Abstract
Background
The influence of survivin isoforms on outcome in glioblastoma is poorly understood. We analyzed the dominant anti-apoptotic transcript variants of survivin using expression data and modeled them in vivo to determine their impact on glioma formation and progression.
Methods
Using data from low- and high-grade glioma knowledge bases, we expressed the anti-apoptotic isoforms of survivin (transcript variants 1 and 2) in vivo using the RCAS/Ntv-a model of murine glioma.
Results
In low-grade gliomas, survivin RNA expression was increased in 22 of 167 (13.2%) of cases and was associated with shortened survival (P = .005). Survivin RNA was preferentially expressed in proneural (PN) relative to mesenchymal high-grade gliomas (P < .0001). In proneural gliomas, survivin was expressed in 94 of 141 (67%) of cases and was associated with shorter disease-free survival (P = .04). In a platelet-derived growth factor subunit B-dependent murine model of PN glioma, ectopic expression of variant 1 yielded tumors in 28 of 30 (93%) of mice, of which 25% were high-grade tumors, whereas ectopic expression of variant 2 yielded tumors in 27 of 28 (96%), of which 81% were high-grade tumors (P < .0001). Microvascular proliferation was significantly more prominent (P < .0001), and tumor-free survival was shorter in mice with variant 2 than variant 1-derived tumors (P = .01).
Conclusions
Survivin expression in low-grade gliomas is associated with poor survival and is preferentially expressed in PN gliomas. Compared with variant 1, variant 2 was associated with poorer survival and promoted malignant progression, angiogenesis, and shorter tumor-free survival in the PN murine model. Inhibiting survivin transcript variant 2, rather than variant 1 (the common isoform), may be an effective treatment strategy for glioma.
Keywords: angiogenesis, apoptosis, glioma, survivin
Genomic profiling of glioblastoma multiforme (GBM) has revealed it to be a heterogeneous disease with disparate subtypes.1,2 These subtypes arise from different molecular origins, but they may also exist as a continuum of disease.3,4 The variability in tumor grade is reflected by the presence or absence of specific histological features including microvascular proliferation, diffuse mitotic activity, and necrosis.5 Low-grade gliomas frequently devolve into high-grade gliomas. The causes of degeneration are unknown, although several mechanisms have been shown to promote this malignant progression. Identifying the cellular programs that drive this progression will be critical to developing novel treatments for high-grade glioma, which remains uniformly fatal.
We showed recently that the expression of genes that are potent suppressors of apoptosis facilitate the transition from low- to high-grade glioma.4,6 Expression of these genes in vivo yields tumors with marked microvascular proliferation and necrosis. The expression of survivin (BIRC5), a member of the inhibitor of apoptosis gene family, has been shown to correlate negatively with survival in patients with high-grade gliomas.7,8 Survivin transcript variants can be pro- or anti-apoptotic and are differentially expressed in various cancers. In addition to having unique biological activities, survivin splice variants appear to correlate with different clinical outcomes in other types of cancer.9–17 Of the 3 dominant isoforms, the common isoform and survivin-ΔEx3 (encoded by transcript variants 1 and 2, respectively) exhibit the most anti-apoptotic activity, and survivin-2B (encoded by transcript variant 3) appears to be proapoptotic.8,12–14,16,18–20
We analyzed expression data for the 2 dominant anti-apoptotic transcript variants to determine their influence on the clinical outcome of human glioblastoma and to correlate the data with the 2 most prevalent phenotypes of this disease, proneural (PN) and mesenchymal (Mes). In addition, we used a mouse glioma model to overexpress these transcript variants in vivo, hypothesizing that they would promote malignant progression of glioma and enhance tumor incidence. Here, we show that survivin expression in low grade gliomas correlates with a significant survival disadvantage, that survivin variant 2 is preferentially expressed in PN high-grade glioma, and that its expression appears to correlate with poorer survival in humans. Moreover, ectopic expression of survivin variant 2 in the mouse model promotes angiogenesis and malignant progression.
Methods
Expression Analysis
We used the open-access portal of the cBIO Cancer Genomics Portal (www.cbioportal.org) to obtain expression data of survivin (BIRC5) in glioblastoma.21 We conducted our analysis between January and March of 2013. During this period, the provisional glioblastoma dataset consisted of 547 cases. Specifically, elevated mRNA expression levels as determined by Agilent microarray or RNASeq V2 RSEM (Illumina) relative to normalized mean expression were obtained in case sets by using the appropriate Onco Query Language syntax as described on the website (http://www.cbioportal.org/public-portal/onco_query_lang_desc.jsp). We used a similar method to analyze survivin expression in the provisional low-grade glioma dataset, which consisted of 167 samples.
The cBIO Cancer Genomics Portal does not provide analysis of splice variants. Therefore, we used The Broad Institute's Firehose (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm) expression data from February 22, 2013, for expression analysis of the survivin transcript variants. RNASeqV2 (IlluminaHiSeq) level 3 data were selected for analysis. Information from 256 cases was available for this analysis. We used the University of California Santa Cruz (UCSC) Genome Bioinformatics tool (genome.ucsc.edu) to identify the unique UCSC identifier for the transcript variants we analyzed. For variant 1, these identifiers were uc002jvi.3 and uc002jvq.3, which correlate with positions 76210277 through 76221716 on chromosome 17. For variant 2, this identifier was uc002jve.1, which correlates with positions 76210277 through 76210973 on chromosome 17.
RCAS Vector Construction
The details of RCAS-platelet-derived growth factor subunit B (PDGFB) construction were described by Dai et al.22 pCMV6-XL4 plasmids containing human survivin variant 1 (NM_001168.2) or variant 2 (NM_001012270.1) cDNA were obtained from OriGene Technologies. To generate Gateway-compatible entry vectors containing the survivin sequences, PCR reactions were performed to isolate the survivin cDNAs containing the sequences required for directional cloning into the pENTR/D-TOPO entry vectors (Invitrogen). The forward primer 5′-CACCATGGGTGCCCCGA-3′ and the reverse primers 5′-TCAATCCATGGCAGCCAG-3′ and 5′-CTAAGACATTGCTAAGGGGCC-3′ were used to generate the blunt-ended variant 1 and variant 2 PCR products, respectively. A Gateway LR recombination reaction between the entry vectors containing our gene of interest and a previously designed RCAS destination vector resulted in RCAS-survivin variants 1 and 2, which were verified by sequencing.
