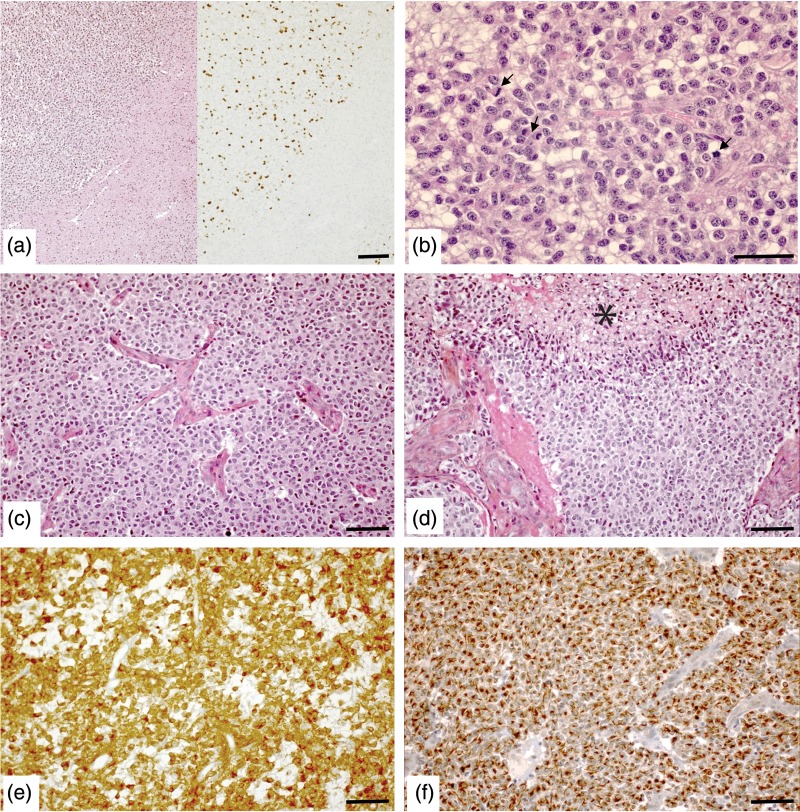
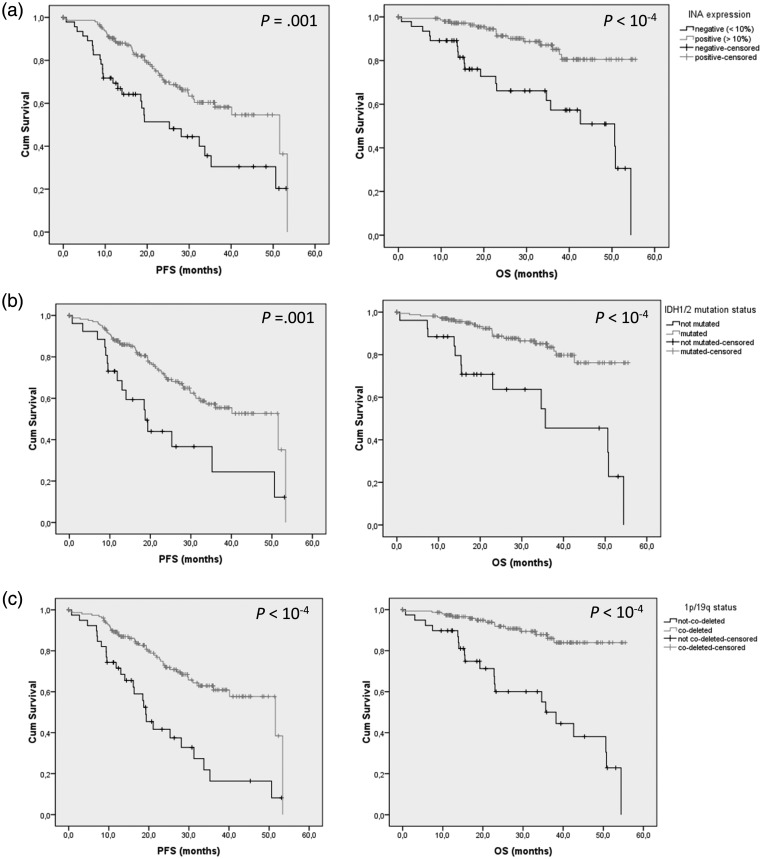
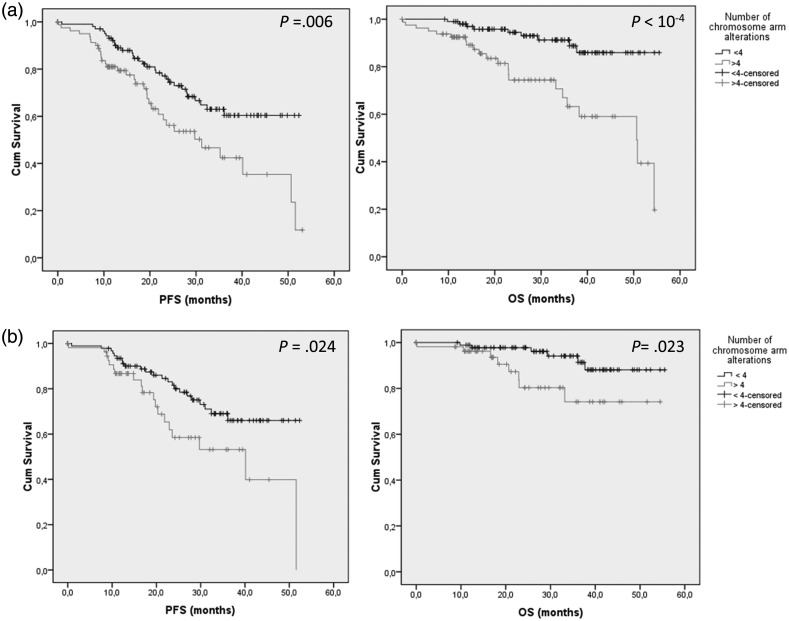
Dominique Figarella-Branger
Dominique Figarella-Branger
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Karima Mokhtari
Karima Mokhtari
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Caroline Dehais
Caroline Dehais
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Anne Jouvet
Anne Jouvet
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Emmanuelle Uro-Coste
Emmanuelle Uro-Coste
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Carole Colin
Carole Colin
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Catherine Carpentier
Catherine Carpentier
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Fabien Forest
Fabien Forest
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Claude-Alain Maurage
Claude-Alain Maurage
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Jean-Michel Vignaud
Jean-Michel Vignaud
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Marc Polivka
Marc Polivka
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Emmanuelle Lechapt-Zalcman
Emmanuelle Lechapt-Zalcman
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Sandrine Eimer
Sandrine Eimer
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Gabriel Viennet
Gabriel Viennet
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Isabelle Quintin-Roué
Isabelle Quintin-Roué
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Marie-Hélène Aubriot-Lorton
Marie-Hélène Aubriot-Lorton
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Marie-Danièle Diebold
Marie-Danièle Diebold
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Delphine Loussouarn
Delphine Loussouarn
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Catherine Lacroix
Catherine Lacroix
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Valérie Rigau
Valérie Rigau
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Annie Laquerrière
Annie Laquerrière
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Fanny Vandenbos
Fanny Vandenbos
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Sophie Michalak
Sophie Michalak
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Henri Sevestre
Henri Sevestre
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Michel Peoch
Michel Peoch
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
