Abstract
The present study was designed to investigate the potential of Fourier transform infrared (FT-IR) spectroscopy to identify Trueperella (T.) pyogenes isolated from bovine clinical mastitis. FT-IR spectroscopy was applied to 57 isolates obtained from 55 cows in a period from 2009 to 2012. Prior to FT-IR spectroscopy these isolates were identified by phenotypic and genotypic properties, also including the determination of seven potential virulence factor encoding genes. The FT-IR analysis revealed a reliable identification of all 57 isolates as T. pyogenes and a clear separation of this species from the other species of genus Trueperella and from species of genus Arcanobacterium and Actinomyces. The results showed that all 57 isolates were assigned to the correct species indicating that FT-IR spectroscopy could also be efficiently used for identification of this bacterial pathogen.
Introduction
Trueperella (T.) pyogenes, according to Yassin et al. [1] reclassified from genus Arcanobacterium to genus Trueperella, is a worldwide known pathogen of domestic ruminants and pigs causing mastitis and a variety of pyogenic infections [2]–[3]. As summarized by Jost and Billington [4] these bacteria are also able to cause diseases in various other animal species. More recently T. pyogenes isolated from reptile and camel origin were characterized in terms of pheno- and genotype [5]–[6]. Infections of human patients with T. pyogenes are rare [7]–[8]. However, conventional methods for identification of T. pyogenes, which are based on biochemical markers such as sugar fermentation, proteolytic activities, haemolytic and CAMP-like haemolytic reactions [2]–[9] are often time consuming. In addition, atypical variants in phenotype might occur [10]–[11]. Alternative methods based on the analysis of species specific regions of the bacterial genome, which could also be used for identification of T. pyogenes [9], can be quite elaborate, expensive and often requires highly skilled staff.
As a high resolving phenotypic technique, FT-IR spectroscopy, analyzing the total composition of components of the bacterial cell, has been established as a method for identification of several bacteria, yeasts and other microorganisms [12]–[13], also including Actinomycetales [14]–[15]. Offering a wide range of application this technique was used in the present study to identify T. pyogenes isolated from bovine mastitis on species level.
Materials and Methods
A total 57 T. pyogenes isolates obtained from milk samples were used in this study. The bacteria were collected from 2009 to 2012 during routine diagnostic of 55 cows with clinical mastitis from 50 farms and 49 locations. The T. pyogenes from milk samples were identified as described previously [6]–[9] by phenotypical and genotypical tests, also including the determination of seven potential virulence factor encoding genes. The studies involved no human or animal participants. The milk samples were taken during routine diagnostic according to national and international guidelines. In all cases from each location was a permission to collect and investigate these samples (Table 1). All samples were taken as part of the hessian udder service to detect reasons for frequently occurring mastitis cases in these holdings.
Table 1. Phenotypical and genotypical properties of Trueperella pyogenes of bovine origin and two T. pyogenes reference strains.
| Phenotypical properties | T. pyogenes (n = 57) | T. pyogenes DSM 20594** | T. pyogenes DSM 20630** |
| Hemolysis on sheep blood agar | + | + | + |
| CAMP-like reaction with*: | |||
| Staphylococcus aureus β-hemolysin | + | + | + |
| Streptococcus agalactiae | − | − | − |
| Rhodococcus equi | + | + | + |
| Reverse CAMP reaction | − | − | − |
| β-Glucuronidase (β-GUR) | +1 , 2 | +1 , 2 | +1 , 2 |
| α-Galactosidase (α-GAL) | − (49)1, (+)(8)1 | −1 | −1 |
| β-Galactosidase (β-GAL) | +(56)2, (+)(1)2 | +2 | +2 |
| α-Glucosidase (α-GLU) | +1 , 2 | +1 , 2 | +1 , 2 |
| β-Glucosidase (β-GLU) | −1 | −1 | −1 |
| N-acetyl- β-Glucosaminidase (β-NAG) | +2 | +2 | +2 |
| α-Mannosidase | − (54)1, (+)(3)1 | −1 | −1 |
| Catalase | − | − | − |
| Serolysis on Loeffler agar | + | + | + |
| Caseinase | + | + | + |
| Starch hydrolysis (amylase) | −(53), +(4) | − | + |
| Cross reaction with streptococcal serogroup G specific antiserum | + | + | + |
| Genotypical properties | |||
| T. pyogenes specific part of gene sodA | + | + | + |
| T. pyogenes specific part of ISR | + | + | + |
| Genes encoding virulence factors | |||
| Pylosin encoding gene plo | + | + | + |
| Collagen-binding protein encoding gene cbpA | + (1), −(56) | − | + |
| Neuraminidase H encoding gene nanH | +(39), −(18) | + | + |
| Neuraminidase P encoding gene nanP | +(48), −(9) | + | + |
| Fimbriae endoding gene fimA | + | + | − |
| Fimbriae endoding gene fimC | +(53), −(4) | + | + |
| Fimbriae endoding gene fimE | +(52), −(5) | + | + |
The reactions are shown as follows:
* = synergistic CAMP-like reaction with indicator strains;
** = results mostly obtained from Hijazin et al., 2011;
+; positive reaction; (+) = weak reaction −; negative reaction;
= tablets containing substrates (Rosco Diagnostica A/S, Taastrup, Denmark);
= 4-methylumbelliferyl conjugated substrates (Sigma, Steinheim, Germany).
