Abstract
Background
As a commonly mutated form of the epidermal growth factor receptor, EGFRvIII strongly promotes glioblastoma (GBM) tumor invasion and progression, but the mechanisms underlying this promotion are not fully understood.
Methods
Through gene manipulation, we established EGFRvIII-, wild-type EGFR-, and vector-expressing GBM cells. We used cDNA microarrays, bioinformatics analysis, target-blocking migration and invasion assays, Western blotting, and an orthotopic U87MG GBM model to examine the phenotypic shifts and treatment effects of EGFRvIII expression in vitro and in vivo. Confocal imaging, co-immunoprecipitation, and siRNA assays detected the focal adhesion-associated complex and their relationships to the EGFRvIII/JAK2/STAT3 axis in GBM cells.
Results
The activation of JAK2/STAT3 signaling is vital for promoting migration and invasion in EGFRvIII-GBM cells. AG490 or WP1066, the JAK2/STAT3 inhibitors, specifically destroyed EGFRvIII/JAK2/STAT3-related focal adhesions and depleted the activation of EGFR/Akt/FAK and JAK2/STAT3 signaling, thereby abolishing the ability of EGFRvIII-expressing GBM cells to migrate and invade. Furthermore, the RNAi silencing of JAK2 in EGFRvIII-expressing GBM cells significantly attenuated their ability to migrate and invade; however, as a result of a potential EGFRvIII-JAK2-STAT3 activation loop, neither EGFR nor STAT3 knockdown yielded the same effects. Moreover, AG490 or JAK2 gene knockdown greatly suppressed tumor invasion and progression in the U87MG-EGFRvIII orthotopic models.
Conclusion
Taken together, our data demonstrate that JAK2/STAT3 signaling is essential for EGFRvIII-driven migration and invasion by promoting focal adhesion and stabilizing the EGFRvIII/JAK2/STAT3 axis. Targeting JAK2/STAT3 therapy, such as AG490, may have potential clinical implications for the tailored treatment of GBM patients bearing EGFRvIII-positive tumors.
Keywords: EGFRvIII, focal adhesion, glioblastoma, invasion, JAK2/STAT3
Glioblastoma (GBM) is an aggressive, highly invasive, vascularized tumor with a long-term survival of <10%.1,2 The tumor carries multiple genetic defects, among which is amplification of the epidermal growth factor receptor (EGFR) in ∼50% of GBMs; half of these GBMs express a truncated EGFR mutant lacking exons 2-7 (EGFRvIII), which includes the extracellular ligand-binding domain.2 The truncated receptor displays the constitutive, ligand-independent activation of receptor tyrosine kinase (RTK) activity, which confers a significant growth advantage, enhanced tumorigenic activity, and a more aggressive tumor phenotype than that associated with the wild-type EGFR.2 EGFRvIII expression is associated with poor prognosis and mediates resistance to EGFR-targeted therapy and chemotherapies in GBM patients.3 Therefore, EGFRvIII-associated signaling events, particularly those for tumor cell migration and invasion, must be elucidated in greater detail to improve treatment strategies.
Aberrant EGFR activation may contribute to tumor progression and invasion through regulation of cell adhesion and migration as well as induction of matrix-degrading proteases.2,4 Recent studies have demonstrated that EGFRvIII expression preferentially upregulates multiple molecules and activates functions responsible for tumor invasion.5–7 For example, EGFRvIII expression in an ovarian cancer cell line results in aberrant cell spreading and focal adhesion complex formation.7 GBM cells exhibit enhanced expression of focal adhesion kinase (FAK) relative to normal brain cells,8 particularly in EGFRvIII overexpressing GBM cells.9 Furthermore, EGFRvIII can promote cell invasion by activating important downstream protein kinases such as c-Src10 and PI3K/Akt signaling, especially in tumors with PTEN loss.11 Moreover, the cross talk between EGFRvIII and FAK12 or Src family kinases (SFK) promotes EGFR-induced tumor invasion and progression in GBM10 and other cancers,6,13 thus providing a rationale for combination therapies.
STAT3 activation is another important contributor to tumor progression and invasion in cancers, including GBM.14 STAT3 can be activated by intrinsic receptor or nonreceptor tyrosine kinases, including EGFR and cytokine receptors, or receptor-associated kinases, such as JAK2 and Src. As a transcription factor, STAT3 regulates the transcription of several genes, including those that function in cell motility.15 STAT3 is constitutively activated in gliomas and coexists with EGFR expression in 27% of primary high-grade gliomas,16 acting as an oncogene in EGFRvIII-expressing cells. STAT3 activation is required for the malignant transformation of EGFRvIII-expressing astrocytes, where it physically interacts with EGFRvIII in the nucleus.17,18 Furthermore, the bidirectional phosphorylation between EGFR and JAK2 leads to activation of both kinases and consequently the activation of downstream signaling effectors such as MAPK and STAT3.19–21 In addition, inhibitors of JAK2, Src, or EGFR have been shown to block STAT3 signaling, inhibit glioma proliferation, and induce apoptosis (alone or in combination).22 Collectively, the cross talk between dysregulated EGFR and JAK2/STAT3 pathways significantly affects intracellular signaling, which leads to more aggressive tumor behaviors.16 However, the mechanisms that underlie this cross talk in malignant gliomas remain unknown.
In this study, we demonstrate for the first time that EGFRvIII is actively involved in a focal adhesion complex in which EGFRvIII, JAK2, and STAT3 form an activation loop that promotes GBM cell invasion in vitro and in vivo. Furthermore, treatment with AG490, or WP1066, the JAK2 kinase inhibitors, or JAK2 gene knockdown efficiently disintegrates the complex and suppresses EGFRvIII/Akt/FAK and JAK2/STAT3 signaling pathways, thereby inhibiting tumor cell invasion and tumor progression.
Materials and Methods
Glioblastoma Cell Culture
The GBM cell lines U87MG-vector, U87MG-EGFRvIII, U87MG-EGFRwt, LN229-vector, and LN229-EGFRvIII were established as previously described23 and maintained in our laboratory. Briefly, full-length cDNAs of EGFRvIII or wild-type EGFR (EGFRwt) were amplified with PSK plasmids (kind gifts from Dr. F. Furnari, Ludwig Institute for Cancer Research, San Diego, California) into the expression vector pcDNA3.1(-) (Invitrogen) before being transfected into either U87MG or LN229 cells with Lipofectamine 2000 (Invitrogen). To establish the GBM cell models (ie, U87-EGFRwt, U87-EGFRvIII, U87-vector, LN229-EGFRvIII, and LN229-vector), stable clones were initially selected using 600–800 μg/mL of the antibiotic G418 (EMD Biosciences) for 2–3 weeks and then maintained in 400 μg/mL G418 in high-glucose Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin/streptomycin at 37°C with 5% CO2. Multiple clones of U87MG and LN229 cells were established that stably expressed EGFRwt, EGFRvIII, or vector control; the expressions were verified by RT-PCR and Western blotting. Clone 1 of the established U87MG-EGFRvIII cells that expressed the highest level of the mutant receptor was used for most of the experiments; some of the major findings were verified in other clones of U87MG or LN229 cells.
Microarrays and Gene Expression Analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). An Agilent Gene Expression array (KangChen Bio-tech Inc.) was used to investigate the transcriptional profiles of the U87MG-vector and -EGFRvIII cells; the array represented more than 41 000 transcripts (http://www.kangchen.com.cn). The microarray datasets were normalized in GeneSpring GX using the Agilent FE one-color scenario (mainly quantile normalization). Differentially expressed genes were identified through fold-change screening. The Gene Ontology (GO) biological process and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using the Database for Annotation, Visualization and Integrated Discovery 6.7, (DAVID; http://david.abcc.ncifcrf.gov/)24 and were ranked by P values.
Antibodies and Reagents
All antibodies were obtained from Cell Signaling Technology except the anti-EGFR and anti-paxillin antibodies, which were purchased from Invitrogen. The molecular-targeted inhibitors and fluorescein isothiocyanate-labeled phalloidin were purchased from Sigma, and WP1066 was purchased from EMD Chemicals. All inhibitors were dissolved in dimethyl sulfoxide (DMSO) and stored at the concentration of 10–100 mM according to their instructions.
Wound-healing Assays
The cells were seeded in 6-well plates at 70%–80% confluence and starved in serum-free medium for 12 hours. A “wounding” line was scratched into the cell monolayer with a sterile 20 μL pipet tip. The cells were washed with phosphate-buffered saline to remove the detached cells. The serum-free growth medium with or without respective inhibitors was added to the plates. The cells were then incubated at 37°C for 24 hours; the migrated cells were monitored with a Nikon ECLIPSE Ti-s inverted microscope and counted in 6 randomly selected fields to quantify the relative cell migration rates with or without treatment. The experiments were performed at least 3 times.