Transfection of DF-1 Cells
DF-1–immortalized chicken fibroblasts (American Type Culture Collection) were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Gemini Bio Products) in a humidified atmosphere of 95% air/5% CO2 at 37°C. Live virus was produced by transfecting the plasmid version of RCAS vectors into DF-1 cells using FuGene6 (Roche) and allowing them to replicate in culture.
Verification of Survivin Expression
To verify survivin variant 1 expression in DF-1 cells, Western blotting was performed for 2 or 3 passages after transfection (data not shown). Whole-cell lysates were prepared from untransfected DF-1 cells as the control and from DF-1 cells transfected with RCAS-survivin variant 1. Protein samples (20 µg) were fractionated by SDS-PAGE using gels containing 15% polyacrylamide, transferred to PVDF membrane, and probed with human-specific antisurvivin antibody (1:10000; ab8113, Abcam). Goat anti-rabbit IgG (1:2500; Santa Cruz Biotechnology) was used as the secondary antibody. Blots were developed with the ECL Plus detection kit (GE Healthcare) following the manufacturer's protocol.
For confirmation of survivin variant 2 expression in DF-1 cells, reverse transcription (RT)-PCR was performed (data not shown). RNA was isolated from 3–4 × 106 DF-1 cells transfected with RCAS-survivin variant 2 using the Qiagen Rneasy mini kit according to the manufacturer's protocol and synthesized into cDNA. Primers for GAPDH (5′-GGTGAAGGTCGGAGTCAACG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′) and for survivin variant 2 (5′-GAATTTGAGGAAACTGCGGAG-3′ and 5′-CTAAGACATTGCTAAGGGGCC-3′) were used in the RT-PCR reactions. PCR products were visualized on a 2% agarose gel.
In vivo Somatic Cell Transfer in Transgenic Mice
The generation of transgenic Ntv-a mice, which are a mix of strains C57BL/6, BALB/c, FVB/N, and CD1, was previously described.23 To transfer genes via RCAS vectors, DF-1 producer cells transfected with a particular RCAS vector or vectors (1 × 104 cells in 1–2 µL of phosphate-buffered saline) were injected bilaterally into the frontal brain lobes of mice from an entry point just anterior to the coronal suture of the skull with a 10-µL gas-tight Hamilton syringe. The mice were injected within 24 to 72 hours after birth, which is the period in which the Nestin+ cells producing TVA receptors proliferate rapidly. Equal amounts of DF-1 cells were injected in the injection sets consisting of 2 RCAS vectors. We coexpressed survivin variants 1 and 2 with PDGFB and independently in Ntv-a mice.
Mice were euthanized by carbon dioxide asphyxiation 90 days post injection, or sooner if symptoms related to tumor burden (eg, hydrocephalus) were present. The brains were excised, fixed in formalin, embedded in paraffin, and sectioned for immunohistochemical analysis. Hematoxylin and eosin staining allowed for analysis of tumor formation. The animal experiments performed in this research were approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center (Protocol No. 08-06-11633).
Immunohistochemical Analysis
Paraffin-embedded, 4-µm-thick brain sections were used for immunohistochemical analysis. The Thermo Scientific PT module with citrate buffer was used for antigen retrieval. Immunoreactive staining was performed using the Lab Vision immunohistochemical autostainer 360 (Thermo Fisher Scientific) and visualized using an avidin-biotin complex technique with diaminobenzidine (Invitrogen) as the chromogenic substrate and hematoxylin as the counterstain. Human survivin variants 1 and 2 expressed by RCAS were detected in tumor sections using antibodies specific for human survivin variant 1 (1:100; AF886; R&D Systems) and variant 2 (1:1000; ab3731; Abcam).
Detection of Survivin Variants in Tumor Sections
In addition to immunohistochemical methods for detecting expression of the survivin transcript variants in tumor-bearing tissue, we also performed protein analysis and RT-PCR on the brains of mice (n = 3) injected with RCAS-survivin variant 1 or RCAS-variant 2. After the mice were killed, the forebrains were removed and frozen in liquid nitrogen. Tissue specimens were homogenized, and RNA was extracted. We confirmed the expression of variant 1 in the tumor samples by Western blot (WB) using a human-specific survivin antibody, and actin was used as loading control. Since this antibody does not recognize the survivin variant 2, we performed RT-PCR analysis using RNA extracted from the tumors to confirm the expression of variant 2, using primer sequences described above. Normal brain was used as a control.
Determination of Tumor Grade
Tumor grade was determined by the study's neuropathologist (GNF) using WHO 2007 criteria.5 Low-grade tumors were identified by the presence of infiltrating tumor cells. High-grade tumors were identified by the presence of microvascular proliferation, foci of necrosis, or brisk mitotic activity.
Apoptotic Assay
Apoptosis was detected and quantified in tumors from the different injection sets by immunostaining of formalin-fixed, paraffin-embedded tumor sections with an antibody against cleaved caspase-3 (CC3) (1:50; 5A1E; Cell Signaling Technology) and cleaved caspase-7 (Cell Signaling Technology). We counted the total number of cells and the number of positively stained cells in the area of highest tumor cell density in 5 nonoverlapping high-power microscopic fields (400x magnification). At least 5 tumors from each injection set were selected for analysis. The expression level was calculated as the percentage of positive cells in each field. For the CC3 analysis from the RCAS-PDGFB + RCAS-survivin variant 1 injection set, the median number of cells counted was 1050 (range, 679–1454); from the RCAS-PDGFB + RCAS survivin variant 2 injection set, the median number of cells counted was 1632 (range, 984–2146). For the CC7 analysis from the RCAS-PDGFB + RCAS-survivin variant 1, the median number of cells counted was 1086 (range, 786–1578); from the RCAS-PDGFB + RCAS survivin variant 2 injection set, the median number of cells counted was 1112 (range, 663–1550).
