Abstract
The cytoskeleton is a dynamic network composed of filamentous polymers and regulatory proteins that provide a flexible structural scaffold to the cell and plays a fundamental role in developmental processes. Mutations that alter the spatial orientation of the cortical microtubule (MT) array of plants are known to cause important changes in the pattern of cell wall synthesis and developmental phenotypes; however, the consequences of such alterations on other MT-network-associated functions in the cytoplasm are not known. In vivo observations suggested a role of cortical MTs in the formation and movement of Tobacco mosaic virus (TMV) RNA complexes along the endoplasmic reticulum (ER). Thus, to probe the significance of dynamic MT behavior in the coordination of MT-network-associated functions related to TMV infection and, thus, in the formation and transport of RNA complexes in the cytoplasm, we performed an evolution experiment with TMV in Arabidopsis thaliana tor1/spr2 and tor2 mutants with specific defects in MT dynamics and asked whether TMV is sensitive to these changes. We show that the altered cytoskeleton induced genetic changes in TMV that were correlated with efficient spread of infection in the mutant hosts. These observations demonstrate a role of dynamic MT rearrangements and of the MT-associated protein TORTIFOLIA1/SPIRAL2 in cellular functions related to virus spread and indicate that MT dynamics and MT-associated proteins represent constraints for virus evolution and adaptation. The results highlight the importance of the dynamic plasticity of the MT network in directing cytoplasmic functions in macromolecular assembly and trafficking and illustrate the value of experimental virus evolution for addressing the cellular functions of dynamic, long-range order systems in multicellular organisms.
Introduction
Microtubules (MTs) are involved in a multitude of cellular processes such as intracellular transport and localization of organelles, determination of cell shape, or the perception and response to mechanical stimulus [1]. The plant MT network is highly dynamic and continuously remodeled into new arrangements in response to environmental and developmental information. In contrast to animal cells, where MTs are attached to the centrosome and extend with their polymerizing plus ends towards the cell periphery, cortical plant MTs are localized underneath the plasma membrane (PM) and form a barrel-shaped interphase array of dispersed MTs that do not share a common nucleation site. New MTs nucleate from mobile γ-tubulin-containing complexes that are, in most cases, recruited to existing MTs. The new MTs emerge either at a 40° angle or in parallel to the associated MTs and thus form either branched/crossover or interbundle arrangements, respectively. Upon nucleation, the new MTs may be severed away from their minus ends, thus creating free minus ends. The liberated minus ends of the severed MTs are now free to depolymerize which, if balanced by polymerization at the plus end, results in treadmilling and the translocation of the MTs along the PM [2]–[4]. MT severing also creates new plus ends that can regrow to elongated MTs at the crossover site [5]. Recent studies indicate that the severing activity of katanin at MT crossover sites is inhibited by the presence of TORTIFOLIA1/SPIRAL2 (TOR1), a MT-associated protein that promotes MT growth and stabilizes MT crossovers [6], [7]. By controlling MT severing, TOR1 appears to play a central role in regulating local MT patterning within the cortical array. Consistently, katanin and tor1 mutants show alterations in the MT array. These and other mutations that affect the dynamic plasticity of the MT cytoskeleton also cause a wide range of developmental phenotypes thus illustrating the important role of MT array patterning during plant development [8]. However, although even slight changes in plant MT alignment caused by such mutations are known to affect growth, barely anything is known about the global consequences of such mutations on localized MT network-associated functions in the cytoplasm. It appears likely that the local patterning of MTs within the cortical array directs the local scaffolding for localized cellular functions and thus the functional and spatial organization of the cellular cortex. A role of MT patterning in directing localized functions of the cell is supported by specific local MT arrangements directing the patterns of cell wall synthesis in xylem and pavement cells [9]. Moreover, recent observations indicate that MTs are associated with endosomes and therefore could influence the abundance of membrane proteins such as PIN2 [10]. MTs linked to an endosomal pathway may also play a role in the targeting of non-cell-autonomous proteins to plasmodesmata (PD) [11]. MTs may also provide a framework for localized protein turnover processes such as ERAD (ER-associated degradation) [12], [13] and autophagy [14]. These and other observations suggest that locally organized MTs may facilitate the localized formation, maintenance, and turnover of PM domains and also of membrane-associated macromolecular complexes that are destined for transport to PD [15].