Transwell Invasion Assays
Transwell filters (8 μm pore size; Corning) were precoated with Matrigel (BD Biosciences) in a 24-well plate. After the cells were serum-starved for 12 hours, 4 × 104 GBM cells in 100 μL of serum-free medium were seeded into the upper chamber. The lower chamber was filled with 600 μL of medium containing 5% fetal bovine serum supplemented with or without inhibitor drugs. After 24 hours of incubation, the cells were removed from the upper surface.25 The cells on the lower surface were stained with 5% crystal violet (Sigma) and counted in 6 randomly selected fields to quantify the cell invasion rates with or without treatment. All experiments were performed independently at least 3 times.
siRNA Transfection and Gene Knockdown Assays
The cells were seeded in 6-well plates at 70%–80% confluence. A nontargeting siRNA scramble control and siRNA targeting at either of EGFRvIII (Invitrogen), JAK2 (Genepharma), or STAT3 (Genepharma) were transfected into U87MG-EGFRvIII cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were harvested for real-time RT-PCR, Western blotting assays, or transwell invasion assays 72 hours post transfection. All experiments were performed independently at least 3 times.
cDNA Preparation and Real-time PCR
Total RNA was extracted from the cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNA was reverse-transcribed from 1 μg RNA with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). To detect the mRNA levels of MMP2 and MMP9, real-time RT-PCR was performed with SYBR Green master mix (Roche). Fold-changes in relative gene expression were calculated by the comparative Ct method (fold change = 2-△△Ct). All experiments were performed independently at least 3 times.
Protein Preparation, Immunoprecipitation, and Western Blotting
To prepare whole-cell lysates, cells were harvested using cell lysis buffer (CST) with PMSF (Sigma), pepstatin A (Sigma) and PhosSTOP (Roche). A total of 40 μg of protein from each sample was used for Western blotting analysis. Cytoplasmic and nuclear proteins were extracted using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime) according to the manufacturer's instructions. For immunoprecipitation, GBM cells (1 × 106) were treated with ice-cold radio immunoprecipitation assay (RIPA) buffer (Sigma), and the cells were disrupted by repeated pipetting. One μg of antibody was added to 1 mg of the protein lysate; after 4 hours incubation at 4°C, 20 μL of protein A-agarose (Santa Cruz Biotechnology) was added. The samples were then incubated overnight at 4°C. After washing with cold RIPA buffer, the immunoprecipitated proteins were eluted for Western blotting. The proteins were separated on an 8%–10% SDS-polyacrylamide gel and electrophoretically transferred onto a nitrocellulose membrane (Millipore). The membranes were blocked with Tris-buffered saline Tween-20 (TBST) containing 5% (w/v) skim milk for 1 hour before incubating overnight at 4°C with the primary antibody. After washing with TBST, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 1 hour at room temperature. The signals were visualized using enhanced chemiluminescence (Roche) and exposed to x-ray film. The lanes were quantitated by using Image J.
Immunostaining With Laser-scanning Confocal Imaging
Immunostaining was performed as previously reported.26 In brief, the cells were grown on glass coverslips in 12-well plates and fixed with 4% paraformaldehyde. The coverslips were then incubated in blocking solution, followed by incubation with the primary antibody at a 1:100 dilution overnight at 4°C. FITC- or Alexa fluor-labeled anti-rabbit or anti-mouse antibody was added to the incubation. The nuclei were stained with DAPI (Sigma), and the slides were examined immediately with a Zeiss LSM 510 Meta confocal microscope (Carl Zeiss), and all images were captured using a 63 × oil immersion objective (Plan-Apochromat 63 × /1.40 Oil DIC M27) and processed using LSM Image browser software.
Nude Mouse Glioma Intracranial Models and Treatment
To construct intracranial glioma models, U87MG-vector and -EGFRvIII cells were infected with a luciferase lentivirus (Genepharma) alone or cotransfected with JAK2 shRNA lentivirus (or scramble oligonucleotide lentivirus) and luciferase lentivirus. After a 4-day infection, 5 × 105 cells were collected and injected into the intracranial striatum of 5-week-old female nu/nu-nude mice with a stereotactic instrument using a previously described guide-screw system.27,28 The animals were randomly divided into 8 groups, with 13 mice in each group. Starting on day 4 after tumor cell implantation, the mice injected with U87MG-vector or -EGFRvIII cells alone were treated with 5 mg/kg AG490 in DMSO or with an equal volume of DMSO alone (vehicle control) every other day until 10 doses were given. To acquire tumor growth status in live animals with or without treatment by bioluminescent imaging, the mice were anesthetized, injected intraperitoneally with 50 mg/mL of D-luciferin (Promega), and imaged with the IVIS Imaging System (Caliper Life Sciences) for 10–120 seconds. Fifteen days after tumor cell implantation, 3 animals from each group were euthanized, and the tumor samples were taken for immunohistochemistry (IHC), hematoxylin and eosin (HE) staining, and Western blotting. The remaining 10 mice in each group were used for survival analysis.
Hematoxylin and Eosin Stain and Immunohistochemistry
Histological sections of the tumor tissue were fixed in 10% neutral buffered formalin for HE staining and IHC analysis. Following paraffin embedding, 5 μm sections were cut and dried, deparaffinized, and rehydrated before the nonspecific binding sites were blocked. For HE staining, the sections were brought to distilled water before the nuclei were stained with hematoxylin; the sections were then rinsed in running tap water and stained with eosin before being dehydrated and mounted. For IHC analysis, the sections were incubated at 4°C overnight in a 1:100 dilution with primary antibodies against matrix metalloproteinase-9 (MMP9), phosphorylated JAK2, STAT3, or FAK, before being incubated with a biotin-labeled secondary antibody (1:100 dilution) for 1 hour at 37°C, followed by incubation with ABC-peroxidase and diaminobenzedine; the sections were then counterstained with hematoxylin and mounted. The images were captured with Olympus IX81 microscopy. All experiments were performed independently at least 3 times.
Statistical Analysis
All experiments were repeated independently at least 3 times, and the data were expressed as the mean ± standard error of the mean using SPSS. For the survival analysis, the Kaplan-Meier method was used to generate survival curves by GraphPad Prism 5. The log-rank test was used to test the differences in survival times of the different groups. The significance of the differences between groups was determined using a 2-tailed Student' t test at P < .05*, <.01**, and <.001***.
Results
Characterized by Alteration of Cell Motility and Inflammation-related Molecules, EGFRvIII Expression Promoted Enhanced Glioblastoma Cell Invasion and Tumor Progression
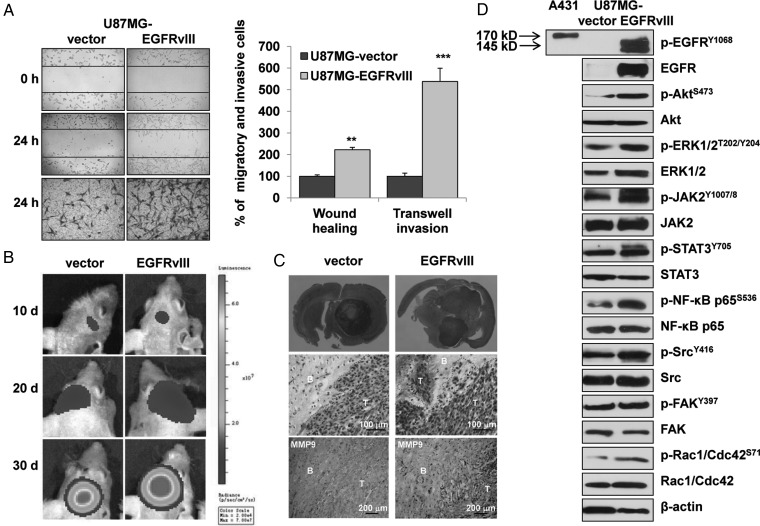
Previous reports clearly demonstrated that EGFRvIII expression produced an invasive and malignant phenotype in patients with GBM.5,6 We previously demonstrated that U87MG-EGFRvIII cells had significantly higher proliferation rates than vector cells.23 In the present study, wound healing and transwell invasion assays demonstrated that the migration and invasion rates of U87MG-EGFRvIII and LN229-EGFRvIII GBM cells, which express EGFRvIII, were significantly increased compared with those of the respective vector cells (Fig. 1A, S1A). Moreover, the U87MG-EGFRvIII orthotopic models displayed significantly accelerated tumor progression (Fig. 1B and C), increased invasion with deeper and advanced infiltration of the tumor fronts (Fig. 1C), and enhanced expression of the proteases, such as MMP9 (one of the most recognizable markers for tumor progression and invasion)29 (Fig. 1C).
Fig. 1.