François Labrousse
François Labrousse
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Christo Christov
Christo Christov
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Jean-Louis Kemeny
Jean-Louis Kemeny
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Marie-Pierre Chenard
Marie-Pierre Chenard
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Danchristian Chiforeanu
Danchristian Chiforeanu
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
François Ducray
François Ducray
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Ahmed Idbaih
Ahmed Idbaih
1APHM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Aix-Marseille Université, Inserm, CRO2 UMR_S 911, Marseille, France (D.F.-B., C.Co.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuropathologie Raymond Escourolle, Paris, France (K.M., A.I.); Université Pierre et Marie Curie – Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière (CRICM), UMRS 975, Paris, France (K.M., C.Ca., A.I.); Inserm U975, Paris, France (K.M., C.Ca., A.I.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2 - Mazarin, Paris, France (C.D.); Centre de Pathologie et de Neuropathologie Est, Bron, France (A.J.); CHU Toulouse, Hôpital Rangueil, Service d'Anatomie Pathologique et Histologie-Cytologie, Toulouse, France (E.U.-C.); Inserm U1037, Centre de Recherche en Cancérologie de Toulouse, Université de Toulouse, France (E.U.-C.); CHU Saint-Etienne, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Saint-Etienne, France (F.F., M.P.); CHU Lille, Pôle Pathologie Biologique, Service Anatomie Pathologique, Lille, France (C.-A.M.); CHU Nancy, Hôpital Central, Laboratoire d'Anatomie Pathologique, Nancy, France (J.-M.V.); AP-HP, Hôpital Lariboisière, Service d'Anatomie et Cytologie Pathologique, Paris, France (M.P.); CHU Caen, Hôpital de la Côte de Nacre, Service d'Anatomie Pathologique, Caen, France (E.L.-Z.); CNRS, UMR 6301 ISTCT, CERVOxy, GIP CYCERON, Caen, France (E.L.-Z.); CHU Bordeaux, Hôpital Pellegrin, Service de Pathologie – Neuropathologie, Bordeaux, France (S.E.); EA2406, Histologie et Pathologie Moléculaire des Tumeurs, Université Bordeaux Segalen, Bordeaux, France (S.E.); CHU Besançon, Hôpital Jean Minjoz, Service Anatomie et Cytologie Pathologiques, Besançon, France (G.V.); CHU Brest, Hôpital de la Cavale Blanche, Service Anatomie Pathologique, Brest, France (I.Q.