Proliferation Index
To determine the extent of tumor cell proliferation in the different injection sets, we analyzed formalin-fixed, paraffin-embedded tumor-bearing tissue sections with an antibody against pHH3, which has been described as a useful marker for determining the mitotic index in gliomas.24 We counted the total number of cells and the number of positively stained cells in the area of highest tumor cell density in 10 nonoverlapping high-power microscopic fields (400 × magnification) in tumor-bearing brains taken from 5 mice in each injection set. The mitotic index was calculated as the percentage of positive cells in each field. The median total number of cells counted from the RCAS-PDGFB + RCAS-survivin variant 1 injection set was 1258 (range, 286–2022); from the RCAS-PDGFB + RCAS survivin variant 2 injection set, the median number of cells counted was 1267 (range, 754–2452).
Microvascular Proliferation Analysis
To determine the extent of endothelial proliferation, we quantified the expression of cluster of differentiation 31 (CD31) in tumor samples. Tissue sections were treated and examined as in the apoptotic assay except that antibody against CD31 (1:400; AF3628; R&D Systems) was used. We also used the methods to quantify microvessel density and microvessel count that have been described for characterizing vascular proliferation in brain tumors and other cancers.25,26 Microvessel densitywas performed by grading tumor vascularization on a scale of 1 to 4. Microvessel counts were performed by counting the number of discrete intratumoral blood vessels in 10 noncontiguous high-power fields (400x) for 5 tumors from each injection set.
Endothelial Tube Formation
To determine the ability of either survivin variant to facilitate endothelial tube formation, human umbilical vein endothelial cells (HUVECs) were seeded with Matrigel basement membrane matrix into 96-well plates, as described previously,27 and incubated with conditioned media collected from A375/TVA cells transfected with virus from DF-1 cells expressing either survivin variant 1 or variant 2. After a 24 hour incubation period at 37°C in a humidified 5% CO2 atmosphere, capillary tube formation was observed, and images were taken with a phase contrast microscope.
Statistical Analysis
Statistical analysis was performed using Graphpad Prism v5.04 software. The chi-square test was used to compare tumor formation rates between different injection sets. A Kaplan–Meier curve was generated to estimate the time to symptomatic tumor development. The log-rank test was used to determine statistical significance for overall and tumor-free survival. The Student' t test was used to determine differences in CD31, CC3, CC7, and pHH3 expression between injection sets. The P value was set at <.05 to indicate statistical significance.
Results
Expression Data Analysis Shows Survivin Negatively Influences Survival in Low-grade Gliomas
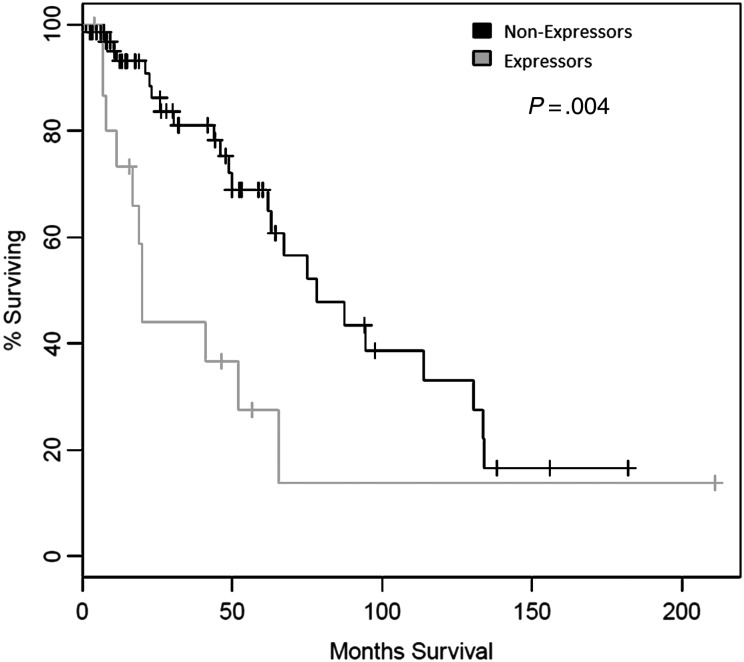
In our analysis of the provisional low-grade glioma dataset available from cBio Cancer Genomic Portal (n = 84 cases with data available), increased survivin mRNA expression had been observed in 17 cases (20%). Median survival was 19.9 months in expressors and 78.1 months in nonexpressors (ie, patients in whom expression was ≤ mean expression) (log rank test, P = .004) (Fig. 1A). We then analyzed the provisional glioblastoma multiforme dataset from cBio Cancer Genomic Portal (n = 547 cases). Increased survivin mRNA expression had been observed in 258 cases (47%). Median survival was 13.9 months for expressors and 14.1 months for nonexpressors (log rank test, P = .496) (Fig. 1B), and progression-free survival was 6.7 and 7.6 months in expressors and nonexpressors, respectively (log rank test, P = .64) (Fig. 1C).
Fig. 1.
Kaplan–Meier curve of 167 patients with low-grade glioma with elevated mRNA expression of survivin (BIRC5) relative to nonexpressors.