EGFRvIII expression induced an invasive phenotype in glioblastoma cells in vivo and in vitro. (A) Representative results (left) and quantitative analysis (middle) of wound healing and transwell invasion assays in U87MG-vector and -EGFRvIII cells in vitro. The data are shown as the mean ± standard error of the mean of at least 3 independent experiments. ** P < .01, *** P < .001. (B) Fluorescence imaging of the tumor growth status in intracranial tumor models with U87MG-vector and -EGFRvIII cells in vivo. (C) Comparison of effect in vivo on tumor growth and invasion between the U87MG-vector and -EGFRvIII models. Fifteen days after tumor cell implantation, the full brain slice section (upper panel) showed significantly enhanced tumor growth and spreading in EGFRvIII models. Hematoxylin and eosin stain (middle panel) displayed significantly increased infiltration and invasive tumor fronts in EGFRvIII models compared with the vector models. IHC analysis (lower panel) showed an increased expression of MMP9 and disordered infiltration and invasive tumor fronts in EGFRvIII tumors. (B, brain; T, tumor). (D) The results of Western blotting showed greatly increased activity of EGFRvIII (140 KD) and its downstream effectors, including ERK, Akt, inflammation-related JAK2/STAT3, NF-κB, and the motility-related Src, FAK in EGFRvIII-cells in vitro. As a wild-type EGFR positive control (170 kD), the protein used was from the epithelial carcinoma cell line A431 and had been stimulated with 50 ng/mL EGF for 15 minutes prior to protein extraction.
The genomic microarray was used to analyze the transcriptome profiles induced by EGFRvIII expression in U87MG cells. A total of 2598 genes were differentially expressed by at least 2-fold between U87MG-EGFRvIII and -vector cells. The differential expression was verified by real-time PCR assays for 78 of the 100 randomly selected genes (data not shown). We next analyzed the biological processes of these 2598 genes using GO or the KEGG pathway database with bioinformatics methods. Among the top 12 most significantly enriched GO terms and/or pathways, 6 GO terms and 4 pathways were related to cell motility. Furthermore, cell responses to environmental stimuli and inflammation-associated events (2 GO terms and 4 pathways), and angiogenesis (3 GO terms) were also prominent (Table 1). Together, the mRNA profiling revealed that EGFRvIII expression significantly altered cell motility, response to environmental stimuli, and inflammatory properties, as well as other advanced cancer features in U87MG cells.
Table 1.
Gene Ontology* and KEGG pathway enrichment analysis of differentially expressed genes between U87MG-EGFRvIII and -vector cells
| Rank | GO ID | GO term | P value | Pathway ID | Definition | P value |
|---|---|---|---|---|---|---|
| 1 | GO:0006928 | Cell motion | <.001 | hsa03010 | Ribosome | .001 |
| 2 | GO:0043062 | Extracellular structure organization | <.001 | hsa04512 | ECM-receptor interaction | .002 |
| 3 | GO:0051674 | Localization of cell | <.001 | hsa04510 | Focal adhesion | .003 |
| 4 | GO:0048870 | Cell motility | <.001 | hsa05200 | Pathways in cancer | .003 |
| 5 | GO:0016477 | Cell migration | <.001 | hsa05322 | Systemic lupus erythematosus | .004 |
| 6 | GO:0070482 | Response to oxygen levels | <.001 | hsa04115 | p53 signaling pathway | .006 |
| 7 | GO:0010033 | Response to organic substance | <.001 | hsa04060 | Cytokine-cytokine receptor interaction | .014 |
| 8 | GO:0051270 | Regulation of cell motion | <.001 | hsa04621 | NOD-like receptor signaling pathway | .024 |
| 9 | GO:0001568 | Blood vessel development | <.001 | hsa04610 | Complement and coagulation cascades | .031 |
| 10 | GO:0001944 | Vasculature development | <.001 | hsa04710 | Circadian rhythm | .038 |
| 11 | GO:0042127 | Regulation of cell proliferation | <.001 | hsa04810 | Regulation of actin cytoskeleton | .041 |
| 12 | GO:0001525 | Angiogenesis | <.001 | hsa04520 | Adherens junction | .043 |
Abbreviation: NOD, nucleotide binding oligomerization domain; EMC, extracellular matrix; ID, identification. *GO term category, biological process (BP).
Western blotting assays confirmed the above findings. Compared with the vector cells, the activation of Akt, ERK1/2, inflammation-related signaling molecules (including JAK2, STAT3, and NF-κB), and cell motility-related molecules (including FAK, Src, and Rac1/Cdc42) was significantly increased in U87MG cells, which expressed different levels of EGFRvIII (Fig. 1D, S1B). Furthermore, consistent results were obtained in LN229-EGFRvIII as compared with those in LN229-vector cells (Fig. S1C). However, the p-Akt levels were unchanged in LN229-EGFRvIII cells, which probably occurred because LN229 cells inherently expressed a wild-type PTEN, whereas the U87MG cells harbored mutant PTEN, which may affect Akt activity.30 Collectively, these data indicated that the expression of mutant EGFRvIII in GBM cells resulted in significant alterations in cancer-related properties such as cell motility and inflammation.
The JAK2/STAT3 Inhibitor Significantly Prevented Glioblastoma Cell Invasion in EGFRvIII-expressing Cells
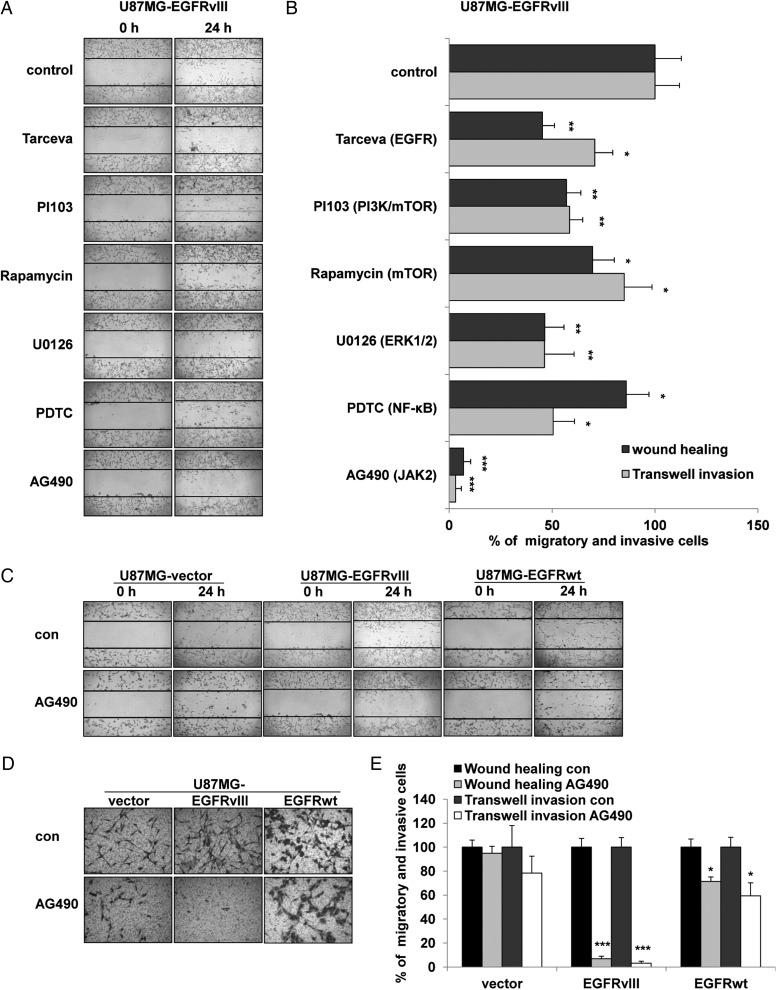
To select an optimal drug to effectively inhibit cell motility, multiple kinase inhibitors were tested on U87MG-EGFRvIII cells. Among the tested inhibitors, AG490, which targets JAK2/STAT3,25 was the most efficient at inhibiting cell migration and invasion (Fig. 2A and B). Other inhibitors included Tarceva, PI103, rapamycin, U0126, and ammonium pyrrolidine dithiocarbamate (PDTC) and targeted EGFR, PI3K, mTOR, ERK1/2, and NF-κB, respectively. AG490 consistently and efficiently inhibited cell motility in LN229-EGFRvIII and U87MG-EGFRvIII clone 2 cells (Fig. S2). In contrast, AG490 treatment affected cell migration and invasion only modestly in either U87MG-vector (Fig. 2C–E, P > .05) or LN229-vector (Fig. S2A, S2B, P > .05) cells, which lack EGFRvIII expression. In addition, the same treatment inhibited cell motility to a lesser extent in U87MG-EGFRwt cells, which express wild-type EGFR (Fig. 2C–E). Furthermore, treatment with WP1066, another JAK2/STAT3 inhibitor and a secondary generation drug of AG490,31 verified this effect on the inhibition of cell invasion by AG490 in 2 clones of U87MG cells that expressed different levels of EGFRvIII (Fig. S2C, S2D). Together, these results indicated that the significantly enhanced cell motility induced by EGFRvIII expression in GBM cells may be the most relevant to the cell inflammation-related JAK2/STAT3 signaling pathways.