Consistent with the above-mentioned examples, cortical MTs are also implicated in the interaction of plants with cytoplasmic viruses and their targeting to PD. Among the plant viruses that have been reported to interact with MTs, Tobacco mosaic virus (TMV) is the best characterized [16]–[19]. TMV replicates its RNA genome in association with MT-associated sites of the cortical endoplasmic reticulum (ER) that together with the underlying actin network provides the structure along which the viral RNA is transported to PD and into adjacent cells. The virus moves its genome between cells in a non-encapsidated form and thus may rely on cellular mechanisms usually supporting the intra- and intercellular transport of endogenous RNA complexes. The virus-encoded movement protein (MP) that binds MTs [16], [20], [21] is essential for cell-to-cell movement of this virus [22]–[24], and the ability of the virus to spread between cells is correlated with the ability of MP to interact with MTs [17], [18]. Similar to RNA transport processes in other systems such as neurons or Drosophila oocytes [25]–[27], the transport of TMV RNA in plants is associated with the formation of mobile MP and RNA-containing particles/granules that are transported in a stop-and-go manner [18], [28]. In vivo studies with conditional mutations in MP demonstrated that the formation of the granules is functionally linked to the ability of the protein to bind MTs and to its function in viral RNA movement [18], [28]. Consistent with a role of MT binding, the granules move along the cortical ER and pause their movements at MT-associated ER sites [28]. The granules observed in cells at the infection front of the virus are proposed to represent early viral replication complexes (VRCs) and either spread along the ER-actin network to reach PD or remain in the infected cell to give rise to large VRC clusters or “virus factories” that are observed during later infection stages [19]. The virus factories contain replicase, viral RNA, as well as MP, and accumulate the viral coat protein (CP) and virions on the surface [29]. They are associated with the ER-actin network as well as with numerous branching MTs suggesting that the assembly and growth of VRCs may involve local rearrangements in the cortical MT array [19]. The local MT rearrangements may reflect a function of MP since this protein appears to be capable to hijack the MT nucleation machinery [17], [19], [28], [30], [31]. By inducing such local MT rearrangements, the MP may support the formation, trafficking, and further maturation of VRCs by creating a local scaffold for VRC anchorage and the recruitment of host factors and ER membranes [15], [19].
However, although the in vivo observations suggest a role of dynamic plasticity in the MT cytoskeleton for virus replication and movement, additional tools are required to support this model. Here, we decided to make use of the high mutation rate and evolvability of RNA viruses [32] to determine the importance of MT dynamics and patterning in the cortical MT array for virus movement by experimental virus evolution. Indeed, if the above model is valid, the dynamic plasticity of the MT cytoskeleton should be vital for the virus and any important change in the dynamic plasticity of the MT cytoskeleton should produce a selective pressure for the virus to evolve. Such evolutionary response would demonstrate the role of MT rearrangements during infection and would highlight the value of TMV evolvability as a system for testing the role of the MT cytoskeleton and its dynamic plasticity in directing cytoplasmic functions, particularly in the formation and trafficking of RNA complexes.
To test the functional intimacy between TMV and the MT cytoskeleton and particularly the role of the dynamic MT cytoskeleton in directing cytoplasmic functions related to the development and transport of viral RNA complexes, we challenged TMV with tortifolia 1/spiral 2 (tor1) and tortifolia 2 (tor2) mutants with specific defects in the dynamic behavior of MTs within the cortical MT array. Whereas the tor1 mutant is a knock-out of the MT-associated protein TOR1 and is inhibited in efficient MT polymerization and in the formation and maintenance of MT crossovers [6], tor2 carries a conservative arginine to lysine change at position 2 of the primary amino acid sequence (R2K mutation) of the α-tubulin 4 protein that is proposed to interfere with contacts between R2 of α-tubulin and the GTPase domain of β-tubulin and thus to reduce the rate of MT polymerization [33]. In this work, we demonstrate that the virus is rather well adapted to the A. thaliana wild type. However, the virus is highly sensitive to tor1 and tor2 mutations and responds to the respective changes in the dynamic plasticity of the MT cytoskeleton with mutations that are correlated with efficient spread within the mutant hosts without significantly increasing the efficiency of virus replication. The results demonstrate the importance of dynamic MT network plasticity for the establishment of cytoplasmic functions required for efficient TMV movement and that MT dynamics and MT-associated proteins represent constraints for virus evolution and adaptation. The results suggest an important role of TOR1, which may stabilize MT crossovers to support the formation and intercellular spread of the viral RNA complexes. We propose that these findings reflect the importance of the dynamic MT cytoskeleton in the formation and transport of viral as well as endogenous RNA complexes.
