Abstract
Structures of integral membrane receptors provide valuable models for drug–receptor interactions across many important classes of drug targets and have become much more widely available in recent years. However, it remains to be determined to what extent these images are relevant to human receptors in their biological context and how subtle issues such as subtype selectivity can be informed by them. The high precision structural modifications enabled by unnatural amino acid mutagenesis on mammalian receptors expressed in vertebrate cells allow detailed tests of predictions from structural studies. Using the Cys-loop superfamily of ligand-gated ion channels, we show that functional studies lead to detailed binding models that, at times, are significantly at odds with the structural studies on related invertebrate proteins. Importantly, broad variations in binding interactions are seen for very closely related receptor subtypes and for varying drugs at a given binding site. These studies highlight the essential interplay between structural studies and functional studies that can guide efforts to develop new pharmaceuticals.
1. Introduction
Recent years have seen a remarkable increase in our knowledge of the structures of integral membrane proteins such as ion channels, transporters, and GPCRs. Of course, these proteins are major targets of the pharmaceutical industry, and the expectation is that structural information will provide valuable guidance to efforts to develop new drugs. The structural insights do, however, come with some caveats. Often the protein is of bacterial origin or from some other, nonmammalian source. The structures are frequently of fragments or homologues of the true receptor, and/or they are heavily modified to enable crystallization. Even in the rare case of an unmodified mammalian receptor succumbing to crystallographic study with a relevant drug bound, the snapshot provided by X-ray crystallography may be ambiguous with regard to the state of the receptor being imaged and may be opaque with regard to the signaling process the receptor initiates. Also, a crucial issue in drug development (the targeting of small molecules to specific subtypes of a family of very closely related receptors) is not well addressed by a single image. To be clear, the structures are extraordinarily valuable, but they do not tell the whole story.
In principle, functional studies of intact mammalian receptors can provide powerful tests of predictions based on structural studies. However, often the tools are too crude to be convincing. How does one establish that a particular hydrogen bond is essential to receptor function? Pharmacology (varying the drug) provides one effective strategy. Mutagenesis (varying the protein) complements the pharmacology, and the two in combination can produce compelling insights. Still, it is often difficult to provide convincing evidence for a particular noncovalent interaction and even more difficult to provide a measure of the strength of a particular noncovalent interaction.
Over the past 20 years our group has conducted in vivo studies on receptors and ion channels, employing unnatural amino acid mutagenesis to gain chemical-scale insights into receptor binding sites.1−3 The advantage of the unnatural amino acid methodology is that it allows the protein scientist to exercise the same level of precision in structurally modifying the protein that the medicinal chemist routinely applies to the small molecule. These studies can provide evidence for (or against) proposed ligand binding interactions and in favorable cases can provide semiquantitative information on noncovalent interactions. In addition, variations in binding among receptor subtypes, a common and especially important theme in receptor pharmacology, can be probed by this approach.
Here we summarize a large number of studies that have evaluated drug–receptor interactions across a family of related receptors, the Cys-loop ligand-gated ion channels. Our focus is on the region of the agonist binding site, leaving a discussion of the fascinating process of channel gating for another time. The more recent work is certainly guided/inspired by the structural studies. In many cases, key noncovalent interactions seen in crystal structures are found to be functionally important. However, we also see instances in which a prediction from the structure is not supported by functional studies, and cases where a very different model is indicated. Our results suggest that variations abound in ligand binding modes, even among very closely related receptors, and that extrapolations from model structures to the mammalian proteins of interest should be made with caution.
2. Cys-Loop Receptors
2.1. Background and Biological Function
Fast synaptic transmission relies on membrane proteins that couple exquisite molecular recognition of neurotransmitters to microsecond allosteric transitions, permitting ion passage across the postsynaptic membrane. Ligand-gated ion channels (LGICs) accomplish this feat,4 with central players being the pentameric receptors of the Cys-loop superfamily.5,6 Other LGICs include the trimeric P2X receptors and the tetrameric ionotropic glutamate receptors. These include excitatory, cation-selective channels that are opened by the ligands acetylcholine (the nicotinic acetylcholine receptors, nAChRs) and serotonin (5-HT3 receptors), along with inhibitory, anion-selective channels that are opened by γ-aminobutyric acid (GABA) and by glycine. As critical mediators of neurotransmission, Cys-loop receptor dysfunction unsurprisingly associates with disease,7,8 including myasthenic syndromes, epilepsy, schizophrenia, Parkinson’s disease, and Alzheimer’s disease. Currently prescribed therapeutics targeting Cys-loop receptors include muscle relaxants, smoking cessation aids, antiemetics, and anxiolytics.
2.2. Structural Studies Related to Cys-Loop Receptors
The global architecture of Cys-loop receptors is well established, based on cryo-EM studies of the Torpedo ray nAChR,9 X-ray structures of the related prokaryotic receptors ELIC and GLIC,10−12 and most recently the first X-ray structure of a true Cys-loop receptor, the anion-selective channel GluCl from C. elegans(13) (Figure 1A). Receptors are pentamers, with each subunit containing four membrane-spanning α-helices and a large, primarily β-sheet, N-terminal extracellular domain. The five subunits arrange pseudo-symmetrically around a pore lined by the second transmembrane helix, and ligand-binding sites are found at subunit interfaces in the extracellular domain.
Figure 1.

(A) Cryo-EM structure of the Torpedo nAChR (PDB 2BG9). (B) Acetylcholine binding protein (AChBP) from Lymnaea stagnalis, with loops colored that form the principal (A–C) and complementary (D–F) faces of the binding site (PDB code 1UW6). (C) Aromatic box of the AChBP binding site comprising Trp and Tyr residues conserved across nAChRs.
Our structural understanding of ligand binding to nAChRs in particular, and to some extent Cys-loop receptors in general, has been greatly enhanced by structures of invertebrate acetylcholine binding proteins (AChBPs).14−17 These soluble proteins have proven to be amenable to crystallization, and they share structural homology and 20–25% sequence identity with the nAChR extracellular domain. Each binding site comprises six “loops” labeled A–F, with A–C forming the “principal face” and D–F contributed by the adjacent subunit and forming the “complementary face” (Figure 1B). The C loop is thought to be mobile in nAChRs, and it wraps over the ligand in the active receptor.
AChBP structures reveal a confluence of aromatic residues that are highly conserved in nAChRs and that are arranged into an “aromatic box” (Figure 1C). These aromatics have been named by the loop on which they reside: TyrA, TrpB, TyrC1, TyrC2, and TrpD. Other Cys-loop receptor binding sites are also rich in Phe, Tyr, and Trp, often seen at these conserved aromatic box sites. AChBP is believed to approximate an agonist-bound conformation of the binding site, corresponding to an active or a desensitized state. Over 60 AChBP structures have been reported, many in complex with pharmacologically relevant ligands.16 Additionally, AChBPs have been modified to more closely mimic the extracellular domain residues and binding sites of the α7 nAChR extracellular domain18,19 and of the 5-HT3 receptor binding site.20 It should be appreciated, however, that AChBP is merely a homologue of the nAChR extracellular domain and further that ligand binding does not gate a transmembrane channel.
Importantly, all these structural studies confirmed conclusions that were reached on the basis of prior biochemical studies, including an interfacial binding site, the loop regions A–F, the preponderance of aromatic amino acids at the agonist binding site, and other features. This engenders confidence that these structures, including those of proteins that are only obliquely related to the mammalian receptors, are highly relevant. Key questions remain, however. Which structural features have functional significance? How does ligand binding lead to channel gating? How is selectivity achieved among very closely related receptors? Our view is that functional studies, guided by these structural results, provide the best route to answering these and other key questions.
3. Unnatural Amino Acid Mutagenesis Strategies To Probe Binding Interactions
Ligand binding is mediated by various noncovalent interactions, such as hydrogen bonds, ion pairs, and cation−π interactions. Establishing the importance of such weak interactions can be challenging using conventional methods. As such, we have developed and applied a number of strategies based on unnatural amino acids that can probe noncovalent interactions with high precision.1−3 The key is that unnatural amino acid mutagenesis allows subtle and systematic changes to a potential binding interaction, thus producing convincing evidence for its significance (or insignificance). Here we describe general strategies that we have employed across the entire Cys-loop receptor family and to many other proteins. All studies involved receptors expressed in Xenopus oocytes, and our primary characterization was through a dose–response curve generated from two electrode-voltage clamp recordings of ion channel activity.