Survivin Expression Is Predominant in Proneural Gliomas
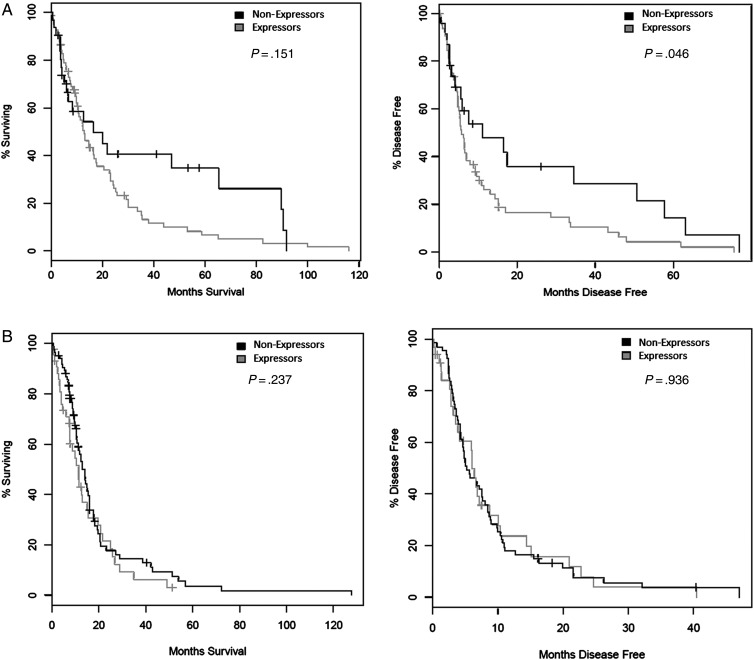
We evaluated survivin mRNA expression in the PN and Mes glioma subtypes, as they exhibit the most disparate survival profiles.1 The percentage of survivin expressors between PN and MES cases was significantly different (PN, 94 of 141 [67%]; Mes, 60 of 160 [38%]; chi-square test, P < .0001). For PN cases, median overall survival was 13 months in expressors and 16.5 months in nonexpressors (log rank test, P = .151). However, disease-free survival was 5.8 months for expressors and 11.2 months in nonexpressors (log rank test, P = .046) (Fig. 2A). For Mes cases, the corresponding values were 11.3 and 13.9 months for expressors (log rank test, P = .237) and 6.0 and 5.2 months for nonexpressors, respectively (log rank test, P = .936) (Fig. 2B).
Fig. 2.
Kaplan-Meier curves of overall survival (left) and disease-free survival (right) in patients with (A) proneural glioblastoma multiforme (GBM) or (B) mesenchymal GBM with elevated expression of survivin (BIRC5) relative to nonexpressors.
Because the preponderance of survivin expressors was in the PN group, we focused on the number of cases from this subtype that expressed variants 1 and 2. We used the unique UCSC identifiers for these variants and identified the cases using Broad Institute Firehose expression data from February 22, 2013. Of the 94 cases of PN expressors, 76 (81%) expressed variant 1, and 8 (9%) expressed variant 2 (the remaining 10 cases were indeterminate). The median survival of variant 2 expressors in the PN subtype was 394 days which, although shorter than the 447 days of variant 1 expressors, was not statistically significant (log rank test, P = .23). Similarly, the median survival of variant 2 expressors in the PN group was shorter than nonexpressors in the PN group (507 days), although not statistically significant (log rank test, P = .19).
In Vivo Modeling of Survivin Variant 1 and 2 Expression
We modeled the overexpression of survivin variants 1 and 2 in mice to determine their biological effect on tumor progression and tumor-free survival. We created RCAS-survivin variant 1 and variant 2 vectors. Because we found that survivin is expressed predominantly in the PN phenotype, we modeled overexpression using a murine model system that is dependent on activation of the platelet-derived growth factor receptor pathway, which is known to be upregulated in PN tumors.28 In the RCAS/Ntv-a system, overexpression of the ligand PDGFB is sufficient to induce tumors with a PN phenotype.29 We also expressed these transcript variants independently but did not observe any tumors.
Survivin Variant 2 Promotes Malignant Progression in Vivo
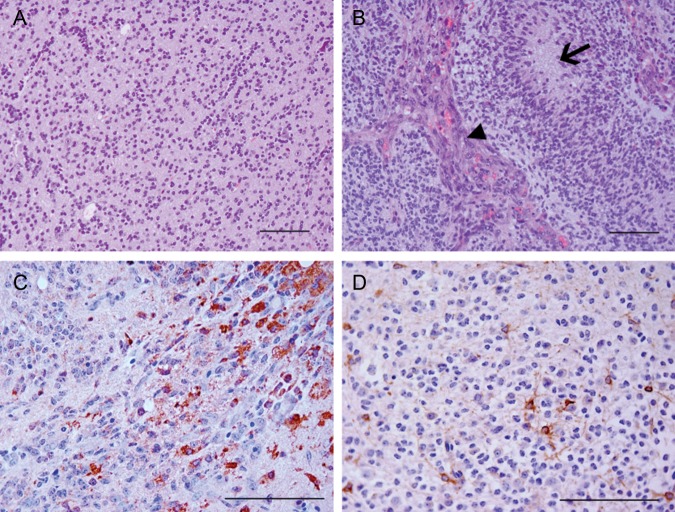
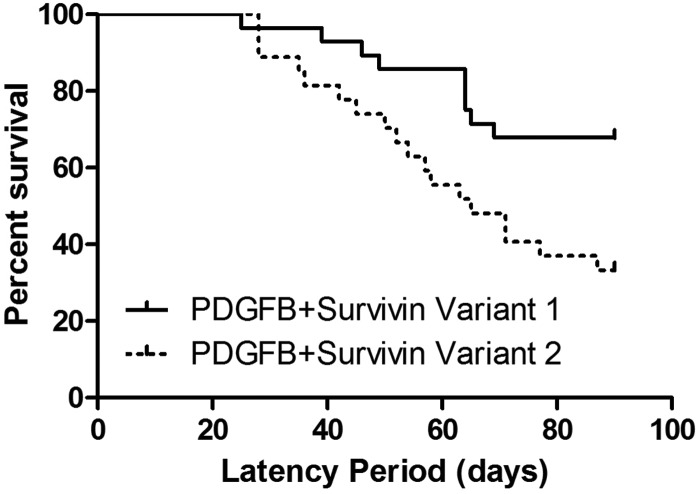
The incidence of tumors in RCAS-PDGFB + RCAS-survivin variant 1 was 93%, with 25% of these demonstrating histological features consistent with high-grade glioma. The tumor incidence in variant 2 was 96%, with 81% of these demonstrating high-grade features (Fig. 3 and Table 1). The difference in high-grade tumor formation was significant (chi-square test, P < .0001). Independent expression of RCAS-survivin variant 1 or 2 was insufficient to induce any tumors by 90 days. Median tumor-free survival of the mice was 90 days for the variant 1 injection set and 65 days for the variant 2 injection set (log rank test, P = .01) (Fig. 4). Expression of each survivin variant was verified (Fig. 3C and D). We also confirmed expression of the variants in tumors using WB and RT-PCR (Supplementary Fig. 1).