In addition T. pyogenes DSM 20594, T. pyogenes DSM 20630T, T. pyogenes CVUAS 0222, Trueperella abortisuis DSM 19515T, Trueperella bernardiae DSM 9152T, Trueperella bialowiezensis DSM 17162T, Trueperella bonasi DSM 17163T, Arcanobacterium haemolyticum DSM 20595T, Arcanobacterium canis DSM 25104T, Arcanobacterium hippocoleae DSM 15539T, Arcanobacterium phocae DSM 10002T, Arcanobacterium phocae DSM 10003, Arcanobacterium phocisimile DSM 26142, Arcanobacterium pluranimalium DSM 18483T, Actinomyces bovis DSM 43014T, Actinomyces hyovaginalis CVUAS 4295, Actinomyces weissii DSM 24894T and Actinomyces canis DSM 15536T, were included and used for comparative purposes.
For FT-IR spectroscopy all isolates were cultivated on sheep blood agar for 48 h (+/−0.5 h) at 37°C in 6–10 replicates under microaerobic conditions (GasPak EZ Campy Container System; Becton, Dickinson and Company, Heidelberg, Germany). Harvesting bacterial biomass and preparation of bacterial films on zinc selenide (ZnSe) plates were performed as described previously [16], using an aliquot of 25 µl in a sample zone of a 96 well format ZnSe-plate. Every isolate for the database was measured at least six times using a TENSOR 27 FT-IR spectrometer supplemented with a HTS-XT module (Bruker Optik GmbH, Ettlingen, Germany) in transmission mode from 500 to 4000 cm−1 with the coupled software (OPUS 6.5).
The data set for the isolates used for the construction of the differentiation method was divided into two equal parts, as described by Kuhm et al. [16]. The first one, called creation set, was used to create the method. The second one was used to verify the created method to gain the recovery rates (internal recovery set). An internal validation was performed with this internal recovery set.
The 57 well described T. pyogenes isolates from bovine mastitis were used for external validation. Results were given as probability for repeated determination, based on the results of the respective internal and external recovery sets (Table 2).
Table 2. Validation of genus and species classification by FT-IR, given as probability for correct identification of strains (repeated determinations).
| Organism | Isolates | Spectra | nref | nval | Identification (%)a | |
| Correct | Incorrect | |||||
| Actinomyces (genus level) | 4 | 36 | 13 | 23 | 95.8 | 0 |
| Arcanobacterium (genus level) | 7 | 87 | 35 | 52 | 92.6 | 0 |
| Trueperella (genus level) | 7 | 89 | 31 | 58 | 95.8 | 0 |
| T. abortisuis | 1 | 15 | 5 | 10 | 96.0 | 0 |
| T. bernardiae | 1 | 13 | 4 | 9 | 100 | 0 |
| T. bialowiezensis | 1 | 19 | 5 | 14 | 91.8 | 0 |
| T. bonsai | 1 | 12 | 3 | 9 | 88.9 | 0 |
| T. pyogenes | 3 | 30 | 14 | 16 | 97.9 | 0 |
| T. pyogenes from milk-samples | 57 | 342 | 0b | 342 | 98.8b | 0 |
n ref, number of spectra used for reference; n val, number of spectra used for validation. For internal validation, all spectra which had not been included in the reference data set were used.
The probability of obtaining uncertain results during repeated determinations is given by the residual to 100%.
For identification of T. pyogenes, an external validation was applied using all isolates not included in the reference.