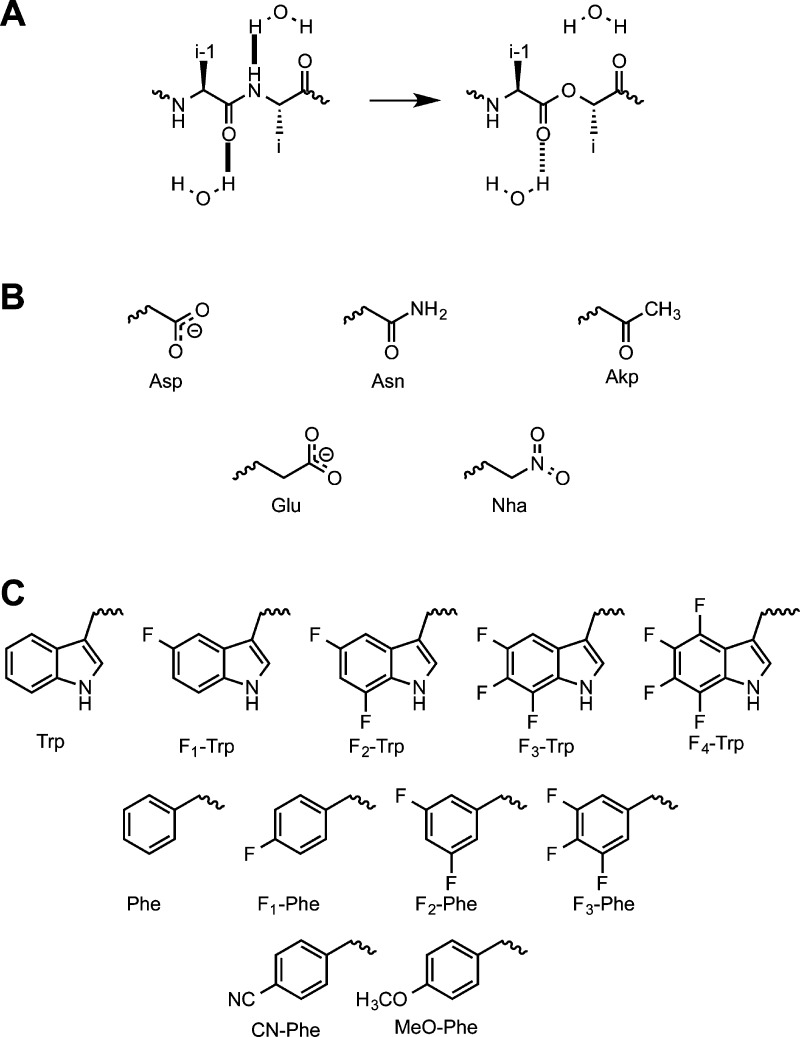
Hydrogen bonding is universally employed in drug–receptor interactions. When the protein side chain is involved, both conventional and unnatural amino acid mutagenesis provide strategies to evaluate the interaction, although the unnatural amino acid approach can provide more compelling evidence. However, often it is the protein backbone that is the hydrogen bond partner for the drug, and such an interaction cannot be probed by conventional mutagenesis. Fortunately, unnatural amino acid mutagenesis provides a powerful approach. Not only can a large, diverse range of unnatural α-amino acids be incorporated by our methodology, but α-hydroxy acids can also be employed. As shown in Figure 2A, this amide-to-ester mutagenesis allows us to probe both hydrogen bond donor and acceptor roles for the protein backbone. Interestingly, both model studies21,22 and our own experience have shown that the two perturbations (removing the backbone NH and attenuating the backbone CO) can have comparable impacts, and we have used both extensively.
Figure 2.
(A) α-Hydroxy acid strategy to evaluate backbone hydrogen bonding. The backbone NH group is removed and the backbone CO becomes a weaker hydrogen bond acceptor (dashed line). (B) Asp and Glu analogues. (C) Trp and Phe analogues.
Ion pairs involving Asp and Glu are conventionally probed with Asn and Gln, respectively, but unnatural amino acid mutagenesis provides more subtle probes. For example, nitrohomoalanine (Nha, Figure 2B) is isosteric and isoelectronic to Glu but lacks the negative charge.23 Also, structures like Akp provide alternatives to Asn and Gln.
Another noncovalent interaction that is important in many drug–receptor interactions, and which was long anticipated to be important in nicotinic receptors,24 is the cation−π interaction.25−27 The primary attraction in a cation−π interaction is between a positive charge (typically on the drug) and the negative electrostatic potential associated with the face of the aromatic rings of Phe, Tyr, and Trp. This attraction can be systematically modulated by progressive fluorination of the aromatic ring (Figure 2C), as fluorines withdraw electron density from the ring. This fluorination strategy has been used to establish cation−π interactions for dozens of drug–receptor pairs,27 and it will figure prominently in our evaluation of Cys-loop receptors.
It should be noted from the start that, technically, the studies described here do not evaluate whether a particular noncovalent interaction is present or not. We are probing whether a particular interaction is functionally significant. That is, we determine whether perturbing or in some cases removing a noncovalent interaction impacts receptor function in a meaningful way. Our typical measure of function is EC50, a composite measurement that reflects both agonist binding and receptor gating and, as such, involves multiple equilibria. As discussed in detail elsewhere,28 when mutating residues that are expected to be in direct contact with the agonist, it is clear that we are perturbing a noncovalent binding interaction, although there may be ambiguity about which particular equilibrium is being perturbed by the change in binding. And of course in any structure–function study, the structure of the receptor must be changed, and it is possible that unanticipated, large structural changes are induced by a mutation. Without structural corroboration, this cannot be ruled out, but we would argue that the very subtle mutations enabled by unnatural amino acid mutagenesis make such a complication less likely.
4. Nicotinic Acetylcholine Receptors (nAChRs)
nAChRs are the most thoroughly studied Cys-loop receptors and will be the primary focus of this review. Subunits are primarily classified as α (10 variants) or β (4 variants), with the former contributing the principal face of the agonist binding site and the latter generally contributing the complementary face. Dozens of receptors subtypes, formed by differing combinations of α and β subunits, have been established to be active in humans.7,8 In the brain, homomeric α7 receptors and receptors containing α4 and β2 subunits are the dominant subtypes expressed.29 The receptor of the neuromuscular junction, a close homologue of Torpedo electroplax nAChR, has a unique subunit composition of (α1)2β1γδ (fetal form; in adults the ε subunit substitutes for γ), with binding sites found at α/γ and α/δ interfaces. Here we consider ligand binding to the α4β2, α4β4, and α7 neuronal receptors and to the muscle-type receptor.
4.1. A Model of nAChR Ligand Binding Suggested by AChBP Structures
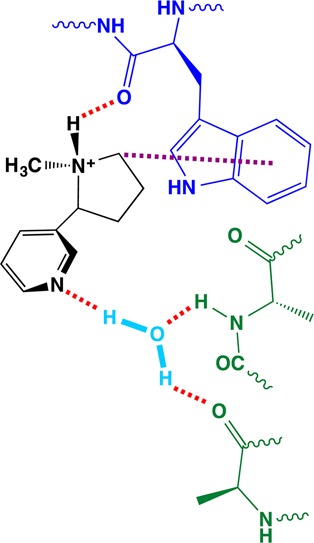
Over 40 years ago it was recognized that nAChR agonists share a pharmacophore comprising a cationic nitrogen separated by approximately 4–6 Å from a hydrogen bond-accepting group.30 A subset of nicotinic agonists is shown in Figure 3, highlighting these common structural features. Over the past decade, AChBP structures in complex with nicotinic agonists (including carbamylcholine, nicotine, and epibatidine) have suggested binding partners for these groups at the receptor.15,31 AChBP structures from different organisms and in complex with different agonists all show similar side chain conformations at the binding site. In all of these structures, the cationic group of the agonist is oriented toward the principal face of the binding site, and three potential noncovalent interactions are evident (Figure 4): (1) a cation−π interaction with TrpB; (2) a cation−π interaction with TyrC2; (3) a hydrogen bond between the agonist N+H group (for agonists with this moiety) and the TrpB backbone CO. The hydrogen bond acceptor of the agonist faces the complementary subunit. In AChBP structures the hydrogen bond donor is a water molecule, which in turn binds to the protein, establishing a network of three hydrogen bonding interactions: (4) agonist hydrogen bond acceptor to water; (5) the backbone NH of a conserved Leu on loop E (“LeuE”) to water; (6) the backbone CO of a conserved Asn on loop E (“AsnE”) to water. We will refer to noncovalent interactions by the numbers of Figures 4 and 5 throughout.
Figure 3.
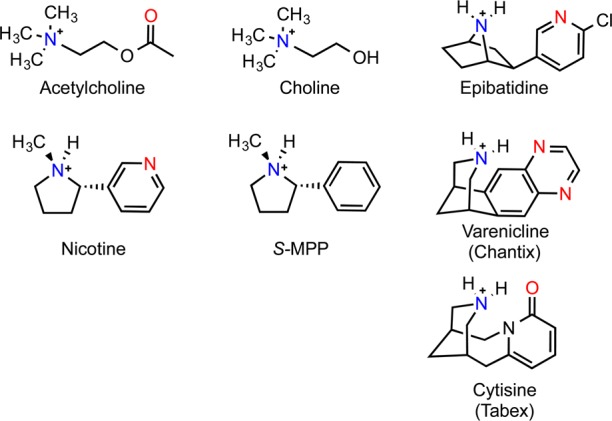
Nicotinic agonists. Cationic nitrogen (blue) and hydrogen bond acceptor (red) correspond to the nicotinic pharmacophore.
Figure 4.
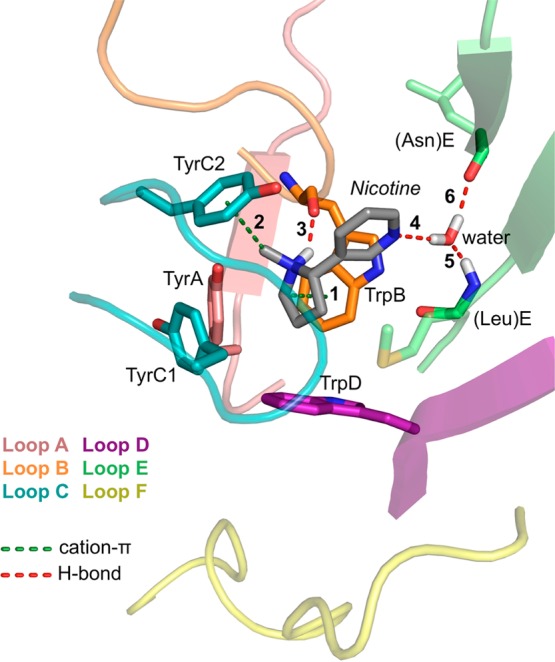
Nicotinic agonist binding interactions suggested by AChBP and key binding site residues. Shown is a structure of AChBP in complex with nicotine (PDB code 1UW6). Numbered interactions are discussed in the text. Explicit hydrogens are displayed for hydrogen bonding groups. AsnE and LeuE are conserved in all nAChRs but are Leu and Met, respectively, in the AChBP structure shown.
Figure 5.