Results
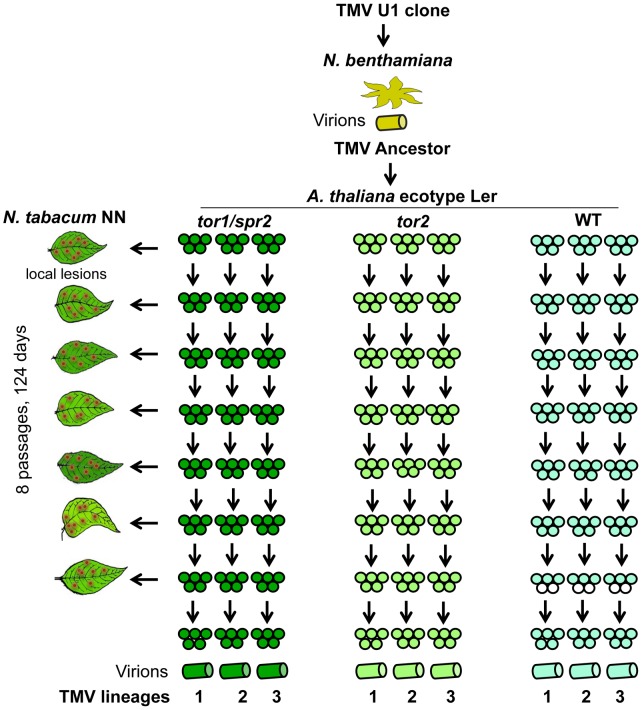
To test the response of TMV to alterations in MT network dynamics, we used a TMV isolate obtained after replication of a TMV cDNA clone [24] in Nicotiana benthamiana plants (Figure 1). Using this isolate as the ancestral virus in our evolution experiment, we inoculated three sets (replicates) of five wild type, five tor1, and five tor2 A. thaliana plants. At 15 days post inoculation (dpi), the non-inoculated, systemically infected, young leaves of all plants of a given set were pooled and used for virus extraction. The respective virus extracts were then used to inoculate new sets of plants. This passaging from one plant generation to the next was performed eight times. The number of infectious virion particles in each inoculum was estimated by quantifying the number of local cell death lesions following inoculation of hypersensitive N. tabacum NN plants (Figure S1). These tests demonstrated that the inoculum carried 150 to 400 infectious virion particles in all passages thus indicating the absence of strong bottlenecks during passages that could favor the accumulation of mutations by genetic drift and hinder fixation of adaptive mutations. After the eighth passage, nine final viral lineages were created (Figure 1): three lineages from tor1 plants (Tor1-1, Tor1-2 and Tor1-3), three lineages from tor2 plants (Tor2-1, Tor2-2 and Tor2-3), and three lineages from wild type plants (WT-1, WT-2 and WT-3).
Figure 1. Layout of the virus evolution experiment.
The initial TMV inoculum (ancestor) was obtained from N. benthamiana plants inoculated with a cDNA clone of TMV and was used for inoculation of tor1/spr2, tor2, and wild type (WT) A. thaliana plants (three sets of five plants each). Following eight passages, nine independent TMV lineages were obtained. The number of infectious particles used for each passage was controlled by local lesion assays with hypersensitive N. tabacum NN plants.
To evaluate adaptation of TMV to each host genotype, the fitness of each experimentally evolved viral lineage was compared with that of the ancestral virus. Absolute fitness, W, was determined by measuring the over-time accumulation of viral RNA in the upper, non-inoculated leaves as described in the Materials and Methods section. Whereas the level of viral accumulation in plants, and thus W, results from intracellular replication and the number of new cells infected by intercellular movement, viral accumulation in protoplasts is determined only by intracellular replication. Thus, to distinguish the contribution of intercellular RNA movement from that of RNA replication, viral accumulation was measured in three sets (replicates) of the wild type and mutant plants as well as in protoplasts upon inoculation with the respective viral lineage or the ancestral virus. The results indicate that relative to the ancestral virus the tor1-derived lineages developed a significant decrease in W in wild type plants (13%; P = 0.035) and a slight, yet statistically significant reduction of W in BY-2 protoplasts (0.53%; P = 0.031). Nevertheless, the same tor1-derived lineages showed a trend of higher W in tor1/spr2 plants (8.4%) (Figure 2). These results indicate that the mutant cytoskeleton in tor1/spr2 plants triggered changes in TMV that allowed TMV to adapt and thus to maintain or even increase fitness, i.e. viral RNA movement efficiency, in this environment. However, as shown by the decrease of W in BY-2 protoplasts, the adaptive changes induced by the mutant cytoskeleton in tor1 plants caused a fitness trade-off for replication. The reduced RNA replication efficiency of the tor1-derived lineages likely contributed to the decrease of W in wild type plants and may have prevented a more significant increase of W in tor1 plants. The ability of tor1 to trigger adaptation in TMV indicates a strong impact of dynamic MT behaviour and of TOR1 on virus replication and movement.
Figure 2. Average absolute fitness (W) of the TMV ancestor and evolved lineages.
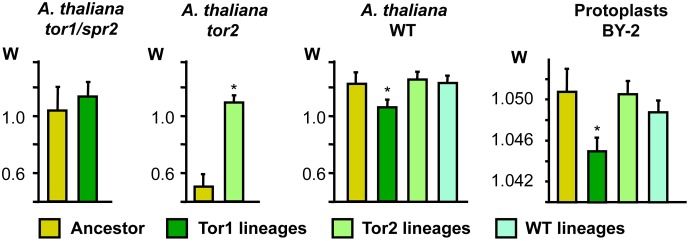
W is shown for infection in A. thaliana tor1/spr2, tor2 and WT plants, and in BY-2 protoplasts. Tor2 lineages show adaptation for efficient systemic movement in tor2 plants without consequences on replication efficiency (lack of a change in W compared to ancestor in protoplasts). Tor1 lineages are affected in replication efficiency and show slight adaptation for efficient systemic movement in tor1/spr2 plants. Asterisks indicate statistically significant differences from the ancestor.