Fig. 3.
Photomicrographs of tumors induced by RCAS-platelet-derived growth factor subunit B + RCAS-survivin variants 1 and 2. The majority of tumors induced by variant 1 were low grade (A), whereas the majority of tumors induced by variant 2 were high grade with microvascular proliferation (arrowhead) and necrosis (arrow) (B). Survivin variant 1 expression was detected in tumors produced in mice injected with variant 1 (C), as was variant 2 in tumors produced in mice injected with variant 2 (D). A and B: hematoxylin and eosin staining. C and D: diaminobenzidine and hematoxylin staining. Magnification x200, scale bar = 100 µm.
Table 1.
Incidence of tumor formation in Ntv-a mice
| Overall tumor formation | Low-grade tumor formation | High-grade tumor formation | |
|---|---|---|---|
| PDGFB + Survivin variant 1 | 28/30 (93%) | 21/28 (75%) | 7/28 (25%) |
| PDGFB + Survivin variant 2 | 27/28 (96%) | 5/27 (19%) | 22/27 (81%) |
Abbreviations: PDGFB, platelet-derived growth factor subunit B.
Fig. 4.
Kaplan–Meier curve of tumor-free survival of RCAS-PDGFB + RCAS-survivin variant 1 and variant 2 injection sets.
Survivin Variant 2 Drives Angiogenesis
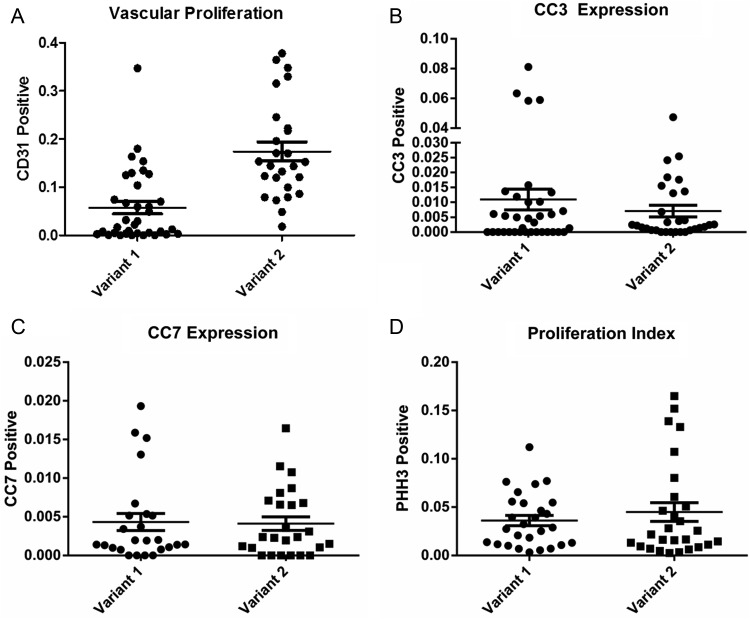
To compare microvascular proliferation between tumors generated from the RCAS-PDGFB + RCAS- survivin variant 1 and variant 2 injection sets, we immunostained representative sections with an antibody against CD31. The median number of cells counted was 1209 (range, 872– 2044) and 1548 (range, 984–2146), respectively. The mean percentage expression of CD31 was 6% (SEM, 1%) for tumors derived from variant 1 and 17% (SEM, 2%) for those derived from variant 2 (P < .0001 by t test) (Fig. 5A). We also counted the number of microvessels per high-power field and scored microvessel density. The mean number of microvessels was 9.906 (± SEM, 1.014) and 19.65 (± SEM, 1.458) for the RCAS-PDGFB + RCAS- survivin variant 1 and RCAS-PDGFB + RCAS-survivin variant 2 injection sets, respectively (P < .0001 by t test). The microvessel density scores were 1.6 (± SEM, 0.12) and 2.6 ± (SEM, 0.16) for the RCAS-PDGFB + RCAS- survivin variant 1 and RCAS-PDGFB + RCAS-survivin variant 2 injection sets, respectively (P < .0001 by t test). The effect of survivin variants on angiogenesis was also determined via in vitro angiogenesis assays. We checked whether capillary formation is modulated by survivin variant 2. While survivin variant 2 conditioned-medium- treated HUVECs formed capillary-like network structures on Matrigel surfaces, this network-forming ability was lost in HUVECs cultured in the presence of survivin variant 1-conditioned medium (Supplementary Fig. 2).
Fig. 5.
Scatter plots of (A) CD31 expression, (B) CC3 expression, (C) CC7 expression, and (D) pHH3 expression in tumors produced in mice injected with RCAS-PDGFB + RCAS-survivin variant 1 or variant 2. Mean expression is represented by the horizontal line. Error bars represent standard error of the mean.
Apoptotic Suppression and Tumor Cell Proliferation are Similar Between Survivin Variants 1 and 2
To compare apoptotic activity between tumors generated from the RCAS-PDGFB + RCAS-survivin variant 1 and variant 2 injection sets, we immunostained representative sections with antibodies against CC3 and CC7. For variant 1-derived tumor samples, the mean expression level of CC3 was 1% (SEM, 0.3%), and for variant 2-derived tumor samples it was 0.7% (SEM, 0.1%) (Fig. 5B) (P = .65 by t test). For variant 1-derived tumor samples the mean expression level of CC7 was 0.43% (SEM, 0.01%), and for variant 2-derived tumor samples it was 0.41% (SEM, 0.008%) (P = .89 by t test). These results indicate that the effect of these transcript variants on suppressing apoptosis was similar.