Fig. 2.
JAK2/STAT3 inhibitors significantly inhibited cell motility in EGFRvIII-expressing glioblastoma cells. (A) Results of wound-healing assays and (B) quantitative analysis of wound-healing and transwell invasion assays for U87MG-EGFRvIII cells treated with respective small-molecule inhibitors. Cells were treated with the EGFR inhibitor Tarceva (20 μM), PI3 K, and mTOR inhibitor PI103 (500 nM), mTOR inhibitor Rapamycin (Rap, 1 nM), ERK inhibitor U0126 (15 μM), NF-κB inhibitor ammonium pyrrolidinedithiocarbamate (PDTC; 5 μM), or JAK2 inhibitor AG490 (100 μM) for 24 hours. The data are shown as the mean ± standard error of the mean of at least 3 independent experiments. * P < .05, ** P < .01, *** P <.001 compared with the control. (C), (D), and (E) Representative wound-healing and transwell invasion assays and quantitative analysis of U87MG-vector, -EGFRvIII, and -EGFRwt cells with or without the treatment of 100 μM AG490. The data are shown as the mean ± standard error of the mean of at least 3 independent experiments. * P < .05, *** P < .001 compared with the control.
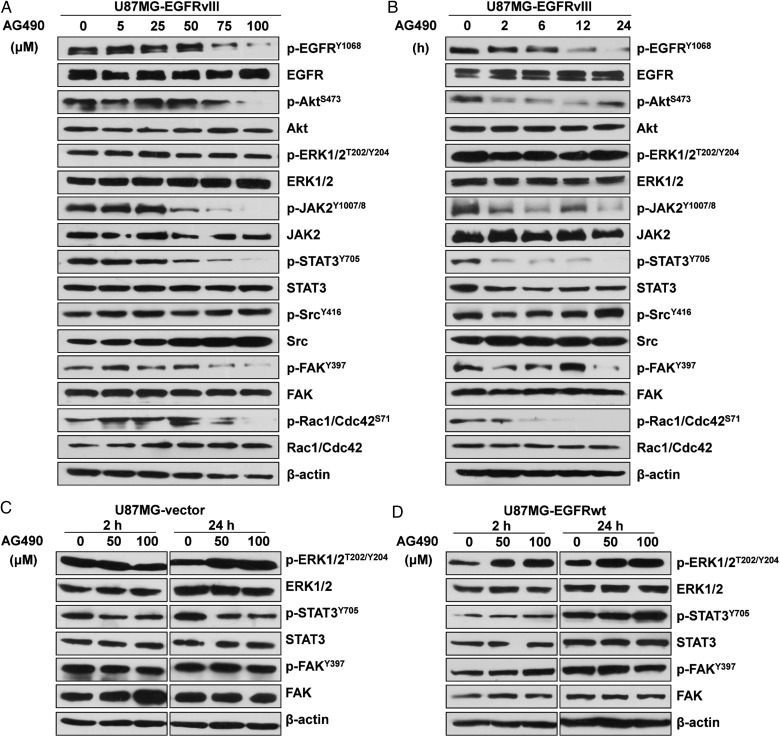
Further Western blotting analysis demonstrated that AG490 had dose-dependent and sustained inhibitory effects on the activity of EGFRvIII, JAK2/STAT3, and motility-related molecules, such as FAK and Rac1/Cdc42 in U87MG-EGFRvIII cells (Fig. 3A). After 24 hours of treatment, 50 μM AG490 inhibited p-JAK2 and p-STAT3. This effect was dose-dependent. In addition, AG490 concentrations of 75 μM or higher efficiently diminished EGFRvIII, FAK, and Rac1/Cdc42 activity, while concentrations up to 100 μM efficiently blocked p-Akt (Fig. 3A). Additional time-course analysis revealed that 100 μM of AG490 stably suppressed the activity of JAK2, STAT3, Akt, FAK, and Rac1/Cdc42 from 2 to 24 hours post treatment, whereas EGFR activity was inhibited after 6 hours of treatment. However, p-Src and p-ERK1/2 were unaffected by treatment, implying that their activities were not affected by inhibition of JAK2/STAT3 signaling (Fig. 3B). Similar effects were observed in LN229-EGFRvIII cells in response to AG490 treatment (Fig. S3A, S3B) and in both U87MG-EGFRvIII and LN229-EGFRvIII cells in response to WP1066 treatment (Fig. S3C).
Fig. 3.
JAK2/STAT3 inhibitor significantly inhibited cell motility-related cellular signaling in EGFRvIII-expressing glioblastoma cells. (A) Concentration-dependent and (B) time-dependent effect of AG490 treatment on the inhibiting activity of EGFRvIII and its downstream effectors that were crucial for cell motility in U87MG-EGFRvIII cells by Western blotting. (C) and (D) The significantly lower inhibition effect of AG490 treatment on EGFRvIII downstream effector in U87MG-vector and -EGFRwt cells compared with that in -EGFRvIII cells in (A) and (B). EGF (50 ng/mL) was added to U87MG-EGFRwt cells for 15 minutes before the cells were harvested for analysis.
Moreover, the effects of AG490 treatment on relevant molecules were examined in U87MG-vector and -EGFRwt cells. Although AG490 treatment moderately decreased p-STAT3 level in U87MG-vector cells, the activation of FAK and ERK1/2 remained unchanged (Fig. 3C). The activity of STAT3, FAK, or ERK1/2 was not affected by AG490 treatment in U87MG-EGFRwt cells (Fig. 3D). These results may explain why cell motility in U87MG-vector and -EGFRwt cells was less affected by AG490 treatment than that in EGFRvIII-expressing cells (Fig. 2C–E).
JAK2/STAT3 Inhibition Disrupted the Focal Adhesion Complex Where EGFRvIII, JAK2, and STAT3 Were Actively Involved in U87MG Cells
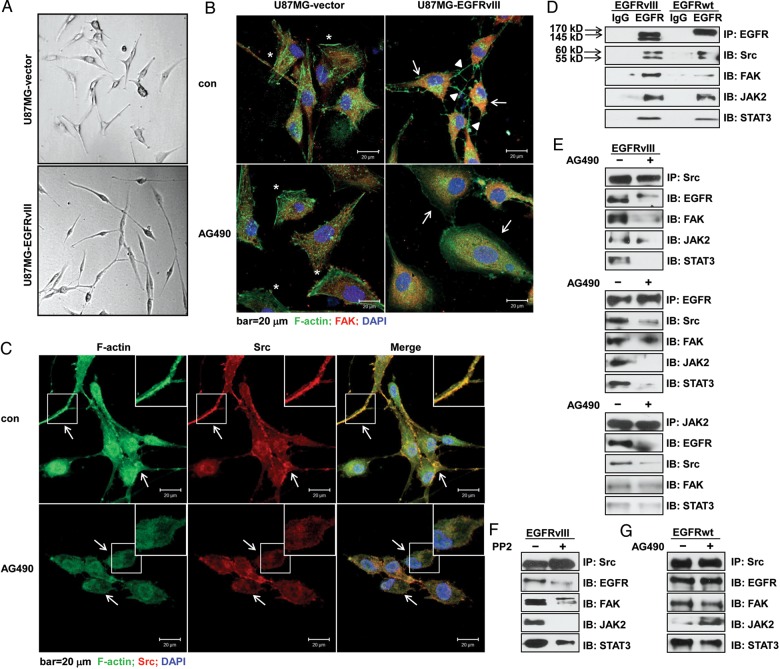
Along with the greatly increased cell motility in U87MG-EGFRvIII cells, we observed a pronounced change in their cell morphology compared with vector cells (Fig. 4A). Confocal imaging analysis demonstrated that the EGFRvIII-expressing cells exhibited pronounced F-actin polymerization in their long, abundant filopodia, resulting in an elongated shape and numerous cell-cell contacts. Conversely, cells treated with AG490 displayed a dramatic decrease in filopodia, stress fibers, cell-cell contacts, and FAK distribution away from the cell edges. By contrast, U87MG-vector cells had a substantially more flattened shape, and AG490 treatment had only a slight effect on cell morphology and FAK distribution (Fig. 4B). The focal adhesion assembly was actively involved in cell shape conformation and motility.32 Thus, we proposed that EGFRvIII expression in U87MG cells might actively participate in this process. The activated Src, FAK, and Src-FAK complex were the focal points for focal adhesion-associated intracellular signaling which controlled cell shape and motility.33 Further analysis demonstrated that F-actin colocalized with Src in U87MG-EGFRvIII cells and that the distribution of these proteins was disturbed when the cells were treated with AG490 (Fig. 4C).
Fig. 4.