The tor2-derived lineages showed a statistically significant average increase in W of 72.2% in tor2 plants (P<0.001) whereas W remained similar to that of the ancestral virus in the wild type plants and protoplasts (Figure 2). Thus, unlike tor1, the presence of tor2 induced TMV adaptation without causing a trade-off in viral replication efficiency. The adaptation of TMV to tor2 supports a role of MT polymerization efficiency in TMV movement. Finally, the fitness of WT-derived lineages remained similar to that of the ancestral virus in wild type plants and protoplast (Figure 2 and Table S1) indicating that the common strain of TMV (U1) is well adapted to A. thaliana.
Sequence analysis for each of the nine lineages revealed 11 nucleotide substitutions in seven lineages with respect to the ancestral virus (Table 1). The tor1/spr2- and tor2- derived lineages showed one or two nucleotide substitutions in the first open reading frame (ORF) of the viral RNA, which encodes the 126 kD and 183 kD subunits of the replicase (Table 1). Nucleotide substitutions in lineages Tor1-1, Tor1-3 and Tor2-3 caused amino acids changes whereas the Tor2-1 and Tor2-2 lineages had synonymous substitutions suggesting that in addition to changes in the protein also the secondary structure of the viral RNA or its translational efficiency may play a role in optimizing viral fitness.
Table 1. Nucleotide substitutions in evolved TMV lineages.
| Repb | MPb | CPb | |||||
| Lineagesa | MTc | HELc | POLc | ||||
| Tor1-1 | - | - | A2542G, T3235C | - | - | - | - |
| Tor1-2 | - | - | - | - | - | - | - |
| Tor1-3 | - | - | - | C3344T | - | - | - |
| Tor2-1 | - | - | T2627C | - | T4293C* | - | - |
| Tor2-2 | - | T1827C* | - | - | - | - | - |
| Tor2-3 | C693Ad | - | - | - | - | - | - |
| WT-1 | - | - | - | - | - | - | - |
| WT-2 | - | T2627C | - | - | - | - | |
| WT-3 | A764G, T798C* | - | - | - | - | - | T180C* |
TMV lineages obtained after eight serial passages of the TMV ancestor through A. thaliana tor1/spr2, tor2, and wild type (WT).
ORF-encoded proteins; Rep, 126 k/183 k replicase; MP, movement protein; CP, coat protein.
Domains of the 126 k/183 k replicase; MT, methyltransferase domain; HEL, helicase domain; POL, RNA-dependent RNA polymerase domain. The shorter 126 k subunit lacks the POL domain.
Nucleotide positions are relative to the first nucleotide of each ORF. Synonymous substitutions are indicated by asterisks.
Discussion
The cortical interphase MT array of plants is well known to drive the insertion [34], [35] and trajectory of the cellulose synthase complex at the plasma membrane [36], [37] and thereby direct the parallel deposition of cellulose microfibrills that mediates anisotropic growth [38], [39]. However, there is only very limited information about the role of the dynamic MT array in guiding other MT-associated functions in the cytoplasm. Our previous studies established that MTs play a role during TMV infection and undergo interactions with the virus-encoded MP, and that these interactions are important for the cell-to-cell movement of the virus [17], [18], [21]. In vivo observations indicated that MTs are associated with VRCs functioning as replication factories during late infection as well as with early VRCs, thus with MP-associated RNA complexes functionally associated with the cell-to-cell movement of the virus [19], [28]. To gain further insight into the role of the MT cytoskeleton and particularly of its dynamic plasticity in guiding cytoplasmic functions related to TMV infection, we challenged TMV with specific A. thaliana mutants affected in MT dynamics. Whereas tor1/spr2 interferes with MT orientation and growth [7], [40] and the stabilization of MT crossovers [6], tor2 affects tubulin polymerization efficiency [33]. We found that TMV is sensitive to these mutations and undergoes adaptive evolution that allows it to enhance its fitness in the respective mutant hosts. The ability of the host mutations to induce adaptive changes in the virus demonstrates the importance of the dynamic plasticity of the MT cytoskeleton for guiding MT network-related functions in the cytoplasm, i.e. functions that are required to support virus infection.
