156A.RP, or ElxO.K156M.FP and ElxO.K156M.RP (Supporting Information Table 5) followed by treatment with DpnI (New England Biolabs) and transformation of Escherichia coli DH5α cells. The correct sequence of the insert was confirmed by sequencing at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana–Champaign. The proteins were expressed and purified using a HisTrap HP column (GE Healthcare) as described elsewhere for His6-ElxO,7 followed by further purification by size exclusion chromatography using an ÄKTApurifier equipped with a HiLoad 16/60 Superdex 200 column (GE Healthcare) and a flow rate of 1.5 mL min–1 of running buffer (50 mM HEPES, 300 mM NaCl, 10% (v/v) glycerol, pH 7.4)
Wild Type and Mutant His6-ElxO Activity Assays
Wild type or mutant His6-ElxO (2 or 10 μM) and purified peptide (0.1 to 5 mM) were incubated with NADPH (2.5 mM) in assay buffer (100 mM HEPES, 500 mM NaCl, pH 7.5) at 25 °C. Reaction progress was monitored by UV spectrophotometry to measure initial rates, measuring the disappearance of NADPH absorbance at 340 nm. Formation of reduced peptides was confirmed by LC-MS using an Agilent 1200 instrument equipped with a single quadruple multimode ESI/APCI ion source mass spectrometry detector and a Synergi Fusion-RP column (4.6 mm × 150 mm, Phenomenex). The mobile phase was 0.1% (v/v) formic acid in water (A) and methanol (B). A gradient of 0–70% (v/v) B in A over 30 min and a flow rate of 0.5 mL min–1 were used.
Production of Dihydrolactocin S
Detailed experimental procedures are described in the Supporting Information.
Bioactivity Assays with Dihydrolactocin S
Agar diffusion bioactivity assays were performed using de Man-Rogosa-Sharpe (MRS) agar media. For each assay, aliquots of agar medium inoculated with overnight cultures of indicator strain (1:100 dilution) were poured into sterile plates. Aliquots of 20 μL of sample were placed into wells made on the solidified agar and the plates were incubated at 37 °C overnight. For determination of critical concentration, the diameter of the inhibition zones was determined and fitted to the equation D = a + b × (C), where D is the diameter of the inhibition zone, C is the concentration of bacteriocin, and a and b are constant parameters.40,46,47 For MIC determinations, serial dilutions of peptides were prepared in MRS broth and aliquots of 50 μL were dissolved in 150 μL of a 1 to 50 dilution of an overnight culture of indicator strain in fresh MRS broth in 96-well plates. The cultures were incubated at 37 °C overnight and the wells with no bacterial growth (OD600 < 0.3) were determined.
Determination of the ElxO X-ray Crystal Structure
Crystallization and structure elucidation conditions and procedures are described in the Supporting Information associated with this article (Supporting Information Table 4).
Acknowledgments
The authors thank W. Tang for helpful experimental suggestions and Prof. J. Vederas (University of Alberta) for kindly supplying a synthetic sample of lactocin S. This work was supported by a grant from the National Institutes of Health (R01 GM 58822) to W.A.v.d.D. M.A.O and J.E.V. were supported by the NIGMS-NIH Chemistry-Biology Interface Training Grant (5T32-GM070421). M.A.O. was supported partially by the Ford Foundation. The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS, National Institutes of Health, or Ford Foundation. The Bruker UltrafleXtreme MALDI TOF/TOF mass spectrometer was purchased in part with a grant from the National Institutes of Health (S10 RR027109).
Supporting Information Available
Descriptions of all molecular biology procedures, protein purifications, determination of ElxP kinetic parameters, synthesis of ElxO peptide substrate analogues, parameters for the production of dihydrolactocin S, crystallographic data summary for ElxO, and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
Atomic coordinates of ElxO have been deposited in the Protein Data Bank (PDB) with the accession code 4QEC for wt ElxO and 4QED for ElxO Y152F.