JAK2/STAT3 inhibition disrupted the focal adhesion complex where EGFRvIII, JAK2, and STAT3 were actively involved in U87MG cells. (A) U87MG-vector and -EGFRvIII cells showed different cell morphologies (100× images of inverted light microscopy). (B) Confocal images showed significantly disturbed cell morphology and focal adhesion contact of F-actin and FAK, and (C) F-actin and Src in U87MG-EGFRvIII cells under the treatment. Triangles indicate filopodia, asterisks indicate lamellipodia, and arrows in (B) indicate alterations in FAK distribution; arrows in (C) show changes in the colocalization of F-actin and Src. U87MG-vector and -EGFRvIII cells were treated with 100 μM AG490 for 24 hours before staining with fluorescein isothiocyanate-labeled phalloidin for F-actin (green), an anti-FAK or anti-Src antibody (red), and DAPI (blue); bar, 20 μm. (D) The analysis of co-immunoprecipitation (co-IP) showed the firm association of EGFRvIII or EGFRwt with the focal adhesion marker FAK, Src, and JAK2, STAT3. The 55-kD band is IgG, and the 60-kD band is Src. (E) Co-IP assays in U87MG-EGFRvIII cells indicated the disruption of the respective focal adhesion contacts that were pulled down by anti-Src, anti-EGFR, or anti-JAK2 antibodies after treatment with 100 μM AG490. (F) Co-IP analysis in U87MG-EGFRvIII cells indicated the disruption of the respective focal adhesion contacts that were pulled down by an anti-Src antibody or treatment with a Src kinase inhibitor PP2 (20 μM). (G) Co-IP analysis in U87MG-EGFRwt cells showed that the treatment with AG490 failed to disrupt the focal adhesion contacts that were pulled down by an anti-Src antibody.
Co-immunoprecipitation with the wild-type EGFR or EGFRvIII revealed a potential connection between either of the receptors and one of the 4 kinases, including Src, FAK, JAK2, and STAT3 (Fig. 4D). While the co-immunoprecipitation also suggested a link between Src or JAK2 and another kinase (Src, FAK, EGFR, JAK2, or STAT3), these connections could be efficiently disrupted by AG490 (Fig. 4E, S4A) or WP1066 treatment (Fig. S4B, S4C) in EGFRvIII-expressing cells. To verify the vital connection between Src-FAK complex and the EGFRvIII-Src-FAK-JAK2-STAT3 focal adhesion complex, U87MG-EGFRvIII cells were treated with the Src kinase inhibitor, PP2. This treatment significantly weakened or abolished the connection between Src and FAK, Src and JAK2, Src and STAT3, respectively (Fig. 4F). By contrast, although wild-type EGFR also formed a complex with Src, JAK2, STAT3 or, to a lesser extent, FAK (Fig. 4D), AG490 treatment failed to disintegrate the essential Src-FAK complex for focal adhesion formation in U87MG-EGFRwt cells (Fig. 4G); such data partially explained why the treatment failed to affect cell motility (Fig. 2C–E). Taken together, these results indicated that EGFRvIII, JAK2, and STAT3 may attach to the focal adhesions, thereby forming a complex to facilitate cell motility in EGFRvIII-expressing cells, whereas the treatment with JAK2/STAT3 inhibitors destroyed the complex.
The Activation of STAT3 Depended on JAK2 Activity in EGFRvIII-expressing Glioblastoma Cells
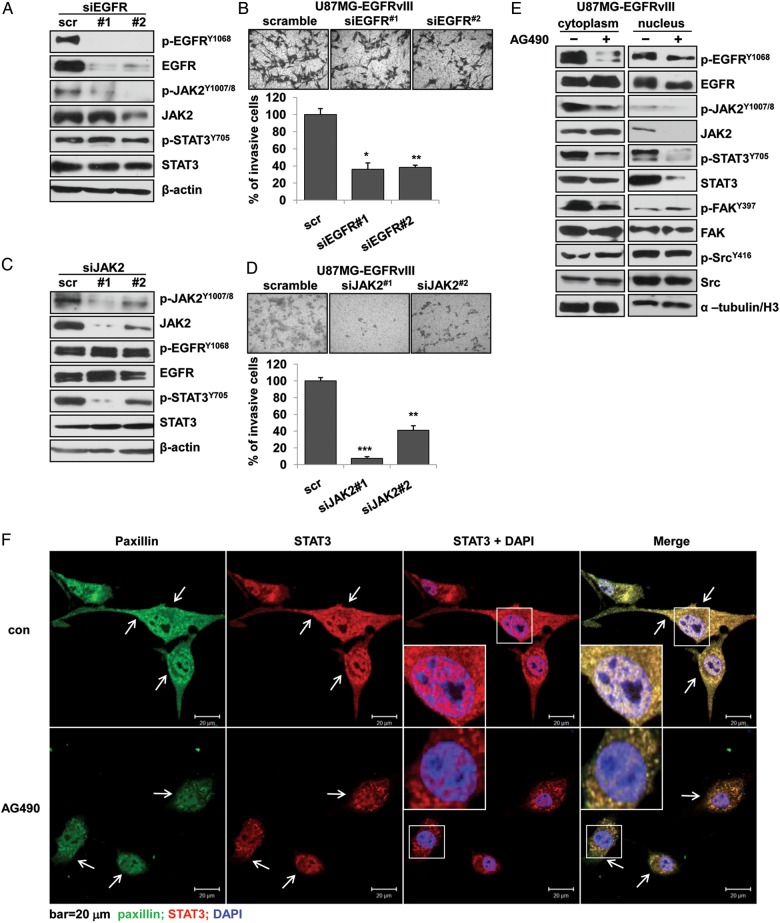
To test whether the activation of STAT3, a key downstream effector of EGFR signaling,2 depended on EGFRvIII activity, the EGFRvIII gene was knocked down by siRNA in U87MG-EGFRvIII cells. Although the effective blockage of EGFRvIII reduced p-JAK2 at Tyr1007/8, the gene knockdown failed to affect STAT3 activity (Fig. 5A), indicating other routes of STAT3 activation. In addition, the siRNA knockdown did not inhibit cell invasion as effectively as AG490 treatment (Fig. 5B, 2B). In comparison, JAK2 knockdown by siRNA in the same cells reduced p-STAT3 and cell invasion in a siRNA interference efficiency-dependent manner (Fig. 5C and D); the effective inhibition was similar to that obtained by treating the same cells with AG490 (Fig. 5D, 2B). These results indicated that JAK2 activity was closely linked to STAT3 activation and cell invasion in U87MG-EGFRvIII cells. Moreover, AG490 not only significantly reduced the levels of p-EGFR, p-JAK2, p-STAT3, and p-FAK in the cytoplasm but also abolished the total and phosphorylated forms of JAK2, STAT3 and, to a lesser extent, EGFR, in the nucleus. However, this treatment did not affect the levels of nuclear FAK, cytoplasmic, and nuclear Src of the phosphorylated and total protein (Fig. 5E). Confocal imaging analysis confirmed that AG490 treatment significantly decreased STAT3 translocation into the nucleus and disturbed the colocalization of STAT3 and the focal adhesion marker paxillin34 in EGFRvIII-expressing cells (Fig. 5F). EGFRvIII has been reported to interact with STAT3 in the nucleus and function as a cotranscription factor.17 Our results suggested that AG490 treatment significantly reduced the expression, and possibly the function, of EGFRvIII and STAT3 in the nucleus, but their interactions in GBM cells require further investigations.
Fig. 5.
The activation of STAT3 depended on JAK2 activity in EGFRvIII-expressing cells. (A) siRNA treatment on EGFRvIII in U87MG-EGFRvIII cells resulted in significant inhibition on JAK2 activity but not STAT3 activity by Western blotting, and (B) inhibition on cell invasion by transwell-invasion assays. (C) siRNA treatment on JAK2 in U87MG-EGFRvIII cells resulted in significant inhibition on STAT3 activity but not EGFRvIII activity by Western blotting, and (B) inhibition on cell invasion by transwell-invasion assays. The data are shown as the mean ± standard error of the mean of at least 3 independent experiments. * P < .05, ** P < .01, *** P < .001 compared with the scramble control. (E) The treatment of AG490 resulted in the significant reduction of nuclear content of total- and phosphorylated-STAT3, JAK2, and EGFRvIII by Western blotting. α-tubulin and H3 were used as internal controls in the cytoplasm and nucleus, respectively. (F) Confocal images showed significantly reduced nuclear content of STAT3 and changes in the colocalization of paxillin and STAT3 (arrows) in U87MG-EGFRvIII cells after AG490 treatment. Paxillin (green), STAT3 (red), DAPI (blue). Bar, 20 μm. U87MG-EGFRvIII cells were treated with 100 μM AG490 for 24 hours.