Given that the mutant host environment induced viral adaption and increases in fitness with respect to systemic spread without involving increases in viral replication efficiency in any of the evolved lineages, the observations indicate a role of dynamic MT network functions in TMV RNA movement. Viral adaptation to tor1 and to tor2 correlated with the fixation of mutations clustering in the replicase-encoding ORF of the virus, thus indicating a role of the replicase subunits in these processes. Although previous evidence supports a role of the replicase in TMV spread [41], an interaction of these proteins with MTs has not been reported. The observation that host mutations affecting the dynamic rearrangements within the MT network caused changes in the replicase is consistent with the role of the 126 k replicase protein in regulating the size of the VRCs [42] and with current models implying a role of MTs in the anchorage and assembly of these complexes [19]. Interestingly, no adaptive mutations were found in the MP, which interacts with MT in vivo and in vitro [21], or in the coat protein, which is required for TMV long distance movement through the phloem [43]. This may suggest that the tor1 and tor2 mutations do not interfere with the interactions of MP with tubulin but rather impair the important role of local MT patterning and scaffolding in the formation, movement and further maturation of VRCs. The CP was reported to control VRC growth by regulating the production of subgenomic RNAs and thus the amount of MP that accumulates in VRCs. However, this protein does not interact with MTs, is dispensable for virus replication, and accumulates outside the VRCs [29] and may therefore not be involved in MT-related mechanisms that interact with the VRCs for movement.
Viral adaptation in response to tor1 and tor2 reveals the importance of local MT patterning and associated functions in the formation and transport of viral and potentially endogenous RNA complexes. Since the mutations in tor1/spr2 and tor2 plants interfere with MT dynamics through different mechanisms, these processes do not necessarily depend on direct interactions with tubulin and SPIRAL2 but rather on their function in controlling the local MT network superstructure. Whereas the viral adaptation to both host mutations reflects the importance of normal MT dynamics, the adaptation to tor1 indicates the specific importance of TOR1 function. The function of TOR1 in maintaining MT crossovers during infection may be consistent with evidence indicating that MP hijacks MT nucleating complexes [30]. According to current hypothesis, the MP recruits MT nucleating complexes to early VRCs to generate a MT scaffold required for VRC maturation and growth, thus leading to the MT crossovers associated with VRCs seen in vivo [19]. TOR1 function may be required to suppress MT severing by katanin [6], which would otherwise act against the MT arrangement induced by MP. That a reduced MT polymerization efficiency and a reduced capacity of the cell to maintain MT crossovers lead to changes in the replicase suggests that these changes in MT activities affect VRC structure, movement, or maturation and that the deficiencies imposed by the mutations can be complemented by the structural composition of the VRC, which may be determined by the structure of the replicase or the viral RNA. The range of the genetic changes in the adapted virus lineages suggests that the respective replicase and RNA domains offer several options and plasticity for adaptation to the alteration in MT dynamics. This plasticity in the viral genome may play an important role in allowing the virus to replicate and move between different cell types and hosts.
Our results demonstrate that the U1 strain of TMV is well adapted to the model plant A. thaliana and that the interaction of TMV with the altered hosts exerted a positive selection of gain-of-function mutations in the viral genome which allowed efficient viral RNA spread. The capacity of TMV to undergo adaptive evolution (evolvability) in response to altered host components commends its suitability for experimental viral evolution experiments to identify and determine the significance of specific gene functions in relation to dynamic membrane and cytoskeleton-related processes that are involved in infection and that may as well also play fundamental roles during plant development and disease.
Materials and Methods
Virus production
To generate the initial inoculum for the virus evolution experiment, a full-length cDNA clone of TMV strain U1 [24] was linearized with Acc65I and capped infectious TMV RNA was in vitro transcribed using the RiboMax Large Scale RNA production system-T7 (Promega). Three N. benthamiana plants (3 to 4 weeks old) were infected by mechanical inoculation of the second true leaf with 4 µg viral RNA per plant, in the presence of Celite 545 (Prolabo). Following inoculation, the plants were maintained in a greenhouse at 16 h light/8 h dark cycles at 22/18°C. Virion particles were purified and quantified from symptomatic tissue at 7 dpi using published methods [12].
Arabidopsis plants and inoculation
Wild type, tor1 [40] and tor2 [33] A. thaliana (ecotype Landsberg erecta) plants were germinated and cultivated in growing chambers with 16 h light/8 h dark cycles at 22/18°C. The 5th and 6th leaves of 4 weeks old plants were rub-inoculated with virions in the presence of Celite 545 (Prolabo). Inoculated plants were maintained under greenhouse conditions, with 16 h light/8 h dark cycles at 22/18°C.
Viral passages in Arabidopsis
The virus evolution experiment involved eight viral passages between generations of plants, each consisting of three sets (replicates) of five plants for each plant genotype (Figure 1). For each passage at 15 dpi, three upper leaves of the systemically infected plants in each set were harvested and pooled. Viruses were extracted from these pools by addition of 3 ml of 10 mM sodium phosphate buffer (pH 7.0) per 1 g of infected plant tissue, followed by homogenization using a Precellys tissue lyser (Bertin). The extracts were clarified by centrifugation for 10 min at 12000×g and used for inoculation of new plants. The titer of infectious viral particles in the inoculum of each passage was determined by counting the number of local cell death lesions following inoculation of hypersensitive N. tabacum NN plants.