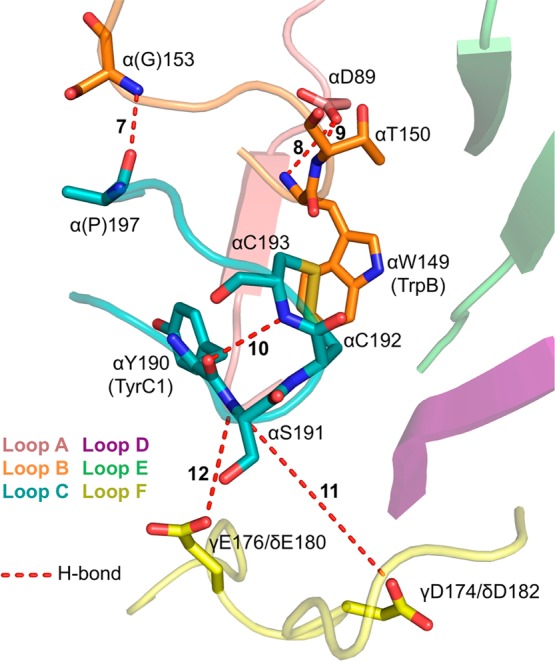
Interactions shaping the nAChR binding site, as seen in AChBP. AChBP structure and view of the binding site are identical to those in Figure 4 (PDB code 1UW6). Residue numbering is for the aligning residues of the muscle-type nAChR. Muscle-type nAChR side chains labeled are identical in AChBP, with the exceptions of G153 and P197, which are Ser and Ala, respectively, in AChBP.
These interactions form a binding model for these agonists with AChBP, interactions that can be tested at the actual nAChRs of interest. AChBP pharmacology differs from that of nAChRs (it is most similar to that of the α7 receptor),16 and of course, AChBPs serve a distinct functional role and do not gate a channel. Further, while AChBP structures with varying ligands bound generally show similar conformations, pharmacology varies considerably across nAChRs. Notably, nicotine is potent at the α4β2 receptor but not at muscle-type or α7 receptors, and in general the α7 receptor displays unique pharmacology from the other neuronal nAChRs.32
4.2. Ligand Binding to the α4β2 Neuronal nAChR
α4β2 receptors are one of the dominant nAChR subtypes expressed in the brain and can assemble in two different stoichiometries: (α4)2(β2)3 and (α4)3(β2)2. This receptor binds nicotine with high affinity and is established to play a key role in nicotine dependence from smoking. Consequently, it has been targeted by smoking cessation therapeutics, including the compounds varenicline33 and cytisine,34 which are marketed commercially as Chantix and Tabex, respectively. We have evaluated ligand binding interactions for the native agonist acetylcholine, for nicotine, and for these two smoking cessation agents at both stoichiometries of this receptor (Tables 1 and 2).
Table 1. EC50 Shifts for Mutations Probing Ligand–Receptor Cation−π Interactions in Cys-Loop Receptorsc.
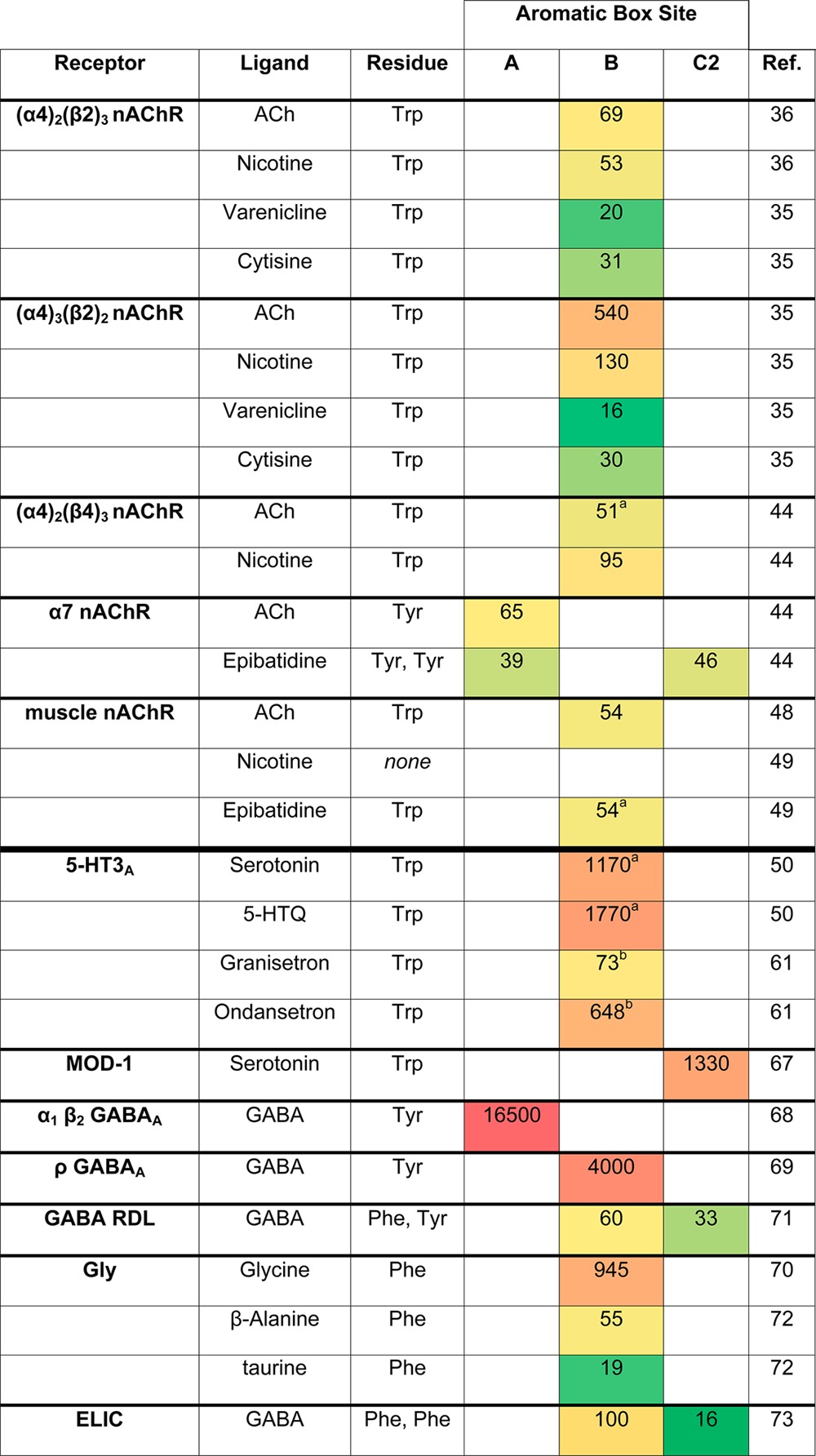
F4Trp data unavailable; value is an extrapolation of the fit of EC50 shifts for other deactivated Trp analogues.
Ratio of F4Trp/Trp IC50 values for these antagonists.
Values are EC50 fold shifts for F4Trp (Trp sites) or F3Phe (Phe or Tyr sites) relative to wild type, and values of >1 indicate a loss of function (increase in EC50 for the mutant). Shading ranges from green (smallest value) to red (largest value), with colors assigned to the logarithm of each value to emphasize differences in free energy.
Table 2. EC50 Shifts for α-Hydroxy Acid Mutations Probing Agonist–Receptor Hydrogen Bonds in nAChRsa.

For TrpB CO and AsnE CO (probing interactions 3 and 6, respectively), values are EC50 fold shift from wild type for the α-hydroxy acid mutation i + 1 to the site of interest. For LeuE NH (probing interaction 5), values are EC50 fold shift from wild type for α-hydroxy leucine at the LeuE site. Values of >1 indicate a loss of function (increase in EC50 for the mutant). Shading ranges from green (smallest value) to red (largest value), with colors assigned to the logarithm of each value to emphasize differences in free energy. ND = not determined.
All agonists, at both stoichiometries, form a cation−π interaction with TrpB (Table 1, interaction 1), as evidenced by responses to fluorotryptophan mutations.35,36 A plot of EC50 vs the cation−π binding ability of the side chain shows a compelling correlation. Single channel studies confirm that the perturbation to EC50 results from a change in the ligand binding step of receptor activation.36
Notably, mutagenesis does not corroborate the cation−π interaction to TyrC2 suggested by AChBP structures (interaction 2): this site accepts the highly deactivating CN-Phe mutation with no shift in receptor EC50 for both ACh and for nicotine.36 At other Cys-loop receptors we have been able to see evidence for cation−π interactions to two aromatics at the same time in a given receptor (see below), so a significant cation−π interaction to TyrC2 could have been detected. The TrpB cation−π interaction provides critical experimental evidence for the positioning of these agonists in the binding site, but the TyrC2 data suggest that we should expect differences between the AChBP binding site and that of the α4β2 receptor.
