Abstract
Insertion and deletion (INDEL) is one of the main events contributing to genetic and phenotypic diversity, which receives less attention than SNP and large structural variation. To gain a better knowledge of INDEL variation in chicken genome, we applied next generation sequencing on 12 diverse chicken breeds at an average effective depth of 8.6. Over 1.3 million non-redundant short INDELs (1–49 bp) were obtained, the vast majority (92.48%) of which were novel. Follow-up validation assays confirmed that most (88.00%) of the randomly selected INDELs represent true variations. The majority (95.76%) of INDELs were less than 10 bp. Both the detected number and affected bases were larger for deletions than insertions. In total, INDELs covered 3.8 Mbp, corresponding to 0.36% of the chicken genome. The average genomic INDEL density was estimated as 0.49 per kb. INDELs were ubiquitous and distributed in a non-uniform fashion across chromosomes, with lower INDEL density in micro-chromosomes than in others, and some functional regions like exons and UTRs were prone to less INDELs than introns and intergenic regions. Nearly 620,253 INDELs fell in genic regions, 1,765 (0.28%) of which located in exons, spanning 1,358 (7.56%) unique Ensembl genes. Many of them are associated with economically important traits and some are the homologues of human disease-related genes. We demonstrate that sequencing multiple individuals at a medium depth offers a promising way for reliable identification of INDELs. The coding INDELs are valuable candidates for further elucidation of the association between genotypes and phenotypes. The chicken INDELs revealed by our study can be useful for future studies, including development of INDEL markers, construction of high density linkage map, INDEL arrays design, and hopefully, molecular breeding programs in chicken.
Introduction
Chicken as one of the most important domestic animals not only provides essential proteins for human food industry, but also serves as an excellent biological model for many scientific researches [1]. Identifying genetic determinants of economically important traits or diseases is one of the main focuses of chicken genetic studies, which requires a comprehensive knowledge of DNA sequence variations as well as the development of numerous informative genetic markers. The near-complete chicken genome has made it possible to systematically study genetic variations. Up to now, several types of genetic variations have been identified across genomes, i.e. single nucleotide polymorphism (SNP), insertion and deletion (INDEL) and structural variation (SV). Studies in human show that INDEL is one of the main forms of genomic variations, with its occurrence in genome only second to SNP and even comparable to SNP in terms of affected bases [2], [3]. INDELs contribute substantially to genetic divergence both within and between species [4]–[7]. Besides, INDELs generally have a greater impact on gene functions than SNPs. Nearly 24% of the heritable disease mutations in human gene mutation database (HGMD) (http://www.hgmd.cf.ac.uk/ac/index.php) are INDELs. Many common human diseases are frequently caused by INDELs, such as cystic fibrosis [8] and Huntington's diseases [9]. In domestic animals, INDELs are also found to be responsible for a number of traits and diseases, such as double muscle trait [10] and factor XI deficiency [11] in cattle, immotile short-tail sperm defect in pig [12], and muscle mass in dog [13]. In chicken, INDELs of 9–15 bp in PMEL17 gene are causative mutations for plumage color (Dominant white, Dun and Smoky) [14] and an INDEL mutation in the growth hormone receptor (GHR) gene causes sex-linked dwarfism [15]. Therefore, INDEL is gaining an increasing attention recently and has been extensively discovered and studied in a variety of species [3], [6], [16]–[20].
With the rapid advance of sequencing technology, considerable progresses have been made in INDEL discovery in chicken genome. Wong et al. [21] partially sequenced three chicken breeds by capillary sequencing and identified 2.8 million SNPs by aligning the resultant reads to the reference genome, and about 10% of these variations are actually INDELs. Based on these results, Brandstrom and Ellegren [5] estimated that INDELs in unique sequence were as 5% abundance as SNPs in chicken genome. In their efforts to detect selective sweeps, Rubin et al. [22] contributed almost 1,300 novel large deletions. Currently, more than 9 million variants have been deposited into the chicken SNP database, 438,865 of which are INDELs, accounting for 4.7% of all variants (ftp://ftp.ncbi.nih.gov/snp/organisms/chicken_9031/VCF/, updated in June 11, 2013). However, previous studies with human and other model organisms showed that INDELs accounted for 9–14% of all genetic polymorphisms [3], [23], . Therefore, the relatively small proportion of INDEL in chicken SNP database indicates that a large number of INDELs in chicken genome may not have been discovered.
Recently, Fan et al. [25] sequenced two chickens and identified over 600,000 INDELs per individual, which was a great contribution to the current variation database. However, for detailed examination and validation of INDEL variation in chicken genome, a larger and more representative collection of INDEL is still desired. To this end, we performed next generation sequencing (NGS) to detect genome-wide INDELs in 12 diverse chickens, representatives of both commercial and Chinese indigenous breeds. We focused on the identification of short INDELs (1–50 bp), which are the predominant forms of INDEL in the genome [5], [17], [26], [27]. We also examined the distribution of INDELs in chicken genome and their potential influence on gene functions, which would be helpful in deepening our understanding of chicken genome variation, developing INDEL markers, and elucidating the association between genetic variations and phenotypes in the future.
Materials and Methods
Ethics statements
The whole blood samples were collected from brachial veins of chickens by standard venipuncture. The whole procedure was performed according to the protocol approved by the Animal Care and Use Committee of China Agricultural University.
Sample selection
Twelve female birds from 12 different chicken breeds were used in this study. Seven breeds, Beijing You (BY), Dongxiang (DX), Luxi game (LX), Shouguang (SG), Silkie (SK), Tibetan (TB) and Wenchang (WC), were Chinese indigenous. Four were commercial breeds, i.e., Cornish (CS), Rhode Island Red (RIR), White Leghorn (WL) and White Plymouth Rock (WR). A Red Jungle Fowl (RJF), the wild ancestor of domestic chickens, was also used. Birds from these breeds exhibit significant differences in appearance (comb type, skin and plumage color, etc.) and production performance (growth, egg production, feed consumption, etc.), and are believed to harbor extensive genetic diversity.