Author Contributions
∥ These authors contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K.-D.; Fischbach M. A.; Garavelli J. S.; Göransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Müller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J. T.; Rebuffat S.; Ross R. P.; Sahl H.-G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Süssmuth R. E.; Tagg J. R.; Tang G.-L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell N.; Entian K.-D.; Schneider U.; Götz F.; Zahner H.; Kellner R.; Jung G. (1988) Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333, 276–278. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R.; Rollema H. S.; Siezen R. J.; Beerthuyzen M. M.; Kuipers O. P.; de Vos W. M. (1994) Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J. Biol. Chem. 269, 3555–3562. [PubMed] [Google Scholar]
- Li B.; Yu J. P.; Brunzelle J. S.; Moll G. N.; van der Donk W. A.; Nair S. K. (2006) Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311, 1464–1467. [DOI] [PubMed] [Google Scholar]
- McClerren A. L.; Cooper L. E.; Quan C.; Thomas P. M.; Kelleher N. L.; van der Donk W. A. (2006) Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc. Natl. Acad. Sci. U.S.A. 103, 17243–17248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekkelenkamp M. B.; Hanssen M.; Danny Hsu S. T.; de Jong A.; Milatovic D.; Verhoef J.; van Nuland N. A. (2005) Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 579, 1917–1922. [DOI] [PubMed] [Google Scholar]
- Velásquez J. E.; Zhang X.; van der Donk W. A. (2011) Biosynthesis of the antimicrobial peptide epilancin 15X and its unusual N-terminal lactate moiety. Chem. Biol. 18, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knerr P. J.; van der Donk W. A. (2012) Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81, 479–505. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Yu Y.; Velásquez J. E.; van der Donk W. A. (2012) Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 109, 18361–18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein L. S.; Diep D. B.; Nes I. F. (1995) A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16, 229–240. [DOI] [PubMed] [Google Scholar]
- Oman T. J.; van der Donk W. A. (2010) Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 6, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirix G.; Monsieurs P.; Dombrecht B.; Daniels R.; Marchal K.; Vanderleyden J.; Michiels J. (2004) Peptide signal molecules and bacteriocins in Gram-negative bacteria: A genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides 25, 1425–1440. [DOI] [PubMed] [Google Scholar]
- Uguen P.; Hindré T.; Didelot S.; Marty C.; Haras D.; Le Pennec J. P.; Vallee-Rehel K.; Dufour A. (2005) Maturation by LctT is required for biosynthesis of full-length lantibiotic lacticin 481. Appl. Environ. Microbiol. 71, 562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S.; Yano T.; Hayashi H. (2006) Expression and characterization of the peptidase domain of Streptococcus pneumoniae ComA, a bifunctional ATP-binding cassette transporter involved in quorum sensing pathway. J. Biol. Chem. 281, 4726–4731. [DOI] [PubMed] [Google Scholar]
- Furgerson Ihnken L. A.; Chatterjee C.; van der Donk W. A. (2008) In vitro reconstitution and substrate specificity of a lantibiotic protease. Biochemistry 47, 7352–7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishie M.; Sasaki M.; Nagao J.; Zendo T.; Nakayama J.; Sonomoto K. (2011) Lantibiotic transporter requires cooperative functioning of the peptidase domain and the ATP binding domain. J. Biol. Chem. 286, 11163–11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völler G. H.; Krawczyk B.; Ensle P.; Süssmuth R. D. (2013) Involvement and unusual substrate specificity of a prolyl oligopeptidase in class III lanthipeptide maturation. J. Am. Chem. Soc. 135, 7426–7429. [DOI] [PubMed] [Google Scholar]
- Siezen R. J.; Rollema H. S.; Kuipers O. P.; de Vos W. M. (1995) Homology modeling of the Lactococcus lactis leader peptidase NisP and its interaction with the precursor of the lantibiotic nisin. Protein Eng. 8, 117–125. [DOI] [PubMed] [Google Scholar]
- Kuipers A.; de Boef E.; Rink R.; Fekken S.; Kluskens L. D.; Driessen A. J.; Leenhouts K.; Kuipers O. P.; Moll G. N. (2004) NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J. Biol. Chem. 279, 22176–22182. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R.; Polman J.; Beerthuyzen M. M.; Siezen R. J.; Kuipers O. P.; de Vos W. M. (1993) Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175, 2578–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plat A.; Kluskens L. D.; Kuipers A.; Rink R.; Moll G. N. (2011) Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl. Environ. Microbiol. 77, 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers O. P.; Rollema H. S.; de Vos W. M.; Siezen R. J. (1993) Biosynthesis and secretion of a precursor of nisin Z by Lactococcus lactis, directed by the leader peptide of the homologous lantibiotic subtilin from Bacillus subtilis. FEBS Lett. 330, 23–27. [DOI] [PubMed] [Google Scholar]