The remaining aromatic box residues of the principal face, TyrA and TyrC1, were also evaluated for cation−π interactions by unnatural amino acid mutagenesis. No such interaction was found at TyrA. TyrC1, well-known to have a critical role in nAChR gating,37,38 was extremely sensitive to substitution, preventing extensive evaluation. The role of a hydrogen bond donor appears to be especially critical at this site: both Phe (which removes the side chain −OH) and MeO-Phe (which retains hydrogen bond acceptor but not donor ability) produced EC50 shifts of approximately 100-fold for acetylcholine.36
We see strong evidence for hydrogen bonds between agonist N+H groups and the backbone CO of TrpB (interaction 3). Converting the residue i + 1 to TrpB to its α-hydroxy analogue weakens the hydrogen bond acceptor strength of this CO. This mutation produced a 19-fold shift in EC50 for nicotine at both receptor stoichiometries but critically had no effect on the EC50 for ACh (which cannot form this interaction).35,36
We have also probed the water-mediated hydrogen bonds seen in AChBP by converting LeuE to its α-hydroxy analogue, thus disrupting interaction 5.39 Significant impacts were seen for ACh and nicotine (Table 2). To convincingly assign an interaction between the LeuE backbone NH and the agonist hydrogen bond acceptor group, we performed a nontraditional “mutant cycle analysis” involving both the receptor and the agonist. We prepared and evaluated the nicotine analogue S-MPP, which replaces the pyridine ring with a phenyl group (Figure 3). We evaluated the additivity of the LeuE α-hydroxy mutation and the nicotine-to-(S-MPP) “mutation”. The mutations were strongly nonadditive, producing a 2.6 kcal/mol coupling energy. This clearly establishes a strong interaction between the agonist hydrogen bond acceptor and the LeuE backbone NH. Unfortunately, efforts to probe the other component of the water-mediated hydrogen bonding network (interaction 6) by modulating the AsnE backbone CO were unsuccessful for technical reasons (but see results below from other nAChRs).
For the three interactions identified above (1, 3, and 5), we see variations among different agonists and between the two receptor stoichiometries. Cation−π interactions to TrpB (interaction 1) are seen for all agonists tested, but the strength of this interaction varies. As discussed elsewhere,28 we consider the EC50 ratio of the tetrafluorotryptophan (F4-Trp) mutant to wild type Trp as a measure of cation−π strength. The cation−π effect ranges from a maximum of 540-fold for ACh at the (α4)3(β2)2 receptor, corresponding to 3.7 kcal/mol, to a minimum shift of 16-fold, 1.6 kcal/mol, for varenicline at this same stoichiometry (Table 1).
It is interesting that ACh, the only agonist evaluated with a quaternary ammonium group, shows the strongest cation−π interaction among this panel of agonists. Intrinsic cation−π binding affinity is greater for protonated amines than for quaternary amines.40,41 The strong binding energy for ACh suggests that the receptor has evolved to optimize the cation−π interaction when binding its native agonist. Also, for agonists such as nicotine it may not be possible to achieve a geometry that is simultaneously optimal for both the hydrogen bond interaction 3 and the cation−π interaction 1, and perhaps these agonists sacrifice some cation−π binding ability to strengthen the hydrogen bond.15,17,42
The interaction involving the agonist hydrogen bond acceptor group and the LeuE backbone NH also varies across the agonists assayed (Table 2). Notably, no interaction was seen for varenicline at either receptor stoichiometry. Two factors could account for this result. First, the hydrogen bond acceptor group and positively charged nitrogen have a greater separation in varenicline than in the other agonists investigated, raising the possibility that the geometry is no longer appropriate for this hydrogen bond, although the interaction is present in a structure of AChBP in complex with varenicline.42 The second possibility is that the agonist hydrogen bond acceptor could indeed be appropriately positioned, but the interaction is much weaker than for the other agonists considered, and thus perturbation of this hydrogen bond has no impact on our functional assay. Indeed, varenicline is expected to be a much poorer hydrogen bond acceptor than nicotine. It is known that pKa can be a reliable predictor of hydrogen bonding strength when considering closely related systems. On this basis, the quinoxaline N of varenicline (pKa = 0.8) is expected to be a significantly weaker hydrogen bond acceptor than the pyridine N of nicotine (pKa = 5.2).
In contrast, evidence for an unusually strong interaction was seen for cytisine: this agonist had the largest shift for the LeuE mutation at each receptor stoichiometry, with a remarkable 62-fold shift seen for (α4)2(β2)3. The hydrogen bond acceptor in cytisine is an amide oxygen, which is well-established to be a stronger hydrogen bond acceptor than a heterocyclic N such as is seen in nicotine or varenicline. Data like these indicate that the unnatural amino acid methodology not only can identify key interactions but also can give semiquantitative guidance as to the strength of a given interaction.
4.3. Ligand Binding to the α4β4 Neuronal nAChR
The α4β4 neuronal nAChR presents a different complementary face to the binding site than does α4β2, resulting in a distinct pharmacology.43 As in all nAChRs, residues of the aromatic box are identical, suggesting that interaction with the complementary face may play a greater role in establishing subtype specificity. Nevertheless, the side chains at the positions corresponding to LeuE and AsnE are very highly conserved. The same agonist–receptor interactions probed in the α4β2 receptor were also tested in the (α4)2(β4)3 stoichiometry of this receptor.
For α4β4, we found that ACh and nicotine bind the receptor using a cation−π interaction with TrpB (interaction 1), with EC50 shifts for TrpB mutants suggesting similar interaction strengths as in the (α4)2(β2)3 receptor (Table 1). Again, no cation−π interaction to TyrC2 (interaction 2) was seen. Also suggesting similar interactions with the α4 residues forming the binding site’s principal face, we observe a hydrogen bond between nicotine’s N+H and the TrpB backbone CO (interaction 3). Other aromatic residues of the principal face (TyrA, TyrC1) also appear to play a similar role in this receptor as in α4β2.44
Interesting contrasts to α4β2 were seen at the complementary (β4) face. The mutation to the LeuE NH (interaction 5) produced only small EC50 shifts, 2- to 3-fold smaller for ACh, nicotine, and epibatidine than in α4β2 and at the margin of being detectable in our assay (Table 2). This interaction is viable for β4, however: with cytisine a 14-fold shift was observed, though this is still 4-fold smaller than in (α4)2(β2)3.45 In the α4β4 receptor we were able to probe the AsnE backbone CO that additionally participates in the water-mediated hydrogen bond to the agonist’s hydrogen bond acceptor group in AChBPs (interaction 6). Here the α-hydroxy mutation was applied i + 1 from this site to weaken the CO’s hydrogen bond acceptor strength, as was done with hydrogen bonding interaction 3 (Figure 2A). The mutation of AsnE did not produce a meaningful effect for a panel of agonists: ACh, nicotine, epibatidine, varenicline, cytisine, and choline (EC50 shifts of 2-fold or less).
Overall, receptors with β4 rather than β2 at their complementary face have reduced binding affinities for a variety of agonists.43 Our data suggest that a weaker interaction involving the agonist hydrogen bond acceptor could contribute to this effect.
4.4. Ligand Binding to the α7 Neuronal nAChR
Several features of the α7 nAChR distinguish it from the other principal neuronal subtypes. It assembles as a homopentamer, and it is phylogenetically more ancestral than the heteromeric receptor subunits.46 In addition, α7 shows broad pharmacology and thus an apparently less specialized binding site.32 Clearly this is a different sort of neuronal receptor, though again, the critical aromatic box residues of the binding site are conserved. Chimeras of the α7 extracellular domain and AChBP have been crystallized, and they show remarkably similar binding sites to other AChBP structures with regard to the aromatic residues, LeuE, and AsnE.18,19
Consistent with this receptor’s distinct pharmacology, we observe a distinct pattern of ligand binding interactions for α7. TrpB is no longer the cation−π binding site for any of the agonists evaluated (ACh, epibatidine, and varenicline). ACh forms a cation−π interaction with TyrA (not seen in AChBPs, where it is not structurally feasible), while epibatidine forms cation−π interactions with both TyrA and TyrC2 (interaction 2), the stronger interaction being with TyrC2 (Table 1).28,44 Note that these findings for TyrA and TyrC2 are in stark contrast to our data from all other nAChRs probed, for which fluorinated Phe and Tyr analogues revealed that side chain electrostatics were relatively unimportant at these sites. Also in contrast to the other nAChRs, wild type function of the TyrA MeO-Phe mutant for ACh and varenicline reveals no functionally meaningful hydrogen bond donor role for this side chain, although a steric placeholder at the 4-position appears to be important. Interestingly, some agonist-specific behavior is observed at TyrC2: a steric placeholder at the 4-position is important for ACh and epibatidine but not for varenicline. In keeping with this receptor’s different utilization of principal face aromatic residues, the TrpB backbone CO hydrogen bond to the agonist N+H (interaction 3) appears to be weak in this receptor.
Hydrogen bonds between the agonist hydrogen bond acceptor and the AsnE backbone CO and LeuE backbone NH groups on the binding site’s complementary face appear to be weak or absent in the α7 nAChR (Table 2). Of the agonists evaluated (ACh, epibatidine, and varenicline) we see a modest shift (2.6-fold) only for epibatidine with the LeuE NH mutation. The α7 receptor is also distinctive in that it is the only one for which we have seen a meaningful interaction with the Asn CO (interaction 6): a 4.3-fold shift is seen for varenicline and only for varenicline. All other shifts were less than 2-fold.28 It is possible that an agonist H-bond acceptor group is not critical to binding and receptor activation for α7. Some α7-specific agonists lack the canonical hydrogen bond acceptor group. Notably, the structurally simple agonist tetramethylammonium has equivalent potency and efficacy to ACh at this receptor, in contrast to its much weaker activity at other nAChRs.32,47
4.5. Ligand Binding to the Muscle-Type nAChR
The extensively characterized (α1)2β1γδ receptor found at the neuromuscular junction is a “low affinity” nAChR, especially with regard to nicotine. Here TrpB is engaged in a cation−π interaction (interaction 1) with acetylcholine.48 We note that the muscle-type nAChR was actually the first receptor to which the unnatural amino acid methodology was applied, and the cation−π interaction to TrpB was established a full 3 years before the first AChBP crystal structure revealed the aromatic box motif.