JAK2/STAT3 Signaling Activation Was Vital for Maintaining the EGFRvIII-induced Network and Invasion in Glioblastoma Cells
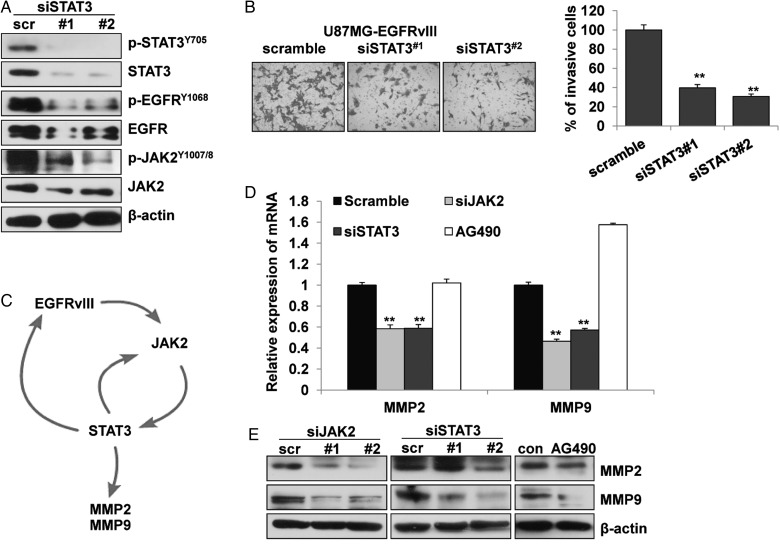
Further siRNA assays in U87MG-EGFRvIII cells demonstrated that the effective blockage of STAT3 significantly reduced the levels of phosphorylated and total EGFR and JAK2 (Fig. 6A); however, the STAT3 knockdown was not as effective as either the AG490 treatment or the JAK2 knockdown for inhibiting cell invasion (Fig. 6B, 2B, 5D). These data, together with the results from the EGFR and JAK2 knockdown assays (Fig. 5A–D), implied that EGFRvIII, JAK2, and STAT3 may form an activation loop to promote effective cell invasion (Fig. 6C). To verify this finding, we compared the effects of JAK2 and STAT3 siRNA as well as AG490 treatment on the mRNA and protein expression of matrix metalloproteinase-2, (MMP2) and MMP9, which were key proteases for effective tumor invasion.29 JAK2 siRNA reduced the levels of these extracellular matrix (ECM)-degrading enzymes significantly, and did STAT3 siRNA and AG490 reduced them to a lesser extent (Fig. 6D and E). Furthermore, the EGFRvIII/JAK2/STAT3 axis also promoted cellular signaling that drove tumor cell invasion by forming a focal adhesion complex with FAK and Src (Fig. 4D and E). Taken together, we concluded that the activity of JAK2/STAT3 was important for maintaining an EGFRvIII-induced network and cell invasion.
Fig. 6.
JAK2/STAT3 were key players in EGFRvIII-induced network and cell invasion in glioblastoma cells. (A) siRNA treatment on STAT3 in U87MG-EGFRvIII cells resulted in significant inhibition on total- and phosphorylated-JAK2 and EGFRvIII by Western blotting and (B) inhibition on cell invasion by transwell invasion assays. The data are shown as the mean ± standard error of the mean of at least 3 independent experiments. ** P < .01 compared with the scramble control. (C) Schematic illustration of a potential activation loop of EGFR, JAK2, and STAT3 in promoting MMP expression and cell invasion. (D) The treatment with siRNA on JAK2 and STAT3 but not AG490 (100 μM for 24 h) reduced the expression of MMP2 and MMP9 according to real-time PCR; and (E) treatment with siRNA on JAK2 and STAT3, and AG490 reduced the same expression by Western blotting in U87MG-EGFRvIII cells. Data are shown as the mean ± standard error of the mean of at least 3 independent experiments. ** P < .01, compared with the control.
Because treatment with the JAK2/STAT3 inhibitors, including AG490 and WP1066, inhibited GBM cell motility the most effectively, our next step was testing the effect of a Src kinase inhibitor, PP2, on U87MG-EGFRvIII cells for excluding other targets that may inhibit cell invasion more effectively than JAK2. The results showed that PP2 did not inhibit cell invasion and STAT3 activity as effectively as AG490 (Fig. S5, 2B, 3A), although the inhibition of Src activity by PP2 disrupted EGFRvIII/JAK2/STAT3-associated focal adhesion contacts and, to some extent, the related signaling (Fig. 4F, S5A). Accordingly, the lower efficacy of EGFRvIII siRNA compared with AG490 may be attributed to its inability to inhibit STAT3 activation (Fig. 5A). Moreover, we proposed that treatment with up to 100 mM diethyl bromodifluoromethyl phosphonate (DEP), a specific STAT3 kinase inhibitor, was ineffective for inhibiting STAT3 activity and cell motility because the EGFRvIII-JAK2-STAT3 axis was continuously activated in the tested GBM cells (Fig. S6). While p-STAT3 was significantly diminished, p-FAK and p-ERK1/2 were unchanged in U87MG-EGFRwt cells after 24 hour treatment with 50 μM DEP (Fig. S6A), which resulted in only moderate inhibition of cell invasion (Fig. S6B). In summary, efficacy in cell invasion inhibition by the JAK2/STAT3 inhibitors or JAK2 knockdown was attained by disrupting the EGFRvIIII/JAK2/STAT3 axis and its associated focal adhesion complex in the GBM cells.
Targeting JAK2/STAT3 Therapy Efficiently Suppressed Glioblastoma Invasion and Progression Induced by EGFRvIII Expression in Vivo
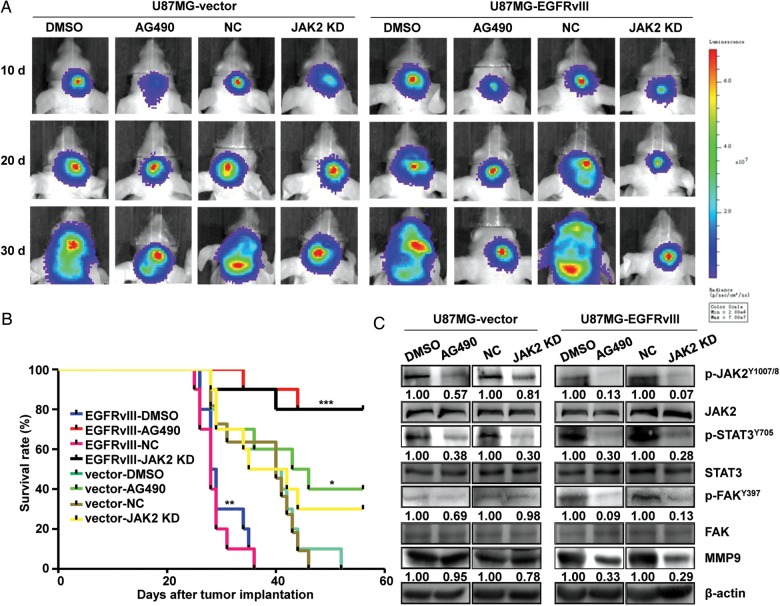
We used GBM orthotopic animal models implanted with U87MG-EGFRvIII or -vector cells to determine whether treatment with the JAK2/STAT3 inhibitor, AG490, efficiently inhibited EGFRvIII-induced tumor invasion and progression in vivo. To verify the best treatment target, we also applied both EGFRvIII-expressing and vector cells that underwent shRNA gene knockdown at JAK2 on the models. During the experiments, the GBM cells were also engineered to stably express firefly luciferase prior to intracranial implantation in order to monitor treatment response and tumor growth status in live animals by bioluminescent imaging. The results indicated that the U87MG-EGFRvIII tumors progressed significantly faster, infiltrated more aggressively, and had a shortened survival time compared with mice bearing vector control tumors (P <.01, Fig. 1B, 1C, 7A and B). The U87MG-EGFRvIII mice in the AG490 treatment group and the JAK2 KD (knockdown) group did not exhibit body weight loss until the late stages of the experiment. In contrast, animals in the nontreatment control group and negative control group exhibited progressive body weight loss from the early stages of the experiment due to advanced disease (data not shown). A survival curve demonstrated that AG490 treatment and JAK2 knockdown in the U87MG-EGFRvIII models greatly enhanced survival compared with nontreatment and negative control of the same tumor model (P < .001, Fig. 7B). Furthermore, the expression levels of key factors involved in active tumor invasion were also examined by Western blotting and IHC. The Western blotting results showed that the level of p-JAK2, p-STAT3, p-FAK, or MMP9 was greatly decreased in the U87MG-EGFRvIII mice models that were either treated with AG490 or experienced JAK2 knockdown compared with the level observed in the respective control groups (Fig. 7C). IHC of GBM models with AG490 treatment or JAK2 knockdown showed consistent results (Fig. S7). In comparison, however, cell proliferation inhibition by Ki67 expression, and apoptosis detected by TUNEL assays, under the same treatments, showed no significant differences between U87MG-vector and -EGFRvIII mice (Fig. S7). Although the U87MG-vector mice also benefitted from AG490 treatment or JAK2 knockdown (P < 0.05, Fig. 7B), the results showed no significant differences between AG490 and the nontreatment control groups or between the JAK2 knockdown and control groups on the expression of those tumor invasion markers (Fig. 7C). These results greatly corroborated our in vitro findings.