Analysis of viral fitness
The fitness of each viral lineage and the ancestral virus was analyzed by inoculation of A. thaliana wild type, tor1/spr2 and tor2 plants (10–13 plants each). Plants were inoculated with 1 µg of purified virions and non-inoculated upper leaves were harvested at 7, 12 and 15 dpi to evaluate systemic infection. Harvested samples were used for TMV detection by a commercial DAS-ELISA kit (DSMZ RT-0041, Germany, using the manufacturer's instructions) and for analysis of viral RNA titer in ELISA-positive plants. RNA titer was determined by quantitative RT-PCR [44] using the TaqMan probe 6-FAM-AGACAGCCCACATGTTTTGGTCGCA and primers TMV-3093F ( CCGATCTCAAAACCCTTGCA) and TMV1R (CGAACAGGTGTGCCTTGACA). Viral RNA titers (Table S1) were used to calculate absolute fitness, W, defined as W = er, where r, the Malthusian growth rate, was estimated by regression of the logarithm of the number of viral RNA molecules that accumulate over time (7, 12 and 15 dpi). W was analyzed by using a Generalized Linear Model [45]. All W calculations refer to times after infection during which the virus was actively spreading into non-inoculated leaves.
To determine replication efficiency in the absence of virus movement, W was measured in infected tobacco Bright Yellow-2 (BY-2) protoplasts [46]. Protoplasts (1×106 cells) were inoculated with 1 µg of purified virions using electroporation [47]. Samples were collected at 0, 6, 12, 24, 48 and 72 hours post inoculation and used for total RNA extraction and subsequent analysis by quantitative RT-PCR. Three independent experiments (biological replicates) were performed for each viral lineage and the ancestor. The results of these quantifications are shown in Table S2.
Nucleotide sequencing
The consensus nucleotide sequences of the TMV ancestor and the nine TMV lineages were obtained by sequencing of reverse-transcribed and PCR-amplified viral RNA extracted from virions (purified as above) belonging to the 8th passage. Viral RNA was purified from virions by a protocol involving phenol/chloroform/isoamyl alcohol extraction followed by isopropanol precipitation [48]. Nine pairs of primers mapping along the TMV genome were used (Table S3). Reverse transcription was performed at 42°C for 1 h with SuperScriptIII Reverse Transcriptase (Invitrogen), followed by PCR amplification using iProof High-Fidelity DNA Polymerase (BIO-RAD) in a GeneAmp PCR System 9700 (Applied Biosystems) and applying 2 min at 94°C, 35 cycles of 15 s at 94°C, 15 s at 55°C and 40 s at 72°C, and a final incubation for 5 min at 72°C. PCR products were purified using Illustra GFX PCR DNA and Gel Band Purification kit (GE Healthcare). Nucleotide sequences were determined with a 3130xL Genetic Analyzer (Applied Biosystems) and assembled with the program STADEN 2.0.0b9 [49]. Nucleotide sequences were deposited at GenBank under accession numbers KF972427-KF972436.
Accession numbers
Nucleotide sequences were deposited at GenBank under accession numbers KF972427-KF972436.
Supporting Information
Local necrotic lesion assay using hypersensitive tobacco NN plants to estimate the number of infectious particles in a plant extract. The left half of the leaf was inoculated with a control extract from a healthy A. thaliana plant. The right half of the leaf was inoculated with extracts from a TMV-infected A. thaliana plant (WT-1 from the fourth passage). The number of lesions reflects the number of particles in the inoculum.
(JPG)
Accumulation of the TMV ancestor and evolved lineages in non-inoculated, systemic leaves in tor1/spr2 , tor2 , and WT plants at 7, 12 and 15 dpi.
(DOCX)
Accumulation of the TMV ancestor and evolved lineages in tobacco BY-2 protoplasts at 6, 12, 24, 48 and 72 hpi.
(DOCX)
Forward (F) and reverse (R) primers used for TMV genome amplification by RT-PCR and nucleotide sequencing.
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. Nucleotide sequences were deposited at GenBank under accession numbers KF972427-KF972436.