There is also a cation−π interaction to TrpB with the relatively potent agonist epibatidine, but for nicotine the interaction is absent, in contrast to α4β2 and α4β4 (Table 1).48−50 Also in contrast to the α4-containing receptors, the agonist N+H–TrpB CO hydrogen bond (interaction 3) appears to be weak or absent for nicotine (Table 2). A modest interaction is detected for epibatidine.50 Electrostatics of TyrA and TyrC2 were unimportant when probing with ACh and with nicotine.51 In another demonstration of ligand-specific behavior at this receptor suggesting high specialization for its native agonist, the ACh analogue NorACh, which lacks a single methyl from its ammonium group, does not form a cation−π interaction with TrpB.49
At the complementary face, the LeuE NH mutation (interaction 5) has a dramatic effect for ACh (29-fold), a large effect for nicotine (10-fold), and as expected, no effect for choline (Table 2). Interestingly, no shift was seen for epibatidine.45 A distinct positioning of this agonist could account for this observation, or alternatively it may reflect the inherently weaker hydrogen bond acceptor strength of epibatidine’s 2-chloropyridine N compared to nicotine’s pyridine N (pKa of 0.5 vs 5.2, respectively).52 No meaningful shifts were detected for mutation of the AsnE CO group (interaction 6),45 as generally seen at other nAChRs evaluated (α4β4 and α7).
4.6. Interactions Shaping the nAChR Binding Site
Differences in ligand binding behavior across nAChRs are well established on the basis of pharmacology and have been elucidated in detail by the unnatural amino acid mutagenesis experiments described above. Such variety is not seen in the dozens of AChBP structures, for which little variation is seen in the relative positions of key residues. The aromatic box residues, TrpB CO, LeuE NH, and AsnE CO, while completely conserved, clearly engage differently with agonists in different receptor subtypes. Hence, peripheral interactions are likely responsible for the differences we observe. We have primarily probed peripheral interactions in the muscle-type receptor, and we will use residue numbering corresponding to the muscle-type receptor here. The relevant interactions are shown in Figure 5.
One distinguishing feature of high affinity nAChRs (such as α4β2) is a loop B lysine four residues from TrpB of the ligand binding site. The aligning residue is glycine in low affinity receptors such as muscle-type and α7. In AChBPs, the backbone NH of this residue forms a hydrogen bond to the backbone carbonyl i + 1 to TyrC2 on loop C (interaction 7, Figure 5). Molecular dynamics simulations suggested that mutations introducing a side chain to the loop B glycine site favor formation of this hydrogen bond.53 We studied this interaction in the muscle-type receptor and found that mutation of this glycine (α1 G153) to lysine (as found in the high affinity α4 subunit) dramatically increases the potency of nicotine at this receptor. The increase in affinity occurs because nicotine now forms a cation−π interaction with TrpB (interaction 1) and a strong hydrogen bond with the TrpB backbone CO (interaction 3).36,54 Such enhanced binding affinity has meaningful functional consequences in humans: a single nucleotide polymorphism producing the α1 G153S mutant induces a slow-channel myasthenic syndrome.55
The loop B Gly to Lys mutation in α7 also increases agonist potency but notably does not induce a cation−π interaction with TrpB; it instead strengthens the existing cation−π interaction with TyrC2 for ACh and for Epi. Hence the G153 mutation appears to effect global repositioning of the binding site, with different ligand binding implications at each receptor. The “reverse mutations” in the α4β2 receptor, Lys-to-Gly or α-hydroxy acid mutations to the proposed loop B–loop C backbone hydrogen bond (interaction 7), had only small effects, suggesting that additional factors may support the high affinity binding evolved at this receptor.35
A conserved aspartate on loop A (α1 D89 in the muscle-type receptor) is also involved in shaping the nAChR binding site. Structural studies of AChBPs revealed that this residue is positioned behind TrpB, and several investigators have proposed an essential role for the negative charge of this Asp in agonist recognition.15,56 Unnatural amino acid mutagenesis in the muscle-type receptor established that side chain charge is not critical, as mutation to neutral analogues such as Nha and Akp produced only modest effects. Instead this Asp side chain participates in a network of functionally significant hydrogen bonds. The hydrogen bond partners are two backbone NH groups on loop B: those of TrpB and of Thr150 (TrpB + 1), confirmed by α-hydroxy acid mutagenesis of these backbone groups (Figure 5, interactions 8 and 9).23
Another interesting motif of the nAChR agonist binding site is the C loop vicinal disulfide of the principal binding face, the defining structural feature of nAChR α subunits. In most AChBP structures, the vicinal disulfide participates in a type I β-turn of the C loop involving a hydrogen bond between the C193 backbone NH and the Y190 (TyrC1) backbone CO (Figure 5, interaction 10). In the muscle-type receptor, backbone mutations and mutant cycle analyses that probe this hydrogen bond establish a strong interaction. These include N-methylcysteine or α-hydroxycysteine mutants of C193 or an α-hydroxy mutation to S191, which modulates the Y190 CO. Interestingly, coupling to the Y190 CO is seen even for the C193A side chain mutation, which preserves the C193 backbone but eliminates the vicinal disulfide. Thus, the primary role of the nAChR α subunit vicinal disulfide appears to be establishing an optimal position of the C193 backbone for hydrogen bonding.57
Further studies of the α-hydroxy acid mutation at S191 established a specific role for the backbone NH as part of an extended network of hydrogen bonds, connecting this NH to the side chain of γD174/δD180, a conserved aspartate located across the subunit interface on loop F (Figure 5, interaction 11).58 The γD174/δD180 aspartate had been shown in a classic cross-linking experiment to lie near the vicinal disulfide.59 However, AChBP structures place γD174/δD180 quite far from the agonist binding site, hence the imagery of Figure 5. Instead, in AChBPs a different loop F aspartate (aligning to γE176/δE182 in the muscle receptor) forms a hydrogen bond to the S191 NH (interaction 12). Conventional mutagenesis of the γE176/δE182 side chain convincingly rules out a significant functional role for this residue. These findings thus establish a substantive difference between AChBP and the full receptors. The AChBP structures show a hydrogen bond corresponding to 12 in Figure 5, but our studies of the full receptor rule out interaction 12 and establish hydrogen bond 11 as playing an important functional role instead, consistent with the earlier biochemical studies. The long-range nature of the interactions probed here was established by mutant cycles between the termini of this hydrogen bonding network, the C193 NH and the γD174/δD180 side chain (spanning interactions 10 and 11), which show a robust coupling energy (2.2 kcal/mol).57 Evidently, the characteristic vicinal disulfide of the α subunit positions the S191 backbone NH for its intersubunit hydrogen bond, via the carbonyl of TyrC1. Presumably hydrogen bond 11 forms in the ligand-bound, open channel and could accompany C loop closure upon ligand binding. Such rearrangements could underlie the well-known gating significance of TyrC1, a critical binding site residue of the aromatic box.37,38
4.7. Overview of nAChR Studies
The family of nAChRs represent a classic example of a common situation in drug development: a family of closely related receptors that show specific distribution patterns, differing pharmacologies, and distinct physiological roles. The large number of AChBP structures with relevant small molecules bound have provided invaluable insights into possible drug–receptor interactions. However, very little variation is seen in protein structure, and so little insight into subtype specificity can be obtained.
Using unnatural amino acid mutagenesis, we have seen substantial variations in drug–receptor interactions in specific receptor subtypes. The anticipated cation−π interaction is evident in all receptors, with TrpB being the aromatic in most subtypes. However, the α7 receptor rejects TrpB and instead makes cation−π interactions to TyrA and TyrC2, with the TyrA interaction being quite incompatible with AChBP structures. This inconsistency arises despite the fact that AChBP pharmacology is closest to that of α7 receptors.
Along with the cation−π interaction, the other key component of the nicotinic pharmacophore is the hydrogen bond acceptor, defined by the CO of ACh and the pyridine N of nicotine. Certainly, it was very difficult to think about probing this interaction before the AChBP structures, and a novel model arose from them. Instead of a direct interaction with the protein, the agonist binds to a water molecule. This water in turn makes two hydrogen bonds to the protein backbone of the complementary face. We find many instances where the LeuE NH is important for receptor function (interaction 5), and using S-MPP as a probe, we have directly linked the NH to the pyridine N of nicotine. There is some variability that could be quite relevant to subtype specificity issues, the α4β4 and α7 receptors showing weaker interactions. Still, the LeuE NH interaction is clearly important in the family. In contrast, we find scant evidence for involvement of the AsnE CO (interaction 6). We probed this interaction in 13 different drug–receptor combinations (Table 2). In 11 cases, we saw no effect; one produced a factor of 2 change, which we consider to be borderline meaningful, and one produced a factor of 4. It may well be that this interaction is not functionally relevant in nAChRs, which raises an interesting possibility. If the water molecule seen in AChBP structures was absent in nAChRs, the LeuE NH can hydrogen-bond directly to the drug, and our studies would show an important effect of mutation. The AsnE CO can only engage in a water-mediated hydrogen bond to the drug, and if the water is absent, our studies would show negligible/small effects.
In other studies we have probed interactions that are peripheral to the agonist binding site but that have strong influences on receptor pharmacology. We characterized several hydrogen bonding interactions that influence receptor function, and in one case, we see a significance difference between predictions from the AChBP structures and the results of our functional studies (interactions 11 and 12).
5. Other Cys-Loop Receptors
We have conducted numerous studies of other members of the Cys-loop family. Here we highlight studies that parallel our work on nAChRs.
5-HT3 receptors, serotonin receptors of the Cys-loop superfamily, are most closely related to the nAChRs and also are cation channels. 5-HT3 receptors are widely distributed in the CNS and also play important roles in the peripheral nervous system. 5-HT3 receptor antagonists are currently prescribed for management of nausea, vomiting, and irritable bowel syndrome.60
As in nAChRs, the binding site of 5-HT3A receptors is rich in aromatic residues. TrpB, TyrC2, and TrpD are conserved, and additionally there are three tyrosines on loop E of the complementary face: Y141 (aligning to the nAChR AsnE site), Y143, and Y153 (aligning to the nAChR LeuE site). TyrA is absent, but an additional glutamate is found on loop A. A structure of a modified AChBP in complex with serotonin has been reported. The construct contains four point mutations that significantly increase serotonin’s binding affinity (by almost 40-fold),20 although some important binding residues are absent.