- Mavaro A.; Abts A.; Bakkes P. J.; Moll G. N.; Driessen A. J.; Smits S. H.; Schmitt L. (2011) Substrate recognition and specificity of the NisB protein, the lantibiotic dehydratase involved in nisin biosynthesis. J. Biol. Chem. 286, 30552–30560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abts A.; Montalban-Lopez M.; Kuipers O. P.; Smits S. H.; Schmitt L. (2013) NisC binds the FxLx motif of the nisin leader peptide. Biochemistry 52, 5387–5395. [DOI] [PubMed] [Google Scholar]
- Khusainov R.; Moll G. N.; Kuipers O. P. (2013) Identification of distinct nisin leader peptide regions that determine interactions with the modification enzymes NisB and NisC. FEBS Open Bio 3, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusainov R.; Heils R.; Lubelski J.; Moll G. N.; Kuipers O. P. (2011) Determining sites of interaction between prenisin and its modification enzymes NisB and NisC. Mol. Microbiol. 82, 706–718. [DOI] [PubMed] [Google Scholar]
- Yang X.; van der Donk W. A. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: new insights into the role of leader and core peptides during biosynthesis. Chem.—Eur. J. 19, 7662–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar D.; Pierce E.; McIntosh J. A.; Schmidt E. W. (2014) Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth. Biol. 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta C.; Entian K. D.; Kellner R.; Jung G.; Reis M.; Sahl H. G. (1989) Pep5, a new lantibiotic: Structural gene isolation and prepeptide sequence. Arch. Microbiol. 152, 16–19. [DOI] [PubMed] [Google Scholar]
- Meyer C.; Bierbaum G.; Heidrich C.; Reis M.; Süling J.; Iglesias-Wind M. I.; Kempter C.; Molitor E.; Sahl H.-G. (1995) Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis of PepP and PepC. Eur. J. Biochem. 232, 478–489. [DOI] [PubMed] [Google Scholar]
- Ryan M. P.; Jack R. W.; Josten M.; Sahl H. G.; Jung G.; Ross R. P.; Hill C. (1999) Extensive post-translational modification, including serine to d-alanine conversion, in the two-component lantibiotic, lacticin 3147. J. Biol. Chem. 274, 37544–37550. [DOI] [PubMed] [Google Scholar]
- Begley M.; Cotter P. D.; Hill C.; Ross R. P. (2009) Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 75, 5451–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dischinger J.; Josten M.; Szekat C.; Sahl H. G.; Bierbaum G. (2009) Production of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13. PLoS One 4, e6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkarev Z. O.; Finkina E. I.; Nurmukhamedova E. K.; Balandin S. V.; Mineev K. S.; Nadezhdin K. D.; Yakimenko Z. A.; Tagaev A. A.; Temirov Y. V.; Arseniev A. S.; Ovchinnikova T. V. (2010) Isolation, structure elucidation, and synergistic antibacterial activity of a novel two-component lantibiotic lichenicidin from Bacillus licheniformis VK21. Biochemistry 49, 6462–6472. [DOI] [PubMed] [Google Scholar]
- Caetano T.; Krawczyk J. M.; Mosker E.; Süssmuth R. D.; Mendo S. (2011) Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem. Biol. 18, 90–100. [DOI] [PubMed] [Google Scholar]
- Li B.; Sher D.; Kelly L.; Shi Y.; Huang K.; Knerr P. J.; Joewono I.; Rusch D.; Chisholm S. W.; van der Donk W. A. (2010) Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 10430–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau P.; Bure C.; Delmas A. F. (2005) Efficient synthesis of C-terminal modified peptide ketones for chemical ligations. Bioorg. Med. Chem. Lett. 15, 5442–5445. [DOI] [PubMed] [Google Scholar]
- Mortvedt C. I.; Nissen-Meyer J.; Sletten K.; Nes I. F. (1991) Purification and amino acid sequence of lactocin S, a bacteriocin produced by Lactobacillus sake L45. Appl. Environ. Microbiol. 57, 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. C.; Liu H.; Pattabiraman V. R.; Vederas J. C. (2010) Synthesis of the lantibiotic lactocin S using peptide cyclizations on solid phase. J. Am. Chem. Soc. 132, 462–463. [DOI] [PubMed] [Google Scholar]
- Delgado A.; Brito D.; Fevereiro P.; Tenreiro R.; Peres C. (2005) Bioactivity quantification of crude bacteriocin solutions. J. Microbiol. Methods 62, 121–124. [DOI] [PubMed] [Google Scholar]
- Blaesse M.; Kupke T.; Huber R.; Steinbacher S. (2000) Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate. EMBO J. 19, 6299–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaesse M.; Kupke T.; Huber R.; Steinbacher S. (2003) Structure of MrsD, an FAD-binding protein of the HFCD family. Acta Cryst. Section D Biol. Cryst. D59, 1414–1421. [DOI] [PubMed] [Google Scholar]
- Kavanagh K. L.; Jörnvall H.; Persson B.; Oppermann U. (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H.; Persson B.; Krook M.; Atrian S.; Gonzàlez-Duarte R.; Jeffery J.; Ghosh D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013. [DOI] [PubMed] [Google Scholar]
- Tanaka N.; Nonaka T.; Nakamura K. T.; Hara A. (2001) SDR: Structure, mechanism of action, and substrate recognition. Curr. Org. Chem. 5, 89–111. [Google Scholar]
- Wolf C. E.; Gibbons W. R. (1996) Improved method for quantification of the bacteriocin nisin. J. Appl. Bacteriol. 80, 453–457. [DOI] [PubMed] [Google Scholar]
- Parente E.; Brienza C.; Moles M.; Ricciardi A. (1995) A comparison of methods for the measurement of bacteriocin activity. J. Microbiol. Methods 22, 95–108. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.