The infrared spectra of the creation set were used in development of the differentiation methods with NeuroDeveloper software (Synthon GmbH, Heidelberg, Germany), which is based on an artificial neural network strategy (ANN) [17]. The second derivatives of the vector-normalized, five-point smoothed spectra of the creation set in the wave number ranges from 2800–3000 cm−1 and 500 to 1800 cm−1 were used for data analysis in covar mode with a significance of 95%. Four-fifth randomly assorted spectra of the creation set were used as the “training set” of the developer module. With the remaining one-fifth of the spectra, put in the “validation set”, the internal method optimization of wavelength combinations was done [18]. In this way a hierarchical classification scheme is build, consisting of a top level dividing the three genera (Actinomyces, Arcanobacterium and Trueperella), and a subsequent classification level, differentiating the five Trueperella-species.
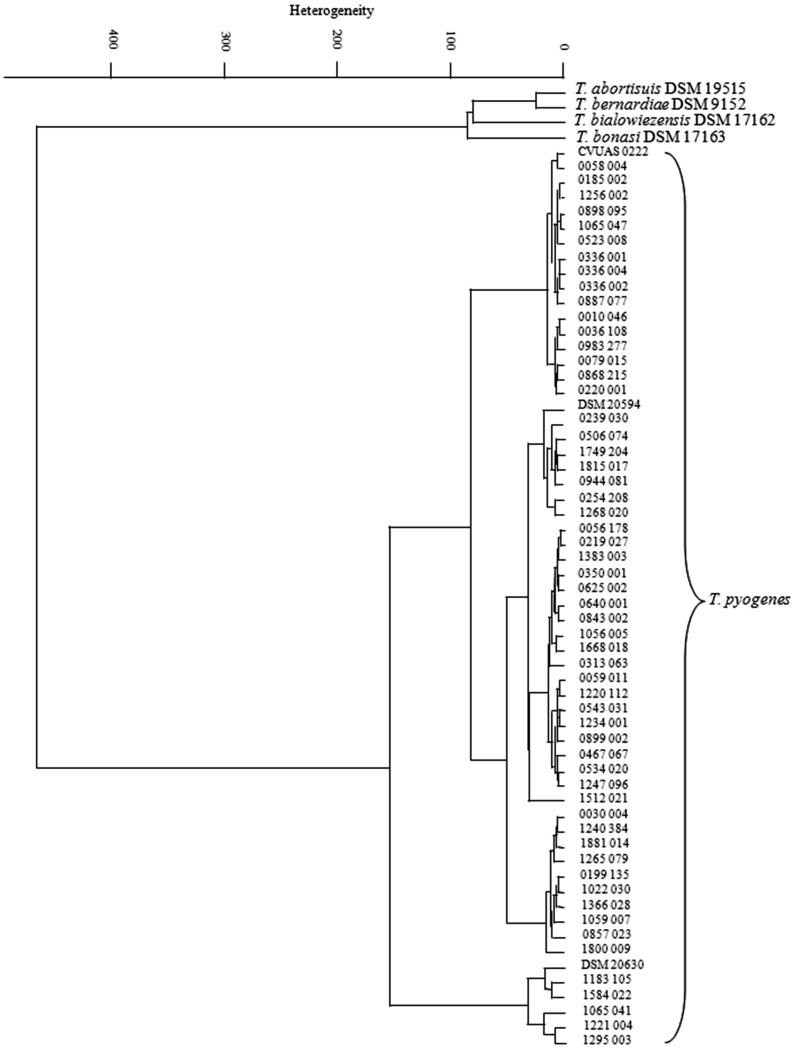
Single infrared spectra of all Trueperella isolates were compared by cluster analysis as described [15]. For this collation, the second derivatives of vector normalized spectra in the wave number ranges of 500–1200 cm−1 and 2800–3000 cm−1 were used for calculation with Ward's algorithm (OPUS 4.2) [19]. The dendrogram obtained depicts the arrangement of isolates according to their spectral differences (Fig. 1).
Figure 1. Dendrogram of infrared spectra of 57 T. pyogenes isolates from milk samples in comparison with reference strains from the same genus.
Cluster analysis was performed by using the second derivatives of the spectra in the spectral ranges of 500 to 1200 cm−1 and 2800 to 3000 cm−1. Ward's algorithm was applied. The arrows show three independent isolates from the same cow.