Library construction and sequencing
Genomic DNAs were extracted from blood samples using standard phenol/chloroform extraction method. DNA concentration and purity were accessed on NanoDrop (Thermo Fisher Scientific Inc. Waltham, MA, USA), and the qualified DNAs were used for library construction. Two paired-end libraries were constructed for each individual, with an intended 10-fold depth (5-fold for each library). Genomic DNAs were sheared to yield an average size of 500 bp and then ligated to Illumina paired-end adaptors. After PCR amplification and purification, the resulting libraries were sequenced on an Illumina Hiseq 2000 sequencer (Illumina Inc., San Diego, CA, USA). Raw reads of 2×100 bp were generated for downstream analysis.
Read mapping and variant calling
Chicken genome assembly (galGal4) was downloaded from UCSC Genome Browser website (http://hgdownload.soe.ucsc.edu/goldenPath/galGal4/bigZips/) [28]. In order to minimize mapping errors, we remove low quality reads with the help of NGS QC Toolkit [29] with default parameters. Considering the increasing error rate towards the end of reads due to the decay of signal intensity [30], we trimmed the last 10 bases of reads.
Mapping reads to reference genome was performed using Burrows-Wheeler Alignment tool (BWA ver 0.6.2) [31], with mainly default parameters. SAMtools (ver.0.1.19) [32] was used to convert the alignment results (in SAM format) to BAM format. Duplicated reads were removed using Picard package [32] and then the two BAM files from two libraries for each individual were merged by SAMtools. Reads were realigned around INDELs and base qualities were recalibrated before calling variants using the Genome Analysis Toolkit (GATK, ver 2.4.9) [33], which can greatly improve the sensitivity and specificity in variant calling [34].
Variant calling
The chicken SNP information was downloaded from SNP database in NCBI (ftp://ftp.ncbi.nih.gov/snp/organisms/chicken_9031/VCF/, updated in June 11, 2013) [35]. To exclude most false positive variants, we applied a conservative strategy for both INDEL and SNP calling. First, we require a minimum quality score of 20 for both mapped reads and bases to call variants [36]. Then, the SAMtools mpileup and GATK UnifiedGenotyper module were used to call variants independently. The samples were analyzed together. The variants called by both algorithms were retained for further analysis.
Post filtering
Stringent filtering criteria were applied to the concordant part of variants using GATK VariantFiltration module. For INDELs, only those meeting all the following criteria were retained: a) read depth between 5 and 31; b) quality by depth (QD)>5.0; c) ReadPosRankSum >−20.0 and d) FS<200.0. For SNPs, the criteria were: a) read depth between 5 and 31; b) QD>5.0; c) MQ>40.0; d) HaplotypeScore <13.0; e) MQRankSum >−12.5; f) ReadPosRankSum >−8.0 and g) FS<60.0. Besides, if more than 3 SNPs were clustered in a 10 bp window, they were all considered as false positives and removed [36]. We also eliminated all the heterozygous variants on sex chromosomes, because in the genome of female chickens, these heterozygous variants can either be caused by error or are within the pseudo-autosomal regions.
PCR assay validation
To evaluate the reliability of our data, we randomly selected a subset of putative variants for PCR validation in the positive chickens. PCR primers were designed using Primer Premier5 (http://www.premierbiosoft.com/primerdesign/) [37] to amplify the genomic sequences of 250–600 bp containing the variant. Purified PCR products were analyzed with Sanger sequencing as the gold standard.
Functional annotation
The Ensembl chicken gene set (Ensembl release 74) was downloaded from the Ensembl website (ftp://ftp.ensembl.org/pub/release-74/fasta/gallus_gallus/cdna/) [38] and gene-based annotation of putative INDELs were conducted using ANNOVAR [39]. The chicken QTL database was downloaded from Animal QTL database website (http://www.animalgenome.org/cgi-bin/QTLdb/GG/index, updated in July 8, 2013) [40]. Originally, there were 3807 QTLs in chicken QTL database, but not all of them are suitable for analysis because the confidence intervals of some QTLs were too large to be used efficiently in post-processing. We discarded QTLs with confidence intervals greater than 10 Mb and merged any two or more QTLs with overlapped confidence intervals greater than 50% into one larger QTL. An in-house PERL script was generated to perform the QTL-based annotation. We performed Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis on genes affected by INDELs with the DAVID tool (ver 6.7) [41]. We used the default population background for enrichment calculation. Statistical significance was assessed by using P value (P<0.05) of a modified Fisher's exact test and Benjamini correction for multiple testing.
Data availability
All raw sequence data had been deposited in NCBI Sequence Read Achieve (SRA) under the Bioproject number PRJNA232548. The experiment numbers for the 12 chickens are SRX408161-SRX408172. The whole variant information was provided in the supplementary files.
Results
Sequencing and mapping summary
On average, about 140 million raw reads were generated for each individual, 90% of which were aligned to the reference genome (Table 1). Here, we defined the “effective depth” as the read depth calculated from the reads with mapping quality greater than 20 (Q20). The effective depth ranged from 6.8 in CS to 10.5× in DX, with an average of 8.6×, which was sufficient for further analysis. The overall genome coverage ranged from 94.42 in SG to 95.42% in WL and WR, with an average of 95.02% (Table 1).
Table 1. Summary of sequencing and mapping statistics.
| Chicken breeds* | Raw reads | Mapped reads (Ratio,%) | Q20 Reads (Ratio,%) | Effective Depth (X) | Coverage (%) |
| BY | 122,734,374 | 108,899,430(89) | 98,055,844(80) | 8.2 | 94.78 |
| CS | 113,814,596 | 102,118,711(90) | 81,636,500(72) | 6.8 | 94.67 |
| DX | 160,799,966 | 146,498,490(91) | 125,254,482(78) | 10.5 | 95.26 |
| LX | 146,127,228 | 129,219,851(88) | 100,947,015(69) | 8.4 | 95.03 |
| RIR | 168,078,474 | 151,117,490(90) | 98,330,708(59) | 8.2 | 95.12 |
| RJF | 161,325,436 | 144,056,310(89) | 100,985,637(63) | 8.4 | 94.92 |
| SG | 141,222,608 | 124,890,623(88) | 82,966,840(59) | 6.9 | 94.42 |
| SK | 115,578,334 | 104,212,249(90) | 91,427,763(79) | 7.6 | 94.83 |
| TB | 132,544,720 | 121,217,370(91) | 103,736,982(78) | 8.7 | 95.07 |
| WC | 143,636,242 | 132,110,332(92) | 114,868,135(80) | 9.6 | 95.24 |
| WL | 131,298,592 | 120,759,421(92) | 112,326,911(86) | 9.4 | 95.42 |
| WR | 143,375,106 | 132,693,886(93) | 123,918,088(86) | 10.4 | 95.42 |
| Average | 140,044,640 | 126,482,847(90) | 102,871,242(73) | 8.6 | 95.02 |
*Chicken abbreviations: BY, Beijing You; CS, Cornish; DX, Dongxiang; LX, Luxi Game; RIR, Rhode Island Red; RJF, Red Jungle Fowl; SG, Shouguang; SK, Silkie; TB, Tibetan; WC, Wenchang; WL, White Leghorn; WR, White Plymouth Rock.