Fluorination studies revealed a strong cation−π interaction to TrpB in the 5-HT3A receptor (Table 1).49 Cation−π interactions involving TrpB were also detected for the antagonists granisetron and ondansetron, suggesting a conserved ligand binding mode that includes these actively prescribed antiemetic compounds (marketed as Kytril and Zofran, respectively).61 Tyrosines of the binding site were also evaluated by unnatural amino acid mutagenesis, revealing a critical hydrogen bond donor role for Y143 and a hydrogen bond acceptor role for Y153 on loop E, both of which may contribute to receptor gating.62
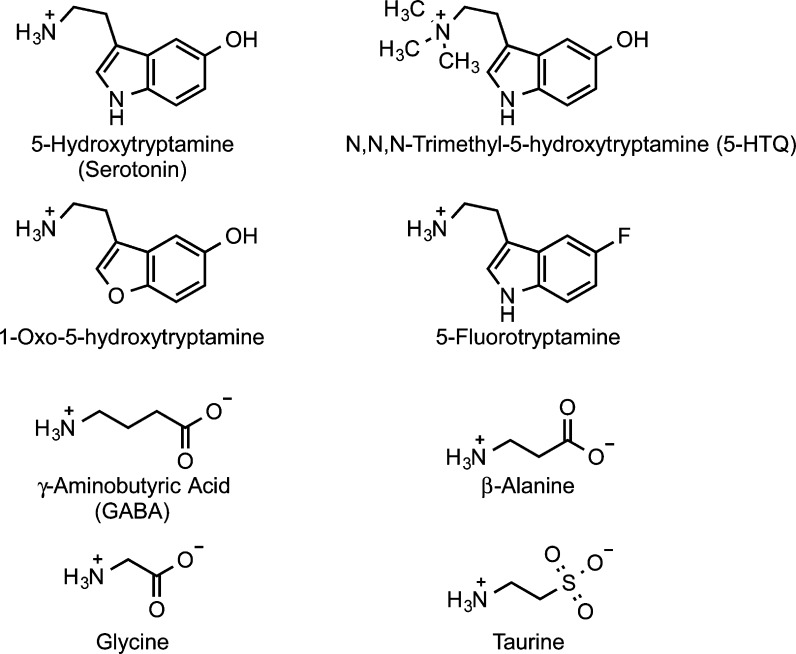
Serotonin analogues have clarified receptor recognition of this ligand’s polar groups (Figure 6). Interestingly, the quaternary trimethylammonium analogue of serotonin (5-HTQ) is as potent as serotonin at this receptor and also forms a strong cation−π interaction with TrpB.49 Surprisingly, 1-oxo-5-hydroxytryptamine, in which an O replaces the indole NH of serotonin, is equipotent to serotonin and a full agonist, suggesting that this NH group is not essential to receptor activation.63 In contrast, serotonin analogues that replace the 5-hydroxy group, such as 5-fluorotryptamine, have notably reduced affinities and low efficacies.64 Conventional and unnatural amino acid mutagenesis studies suggest that the hydrogen bonding partner for the 5-hydroxy group is E129 on loop A. At this site hydrogen bond acceptor ability, but not charge, is critical to ligand binding.65
Figure 6.
Agonists of serotonin, GABA, and glycine receptors.
Hence, both key interactions for serotonin recognition by the 5-HT3A receptor apparently lie on the binding site’s primary face: a cation−π interaction with TrpB and a hydrogen bond between E129 (corresponding to TyrA) and the ligand’s 5-hydroxy group. As in the nAChRs, a conserved aspartate lies behind TrpB on loop A, and double mutant cycles between this side chain (D124) and α-hydroxy acid mutations that delete the backbone NH of either TrpB (W183) or L184 demonstrate important hydrogen bonds between the Asp and these loop B groups. Triple mutant cycles that expand to include the critical E129 side chain demonstrate coupling between E129 and the loop A–loop B interaction: any single mutation out of the three decouples the other two. These results suggest a network of coupled noncovalent interactions spanning from the TrpB cation−π interaction with serotonin, through a series of hydrogen bonds to loop A, to the proximal loop A side chain E129, which makes a hydrogen bond to the agonist 5-hydroxy group.66
Another receptor gated by serotonin is the C. elegans MOD-1 receptor, a chloride channel that is actually more closely related to the GABA and glycine receptors. MOD-1 has the B and C2 side chains of the aromatic box transposed relative to 5-HT3A: a Tyr is at the B site, and a Trp is at the C2 site. Remarkably, the cation−π interaction for serotonin follows the tryptophan to the C2 site (Table 1), providing a case of the same agonist binding to two homologous binding sites with differing orientations.67 The switch could reflect the inherently stronger cation−π binding ability of a Trp vs a Tyr.
GABAA and glycine receptors are anion-selective Cys-loop receptors that mediate fast inhibitory neurotransmission in adult neurons. These receptors have high homology and ∼30–35% sequence identity to the crystallized C. elegans GluCl receptor, making this structure a useful template for these receptors’ binding sites.13 In the GluCl structure aromatic residues are found at the nAChR aromatic box A, B, and C2 sites (Phe, Tyr, and Tyr, respectively), and the structure implies a strong cation−π interaction between glutamate and TyrC2, with perhaps a weaker one to TyrB (Figure 7).
Figure 7.
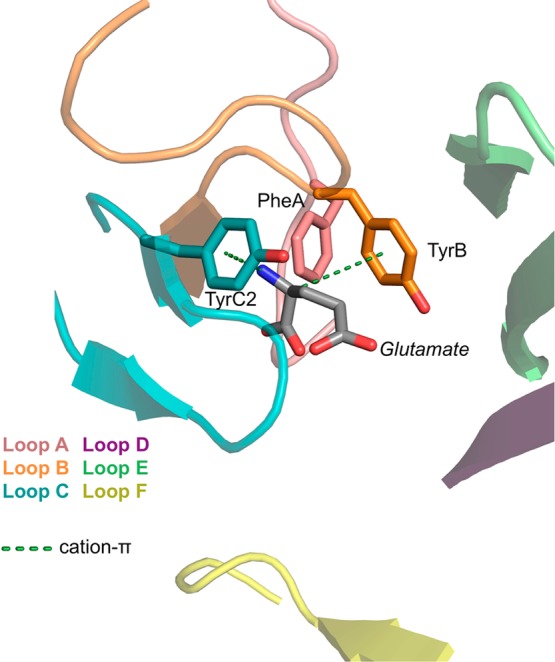
Ligand binding site of C. elegans GluCl in complex with glutamate (PDB code 3RIF). The cationic group of glutamate participates in a cation−π interaction with the conserved aromatic box site TyrC2 and perhaps the TyrB site as well. The carboxylates of glutamate are paired with Arg side chains from the complementary face of the binding site.
Unnatural amino acid mutagenesis has been used to identify cation−π interactions in GABAA and glycine receptors. In all cases studied, such interactions have been localized to aromatic residues on the principal face of the binding site, but the residue(s) involved is surprisingly variable. In the GABAA receptor comprising α1 and β2 subunits, the cation−π site for GABA is TyrA (β2Tyr97),68 while in the GABAA ρ1 homomeric receptor (also known as GABAC), the site is TyrB (ρ1Tyr198).69 In the glycine receptor (α1 homomer) the glycine cation−π site is PheB (α1Phe159).70 In the above examples the entire binding site was investigated, but only a single cation−π site was found. However, in the insect GABA RDL receptor, an anion-selective channel related to the vertebrate GABAA receptors, GABA is bound by two cation−π interactions: both PheB and TyrC2 are involved, consistent with the GluCl structure.71
The variability of cation−π sites among GABA and glycine receptors suggests that while the GluCl structure is no doubt a valuable template for homology modeling, positioning of agonists in the binding site is likely to vary. We also see that the apparent strength of the cation−π interaction varies considerably (Table 1). For cation−π interactions involving Phe and Tyr, we consider the EC50 ratio of the trifluorophenylalanine (F3-Phe) mutant to Phe as a measure of cation−π strength. Among the GABA and glycine receptors, estimated cation−π strengths range from a low of 2.1 kcal/mol for TyrC2 with GABA at the insect RDL receptor71 to a high of 5.8 kcal/mol for GABA at TyrA of the α1, β2 GABAA receptor, the strongest cation−π interaction we have characterized at any Cys-loop receptor.68 Interestingly, variations in cation−π strength are seen among different agonists at the glycine receptor (all of which bind to PheB): for glycine, the interaction strength is 4.1 kcal/mol, while significantly weaker interactions of 2.4 and 1.8 kcal/mol are seen for the partial agonists β-alanine and taurine, respectively (Table 1, Figure 6).72
Finally, there is one bona fide pentameric receptor–agonist complex captured by crystallography that has been functionally interrogated by unnatural amino acid mutagenesis. The prokaryotic receptor ELIC, a homologue of the eukaryotic Cys loop receptors, was crystallized in complex with GABA. The structure shows two cation−π interactions with phenylalanines at the aromatic box B and C2 sites, and mutagenesis confirms both interactions. The stronger interaction properly corresponds to the Phe closest to GABA’s ammonium group in the structure.73
6. Conclusions
This work highlights the fruitful interplay between structural studies of model systems and functional studies on full mammalian receptors. There is no doubt that the structures provide essential guidance as to possible key binding interactions. However, extrapolating one structural model to an entire family of receptors and to a wide range of agonists and antagonists is not warranted. It will always be essential to probe the real receptors to establish the essential binding features. Along with providing crucial tests of predictions based on structural information, the detailed binding interactions revealed by unnatural amino acid mutagenesis present excellent benchmarks for modeling studies that aim to reveal the origins of subtype specificity.