We quantified proliferation in tumors from the injection sets by staining them for pHH3. Although the proliferation index was higher for variant 2-derived tumor samples (4.4%; SEM, 0.09%) than for variant 1-derived tumor samples 3.6% (SEM, 0.05%) the difference was not statistically significant (P = .42 by t test).
Discussion
Gene expression profiling has revealed significant heterogeneity in GBM, and subtypes of the disease have been described by both clinical and genetic characteristics. We evaluated the expression of anti-apoptotic survivin (BIRC5) transcript variants to determine whether their expression varied between the PN and Mes subtypes (2 GBM phenotypes with marked genetic and survival differences). Survivin expression has been associated with a poorer prognosis in patients diagnosed with GBM. However, the effects of survivin transcript variants within GBM subtypes and on tumor progression are less well understood. In this study we analyzed TCGA expression data and determined that survivin expression was significantly more prevalent in the PN glioma subtype. We also found that progression-free survival was significantly shorter in patients with PN GBM with a higher mRNA expression level of survivin relative to normalized mean expression. In addition, after expressing the dominant anti-apoptotic transcript variants of survivin in a PDGFB-dependent murine model of glioma that recapitulates the PN phenotype, we showed that ectopic expression of variant 2 (survivin-ΔEx3) resulted in marked malignant progression of tumors compared with variant 1 (the common survivin isoform) with a commensurate decrease in tumor-free survival.
Survivin is a structurally unique member of the inhibitor of apoptosis family. Both variants 1 and 2 appear to inhibit mitochondrial-dependent apoptosis.18 We previously showed that suppression of apoptosis promoted malignant progression in a murine glioma model.4,6 Thus, we hypothesized that the anti-apoptotic activity mediated by survivin transcript variants would have a similar effect on malignant progression (variant 3, also known as survivin-2B, appears to have had lost its anti-apoptotic effect, and its expression appeared to decrease in a stage-dependent manner).12 In the current study, we analyzed the provisional low-grade glioma TCGA dataset to determine the effect of survivin expression on survival and found that increased mRNA expression levels of survivin correlated with decreased survival in this patient population. A similar effect was not observed for the GBM TCGA dataset but, after subanalysis of the transcript variants, we found that cases in which variant 2 mRNA was overexpressed had a shorter (although not statistically significant) overall survival compared with cases demonstrating variant 1 overexpression or with nonexpressors of survivin. Consistent with this result, we observed that ectopic expression of PDGFB + survivin variant 1 in mice yielded an overall tumor-free survival time similar to that of mice expressing PDGFB alone (based on our previous studies4,6), suggesting that variant 1 may not contribute to malignant progression. In contrast, mice coexpressing PDGFB + survivin variant 2 had a significantly shorter tumor-free survival.
Survivin is expressed in several different cancer types, but its expression is undetectable in normal adult tissues. Several studies have linked the expression of this protein with poor survival in patients with cancer. In GBM, increased protein levels of survivin from human tumor samples have been shown to correlate negatively with overall survival.7 A previous report showed that expression of both survivin variants 1 and 2 was significantly higher in malignant brain tumors relative to benign ones (this correlation did not hold for variant 3).30 A study using nonquantitative RT-PCR techniques showed that survivin variants 1 and 2 were detectable in a significant number of glioblastoma cases and had a strong negative correlation with survival in cases displaying mRNA positivity for variants 1 and 2.8 Our analysis of the TCGA dataset was calibrated to determine how expression above normalized mean expression of these variants correlated with survival. Therefore, our analysis shows that variant 2 is less frequently overexpressed in human glioma. This finding that the common isoform of survivin was expressed in the majority of low- and high-grade glioma cases relative to variant 2 is consistent with observations made for other cancers.9–17,31
In a murine model, we found that coexpression of PDGFB and survivin variant 2 produced a significantly higher number of high-grade gliomas relative to coexpression of PDGFB and survivin variant 1. The presence of microvascular proliferation was the most common histological criterion conferring high-grade status, and we were able to demonstrate a strong increase in microvessel density (as quantified by CD31 expression) in tumors produced by coexpression of PDGFB and survivin variant 2 relative to PDGFB and survivin variant 1, even though the level of anti-apoptotic activity was similar between these injection sets. We previously showed that the expression of other anti-apoptotic genes (ie, BCL2 and STAT3) also resulted in the formation of high-grade gliomas, although this finding was primarily due to the presence of necrotic foci (as well as microvascular proliferation).4,6 In the present study, we did not find the same predilection towards the formation of necrosis after expression of either survivin isoform. Other investigators have observed a strong association between the expression of survivin and tumor grade, demonstrating that increased survivin expression (based on immunohistochemical detection) correlated with increased tumor vascularity.32 The mechanism of angiogenesis mediated by survivin occurs through the expression of vascular endothelial and basic fibroblast growth factors.33–38 Further, it has been shown that survivin is upregulated after various types of vascular injury, resulting in neointima formation.39 Not surprisingly, targeting survivin experimentally has been shown to reduce tumor-associated angiogenesis.40 At least one other group has shown that survivin variant 2 plays a prominent role in angiogenesis,41 which corroborates our finding that this isoform may confer a more profound effect on tumor vascular proliferation. Caldas et al. demonstrated that survivin variant 2 primarily localized to vascular endothelial cells of pediatric brain tumor tissues and that these cells were more prominent in the tumor cells themselves. They also demonstrated that specific suppression of survivin variant 2 reduced angiogenesis.41 Prior studies using nonspecific inhibitors of survivin made it difficult to determine which variant had proangiogenic properties.