Funding Statement
Work of E.P. and of A.N. was supported by grants from the Agence National de la Recherche (ANR, grant ANR-08-BLAN-244) and the Swiss National Science Foundation (SNF, grant 310031A_140694) to M.H. We also acknowledge the funding of an EMBO short-term fellowship to H.B. I.F. was supported by a pre-doctoral fellowship from the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) from Spain. L.R. was supported by grant AGL2005-07754 from the Spanish Ministry of Science and Education and S.F.E. by grant BFU2012-30805 from the Spanish Secretaría de Estado de Investigación, Desarrollo e Innovación. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fletcher DA, Mullins RD (2010) Cell mechanics and the cytoskeleton. Nature 463: 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, et al. (2005) Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol 7: 961–968. [DOI] [PubMed] [Google Scholar]
- 3. Shaw SL, Kamyar R, Ehrhardt DW (2003) Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718. [DOI] [PubMed] [Google Scholar]
- 4. Nakamura M, Ehrhardt DW, Hashimoto T (2010) Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat Cell Biol 12: 1064–1070. [DOI] [PubMed] [Google Scholar]
- 5. Chan J, Sambade A, Calder G, Lloyd C (2009) Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21: 2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wightman R, Chomicki G, Kumar M, Carr P, Turner SR (2013) SPIRAL2 determines plant microtubule organization by modulating microtubule severing. Curr Biol 23: 1902–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yao M, Wakamatsu Y, Itoh TJ, Shoji T, Hashimoto T (2008) Arabidopsis SPIRAL2 promotes uninterrupted microtubule growth by suppressing the pause state of microtubule dynamics. J Cell Sci 121: 2372–2381. [DOI] [PubMed] [Google Scholar]
- 8. Buschmann H, Lloyd CW (2008) Arabidopsis mutants and the network of microtubule-associated functions. Mol Plant 1: 888–898. [DOI] [PubMed] [Google Scholar]
- 9. Oda Y, Fukuda H (2013) Spatial organization of xylem cell walls by ROP GTPases and microtubule-associated proteins. Curr Opin Plant Biol 16: 743–748. [DOI] [PubMed] [Google Scholar]
- 10. Ambrose C, Ruan Y, Gardiner J, Tamblyn LM, Catching A, et al. (2013) CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana . Dev Cell 24: 649–659. [DOI] [PubMed] [Google Scholar]
- 11. Wu S, Gallagher KL (2013) Intact microtubules are required for the intercellular movement of the SHORT-ROOT transcription factor. Plant J 74: 148–159. [DOI] [PubMed] [Google Scholar]
- 12. Niehl A, Amari K, Gereige D, Brandner K, Mely Y, et al. (2012) Control of Tobacco mosaic virus movement protein fate by CELL-DIVISION-CYCLE protein 48. Plant Physiol 160: 2093–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niehl A, Amari K, Heinlein M (2013) CDC48 function during TMV infection: Regulation of virus movement and replication by degradation? Plant Signal Behav 8: e22865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ (2004) Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett 567: 302–306. [DOI] [PubMed] [Google Scholar]
- 15. Peña EJ, Heinlein M (2013) Cortical microtubule-associated ER sites: organization centers of cell polarity and communication. Curr Opin Plant Biol 16: 764–773. [DOI] [PubMed] [Google Scholar]
- 16. Heinlein M, Epel BL, Padgett HS, Beachy RN (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985. [DOI] [PubMed] [Google Scholar]
- 17. Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M (2000) Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2: 826–832. [DOI] [PubMed] [Google Scholar]
- 18. Boyko V, Hu Q, Seemanpillai M, Ashby J, Heinlein M (2007) Validation of microtubule-associated Tobacco mosaic virus RNA movement and involvement of microtubule-aligned particle trafficking. Plant J 51: 589–603. [DOI] [PubMed] [Google Scholar]
- 19. Niehl A, Peña EJ, Amari K, Heinlein M (2013) Microtubules in viral replication and transport. Plant J 75: 290–308. [DOI] [PubMed] [Google Scholar]
- 20. Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, et al. (1998) Changing patterns of localization of the Tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ashby J, Boutant E, Seemanpillai M, Groner A, Sambade A, et al. (2006) Tobacco mosaic virus movement protein functions as a structural microtubule-associated protein. J Virol 80: 8329–8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deom CM, Oliver MJ, Beachy RN (1987) The 30-kilodalton gene product of Tobacco mosaic virus potentiates virus movement. Science 237: 384–389. [DOI] [PubMed] [Google Scholar]
- 23. Deom CM, Schubert KR, Wolf S, Holt CA, Lucas WJ, et al. (1990) Molecular characterization and biological fuction of the movement protein of Tobacco mosaic virus in transgenic plants. Proc Natl Acad Sci USA 87: 3284–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holt CA, Beachy RN (1991) In vivo complementation of infectious transcripts from mutant Tobacco mosaic virus cDNAs in transgenic plants. Virology 181: 109–117. [DOI] [PubMed] [Google Scholar]
- 25. Dohner K, Nagel CH, Sodeik B (2005) Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol 13: 320–327. [DOI] [PubMed] [Google Scholar]