Across the Cys loop family, we see that the details of ligand binding (which interactions are present and which residues are involved) vary across receptors and among different ligands at the same receptor. It has long been appreciated that closely related receptors can have distinct pharmacologies, so these differences should not be surprising. However, rationalizing such distinctions across receptors is no easy task. Among the closely related subtypes of the nAChR family, similar binding sites engage agonists differently, and the differences become more pronounced on moving to other members of the Cys-loop receptor superfamily. Related binding sites must be shaped differently, and extrapolations of ligand binding from one receptor to another cannot readily be made. It is clear that caution is warranted when extrapolating from model structures to receptors of interest in human health.
Acknowledgments
Our work in this area has been supported by the NIH and by the California Tobacco-Related Disease Research Program of the University of California.
Glossary
Abbreviations Used
- LGIC
ligand-gated ion channel
- nAChR
nicotinic acetylcholine receptor
- GABA
γ-aminobutyric acid
- AChBP
acetylcholine binding protein
- Nha
nitrohomoalanine
- F4-Trp
tetrafluorotryptophan
- F3-Phe
trifluorophenylalanine
Biographies
Ethan B. Van Arnam received his Bachelors degree in Chemistry from Bowdoin College, ME. He earned his Ph.D. from California Institute of Technology, CA, in 2013, working with Dennis Dougherty. There he studied ligand binding and conformational changes in ligand-gated ion channels and G-protein-coupled receptors using unnatural amino acid mutagenesis. He is currently a postdoctoral scholar at Harvard Medical School, MA.
Dennis A. Dougherty received a B.S./M.S. from Bucknell University, PA, and a Ph.D. from Princeton University, NJ, working with Kurt Mislow, and then did postdoctoral work with Jerome Berson at Yale University, CT. In 1979 he joined the faculty of California Institute of Technology, CA, where he is now the George Grant Hoag Professor of Chemistry.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Nowak M. W.; Kearney P. C.; Sampson J. R.; Saks M. E.; Labarca C. G.; Silverman S. K.; Zhong W.; Thorson J.; Abelson J. N.; Davidson N.; Schultz P. G.; Dougherty D. A.; Lester H. A. Nicotinic receptor binding site probed with unnatural amino acid incorporation in intact cells. Science 1995, 268, 439–442. [DOI] [PubMed] [Google Scholar]
- Nowak M. W.; Gallivan J. P.; Silverman S. K.; Labarca C. G.; Dougherty D. A.; Lester H. A. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998, 293, 504–529. [DOI] [PubMed] [Google Scholar]
- Dougherty D. A. Cys-loop neuroreceptors: structure to the rescue?. Chem. Rev. 2008, 10851642–2653. [DOI] [PubMed] [Google Scholar]
- Lemoine D.; Jiang R.; Taly A.; Chataigneau T.; Specht A.; Grutter T. Ligand-gated ion channels: new insights into neurological disorders and ligand recognition. Chem. Rev. 2012, 112, 6285–6318. [DOI] [PubMed] [Google Scholar]
- Corringer P. J.; Poitevin F.; Prevost M. S.; Sauguet L.; Delarue M.; Changeux J. P. Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure 2012, 206941–956. [DOI] [PubMed] [Google Scholar]
- Corringer P. J.; Baaden M.; Bocquet N.; Delarue M.; Dufresne V.; Nury H.; Prevost M.; Van Renterghem C. Atomic structure and dynamics of pentameric ligand-gated ion channels: new insight from bacterial homologues. J. Physiol. 2010, 588Part 4565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C.; Zoli M.; Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol. Sci. 2006, 279482–491. [DOI] [PubMed] [Google Scholar]
- Jensen A. A.; Frolund B.; Liljefors T.; Krogsgaard-Larsen P. Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J. Med. Chem. 2005, 48154705–4745. [DOI] [PubMed] [Google Scholar]
- Miyazawa A.; Fujiyoshi Y.; Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature 2003, 4236943949–955. [DOI] [PubMed] [Google Scholar]
- Hilf R. J.; Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 2008, 4527185375–379. [DOI] [PubMed] [Google Scholar]
- Hilf R. J.; Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 2009, 4577225115–118. [DOI] [PubMed] [Google Scholar]
- Bocquet N.; Nury H.; Baaden M.; Le Poupon C.; Changeux J. P.; Delarue M.; Corringer P. J. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 2009, 4577225111–114. [DOI] [PubMed] [Google Scholar]
- Hibbs R. E.; Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 2011, 474734954–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixma T. K.; Smit A. B. Acetylcholine binding protein (AChBP): a secreted glial protein that provides a high-resolution model for the extracellular domain of pentameric ligand-gated ion channels. Ann. Rev. Biophys. Biomol. Struct. 2003, 32, 311–334. [DOI] [PubMed] [Google Scholar]
- Celie P. H.; van Rossum-Fikkert S. E.; van Dijk W. J.; Brejc K.; Smit A. B.; Sixma T. K. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 2004, 416907–914. [DOI] [PubMed] [Google Scholar]
- Rucktooa P.; Smit A. B.; Sixma T. K. Insight in nAChR subtype selectivity from AChBP crystal structures. Biochem. Pharmacol. 2009, 787777–787. [DOI] [PubMed] [Google Scholar]
- Rucktooa P.; Haseler C. A.; van Elk R.; Smit A. B.; Gallagher T.; Sixma T. K. Structural characterization of binding mode of smoking cessation drugs to nicotinic acetylcholine receptors through study of ligand complexes with acetylcholine-binding protein. J. Biol. Chem. 2012, 2872823283–23293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. X.; Huang S.; Bren N.; Noridomi K.; Dellisanti C. D.; Sine S. M.; Chen L. Ligand-binding domain of an alpha7-nicotinic receptor chimera and its complex with agonist. Nat. Neurosci. 2011, 14101253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecz A.; Taylor P. Creating an alpha7 nicotinic acetylcholine recognition domain from the acetylcholine-binding protein: crystallographic and ligand selectivity analyses. J. Biol. Chem. 2011, 2864942555–42565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesters D.; Thompson A. J.; Brams M.; van Elk R.; Spurny R.; Geitmann M.; Villalgordo J. M.; Guskov A.; Danielson U. H.; Lummis S. C.; Smit A. B.; Ulens C. Structural basis of ligand recognition in 5-HT3 receptors. EMBO Rep. 2013, 14149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J. T.; Cornish V. W.; Schultz P. G. An experimental approach to evaluating the role of backbone interactions in proteins using unnatural amino acid mutagenesis. Biochemistry 1997, 36, 11314–11322. [DOI] [PubMed] [Google Scholar]
- Deechongkit S.; Nguyen H.; Powers E. T.; Dawson P. E.; Gruebele M.; Kelly J. W. Context-dependent contributions of backbone hydrogen bonding to beta-sheet folding energetics. Nature 2004, 4306995101–105. [DOI] [PubMed] [Google Scholar]
- Cashin A. L.; Torrice M. M.; McMenimen K. A.; Lester H. A.; Dougherty D. A. Chemical-scale studies on the role of a conserved aspartate in preorganizing the agonist binding site of the nicotinic acetylcholine receptor. Biochemistry 2007, 463630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty D. A.; Stauffer D. A. Acetylcholine binding by a synthetic receptor. Implications for biological recognition. Science 1990, 250, 1558–1560. [DOI] [PubMed] [Google Scholar]
- Dougherty D. A. Cation−π interactions in chemistry and biology. A new view of benzene, Phe, Tyr, and Trp. Science 1996, 271, 163–168. [DOI] [PubMed] [Google Scholar]
- Ma J. C.; Dougherty D. A. The cation−π interaction. Chem. Rev. 1997, 9751303–1324. [DOI] [PubMed] [Google Scholar]
- Dougherty D. A. The cation−π interaction. Acc. Chem. Res. 2013, 464885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arnam E. B.; Blythe E. E.; Lester H. A.; Dougherty D. A. An unusual pattern of ligand–receptor interactions for the alpha7 nicotinic acetylcholine receptor, with implications for the binding of varenicline. Mol. Pharmacol. 2013, 842201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar N. S.; Gotti C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology 2009, 561237–246. [DOI] [PubMed] [Google Scholar]
- Beers W. H.; Reich E. Structure and activity of acetylcholine. Nature 1970, 2285275917–922. [DOI] [PubMed] [Google Scholar]
- Hansen S. B.; Sulzenbacher G.; Huxford T.; Marchot P.; Taylor P.; Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005, 24203635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horenstein N. A.; Leonik F. M.; Papke R. L. Multiple pharmacophores for the selective activation of nicotinic alpha7-type acetylcholine receptors. Mol. Pharmacol. 2008, 7461496–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe J. W.; Brooks P. R.; Vetelino M. G.; Wirtz M. C.; Arnold E. P.; Huang J.; Sands S. B.; Davis T. I.; Lebel L. A.; Fox C. B.; Shrikhande A.; Heym J. H.; Schaeffer E.; Rollema H.; Lu Y.; Mansbach R. S.; Chambers L. K.; Rovetti C. C.; Schulz D. W.; Tingley F. D. 3rd; O’Neill B. T. Varenicline: an alpha4beta2 nicotinic receptor partial agonist for smoking cessation. J. Med. Chem. 2005, 48103474–3477. [DOI] [PubMed] [Google Scholar]
- Etter J.-F.; Lukas R. J.; Benowitz N. L.; West R.; Dresler C. M. Cytisine for smoking cessation: A research agenda. Drug Alcohol Depend. 2008, 921–33–8. [DOI] [PubMed] [Google Scholar]