INDEL discovery
Although both INDELs and SNPs were identified in our study, we focused on INDELs for further analysis and discussion. In total, 1,766,724 and 1,759,849 raw INDELs were called by SAMtools and GATK, respectively. The concordant part contained 1,425,081 INDELs, accounting for 80.66% and 80.98% of the total number called by SAMtools and GATK, respectively (Figure S1A). In terms of SNPs, 16,153,912 and 16,750,183 raw SNPs were called by SAMtools and GATK, respectively, and the 15,470,364 concordant SNPs corresponded to 95.77% and 92.36% of the two datasets, respectively (Figure S1B). Finally, a huge non-redundant set of variants were obtained after stringent filtering, including 1,343,782 INDELs and 13,708,560 SNPs (Table 2; Table S1; File S1; File S2). The number of INDELs detected in each chicken varied from 368,813 in CS to 528,174 in WR, with an average of 442,794. More than 70% of these variants were detected in two or more individuals.
Table 2. Short INDELs detected in 12 diverse chicken breeds.
| Chicken breedsa | INDEL count | Affected bases (bp) | Novel (Ratio,%) | Maximum length (bp) | Indel Rate (kb−1) | |||||
| Total | Insertionb | Deletionb | Totalb | Insertionb | Deletionb | Insertion | Deletion | |||
| BY | 415,540 | 196,981 | 201,852 | 1,176,135 | 526,774 | 649,361 | 370,997(89.28) | 29 | 47 | 0.48 |
| CS | 368,813 | 175,456 | 179,557 | 1,050,011 | 470,460 | 579,551 | 327,938(88.92) | 29 | 47 | 0.52 |
| DX | 497,358 | 233,956 | 241,907 | 1,439,295 | 643,553 | 795,742 | 445,606(89.59) | 29 | 49 | 0.45 |
| LX | 435,935 | 205,138 | 213,611 | 1,266,168 | 562,840 | 703,328 | 390,120(89.49) | 28 | 45 | 0.50 |
| RIR | 421,309 | 200,618 | 203,666 | 1,219,384 | 550,048 | 669,336 | 375,640(89.16) | 29 | 47 | 0.49 |
| RJF | 451,695 | 213,427 | 219,902 | 1,294,215 | 582,830 | 711,385 | 407,594(90.24) | 30 | 47 | 0.51 |
| SG | 383,782 | 182,043 | 186,521 | 1,092,190 | 489,868 | 602,322 | 342,332(89.20) | 29 | 44 | 0.53 |
| SK | 400,982 | 189,927 | 195,966 | 1,146,582 | 512,067 | 634,515 | 356,250(88.84) | 29 | 45 | 0.50 |
| TB | 448,575 | 211,393 | 218,550 | 1,284,935 | 572,294 | 712,641 | 401,789(89.57) | 30 | 45 | 0.49 |
| WC | 476,889 | 223,743 | 233,162 | 1,384,543 | 614,443 | 770,100 | 427,478(89.64) | 29 | 45 | 0.47 |
| WL | 484,471 | 229,597 | 231,861 | 1,371,739 | 620,051 | 751,688 | 431,757(89.12) | 29 | 44 | 0.49 |
| WR | 528,174 | 248,212 | 254,789 | 1,519,228 | 681,962 | 837,266 | 472,901(89.54) | 29 | 45 | 0.49 |
| Unionc | 1,343,782 | 549,806 | 701,623 | 3,794,977 | 1,439,988 | 2,354,989 | 1,242,748(92.48) | 30 | 49 | 1.49 |
Chicken abbreviations: BY, Beijing You; CS, Cornish; DX, Dongxiang; LX, Luxi Game; RIR, Rhode Island Red; RJF, Red Jungle Fowl; SG, Shouguang; SK, Silkie; TB, Tibetan; WC, Wenchang; WL, White Leghorn; WR, White Plymouth Rock.
INDELs that have multiple genotypes were excluded.
Corrected for INDELs called in more than one individual.
Assessment of the variant discovery strategy
We compared our results with the variants in SNP database (NCBI dbSNP, updated in June 11, 2013) and found that the vast majority (92.48%) of our INELs were novel (Table 2). The 101,034 concordant INDELs account for 23.02% of the INDELs in the current SNP database. Similarly, about half of the SNPs (48.01%) in our dataset had not been discovered previously (Table S1) and the 7,127,652 concordant SNPs account for 81.07% of all known SNPs in the SNP database.
In order to evaluate the accuracy of our variant detection strategy, we randomly selected 57 INDELs for validation and 50 of them were successfully amplified and sequenced. These INDELs include 23 insertions and 27 deletions, with their sizes ranging from 1 to 31 bp (Table S2). To address the ambiguity of coordinates of some INDELs in repeat regions, we left aligned these INDELs. For example, if the result of whole genome sequencing is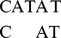 , and the Sanger sequencing result may be like
, and the Sanger sequencing result may be like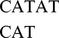 . In such case, we assumed that they were different representations of the same allele, and performed left-alignment of INDELs and considered this INDEL as true variation. Finally, 44 of the 50 sequenced INDELs were consistent with the whole genome sequencing results, corresponding to a validation rate of 88.00%. In addition, we also successfully sequenced 44 SNPs from the selected 53 SNPs and obtained an accuracy of 90.91% (40/44) (Table S3).