Results and Discussion
All T. pyogenes investigated in the present study were identified, comparable to previously described T. pyogenes [5]–[6]–[9], by determination of haemolytic and CAMP-like haemolytic reactions, by using various biochemical tests and genotypically using T. pyogenes specific parts of 16S–23S rDNA intergenic spacer region (ISR) [5] and T. pyogenes specific parts of superoxide dismutase A encoding gene sodA as molecular targets [9]. In addition the amplification of the known and putative virulence factor encoding genes, which had also been previously used to characterize various isolates of this bacterial species [6]–[9], yielded the presence of gene plo encoding pyolysin in 57 isolates, gene cbpA encoding a collagen-binding protein in one isolate, gene nanH encoding neuraminidase H in 39 isolates, gene nanP encoding neuraminidase P in 48 isolates and the fimbriae encoding genes fimA, fimC and fimE in 57, 53 and 52 isolates of the investigated isolates, respectively The phenotypical and genotypical properties are summarized in Table 1.
FT-IR spectroscopy, a promising technique for rapid and reliable identification of bacterial microorganisms, had already been used as tool for classification of Listeria and Yersinia species [16]–, coryneform bacteria [14] and for a large number of other clinically relevant pathogens [15]–[22]–[23]. This spectroscopic technique had also been approved to investigate the most common mastitis-inducing bacteria from genera Staphylococcus [24] and Streptococcus [25]–[26] and to determine the predominant bacterial flora in raw milk [27]. According to the preliminary results of Prunner et al. [28] FT-IR spectroscopy could also be used to detect T. pyogenes in the uterus of cows of Austrian dairy farms.
In order to expand the application of FT-IR spectroscopy for mastitis diagnostics on T. pyogenes, a hierarchically structured method based on reference isolates from genus Trueperella comprising all type-strains, was created. Additionally, to include taxonomically close relatives cultivated on the same conditions, reference isolates from genera Arcanobacterium and Actinomyces were integrated in the first level of the hierarchical method. In this first step the isolates were divided into three classes, representing the genus, which were used as a preliminary filter. Trueperella isolates were differentiated in a second step down to the species level, distinguishing all recently described members of this genus. For T. pyogenes the creation of the FT-IR method succeeded by using three selected isolates (T. pyogenes DSM 20594, T. pyogenes DSM 20630T and T. pyogenes CVUAS 0222). Therewith an adequate segregation of this species from the other species of genus Trueperella and the used taxonomically close members of Arcanobacterium and Actinomyces was achieved (Table 2).
Subsequently FT-IR spectroscopy also allowed the correct classification of the external 57 isolates obtained from bovine clinical mastitis as T. pyogenes, with a probability of 98.8% and no error (Table 2). All other bacterial species comprised mostly single isolates. Therefore, a synoptic appraisal for internal validation on genus level was performed. For more than 92% Arcanobacterium, Actinomyces, and Trueperella isolates were assigned to the respective genus correctly.
As mentioned above, only three isolates were required to create the T. pyogenes-module. This mirrors the limited intra-species variation of infrared spectra of this species, which can also be shown in the cluster analysis of the Trueperella isolates used in this study (Fig. 1). In the dendrogram (Fig. 1) it is shown that the distance of the infrared-spectra of the T. pyogenes isolate-variations are far away from all the other known Trueperella species, to separate the species T. pyogenes unequivocally in this environment (Tab. 2). Noticeable in this context is the clear division of the genus in two branches. One branch comprises the type strains of the four species T. abortisuis, T. bernardiae, T. bialowiezensis, and T. bonasi. The second, very close branch was formed by all T. pyogenes reference strains, and the 57 isolates from the mastitis cases, independent from the observed variations in phenotype and genotype (Table 1). This shows the slight resolution for single isolates in IR analysis in this isolate set. Up to an exception, the T. pyogenes isolates of the present study were obtained from different animals in a wide diversity of farms and locations. Merely three isolates were isolated from one cow. Conspicuously, these three isolates clump together in a special sub-branch (Fig. 1, arrows). Because of the small difference in heterogeneity to the next neighbour isolates, the suitability for a meaningful isolate differentiation cannot be estimated from this limited data-set. In another context, and for other Gram-positive bacteria, like Bacillus cereus or Staphylococcus aureus, contamination route analysis with FT-IR succeeded by use of FT-IR [29]–[30].
Based on the database containing well defined isolates of the various species of genus Trueperella, Arcanobacterium and Actinomyces FT-IR spectroscopy, comparable to previously described Matrix-assisted laser desorption-ionization-time of flight mass spectrometry (MALDI-TOF MS) [31], appears to be a promising tool for rapid and reliable identification of T. pyogenes in routine diagnosis. In several laboratories FT-IR spectroscopy has become the first choice method for differentiation of various bacterial species. However, FT-IR spectroscopy will not replace but complement the classical phenotypical and genotypical diagnostic systems useful for characterization of T. pyogenes.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no funding or support to report.