- 26. Brengues M, Teixeira D, Parker R (2005) Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greber UF, Way M (2006) A superhighway to virus infection. Cell 124: 741–754. [DOI] [PubMed] [Google Scholar]
- 28. Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, et al. (2008) Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic 9: 2073–2088. [DOI] [PubMed] [Google Scholar]
- 29. Asurmendi S, Berg RH, Koo JC, Beachy RN (2004) Coat protein regulates formation of replication complexes during Tobacco mosaic virus infection. Proc Natl Acad Sci U S A 101: 1415–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferralli J, Ashby J, Fasler M, Boyko V, Heinlein M (2006) Disruption of microtubule organization and centrosome function by expression of Tobacco mosaic virus movement protein. J Virol 80: 5807–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brandner K, Sambade A, Boutant E, Didier P, Mély Y, et al. (2008) Tobacco mosaic virus movement protein interacts with green fluorescent protein-tagged microtubule end-binding protein 1. Plant Physiol 147: 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elena SF, Sanjuán R (2007) Virus evolution: Insights from an experimental approach. Annu Rev Ecol Evol Syst 38: 27–52. [Google Scholar]
- 33. Buschmann H, Hauptmann M, Niessing D, Lloyd CW, Schaffner AR (2009) Helical growth of the Arabidopsis mutant tortifolia2 does not depend on cell division patterns but involves handed twisting of isolated cells. Plant Cell 21: 2090–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, et al. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806. [DOI] [PubMed] [Google Scholar]
- 36. Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- 37. Chan J, Crowell E, Eder M, Calder G, Bunnewell S, et al. (2010) The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J Cell Sci 123: 3490–3495. [DOI] [PubMed] [Google Scholar]
- 38. Baskin TI (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222. [DOI] [PubMed] [Google Scholar]
- 39. Lloyd C (2011) Dynamic microtubules and the texture of plant cell walls. Int Rev Cell Mol Biol 287: 287–329. [DOI] [PubMed] [Google Scholar]
- 40. Buschmann H, Fabri CO, Hauptmann M, Hutzler P, Laux T, et al. (2004) Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr Biol 14: 1515–1521. [DOI] [PubMed] [Google Scholar]
- 41. Hirashima K, Watanabe Y (2001) Tobamovirus replicase coding region is involved in cell-to-cell movement. J Virol 75: 8831–8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu J-Z, Blancaflor EB, Nelson RS (2005) The Tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol 138: 1877–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Osbourn JK, Sarkar S, Wilson TM (1990) Complementation of coat protein-defective TMV mutants in transgenic tobacco plants expressing TMV coat protein. Virology 179: 921–925. [DOI] [PubMed] [Google Scholar]
- 44. Mansilla C, Sanchez F, Padgett HS, Pogue GP, Ponz F (2009) Chimeras between Oilseed rape mosaic virus and Tobacco mosaic virus highlight the relevant role of the tobamoviral RdRp as pathogenicity determinant in several hosts. Mol Plant Pathol 10: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nelder JA, Wedderburn RW (1992) Generalized linear models. J R Stat Soc Series A 135: 370–384. [Google Scholar]
- 46. Nagata T, Nernoto Y, Hasezawa S (1992) Tobacco BY-2 cell line as the "HeLa" cell line in the cell biology of higher plants. Int Rev Cytol 132: 1–30. [Google Scholar]
- 47. Herzog E, Hemmer O, Hauser S, Meyer G, Bouzoubaa S, et al. (1998) Identification of genes involved in replication and movement of Peanut clump virus . Virology 248: 312–322. [DOI] [PubMed] [Google Scholar]
- 48. Debreczeni DE, Ruiz-Ruiz S, Aramburu J, López C, Belliure B, et al. (2011) Detection, discrimination and absolute quantitation of Tomato spotted wilt virus isolates using real time RT-PCR with TaqManMGB probes. J Virol Methods 176: 32–37. [DOI] [PubMed] [Google Scholar]
- 49. Bonfield JK, Whitwham A (2010) Gap5—editing the billion fragment sequence assembly. Bioinformatics 26: 1699–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Local necrotic lesion assay using hypersensitive tobacco NN plants to estimate the number of infectious particles in a plant extract. The left half of the leaf was inoculated with a control extract from a healthy A. thaliana plant. The right half of the leaf was inoculated with extracts from a TMV-infected A. thaliana plant (WT-1 from the fourth passage). The number of lesions reflects the number of particles in the inoculum.
(JPG)
Accumulation of the TMV ancestor and evolved lineages in non-inoculated, systemic leaves in tor1/spr2 , tor2 , and WT plants at 7, 12 and 15 dpi.
(DOCX)
Accumulation of the TMV ancestor and evolved lineages in tobacco BY-2 protoplasts at 6, 12, 24, 48 and 72 hpi.
(DOCX)
Forward (F) and reverse (R) primers used for TMV genome amplification by RT-PCR and nucleotide sequencing.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. Nucleotide sequences were deposited at GenBank under accession numbers KF972427-KF972436.