. In such case, we assumed that they were different representations of the same allele, and performed left-alignment of INDELs and considered this INDEL as true variation. Finally, 44 of the 50 sequenced INDELs were consistent with the whole genome sequencing results, corresponding to a validation rate of 88.00%. In addition, we also successfully sequenced 44 SNPs from the selected 53 SNPs and obtained an accuracy of 90.91% (40/44) (Table S3).
Genomic distribution of INDELs
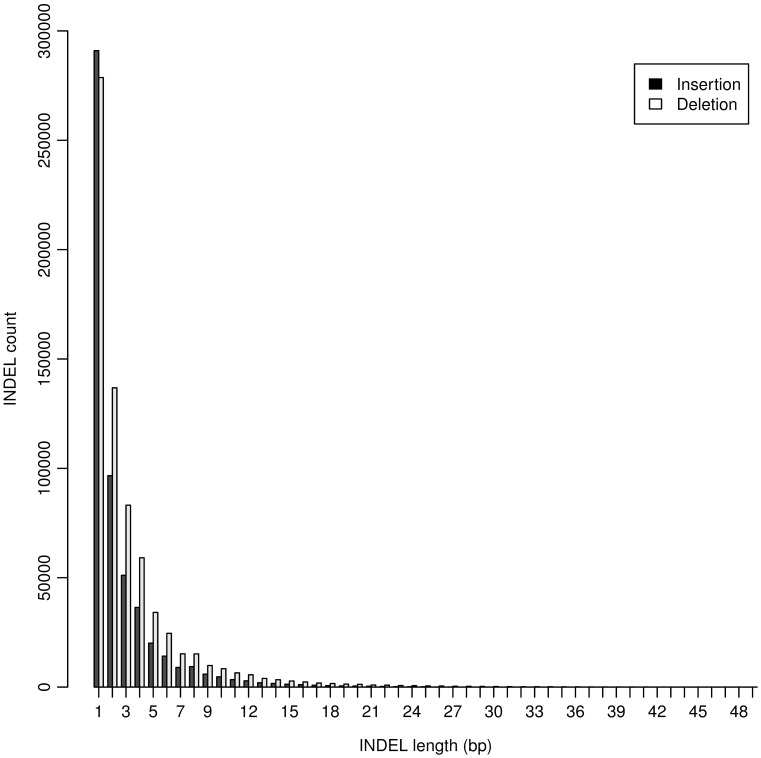
The largest INDEL detected in this study was 49 bp (Table 2), and the majority (95.76%) of INDELs were less than 10 bp (Figure 1). Single base-pair INDEL was the dominant form and accounted for 45.33% of all detected INDELs. Both the detected number and affected bases were larger for deletions than insertions (Table 2). In total, INDELs affected 3.8 million bases, accounting for 0.36% of the chicken genome.
Figure 1. Distribution of INDEL length.
INDELs with multiple genotypes were not included.
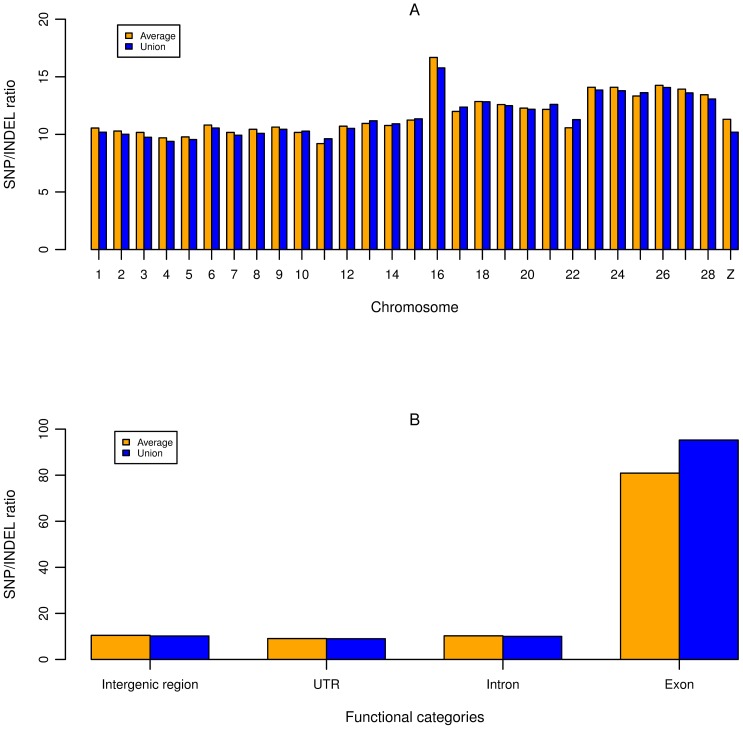
After correcting the read depth in each individual, we observed the average genomic INDEL density was 0.49 per kb, ranging from 0.45 in DX to 0.53 INDELs per kb in SG (Table 2). We calculated the INDEL density for each chromosome and corrected the density by corresponding read depth. INDELs were distributed in a non-uniform fashion across chromosomes (P<2e-16), with INDEL densities of macro-chromosomes (GGA1-5) and intermediate chromosomes (GGA6-10) significantly higher than that of micro-chromosomes (GGA11-28) (0.48, 0.50 vs. 0.38, P = 0.0018) (Figure 2). The Z chromosome tended to have lower INDEL density than most autosomes, with its density 45% lower than the average of autosomes. The chromosome 16 was found to have the lowest INDEL density. The SNP to INDEL ratio was calculated and plotted across each chromosome, based on the union and average data, respectively (Figure 3A). Micro-chromosomes tended to have a higher SNP to INDEL ratio, and notably, GGA16 showed the highest ratio, both on average and union (16.67 and 15.74, respectively).
Figure 2. INDEL and SNP density in each chromosome.
Densities were calculated as the number per 10 kb (INDEL) and kb (SNP), respectively. Densities are averaged by chicken individuals and corrected by read depth. Coverage was calculated based on Q20 reads.
Figure 3. SNP to INDEL ratio.
The ratios were plotted based on the non-redundant (Union) data and the data averaged by chickens (Average), respectively. A: SNP to INDEL ratio across chromosomes. B: SNP to INDEL ratios in functional categories.