References
- 1. Yassin AF, Hupfer H, Siering C, Schumann P, et al. (2011) Comparative chemotaxonomic and phylogenetic studies on the genus Arcanobacterium Collins et al. 1982 emend. Lehnen et al. 2006: proposal for Trueperella gen. nov. and emended description of the genus Arcanobacterium . Int J Syst Evol Microbiol 61: 1265–1274. [DOI] [PubMed] [Google Scholar]
- 2.Lämmler C, Hartwigk H (1995) Actinomyces pyogenes und Arcanobacterium haemolyticum. [German] In: Blobel H, Schlieβer T, editors. Handbuch der bakteriellen Infektionen bei Tieren, Band II/3. Gustav Fischer Verlag, Jena, Stuttgart, Germany. pp. 196–240. [Google Scholar]
- 3.Moore R, Miyoshi A, Pacheco LGC, Seyffert N, Azevedo V, et al. (2010) Corynebacterium and Arcanobacterium. In: Gyles CL, Prescott JF, Glenn Songer J, Thoen CO, editors. Pathogenesis of Bacterial Infections in Animals. Oxford: Wiley-Blackwell. pp. 133–147. [Google Scholar]
- 4. Jost BH, Billington SJ (2005) Arcanobacterium pyogenes: molecular pathogenesis of an animal opportunist. Antonie Van Leeuwenhoek 88: 87–102. [DOI] [PubMed] [Google Scholar]
- 5. Ülbegi-Mohyla H, Hijazin M, Alber J, Lämmler C, Hassan AA, et al. (2010) Identification of Arcanobacterium pyogenes isolated by post mortem examinations of a bearded dragon and a gecko by phenotypic and genotypic properties. J Vet Sci 11: 265–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Al-Tarazi Y, Hijazin M, Alber J, Lämmler C, Hassan AA, et al. (2012) Phenotypic and genotypic characteristics of Trueperella (Arcanobacterium) pyogenes isolated from lung abscesses of one-humped camels (Camelus dromedarius) in Jordan. J Camelid Sci 5: 99–104. [Google Scholar]
- 7. Gahrn-Hansen B, Frederiksen W (1992) Human infections with Actinomyces pyogenes (Corynebacterium pyogenes). Diagn Microbiol Infect Dis 15: 349–354. [DOI] [PubMed] [Google Scholar]
- 8. Kavitha K, Latha R, Udayashankar C, Jayanthi K, Oudeacoumar P, et al. (2010) Three cases of Arcanobacterium pyogenes-associated soft tissue infection. J Med Microbiol 59: 736–739. [DOI] [PubMed] [Google Scholar]
- 9. Hijazin M, Ülbegi-Mohyla H, Alber J, Lämmler C, Hassan AA, et al. (2011) Molecular identification and further characterization of Arcanobacterium pyogenes isolated from bovine mastitis and from various other origins. J Dairy Sci 94: 1813–1819. [DOI] [PubMed] [Google Scholar]
- 10. Hartwigk H, Grund S (1960) Atypische Corynebakterien beim Rind. Zentralbl Bakteriol Orig 179: 499–508. [Google Scholar]
- 11. Hartwigk H, Marcus I (1962) Gelatinaseaktivität und proteolytische Eigenschaft der typischen und atypischen Corynebakterien des Corynebacterium (C.) pyogenes . Zentralbl Bakteriol Orig 186: 544–549. [Google Scholar]
- 12.Naumann D (2000) Infrared Spectroscopy in Microbiology. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. John Wiley & Sons, Ltd. pp. 102–131. [Google Scholar]
- 13. Wenning M, Scherer S (2013) Identification of microorganisms by FTIR spectroscopy: perspectives and limitations of the method. Appl Microbiol Biotechnol 97: 7111–7120. [DOI] [PubMed] [Google Scholar]
- 14. Oberreuter H, Seiler H, Scherer S (2002) Identification of coryneform bacteria and related taxa by Fourier-transform infrared (FT-IR) spectroscopy. Int J Syst Evol Microbiol 52: 91–100. [DOI] [PubMed] [Google Scholar]
- 15. Contzen M, Sting R, Blazey B, Rau J, et al. (2011) Corynebacterium ulcerans from diseased wild boars. Zoonoses Public Health 58: 479–488. [DOI] [PubMed] [Google Scholar]