Our observation that survivin expression correlates with poorer survival in low-grade gliomas is relevant to the observation from our in vivo experiments that demonstrated malignant progression in the PDGFB and survivin variant 2 injection set. The stark difference in median survival in survivin expressors versus nonexpressors (19.9 vs 78.1 months, respectively) for the 84 patients with RNA expression data in the low-grade dataset indicates that the expression of this gene may be critical to degeneration from low-grade to high-grade glioma. However, at the time of our study, the provisional low-grade glioma expression data did not permit the analysis of individual survivin transcript variants. It is not possible to determine whether the disease degenerated to a more malignant phenotype in these patients, although their shortened survival was unusual for low-grade glioma because patients with low-grade glioma typically have a median survival much longer than 20 months. Further, the number of survivin expressors identified with Onco Query Language indicated that only 20% in the low-grade glioma dataset expressed survivin. This is a lower rate than the 47% of glioblastoma samples that displayed survivin overexpression (regardless of variant) using the same query language; only 10% of those cases overexpressed variant 2. Thus, overexpression of variant 2 in low-grade glioma is rare. However, given that PN gliomas are thought to represent secondary GBMs, expression of variant 2 may contribute to disease progression in these cases. Further, our in vivo data demonstrated the importance of variant 2 in driving malignant progression relative to variant 1.
For more than a decade, survivin has been described as a therapeutic target.42 It has been used as an antigen in dendritic cell vaccines created to treat glioma, although the promise of these treatments has yet to be fully realized.43–48 In vitro and in vivo RNA knockdown targeting survivin has been demonstrated to reduce invasion, proliferation, and angiogenesis.43,49 However, the clinical application of inhibiting survivin expression has been limited. Our results suggest that inhibiting survivin transcript variant 2, rather than the common isoform, may be a more effective strategy.
Supplementary Material
Funding
This research was supported by the National Institutes of Health grant K08 NS070928 (G. Rao).
Supplementary Material
Acknowledgements
We are grateful to Elizabeth Hess, PhD, for critical review of the manuscript.
Conflict of interest statement. None declared.
References
- 1.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat KP, Salazar KL, Balasubramaniyan V, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doucette TA, Kong LY, Yang Y, et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012;14(9):1136–1145. doi: 10.1093/neuonc/nos139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doucette T, Yang Y, Zhang W, et al. Bcl-2 promotes malignant progression in a PDGF-B-dependent murine model of oligodendroglioma. Int J Cancer. 2011;129(9):2093–2103. doi: 10.1002/ijc.25869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti A, Noll E, Black PM, et al. Quantitatively determined survivin expression levels are of prognostic value in human gliomas. J Clin Oncol. 2002;20(4):1063–1068. doi: 10.1200/JCO.2002.20.4.1063. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Chen X, Chen N, et al. Expression and prognostic significance of survivin splice variants in diffusely infiltrating astrocytoma. J Clin Pathol. 2011;64(11):953–959. doi: 10.1136/jclinpath-2011-200066. [DOI] [PubMed] [Google Scholar]
- 9.Atlasi Y, Mowla SJ, Ziaee SA. Differential expression of survivin and its splice variants, survivin-DeltaEx3 and survivin-2B, in bladder cancer. Cancer Detect Prev. 2009;32(4):308–313. doi: 10.1016/j.cdp.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 10.De Maria S, Pannone G, Bufo P, et al. Survivin gene-expression and splicing isoforms in oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2009;135(1):107–116. doi: 10.1007/s00432-008-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futakuchi H, Ueda M, Kanda K, et al. Transcriptional expression of survivin and its splice variants in cervical carcinomas. Int J Gynecol Cancer. 2007;17(5):1092–1098. doi: 10.1111/j.1525-1438.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 12.Krieg A, Mahotka C, Krieg T, et al. Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br J Cancer. 2002;86(5):737–743. doi: 10.1038/sj.bjc.6600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XN, Shu Q, Su JM, et al. Differential expression of survivin splice isoforms in medulloblastomas. Neuropathol Appl Neurobiol. 2007;33(1):67–76. doi: 10.1111/j.1365-2990.2006.00782.x. [DOI] [PubMed] [Google Scholar]
- 14.Ryan B, O'Donovan N, Browne B, et al. Expression of survivin and its splice variants survivin-2B and survivin-DeltaEx3 in breast cancer. Br J Cancer. 2005;92(1):120–124. doi: 10.1038/sj.bjc.6602314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suga K, Yamamoto T, Yamada Y, et al. Correlation between transcriptional expression of survivin isoforms and clinicopathological findings in human colorectal carcinomas. Oncol Rep. 2005;13(5):891–897. [PubMed] [Google Scholar]
- 16.Vegran F, Boidot R, Oudin C, et al. Distinct expression of Survivin splice variants in breast carcinomas. Int J Oncol. 2005;27(4):1151–1157. [PubMed] [Google Scholar]
- 17.Wagner M, Schmelz K, Wuchter C, et al. In vivo expression of survivin and its splice variant survivin-2B: impact on clinical outcome in acute myeloid leukemia. Int J Cancer. 2006;119(6):1291–1297. doi: 10.1002/ijc.21995. [DOI] [PubMed] [Google Scholar]
- 18.Caldas H, Jiang Y, Holloway MP, et al. Survivin splice variants regulate the balance between proliferation and cell death. Oncogene. 2005;24(12):1994–2007. doi: 10.1038/sj.onc.1208350. [DOI] [PubMed] [Google Scholar]
- 19.Kanwar JR, Kamalapuram SK, Kanwar RK. Survivin signaling in clinical oncology: a multifaceted dragon. Med Res Rev. 2013;33(4):765–789. doi: 10.1002/med.21264. [DOI] [PubMed] [Google Scholar]