To explore the distribution of INDELs in genic regions, we annotated all detected INDELs using Ensembl gene set (containing 17,954 genes). For each INDEL, its genomic location (intergenic, exonic, intronic, splicing, 5′UTR, 3′UTR, upstream or downstream) and functional role (frameshifting, non-frameshifting and stop gain/loss) were determined. In total, 620,253 (46.15%) INDELs were mapped to genic regions (13,489 genes) (Table 3). Among them, 17,770 INDELs (2.87%) fell in untranslated regions (UTRs), 219 (0.04%) in non-coding transcripts (ncRNAs), 318 (0.05%) in splicing sites, 600,181 (96.76%) in introns, and 1,765 (0.28%) in coding exons. The INDEL densities of intergenic regions, exon, intron and UTR were 0.40, 0.02, 0.45 and 0.09 per kb, respectively. We then examined the SNP to INDEL ratio in these functional regions, and as expected, exon showed the highest ratio (Figure 3B).
Table 3. Statistics of INDELs and SNPs in functional regions.
| Category | INDEL | Category | SNP |
| Intergenic | 690,303 | Intergenic | 7,035,013 |
| Flanking regiona | 33,226 | Flanking regiona | 345,361 |
| Upstream | 13,925 | Upstream | 163,068 |
| Downstream | 18,374 | Downstream | 171,990 |
| Up/downstreamb | 927 | Up/downstreamb | 10,303 |
| Genic | 620,253 | Genic | 6,328,186 |
| 5′URT | 1,388 | 5′URT | 19,814 |
| 3′URT | 16,372 | 3′URT | 139,967 |
| 5′/3′UTRc | 10 | 5′/3′UTRc | 151 |
| Splicing | 318 | Splicing | 543 |
| ncRNAd | 219 | ncRNAd | 1,740 |
| Intronic | 600,181 | Intronic | 5,997,846 |
| Exonic | 1,765 | Exonic | 168,125 |
| Non-frameshift | 720 | Synonymous | 119,816 |
| Frameshift | 1,022 | Non-synonymous | 47,915 |
| Stop gain/losse | 23 | Stop gain/losse | 394 |
Regions that are 1 kb apart from the transcription start site.
Variant located in both upstream and downstream regions (possibly for two different genes).
Variants located in both 5′UTR and 3′UTR regions (possibly for two different genes).
Variants located in the transcripts without coding annotation in the current Ensembl gene annotation.
Variants caused gain or loss of stop codon.
In terms of the potential roles of the coding INDELs, 720 (40.79%) were triplet (non-frameshifting), thus retaining the reading frame, and 23 (1.30%) caused gain or loss of stop codon. The remaining 1,022 (57.90%) were non-triplet INDELs, which were predicted to cause frameshift mutation, and this proportion was significantly lower compared with the genomic level (83.55%, P = 2.2e-16). A large number (1,358, 7.56%) of functionally important genes were covered by coding INDELs, many (284, 20.91%) of which contained two or more coding INDELs (Table S4).
We then examined the distribution of INDELs in quantitative trait loci (QTL) regions (Table S5). According to our filtering criteria, 595 non-overlapping QTL regions were obtained for analysis. A total of 76,387 INDELs fell into these regions, 37,330 (48.87%) of which located in genic regions. INDEL densities varied significantly across QTL regions, ranging from 0.09 to 3.89 per kb. The average INDEL density for all QTL regions was 1.50 per kb, slightly higher than the genomic level. Several QTLs on GGA4, GGA1, GGA6 and GGA12 that govern feather pecking, chicken body composition, body weight, growth, and abdominal fat percentage had the highest INDEL density.
Gene enrichment
GO and KEGG pathway analysis were performed on 1,593 genes that contained more than one hundred INDELs, which we assumed to be under high mutation load of INDELs. GO results showed 211 terms, 76 of which were significant after Benjamini correction. These genes were significantly enriched in the molecular functions of protein kinase activity, enzyme activator activity, molecule binding (including nucleotide binding, ion binding), GTPase regulator activity, channel activity, and substrate specific channel activity (Table S6). The KEGG pathway analysis revealed that the genes were overrepresented in 13 pathways, but only one (gga04070: Phosphatidylinositol signaling system) was significant after Benjamini correction.
Discussion
In this study, we performed NGS on 12 chicken individuals for INDEL discovery to gain a comprehensive understanding of INDEL variation in chicken genome. Although NGS technologies are routinely used to detect genome-wide variations [42], [43], accurately discriminating true variants from false positives from NGS data is still challenging with no easy fix, especially for short INDELs [36], [44], [45]. In this study, we adopted a conservative method to minimize the false positive rate. Several steps that had been proven effective in reducing false positives in variant detection [34], [43], [46]–[48] were adopted in the current study (See Materials and Methods). These measures ensured a significant improvement in detection accuracy compared with a recent study [25] (88.0% vs. 68.4%), even that we had a much lower depth (8.6 vs. 24.9, on average). We anticipate that the combination of advanced sequencing platforms, higher sequencing depth, and superior calling algorithms can further improve the accuracy of INDEL detection in the future. Meanwhile, our method should also suffer a significant false negative rate since we gave much priority to specificity with the sacrifice of sensitivity. For instance, only INDELs called by both algorithms were retained and then subjected to stringent filtering.
To our knowledge, the number of INDELs identified in our study is the highest so far in chicken. Compared with the study in human, this number is smaller than the results from Mills et al. [3], but comparable with a recent study [49], and if we note that the chicken genome is only about one third to human [50], this number will be of great significance. The INDELs accounted for 8.92% of all detected variants and 21.68% in terms of bases involved. These proportions were lower than those observed in other species as described above, and also lower than a recent study in chicken [25], which is probably due to the more stringent filtering criteria and the narrower range of INDEL length in our study. Anyway, our INDELs affected 0.36% of the chicken genome, suggesting that INDELs are widespread in chicken genome and may be an important source of both genetic and phenotypic variation. Over 70% of the 1.2 million INDELs were shared by two or more individuals in spite of their distant genetic relationship, probably representing common variations. Certain unique INDELs may represent the special individual characteristics. The vast majority of detected INDELs were novel, indicating that the discovery of INDELs in chicken, or at least short INDELs, is far from complete. Our results also demonstrated that employing chickens with diverse genetic background for variant detection promoted identifying more variants, as can be seen from the low concordant rate with the INDELs in the SNP database. Hence, for a more comprehensive genetic variation map in the future, multiple individuals and more diverse breeds will be desired.