Fig. 7.
Evaluating the efficiency of targeting JAK2 therapy in U87MG-vector and EGFRvIII intracranial models. (A) Bioluminescent images from control, AG490-treated, negative control (NC), and JAK2 knockdown (JAK2 KD) groups at 10, 20, and 30 days after tumor cell implantation. (B) Survival curves for all groups of intracranial tumor-bearing mice. * P < .05, vector-AG490 group compared with vector-DMSO group; vector-JAK2 KD group compared with vector-NC group; ** P < .01, EGFRvIII-DMSO group compared with vector-DMSO group; EGFRvIII-NC group compared with vector-NC group; *** P < .001, EGFRvIII-AG490 group compared with EGFRvIII-DMSO group, EGFRvIII-JAK2 KD group compared with EGFRvIII-NC group. (C) Comparisons of p-JAK2Tyr1007/1008, p-STAT3Tyr705, p-FAKTyr397, and their total levels, and MMP9 expression levels by Western blotting in the tumors from each group in vivo. Tumor samples for Western blotting analysis were taken 15 days after tumor cell implantation.
Discussion
EGFRvIII promotes growth and invasion by interacting with many induced or pre-existing signaling effectors in cancers including GBM. Thus, development of targeted therapies for cancers that express this oncogenic protein has been challenging.35 In our study, we found that EGFRvIII interacted vigorously with JAK2/STAT3 signaling in the GBM cell U87MG-EGFRvIII, which stably overexpressed the mutant receptor, EGFRvIII. This interaction was facilitated by formation of a focal adhesion complex and activation loop that promoted effective tumor cell invasion and progression. U87MG-EGFRvIII and -vector cells have been analyzed in many other reports.12,36 Specifically, our results showed that treatment with a JAK2 kinase inhibitor, either AG490 or WP1066, efficiently disrupted the cellular network and machinery established by the mutant receptor and prevented the aggressive invasion of GBM cells in vitro and the malignant progression of intracranial tumor models in vivo. This effect was not observed with expression of wild-type EGFR or without EGFRvIII expression GBM cells.
We observed dramatic changes in morphology and motility in U87MG-EGFRvIII cells, which displayed a more motile and invasive phenotype than vector cells. These findings concur with previously published reports.7,37 These features may be strongly related to the cellular machinery linking the dynamic assembly and disassembly of focal adhesions and active cell migration.32 During these processes, the cell adhesion molecule integrins first bind to the ECM at focal adhesions and attract FAK to facilitate its activation. In turn, this interaction forms an active FAK-Src complex that converges as the focal point for activating many intracellular signaling events, including PI3K/Akt and Ras/MAPK.38 Thus, the integrin and growth factor signaling pathways interrelate and overlap significantly. Kim et al. (2011) proposed that integrins colocalized with EGFR at focal adhesions and their associated signaling molecules, such as FAK and Src, to regulate cell functions and activities of downstream effectors such as ERK1/2, Akt, and the Rho GTPases.38 A previous report clearly demonstrated that FAK formed a complex with the activated EGFR and was required for EGF-stimulated cell motility.39 Our data demonstrated for the first time that EGFRvIII, JAK2, and STAT3 attached to the FAK-Src complex and focal adhesions to promote enhanced cell motility in GBM cells. This effect was also observed upon expression of wild-type EGFR. In addition, confocal microscopy demonstrated that the focal adhesion marker colocalized with Src or STAT3 in U87MG-EGFRvIII cells, which is in line with previous reports.34,40 Our genomic microarray data and analysis corroborated these findings: the expression of EGFRvIII resulted in significant alterations of various integrin dimers and components of the ECM and ECM-degrading proteases (Table 2), thereby changing the signaling pathways related to cell adhesion and motility in the GBM cells.
Table 2.
Cell motility-related molecules that were differentially expressed between U87MG-EGFRvIII cells and -vector cells
| ECM | Fold change | Integrins | Fold change | Proteases | Fold change |
|---|---|---|---|---|---|
| COL5A1 | −3.69 | ITGA2 | −6.16 | MMP1 | 3.43 |
| COL6A1 | −2.01 | ITGA3 | 3.81 | MMP12 | 9.34 |
| COL8A2 | −3.46 | ITGA6 | −39.56 | MMP23B | −2.54 |
| COL13A1 | −11.40 | ITGA7 | 4.37 | ADAM8 | 4.96 |
| COL14A1 | −2.03 | ITGAV | −3.73 | ADAM10 | −3.04 |
| LAMA3 | −2.04 | ITGA11 | −232.25 | ADAM12 | −3.91 |
| LAMA4 | 2.07 | ITGB1 | −2.83 | ADAM19 | −4.82 |
| LAMC1 | −2.15 | ITGB3 | 4.06 | ADAM33 | 2.84 |
| FN1 | −4.93 | ITGB5 | −2.54 | CTSH | 2.51 |
| FN3KRP | 3.05 | ITGB7 | −3.38 | CTSO | −2.47 |
| FNDC3B | −2.10 | PLAU | −3.48 | ||
| FNDC4 | 3.24 | PLAUR | −2.39 |
Furthermore, we showed that EGFRvIII expression promoted the phosphorylation of JAK2, which then activated STAT3. This finding concurred with previously published reports.19–21 Nevertheless, our data indicated that STAT3 activation primarily depended on the activation of JAK2 rather than EGFRvIII or Src in the tested GBM cells. We also proved that activated STAT3 further enhanced the expression and activity of both EGFRvIII and JAK2, forming an EGFRvIII-JAK2-STAT3 activation loop to strengthen downstream signaling events for tumor invasion, including gene transcription of the ECM-degrading proteases MMP2 and MMP9.15 Thus, we are the first to show that the ligand-independent activation of EGFRvIII served as a key component of the JAK/STAT system, which functioned to enhance GBM cell invasion.41 Numerous reports indicated important links between the malignant behavior of cancer cells and the complex interaction of the kinases, including EGFRvIII, JAK2, STAT3, FAK, and Src among others.11,42,43 Our results demonstrated that the EGFRvIII-JAK2-STAT3 axis attached to the FAK-Src complex at the focal adhesion and formed a larger complex to facilitate active cellular signaling and cell motility. Moreover, treatment with AG490 or WP1066 efficiently inhibited the activities of these kinases and disrupted the vital complex in GBM cells. However, the exact interaction between EGFRvIII and STAT3 or other kinases requires further investigations.
Recent cancer reports have indicated that JAK2/STAT3 signaling plays a role in promoting EGFR-associated cancer signaling44 and progression in high-grade tumors16,45 or resistance to anti-EGFR agents.16,46 Thus, these reports recommend a combination treatment consisting of anti-EGFR and anti-JAK2/STAT3 signaling agents.16 Specifically, a report on non-small cell lung cancer indicated that JAK2/STAT3 signaling was required for EGFRvIII-mediated oncogenic effects.47 To the best of our knowledge, we are the first to report the interaction between JAK2/STAT3 activation and EGFRvIII. We also demonstrated that the kinase JAK2 was the most effective target for an optimal target therapy to inhibit invasion of EGFRvIII-expressing GBM cells. However, our study has certain limitations. Although treatment with JAK2/STAT3 inhibitors, including AG490 and WP1066, significantly decreased cell motility, this decrease was only validated in 2 EGFRvIII-expressing human GBM cell lines and intracranial tumor models. Due to the heterogeneity of GBM tumors, it is unclear whether AG490 treatment is effective on other types of EGFRvIII-expressing GBM cells. Nevertheless, we proved that the EGFRvIII-null and wild-type EGFR-positive GBM cells were substantially less sensitive to the same treatment than EGFRvIII-expressing cells. In addition, by effective gene knockdown assays at JAK2 in vivo and in vitro, we further verified that the targeted JAK2 therapy most efficiently destroyed the EGFRvIII-induced network in GBM.
In conclusion, we demonstrated that the targeted inhibition of JAK2/STAT3 signaling by JAK2/STAT3 inhibitors significantly suppressed tumor cell invasion and progression by disrupting the EGFRvIII/JAK2/STAT3 axis and related focal adhesion in EGFRvIII-expressing GBM. Our data may have potential clinical implications for the tailored treatment of GBM patients harboring tumors that express the mutant receptor.
Supplementary Material
Funding
This work was supported by the Natural Science Foundation of China (91229112 and 30772238, to H.R.; 91229121, to T.J.), the Key Project Science Foundation of Heilongjiang Province, China (ZD200804-01, to H.R.) and the National High Technology Research and Development Program 863 (2012AA02A508, to T.J. and C.-S.K.; SS2014AA021102, to X.L.), Natural Science Foundation of Tianjin Municipal Science and Technology commission (12ZCDZSY17300, to J.-N.Z. and C.-S.K.).