- Da Silva Tavares X.; Blum A. P.; Nakamura D. T.; Puskar N. L.; Shanata J. A.; Lester H. A.; Dougherty D. A. Variations in binding among several agonists at two stoichiometries of the neuronal, alpha4beta2 nicotinic receptor. J. Am. Chem. Soc. 2012, 1342811474–11480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu X.; Puskar N. L.; Shanata J. A.; Lester H. A.; Dougherty D. A. Nicotine binding to brain receptors requires a strong cation–pi interaction. Nature 2009, 4587237534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit P.; Bruhova I.; Auerbach A. Sources of energy for gating by neurotransmitters in acetylcholine receptor channels. Proc. Natl. Acad. Sci. U.S.A. 2012, 109249384–9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaselli G. F.; McLaughlin J. T.; Jurman M. E.; Hawrot E.; Yellen G. Mutations affecting agonist sensitivity of the nicotinic acetylcholine receptor. Biophy. J. 1991, 603721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A. P.; Lester H. A.; Dougherty D. A. Nicotinic pharmacophore: the pyridine N of nicotine and carbonyl of acetylcholine hydrogen bond across a subunit interface to a backbone NH. Proc. Natl. Acad. Sci. U.S.A. 2010, 1073013206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakyne C. A.; Meot-Ner (Mautner) M. Unconventional Ionic Hydrogen Bonds. 2. NH+ ...π. Complexes of Onium Ions with Olefins and Benzene Derivatives. J. Am. Chem. Soc. 1985, 107, 474–479. [Google Scholar]
- Meot-Ner M.; Deakyne C. A. Unconventional Ionic Hydrogen Bonds. 1. CHδ+...X. Complexes of Quaternary Ions with n- and π-Donors. J. Am. Chem. Soc. 1985, 107, 469–474. [Google Scholar]
- Billen B.; Spurny R.; Brams M.; van Elk R.; Valera-Kummer S.; Yakel J. L.; Voets T.; Bertrand D.; Smit A. B.; Ulens C. Molecular actions of smoking cessation drugs at alpha4beta2 nicotinic receptors defined in crystal structures of a homologous binding protein. Proc. Natl. Acad. Sci. U.S.A. 2012, 109239173–9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. J.; Beck A.; Luetje C. W. Neuronal nicotinic receptor beta2 and beta4 subunits confer large differences in agonist binding affinity. Mol. Pharmacol. 1998, 5461132–1139. [PubMed] [Google Scholar]
- Puskar N. L.; Xiu X.; Lester H. A.; Dougherty D. A. Two neuronal nicotinic acetylcholine receptors, alpha4beta4 and alpha7, show differential agonist binding modes. J. Biol. Chem. 2011, 2861614618–14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A. P.; Van Arnam E. B.; German L. A.; Lester H. A.; Dougherty D. A. Binding interactions to the complementary subunit of nicotinic receptors. J. Biol. Chem. 2013, 288, 6991–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N.; Corringer P. J.; Changeux J. P. The diversity of subunit composition in nAChRs: evolutionary origins, physiologic and pharmacologic consequences. J. Neurobiol. 2002, 534447–456. [DOI] [PubMed] [Google Scholar]
- Papke R. L.; Bencherif M.; Lippiello P. An evaluation of neuronal nicotinic acetylcholine receptor activation by quaternary nitrogen compounds indicates that choline is selective for the alpha 7 subtype. Neurosci. Lett. 1996, 2133201–204. [DOI] [PubMed] [Google Scholar]
- Zhong W.; Gallivan J. P.; Zhang Y.; Li L.; Lester H. A.; Dougherty D. A. From ab initio quantum mechanics to molecular neurobiology: a cation−π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. U.S.A. 1998, 952112088–12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beene D. L.; Brandt G. S.; Zhong W.; Zacharias N. M.; Lester H. A.; Dougherty D. A. Cation–pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry 2002, 413210262–10269. [DOI] [PubMed] [Google Scholar]
- Cashin A. L.; Petersson E. J.; Lester H. A.; Dougherty D. A. Using physical chemistry to differentiate nicotinic from cholinergic agonists at the nicotinic acetylcholine receptor. J. Am. Chem. Soc. 2005, 1271350–356. [DOI] [PubMed] [Google Scholar]
- Kearney P. C.; Nowak M. W.; Zhong W.; Silverman S. K.; Lester H. A.; Dougherty D. A. Dose-response relations for unnatural amino acids at the agonist binding site of the nicotinic acetylcholine receptor: tests with novel side chains and with several agonists. Mol. Pharmacol. 1996, 5051401–1412. [PubMed] [Google Scholar]
- Linnell R. H. Dissociation constants of 2-substituted pyridines. J. Org. Chem. 1960, 252290–290. [Google Scholar]
- Grutter T.; de Carvalho L. P.; Le Novere N.; Corringer P. J.; Edelstein S.; Changeux J. P. An H-bond between two residues from different loops of the acetylcholine binding site contributes to the activation mechanism of nicotinic receptors. EMBO J. 2003, 2291990–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskar N. L.; Lester H. A.; Dougherty D. A. Probing the effects of residues located outside the agonist binding site on drug-receptor selectivity in the nicotinic receptor. ACS Chem. Biol. 2012, 75841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine S. M.; Ohno K.; Bouzat C.; Auerbach A.; Milone M.; Pruitt J. N.; Engel A. G. Mutation of the acetylcholine receptor alpha subunit causes a slow-channel myasthenic syndrome by enhancing agonist binding affinity. Neuron 1995, 151229–239. [DOI] [PubMed] [Google Scholar]
- Lee W. Y.; Sine S. M. Invariant aspartic acid in muscle nicotinic receptor contributes selectively to the kinetics of agonist binding. J. Gen. Phys. 2004, 1245555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A. P.; Gleitsman K. R.; Lester H. A.; Dougherty D. A. Evidence for an extended hydrogen bond network in the binding site of the nicotinic receptor role of the vicinal disulfide of the alpha 1 subunit. J. Biol. Chem. 2011, 2863732251–32258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleitsman K. R.; Kedrowski S. M. A.; Lester H. A.; Dougherty D. A. An intersubunit hydrogen bond in the nicotinic acetylcholine receptor that contributes to channel gating. J. Biol. Chem. 2008, 2835135638–35643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowski C.; Karlin A. Agonist binding site of Torpedo electric tissue nicotinic acetylcholine receptor. A negatively charged region of the delta subunit within 0.9 nm of the alpha subunit binding site disulfide. J. Biol. Chem. 1991, 2663322603–22612. [PubMed] [Google Scholar]
- Lummis S. C. 5-HT3 receptors. J. Biol. Chem. 2012, 287, 40239–40245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy N. H.; Lester H. A.; Dougherty D. A. Ondansetron and granisetron binding orientation in the 5-HT(3) receptor determined by unnatural amino acid mutagenesis. ACS Chem. Biol. 2012, 7101738–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beene D. L.; Price K. L.; Lester H. A.; Dougherty D. A.; Lummis S. C. R. Tyrosine residues that control binding and gating in the 5-hydroxytryptamine(3) receptor revealed by unnatural amino acid mutagenesis. J. Neurosci. 2004, 24419097–9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrowski S. M. A.; Bower K. S.; Dougherty D. A. 1-Oxo-5-hydroxytryptamine: a surprisingly potent agonist of the 5-HT3 (serotonin) receptor. Org. Lett. 2007, 9173205–3207. [DOI] [PubMed] [Google Scholar]
- Bower K. S.; Price K. L.; Sturdee L. E.; Dayrell M.; Dougherty D. A.; Lummis S. C. 5-Fluorotryptamine is a partial agonist at 5-HT3 receptors, and reveals that size and electronegativity at the 5 position of tryptamine are critical for efficient receptor function. Eur. J. Pharmacol. 2008, 5803291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price K. L.; Bower K. S.; Thompson A. J.; Lester H. A.; Dougherty D. A.; Lummis S. C. R. A hydrogen bond in loop a is critical for the binding and function of the 5-HT3 receptor. Biochemistry 2008, 47246370–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles T. F.; Bower K. S.; Lester H. A.; Dougherty D. A. A coupled array of noncovalent interactions impacts the function of the 5-HT3A serotonin receptor in an agonist-specific way. ACS Chem. Neurosci. 2012, 310753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu T. W.; Lester H. A.; Dougherty D. A. Different binding orientations for the same agonist at homologous receptors: a lock and key or a simple wedge?. J. Am. Chem. Soc. 2003, 125236850–6851. [DOI] [PubMed] [Google Scholar]
- Padgett C. L.; Hanek A. P.; Lester H. A.; Dougherty D. A.; Lummis S. C. R. Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation–pi interaction between GABA and beta(2)Tyr97. J. Neurosci. 2007, 274886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummis S. C.; D L. B.; Harrison N. J.; Lester H. A.; Dougherty D. A. A cation–pi binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem. Biol. 2005, 129993–997. [DOI] [PubMed] [Google Scholar]
- Pless S. A.; Millen K. S.; Hanek A. P.; Lynch J. W.; Lester H. A.; Lummis S. C.; Dougherty D. A. A cation–pi interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J. Neurosci. 2008, 284310937–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummis S. C. R.; McGonigle I.; Ashby J. A.; Dougherty D. A. Two amino acid residues contribute to a cation–pi binding interaction in the binding site of an insect GABA receptor. J. Neurosci. 2011, 313412371–12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless S. A.; Hanek A. P.; Price K. L.; Lynch J. W.; Lester H. A.; Dougherty D. A.; Lummis S. C. R. A cation–pi interaction at a phenylalanine residue in the glycine receptor binding site is conserved for different agonists. Mol. Pharmacol. 2011, 794742–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurny R.; Ramerstorfer J.; Price K.; Brams M.; Ernst M.; Nury H.; Verheij M.; Legrand P.; Bertrand D.; Bertrand S.; Dougherty D. A.; de Esch I. J.; Corringer P. J.; Sieghart W.; Lummis S. C.; Ulens C. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl. Acad. Sci. U.S.A. 2012, 10944E3028–E3034. [DOI] [PMC free article] [PubMed] [Google Scholar]