The INDEL density analyzed in 12 chickens was higher than that observed by Brandstrom and Ellegren [5] because we didn't exclude INDELs in tandem repeat sequence, and also higher than that in human [3]. The Z chromosome had lower INDEL density than autosomes, which was also observed by Brandstrom and Ellegren [5]. This difference would be in part due to the lower effective population size of Z chromosome caused by skewed reproductive success among male chickens [51]. In addition, the lower coverage of Z chromosome than autosomes (88.61% vs 93.03%) and the filtering of heterozygous variants on Z chromosome may also contribute to the lower INDEL density. We observed that the micro-chromosomes tend to have lower INDEL densities, which was consistent with previous results [5]. This may be explained partly by their lower coverage of Q20 reads and partly by the fact that micro-chromosomes are extremely gene rich [50], therefore length mutations, like INDELs, are strongly selected against. In our study, the GGA16 was found to have lower INDEL density than other chromosomes, contrary to previous findings [21], [25]. It could be speculated that this may be caused by the poor coverage of Q20 reads, as well as the partial representation of GGA16 in the current chicken genome assembly. The GGA16 has only been sequenced 535.27 kb, whereas its full length is predicted to be between 9 and 11 Mb [52]. In spite of the low density of INDELs and SNPs on GGA16, the SNP to INDEL ratio in GGA16 was the highest among all chromosomes. This may result from the presence of several important gene families, like nucleolus organizer region (NOR) and major histocompatibility complex (MHC), an immune-related gene family, which could impose a greater selection pressure on INDELs than SNPs. As mentioned above, INDELs as a kind of length variants are often deleterious to gene functions, whereas SNPs generally cause little or no effects to gene functions. Besides, the SNP to INDEL ratio was strikingly high in exons, and both the INDEL density and the proportion of frameshifting INDELs was significantly lower than that of genomic level. This indicated that INDELs in exons, frameshifting INDELs in particular, were strongly eliminated by purifying selection. We also found that INDELs were enriched in some QTLs, which was likely due to the recent selection for favorable alleles. These INDELs could be used as candidate markers for fine mapping of causative genes.
Like SNPs and CNVs, INDELs are of great importance for their ability to alter gene functions, especially those frameshifting INDELs locating in exons. In this study, lots of genes were affected by frameshifting INDELs. Some genes are associated with chicken performance traits. For instance, THRSP encodes a small acidic protein that responds to thyroid hormone (TH) stimulation and is thought to play a role in growth. A 9 bp INDEL polymorphism and several SNPs in the exon1 of THRSP were found to be associated with abdominal fat content [53], [54] and body weight [55]. In our study, two novel INDELs within the exon1 were found in several chickens, implicating that this gene was highly polymorphic and the two novel INDELs were worth further studying for their association with economic traits. MUC6 (β-subset of ovomucin) is the homologue of human MUC6 [56] and reported to be involved in determining the gel property of thick egg white [57]. As many as five coding INDELs were identified in MUC6 and we suggested that these INDELs could be used as potential candidates for egg quality. In addition, quite many genes related to the development of chicken embryo or are the homologues of human disease-related genes. The results demonstrated that though strongly selected against, INDELs were common in some functionally important genes, arguing for their incorporation to elucidate the association between genes and traits.
It is increasingly recognized that INDEL polymorphisms can be effectively used as genetic markers [16], [58]–[60]. In fact, INDELs merit as promising genetic markers for many aspects. First, INDELs are diallelic and widespread throughout chicken genome. The density of INDELs in chicken genome is much higher than that of microsatellite [61], which can compensate their shortcoming of lower level of polymorphism. Second, INDELs are relatively easy and cost-effective to genotype, allowing ordinary laboratories to rapidly screen a large number of individuals [16]. Third, the probability of two INDELs of the same length occurring at the same position is very low that the shared INDELs can confidently be related to identity by descent [62]. This can reduce the occurrence of the homoplasy, a common problem in phylogenetic studies using microsatellites as markers. Forth, most INDELs have a minor allele frequency (MAF) greater than 0.05 [3], [17], [58], [60], meeting the criteria of common genetic variations. Finally, most INDELs are in strong linkage disequilibrium (LD) with SNPs of genome-wide association studies (GWASs) [3], suggesting that INDELs are likely to associate with a substantial amount of phenotypic diversity and disease susceptibility. Therefore, INDELs can be efficiently integrated into current genetic variation map to construct a more comprehensive map including SNPs, INDELs and CNVs, which will facilitate the identification of causative mutations and accelerate genetic improvement for complex traits and diseases.
Microarrays are very powerful and essential tools in GWAS and genomic selection (GS). Up to date, medium and high density SNP arrays have been commercially available [63], [64] in chicken, whereas no INDEL array is reported available not only in chicken but also in any other domestic animals. Efforts to design INDEL arrays have been made by Salathia et al. [65] and Mills et al. [3] in Arabidopsis thaliana and human, respectively. Though both arrays contained a relatively small number of INDELs, they shed light on the feasibility of designing INDEL arrays and genotyping large number of individuals. Currently, the paucity of available INDEL resources may hamper the development process since developing INDEL arrays requires a large collection of polymorphic INDELs. The large quantity of INDELs screened in our study enriched the current INDEL database and will be beneficial to future development of INDEL arrays. In addition, researchers can also select a number of informative INDELs and integrate them into SNP arrays to increase their power in GWAS and GS.
Conclusions
We performed whole genome sequencing on 12 diverse chicken breeds and identified the largest number of INDELs in chicken genome so far. Incorporating diverse chicken breeds for variant detection allowed for a larger collection of variants to be discovered. A large number of coding INDELs located in previously reported genes associated with chicken performance traits. We suggest that INDELs are crucial determinants causing genetic and phenotypic diversity and can be promising genetic markers. Our results can be used for a variety of studies in the future, including development of INDEL markers, construction of high density linkage map, INDEL arrays design, and hopefully, molecular breeding programs in chicken.
Supporting Information
The number of raw variants called by SAMtools and GATK, respectively. A: INDELs. B: SNPs.
(TIFF)
SNPs detected in 12 chickens.