Supplementary Material
Acknowledgments
We thank Dr. F Furnari (Ludwig Institute for Cancer Research, San Diego, California) for kindly providing the PSK plasmids.
Conflict of interest statement. None declared.
References
- 1.Chow LM, Endersby R, Zhu X, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19(3):305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2(87):re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 3.Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16(6):748–754. doi: 10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Van Meter TE, Broaddus WC, Rooprai HK, et al. Induction of membrane-type-1 matrix metalloproteinase by epidermal growth factor-mediated signaling in gliomas. Neuro Oncol. 2004;6(3):188–199. doi: 10.1215/S1152851703000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lal A, Glazer CA, Martinson HM, et al. Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Cancer Res. 2002;62(12):3335–3339. [PubMed] [Google Scholar]
- 6.Wheeler SE, Morariu EM, Bednash JS, et al. Lyn kinase mediates cell motility and tumor growth in EGFRvIII-expressing head and neck cancer. Clin Cancer Res. 2012;18(10):2850–2860. doi: 10.1158/1078-0432.CCR-11-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ning Y, Zeineldin R, Liu Y, et al. Down-regulation of integrin alpha2 surface expression by mutant epidermal growth factor receptor (EGFRvIII) induces aberrant cell spreading and focal adhesion formation. Cancer Res. 2005;65(20):9280–9286. doi: 10.1158/0008-5472.CAN-05-0407. [DOI] [PubMed] [Google Scholar]
- 8.Zagzag D, Friedlander DR, Margolis B, et al. Molecular events implicated in brain tumor angiogenesis and invasion. Pediatr Neurosurg. 2000;33(1):49–55. doi: 10.1159/000028975. [DOI] [PubMed] [Google Scholar]
- 9.Liu M, Yang Y, Wang C, et al. The effect of epidermal growth factor receptor variant III on glioma cell migration by stimulating ERK phosphorylation through the focal adhesion kinase signaling pathway. Arch Biochem Biophys. 2010;502(2):89–95. doi: 10.1016/j.abb.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Lu KV, Zhu S, Cvrljevic A, et al. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009;69(17):6889–6898. doi: 10.1158/0008-5472.CAN-09-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwiatkowska A, Symons M. Signaling determinants of glioma cell invasion. Adv Exp Med Biol. 2013;986:121–141. doi: 10.1007/978-94-007-4719-7_7. [DOI] [PubMed] [Google Scholar]
- 12.Cai XM, Tao BB, Wang LY, et al. Protein phosphatase activity of PTEN inhibited the invasion of glioma cells with epidermal growth factor receptor mutation type III expression. Int J Cancer. 2005;117(6):905–912. doi: 10.1002/ijc.21251. [DOI] [PubMed] [Google Scholar]
- 13.Chung BM, Dimri M, George M, et al. The role of cooperativity with Src in oncogenic transformation mediated by non-small cell lung cancer-associated EGF receptor mutants. Oncogene. 2009;28(16):1821–1832. doi: 10.1038/onc.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groner B, Lucks P, Borghouts C. The function of Stat3 in tumor cells and their microenvironment. Semin Cell Dev Biol. 2008;19(4):341–350. doi: 10.1016/j.semcdb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Swiatek-Machado K, Kaminska B. STAT signaling in glioma cells. Adv Exp Med Biol. 2013;986:189–208. doi: 10.1007/978-94-007-4719-7_10. [DOI] [PubMed] [Google Scholar]
- 16.Lo HW, Cao X, Zhu H, et al. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res. 2008;14(19):6042–6054. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Iglesia N, Konopka G, Puram SV, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22(4):449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan QW, Cheng CK, Gustafson WC, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24(4):438–449. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moghal N, Sternberg PW. Multiple positive and negative regulators of signaling by the EGF-receptor. Curr Opin Cell Biol. 1999;11(2):190–196. doi: 10.1016/s0955-0674(99)80025-8. [DOI] [PubMed] [Google Scholar]
- 20.Vignais ML, Sadowski HB, Watling D, et al. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol Cell Biol. 1996;16(4):1759–1769. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamauchi T, Ueki K, Tobe K, et al. Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature. 1997;390(6655):91–96. doi: 10.1038/36369. [DOI] [PubMed] [Google Scholar]
- 22.Heimberger AB, Priebe W. Small molecular inhibitors of p-STAT3: novel agents for treatment of primary and metastatic CNS cancers. Recent Pat CNS Drug Discov. 2008;3(3):179–188. doi: 10.2174/157488908786242489. [DOI] [PubMed] [Google Scholar]
- 23.Xing WJ, Zou Y, Han QL, et al. Effects of epidermal growth factor receptor and phosphatase and tensin homologue gene expression on the inhibition of U87MG glioblastoma cell proliferation induced by protein kinase inhibitors. Clin Exp Pharmacol Physiol. 2013;40(1):13–21. doi: 10.1111/1440-1681.12026. [DOI] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Xiong H, Zhang ZG, Tian XQ, et al. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10(3):287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia X, Hu M, Wang C, et al. Coordinated gene expression of Th17- and Treg-associated molecules correlated with resolution of the monophasic experimental autoimmune uveitis. Mol Vis. 2011;17:1493–1507. [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Han L, Zhang K, et al. VHL regulates the effects of miR-23b on glioma survival and invasion via suppression of HIF-1alpha/VEGF and beta-catenin/Tcf-4 signaling. Neuro Oncol. 2012;14(8):1026–1036. doi: 10.1093/neuonc/nos122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding X, He Z, Zhou K, et al. Essential role of TRPC6 channels in G2/M phase transition and development of human glioma. J Natl Cancer Inst. 2010;102(14):1052–1068. doi: 10.1093/jnci/djq217. [DOI] [PubMed] [Google Scholar]
- 29.Levicar N, Nuttall RK, Lah TT. Proteases in brain tumour progression. Acta Neurochir (Wien) 2003;145(9):825–838. doi: 10.1007/s00701-003-0097-z. [DOI] [PubMed] [Google Scholar]
- 30.Park MJ, Kim MS, Park IC, et al. PTEN suppresses hyaluronic acid-induced matrix metalloproteinase-9 expression in U87MG glioblastoma cells through focal adhesion kinase dephosphorylation. Cancer Res. 2002;62(21):6318–6322. [PubMed] [Google Scholar]
- 31.Iwamaru A, Szymanski S, Iwado E, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26(17):2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 32.Wozniak MA, Modzelewska K, Kwong L, et al. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692(2–3):103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18(5):516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Badgwell DB, Lu Z, Le K, et al. The tumor-suppressor gene ARHI (DIRAS3) suppresses ovarian cancer cell migration through inhibition of the Stat3 and FAK/Rho signaling pathways. Oncogene. 2012;31(1):68–79. doi: 10.1038/onc.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weller M, Stupp R, Hegi M, et al. Individualized targeted therapy for glioblastoma: fact or fiction? Cancer J. 2012;18(1):40–44. doi: 10.1097/PPO.0b013e318243f6c9. [DOI] [PubMed] [Google Scholar]
- 36.Mukherjee B, McEllin B, Camacho CV, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69(10):4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Del Vecchio CA, Giacomini CP, Vogel H, et al. EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene. 2013;32(21):2670–2681. doi: 10.1038/onc.2012.280. [DOI] [PubMed] [Google Scholar]
- 38.Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J Endocrinol. 2011;209(2):139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 39.Sieg DJ, Hauck CR, Ilic D, et al. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol. 2000;2(5):249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 40.Silver DL, Naora H, Liu J, et al. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64(10):3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- 41.Stechishin OD, Luchman HA, Ruan Y, et al. On-target JAK2/STAT3 inhibition slows disease progression in orthotopic xenografts of human glioblastoma brain tumor stem cells. Neuro Oncol. 2013;15(2):198–207. doi: 10.1093/neuonc/nos302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Doucette TA, Kong LY, Yang Y, et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012;14(9):1136–1145. doi: 10.1093/neuonc/nos139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colomiere M, Ward AC, Riley C, et al. Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br J Cancer. 2009;100(1):134–144. doi: 10.1038/sj.bjc.6604794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindemann C, Hackmann O, Delic S, et al. SOCS3 promoter methylation is mutually exclusive to EGFR amplification in gliomas and promotes glioma cell invasion through STAT3 and FAK activation. Acta Neuropathol. 2011;122(2):241–251. doi: 10.1007/s00401-011-0832-0. [DOI] [PubMed] [Google Scholar]
- 46.Harada D, Takigawa N, Ochi N, et al. JAK2-related pathway induces acquired erlotinib resistance in lung cancer cells harboring an epidermal growth factor receptor-activating mutation. Cancer Sci. 2012;103(10):1795–1802. doi: 10.1111/j.1349-7006.2012.02363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alvarez JV, Greulich H, Sellers WR, et al. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. 2006;66(6):3162–3168. doi: 10.1158/0008-5472.CAN-05-3757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.