- 16. Kuhm AE, Suter D, Felleisen R, Rau J, et al. (2009) Identification of Yersinia enterocolitica at the species and subspecies levels by Fourier transform infrared spectroscopy. Appl Environ Microbiol 75: 5809–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Udelhoven T, Novozhilov M, Schmitt J (2003) The NeuroDeveloper: a tool for modular neural classification of spectroscopic data. Chemometr Intell Lab Syst 66: 219–226. [Google Scholar]
- 18. Udelhoven T, Naumann D, Schmitt J (2000) Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl Spectrosc 54: 1471–1479. [Google Scholar]
- 19. Ward JH Jr (1963) Hierarchical grouping to optimize an objective function. J Am Statist Assoc 58: 236–244. [Google Scholar]
- 20. Janbu AO, Moretro T, Bertrand D, Kohler A, et al. (2008) FT-IR microspectroscopy: a promising method for the rapid identification of Listeria species. FEMS Microbiol Lett 278: 164–170. [DOI] [PubMed] [Google Scholar]
- 21. Wortberg F, Nardy E, Contzen M, Rau J, et al. (2012) Identification of Yersinia ruckeri from diseased salmonid fish by Fourier transform infrared spectroscopy. J Fish Dis 35: 1–10. [DOI] [PubMed] [Google Scholar]
- 22. Samuels AC, Snyder AP, Emge DK, Amant D, Minter J, et al. (2009) Classification of select category A and B bacteria by Fourier transform infrared spectroscopy. Appl Spectrosc 63: 14–24. [DOI] [PubMed] [Google Scholar]
- 23. Grunert T, Wenning M, Barbagelata MS, Fricker M, Sordelli DO, et al. (2013) Rapid and reliable identification of Staphylococcus aureus capsular serotypes by means of artificial neural network-assisted Fourier transform infrared spectroscopy. J Clin Microbiol 51: 2261–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spohr M, Rau J, Friedrich A, Klittich G, Fetsch A, et al. (2011) Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 58: 252–261. [DOI] [PubMed] [Google Scholar]
- 25.Horlacher S, Spohr M, Rau J (2009) Einsatz der FT-IR in der Mastitis-Diagnostik [German]. Amtstierärztlicher Dienst und Lebensmittelkontrolle, Sonderausgabe zur 50 Arbeitstagung des Arbeitsgebietes Lebensmittelhygiene der Deutschen Veterinärmedizinischen Gesellschaft. Garmisch-Partenkirchen, Germany, 29.09-02.10.2009.
- 26.Schabauer L, Idris R, Wenning M, Ehling-Schulz M, et al. (2011) Identification and differentiation of mastitic associated Streptococcus spp. and related bacteria by FTIR - spectroscopy. FT-IR Spectroscopy in Microbiological and Medical Diagnostics Workshop. Robert Koch-Institute, Berlin, Germany. [Google Scholar]
- 27. Fricker M, Skanseng B, Rudi K, Stessl B, Ehling-Schulz M, et al. (2011) Shift from farm to dairy tank milk microbiota revealed by a polyphasic approach is independent from geographical origin. Int J Food Microbiol 145: 24–30. [DOI] [PubMed] [Google Scholar]
- 28.Prunner I, Karen W, Pothmann-Reichl H, Ehling-Schulz M, Marc D, et al.. (2013) Bacteriological, vaginoscopical and cytological examination of the bovine uterus in Austrian dairy herds. XIII Middle European Buiatrics Congress, Belgrade, Serbia. pp. 168–174.
- 29. Rau J, Perz R, Klittich G, Contzen M, et al. (2009) Cereulide forming presumptive Bacillus cereus strains from food-differentiating analyses using cultural methods, LC-MS/MS, PCR, and infrared spectroscopy in consideration of thermotolerant isolates. Berl Munch Tierarztl Wochenschr 122: 25–36. [PubMed] [Google Scholar]
- 30. Johler S, Tichaczek-Dischinger PS, Rau J, Sihto HM, Lehner A, et al. (2013) Outbreak of Staphylococcal food poisoning due to SEA-producing Staphylococcus aureus . Foodborne Pathog Dis 10: 777–781. [DOI] [PubMed] [Google Scholar]
- 31. Hijazin M, Hassan AA, Alber J, Lämmler C, Timke M, et al. (2012) Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for species identification of bacteria of genera Arcanobacterium and Trueperella . Vet Microbiol 157: 243–245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.