(DOCX)
Summary of validated INDELs.
(XLSX)
Summary of validated SNPs.
(XLSX)
Coding INDELs in each chromosome.
(XLSX)
Distribution of INDELs in QTL regions.
(XLSX)
Functional enrichment of genes under high mutation load of INDELs.
(XLSX)
The non-redundant set of detected INDELs in 12 chickens. To minimize the file size, only minimal essential information was listed, including CHR (chromosome), POS (position), REF (reference allele), ALT (alternative allele), and BREEDS (chickens in which this INDEL was detected).
(GZ)
The non-redundant set of detected SNPs in 12 chickens. To minimize the file size, only minimal essential information was listed, including CHR (chromosome), POS (position), REF (reference allele), ALT (alternative allele), and BREEDS (chickens in which this SNP was detected).
(GZ)
Acknowledgments
We thank Dr. Xiquan Zhang for sharing some samples.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All raw sequence data had been deposited in NCBI Sequence Read Achieve (SRA) under the Bioproject number PRJNA232548. The experiment numbers for the 12 chickens are SRX408161-SRX408172. The whole variant information was provided in the supplementary files.
Funding Statement
The current research was supported by grants of National High Technology Development Plan of China (2013AA102501), Natural Science Foundation of China (31320103905), Programs for Changjiang Scholars and Innovative Research in University (IRT1191), and China Agriculture Research Systems (CARS-41). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Siegel PB, Dodgson JB, Andersson L (2006) Progress from chicken genetics to the chicken genome. Poult Sci 85: 2050–2060. [DOI] [PubMed] [Google Scholar]
- 2. Mullaney JM, Mills RE, Pittard WS, Devine SE (2010) Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet 19: R131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mills RE, Pittard WS, Mullaney JM, Farooq U, Creasy TH, et al. (2011) Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res 21: 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wetterbom A, Sevov M, Cavelier L, Bergstrom TF (2006) Comparative genomic analysis of human and chimpanzee indicates a key role for indels in primate evolution. J Mol Evol 63: 682–690. [DOI] [PubMed] [Google Scholar]
- 5. Brandstrom M, Ellegren H (2007) The genomic landscape of short insertion and deletion polymorphisms in the chicken (Gallus gallus) genome: a high frequency of deletions in tandem duplicates. Genetics 176: 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan Y, Wang W, Ma G, Liang L, Shi Q, et al. (2007) Patterns of insertion and deletion in mammalian genomes. Curr Genomics 8: 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins FS, Drumm ML, Cole JL, Lockwood WK, Vande Woude GF, et al. (1987) Construction of a general human chromosome jumping library, with application to cystic fibrosis. Science 235: 1046–1049. [DOI] [PubMed] [Google Scholar]
- 9. Ashley CT Jr, Warren ST (1995) Trinucleotide repeat expansion and human disease. Annu Rev Genet 29: 703–728. [DOI] [PubMed] [Google Scholar]
- 10. Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, et al. (1997) A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17: 71–74. [DOI] [PubMed] [Google Scholar]
- 11. Kunieda M, Tsuji T, Abbasi AR, Khalaj M, Ikeda M, et al. (2005) An insertion mutation of the bovine Fii gene is responsible for factor XI deficiency in Japanese black cattle. Mamm Genome 16: 383–389. [DOI] [PubMed] [Google Scholar]
- 12. Sironen A, Thomsen B, Andersson M, Ahola V, Vilkki J (2006) An intronic insertion in KPL2 results in aberrant splicing and causes the immotile short-tail sperm defect in the pig. Proc Natl Acad Sci U S A 103: 5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mosher DS, Quignon P, Bustamante CD, Sutter NB, Mellersh CS, et al. (2007) A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 3: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kerje S, Sharma P, Gunnarsson U, Kim H, Bagchi S, et al. (2004) The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics 168: 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agarwal SK, Cogburn LA, Burnside J (1994) Dysfunctional growth hormone receptor in a strain of sex-linked dwarf chicken: evidence for a mutation in the intracellular domain. J Endocrinol 142: 427–434. [DOI] [PubMed] [Google Scholar]
- 16. Vasemagi A, Gross R, Palm D, Paaver T, Primmer CR (2010) Discovery and application of insertion-deletion (INDEL) polymorphisms for QTL mapping of early life-history traits in Atlantic salmon. BMC Genomics 11: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mills R, Luttig C, Larkins C, Beauchamp A, Tsui C, et al. (2006) An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res 16: 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B, Wang Y, Zhai W, Deng J, Wang H, et al. (2013) Development of InDel markers for Brassica rapa based on whole-genome re-sequencing. Theor Appl Genet 126: 231–239. [DOI] [PubMed] [Google Scholar]
- 19.Zou X, Shi C, Austin R, Merico D, Munholland S, et al.. (2013) Genome-wide single nucleotide polymorphism and Insertion-Deletion discovery through next-generation sequencing of reduced representation libraries in common bean. Molecular Breeding: 1–10.