- 20.Mahotka C, Wenzel M, Springer E, et al. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59(24):6097–6102. [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai C, Celestino JC, Okada Y, et al. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15(15):1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc Natl Acad Sci USA. 1998;95(3):1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colman H, Giannini C, Huang L, et al. Assessment and prognostic significance of mitotic index using the mitosis marker phospho-histone H3 in low and intermediate-grade infiltrating astrocytomas. Am J Surg Pathol. 2006;30(5):657–664. doi: 10.1097/01.pas.0000202048.28203.25. [DOI] [PubMed] [Google Scholar]
- 25.Leon SP, Folkerth RD, Black PM. Microvessel density is a prognostic indicator for patients with astroglial brain tumors. Cancer. 1996;77(2):362–372. doi: 10.1002/(SICI)1097-0142(19960115)77:2<362::AID-CNCR20>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 26.Weidner N, Semple JP, Welch WR, et al. Tumor angiogenesis and metastasis--correlation in invasive breast carcinoma. N Engl J Med. 1991;324(1):1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 27.Ezhilarasan R, Jadhav U, Mohanam I, et al. The hemopexin domain of MMP-9 inhibits angiogenesis and retards the growth of intracranial glioblastoma xenograft in nude mice. Int J Cancer. 2009;124(2):306–315. doi: 10.1002/ijc.23951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hambardzumyan D, Parada LF, Holland EC, et al. Genetic modeling of gliomas in mice: new tools to tackle old problems. Glia. 2011;59(8):1155–1168. doi: 10.1002/glia.21142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada Y, Kuroiwa T, Nakagawa T, et al. Transcriptional expression of survivin and its splice variants in brain tumors in humans. J Neurosurg. 2003;99(4):738–745. doi: 10.3171/jns.2003.99.4.0738. [DOI] [PubMed] [Google Scholar]
- 31.Fangusaro JR, Jiang Y, Holloway MP, et al. Survivin, Survivin-2B, and Survivin-deItaEx3 expression in medulloblastoma: biologic markers of tumour morphology and clinical outcome. Br J Cancer. 2005;92(2):359–365. doi: 10.1038/sj.bjc.6602317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhen HN, Zhang X, Hu PZ, et al. Survivin expression and its relation with proliferation, apoptosis, and angiogenesis in brain gliomas. Cancer. 2005;104(12):2775–2783. doi: 10.1002/cncr.21490. [DOI] [PubMed] [Google Scholar]
- 33.Wang P, Zhen H, Zhang J, et al. Survivin promotes glioma angiogenesis through vascular endothelial growth factor and basic fibroblast growth factor in vitro and in vivo. Mol Carcinog. 2012;51(7):586–595. doi: 10.1002/mc.20829. [DOI] [PubMed] [Google Scholar]
- 34.Coma S, Noe V, Lavarino C, et al. Use of siRNAs and antisense oligonucleotides against survivin RNA to inhibit steps leading to tumor angiogenesis. Oligonucleotides. 2004;14(2):100–113. doi: 10.1089/1545457041526290. [DOI] [PubMed] [Google Scholar]
- 35.Iurlaro M, Demontis F, Corada M, et al. VE-cadherin expression and clustering maintain low levels of survivin in endothelial cells. Am J Pathol. 2004;165(1):181–189. doi: 10.1016/s0002-9440(10)63287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesri M, Morales-Ruiz M, Ackermann EJ, et al. Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am J Pathol. 2001;158(5):1757–1765. doi: 10.1016/S0002-9440(10)64131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connor DS, Schechner JS, Adida C, et al. Control of apoptosis during angiogenesis by survivin expression in endothelial cells. Am J Pathol. 2000;156(2):393–398. doi: 10.1016/S0002-9440(10)64742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran J, Master Z, Yu JL, et al. A role for survivin in chemoresistance of endothelial cells mediated by VEGF. Proc Natl Acad Sci USA. 2002;99(7):4349–4354. doi: 10.1073/pnas.072586399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conte MS, Altieri DC. Survivin regulation of vascular injury. Trends Cardiovasc Med. 2006;16(4):114–117. doi: 10.1016/j.tcm.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Blanc-Brude OP, Mesri M, Wall NR, et al. Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin Cancer Res. 2003;9(7):2683–2692. [PubMed] [Google Scholar]
- 41.Caldas H, Fangusaro JR, Boue DR, et al. Dissecting the role of endothelial SURVIVIN DeltaEx3 in angiogenesis. Blood. 2007;109(4):1479–1489. doi: 10.1182/blood-2006-02-003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3(1):46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 43.George J, Banik NL, Ray SK. Survivin knockdown and concurrent 4-HPR treatment controlled human glioblastoma in vitro and in vivo. Neuro Oncol. 2010;12(11):1088–1101. doi: 10.1093/neuonc/noq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim CH, Woo SJ, Park JS, et al. Enhanced antitumour immunity by combined use of temozolomide and TAT-survivin pulsed dendritic cells in a murine glioma. Immunology. 2007;122(4):615–622. doi: 10.1111/j.1365-2567.2007.02680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang JG, Kruse CA, Driggers L, et al. Tumor antigen precursor protein profiles of adult and pediatric brain tumors identify potential targets for immunotherapy. J Neurooncol. 2008;88(1):65–76. doi: 10.1007/s11060-008-9534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler CJ, Black KL. Vaccines for glioblastoma and high-grade glioma. Expert Rev Vaccines. 2011;10(6):875–886. doi: 10.1586/erv.11.71. [DOI] [PubMed] [Google Scholar]
- 47.Ciesielski MJ, Ahluwalia MS, Munich SA, et al. Antitumor cytotoxic T-cell response induced by a survivin peptide mimic. Cancer Immunol Immunother. 2010;59(8):1211–1221. doi: 10.1007/s00262-010-0845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciesielski MJ, Apfel L, Barone TA, et al. Antitumor effects of a xenogeneic survivin bone marrow derived dendritic cell vaccine against murine GL261 gliomas. Cancer Immunol Immunother. 2006;55(12):1491–1503. doi: 10.1007/s00262-006-0138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendruschk S, Wiedemuth R, Aigner A, et al. RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo. Neuro Oncol. 2011;13(10):1074–1089. doi: 10.1093/neuonc/nor098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.