- 20. Leushkin EV, Bazykin GA, Kondrashov AS (2013) Strong mutational bias toward deletions in the Drosophila melanogaster genome is compensated by selection. Genome Biol Evol 5: 514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong GK, Liu B, Wang J, Zhang Y, Yang X, et al. (2004) A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature 432: 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rubin C, Zody M, Eriksson J, Meadows J, Sherwood E, et al. (2010) Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464: 587–591. [DOI] [PubMed] [Google Scholar]
- 23. Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, et al. (2008) Whole-genome sequencing and variant discovery in C. elegans. Nat Methods 5: 183–188. [DOI] [PubMed] [Google Scholar]
- 24. Tsuda K, Kawahara-Miki R, Sano S, Imai M, Noguchi T, et al. (2013) Abundant sequence divergence in the native Japanese cattle Mishima-Ushi (Bos taurus) detected using whole-genome sequencing. Genomics 102: 372–378. [DOI] [PubMed] [Google Scholar]
- 25. Fan WL, Ng CS, Chen CF, Lu MY, Chen YH, et al. (2013) Genome-wide patterns of genetic variation in two domestic chickens. Genome Biol Evol 5: 1376–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Z, Gerstein M (2003) Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res 31: 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhangale TR, Rieder MJ, Livingston RJ, Nickerson DA (2005) Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Human Molecular Genetics 14: 59–69. [DOI] [PubMed] [Google Scholar]
- 28. Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, et al. (2013) The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41: D64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ledergerber C, Dessimoz C (2011) Base-calling for next-generation sequencing platforms. Brief Bioinform 12: 489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H, Ruan J, Durbin R (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lalitha S (2000) Primer premier 5. Biotech Software & Internet Report: The Computer Software Journal for Scient 1: 270–272. [Google Scholar]
- 38. Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, et al. (2013) Ensembl 2013. Nucleic Acids Res 41: D48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu ZL, Park CA, Wu XL, Reecy JM (2013) Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Research 41: D871–D879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 42. Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, et al. (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12: 499–510. [DOI] [PubMed] [Google Scholar]
- 43. Nielsen R, Paul JS, Albrechtsen A, Song YS (2011) Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 12: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koboldt DC, Ding L, Mardis ER, Wilson RK (2010) Challenges of sequencing human genomes. Brief Bioinform 11: 484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, et al. (2011) Dindel: accurate indel calls from short-read data. Genome Res 21: 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, et al. (2012) Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol 30: 61–68. [DOI] [PubMed] [Google Scholar]
- 47. Yu X, Sun S (2013) Comparing a few SNP calling algorithms using low-coverage sequencing data. BMC Bioinformatics 14: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Neuman JA, Isakov O, Shomron N (2013) Analysis of insertion-deletion from deep-sequencing data: software evaluation for optimal detection. Brief Bioinform 14: 46–55. [DOI] [PubMed] [Google Scholar]
- 49. Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Consortium ICGS (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. [DOI] [PubMed] [Google Scholar]
- 51. Sundstrom H, Webster MT, Ellegren H (2004) Reduced variation on the chicken Z chromosome. Genetics 167: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Delany ME, Gessaro TM, Rodrigue KL, Daniels LM (2007) Chromosomal mapping of chicken mega-telomere arrays to GGA9, 16, 28 and W using a cytogenomic approach. Cytogenet Genome Res 117: 54–63. [DOI] [PubMed] [Google Scholar]
- 53. Wang X, Carre W, Zhou H, Lamont SJ, Cogburn LA (2004) Duplicated Spot 14 genes in the chicken: characterization and identification of polymorphisms associated with abdominal fat traits. Gene 332: 79–88. [DOI] [PubMed] [Google Scholar]
- 54. D'Andre Hirwa C, Yan W, Wallace P, Nie Q, Luo C, et al. (2010) Effects of the thyroid hormone responsive spot 14alpha gene on chicken growth and fat traits. Poult Sci 89: 1981–1991. [DOI] [PubMed] [Google Scholar]
- 55. Cao ZP, Wang SZ, Wang QG, Wang YX, Li H (2007) Association of Spot14alpha gene polymorphisms with body weight in the chicken. Poult Sci 86: 1873–1880. [DOI] [PubMed] [Google Scholar]
- 56. Lang T, Hansson GC, Samuelsson T (2006) An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics 7: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rabouille C, Aon MA, Muller G, Cartaud J, Thomas D (1990) The supramolecular organization of ovomucin. Biophysical and morphological studies. Biochem J 266: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vali U, Brandstrom M, Johansson M, Ellegren H (2008) Insertion-deletion polymorphisms (indels) as genetic markers in natural populations. BMC Genet 9: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maw AA, Shimogiri T, Riztyan, Kawabe K, Kawamoto Y, et al. (2012) Genetic diversity of Myanmar and Indonesia native chickens together with two Jungle Fowl species by using 102 Indels polymorphisms. Asian-Australasian Journal of Animal Science 25: 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maw AA, Shimogiri T, Yamamoto K, Kawabe K, Hamada K, et al. (2013) The genetic diversity of eight chicken populations assessed by 102 indels markers. The Journal of Poultry Science 50: 99–103. [Google Scholar]
- 61. Primmer CR, Raudsepp T, Chowdhary BP, Moller AP, Ellegren H (1997) Low frequency of microsatellites in the avian genome. Genome Res 7: 471–482. [DOI] [PubMed] [Google Scholar]
- 62. Garcia-Lor A, Luro F, Navarro L, Ollitrault P (2012) Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Mol Genet Genomics 287: 77–94. [DOI] [PubMed] [Google Scholar]
- 63. Groenen M, Megens H-J, Zare Y, Warren W, Hillier L, et al. (2011) The development and characterization of a 60K SNP chip for chicken. BMC Genomics 12: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kranis A, Gheyas AA, Boschiero C, Turner F, Yu L, et al. (2013) Development of a high density 600K SNP genotyping array for chicken. BMC Genomics 14: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salathia N, Lee HN, Sangster TA, Morneau K, Landry CR, et al. (2007) Indel arrays: an affordable alternative for genotyping. Plant J 51: 727–737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The number of raw variants called by SAMtools and GATK, respectively. A: INDELs. B: SNPs.
(TIFF)
SNPs detected in 12 chickens.
(DOCX)
Summary of validated INDELs.
(XLSX)
Summary of validated SNPs.
(XLSX)
Coding INDELs in each chromosome.
(XLSX)
Distribution of INDELs in QTL regions.
(XLSX)
Functional enrichment of genes under high mutation load of INDELs.
(XLSX)
The non-redundant set of detected INDELs in 12 chickens. To minimize the file size, only minimal essential information was listed, including CHR (chromosome), POS (position), REF (reference allele), ALT (alternative allele), and BREEDS (chickens in which this INDEL was detected).
(GZ)
The non-redundant set of detected SNPs in 12 chickens. To minimize the file size, only minimal essential information was listed, including CHR (chromosome), POS (position), REF (reference allele), ALT (alternative allele), and BREEDS (chickens in which this SNP was detected).
(GZ)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All raw sequence data had been deposited in NCBI Sequence Read Achieve (SRA) under the Bioproject number PRJNA232548. The experiment numbers for the 12 chickens are SRX408161-SRX408172. The whole variant information was provided in the supplementary files.
All raw sequence data had been deposited in NCBI Sequence Read Achieve (SRA) under the Bioproject number PRJNA232548. The experiment numbers for the 12 chickens are SRX408161-SRX408172. The whole variant information was provided in the supplementary files.