-R.); CHU Dijon, Plateau Technique de Biologie G. Mack, Service Anatomie et Cytologie Pathologiques, Dijon, France (M.-H.A.-L.); CHU Reims, Hôpital Robert Debré, Laboratoire d'Anatomie et Cytologie Pathologiques, Reims, France (M.-D.D.); CHU Nantes, Hôpital Laennec, Service d'Anatomie Pathologique B, Nantes, France (D.L.); AP-HP, Hôpital Bicêtre, Service Anatomie et Cytologie Pathologiques, Kremlin-Bicêtre, France (C.L.); CHU Montpellier, Hôpital Gui de Chaulliac, Laboratoire d'Anatomie et Cytologie Pathologiques, Montpellier, France (V.R.); CHU Rouen, Hôpital Charles Nicolle, Laboratoire de Pathologie, Rouen, France (A.L.); CHU Nice, Hôpital Pasteur, Laboratoire d'Anatomie et Cytologie Pathologiques, Nice, France (F.V.); CHU Angers, Département Pathologie Cellulaire et Tissulaire, Angers, France (S.M.); CHU Amiens, Hôpital Nord, Service d'Anatomie et Cytologie Pathologiques, Amiens, France (H.S.); CHU Limoges, Hôpital Dupuytren, Service Anatomie Pathologique, Limoges, France (F.L.); AP-HP, Hôpital Henri Mondor, Service Histologie-Embryologie, Creteil, France (C.Ch.); CHU Clermont-Ferrand, Hôpital Gabriel Montpied, Service d'Anatomie et Cytologie Pathologiques, Clermont-Ferrand, France (J.-L.K.); CHU Strasbourg, Hôpital Hautepierre, Service d'Anatomie Pathologique, Strasbourg, France (M.-P.C.); CHU Rennes, Hôpital Pontchaillou, Service d'Anatomie et Cytologie Pathologiques, Rennes, France (D.C.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-oncologie, Bron, France (F.D.); INSERM U1028, CNRS UMR5292, Bron, France (F.D.)
1,
Christine Desenclos,
Philippe Menei,
Edmond Al Nader,
Joel Godard,
Stéphanie Servagi-Vernat,
Antoine Carpentier,
Hugues Loiseau,
Phong Dam-Hieu,
Jean Sebastien Guillamo,
Evelyne Emery,
Pierre Verelle,
Xavier Durando,
Thierry Faillot,
Caroline Le Guerinel,
François Ghiringhelli,
Fabrice Parker,
Clovis Adam,
François Dubois,
Carole Ramirez,
Edouard Marcel Gueye,
Jerome Honnorat,
Olivier Chinot,
Luc Bauchet,
Patrick Beauchesne,
Mario Campone,
Jean Sébastien Frenel,
Denys Fontaine,
Chantal Campello,
Pascal Roger,
Anne Heitzmann,
Mélanie Fesneau,
Jean Yves Delattre,
Selma Elouadhani-Hamdi,
Damien Ricard,
Philippe Colin,
Elodie Vauléon,
Olivier Langlois,
Marie Janette Motsuo Fotso
Marie Janette Motsuo Fotso
,
Marie Andraud,
Servane Mouton,
Georges Noel,
Nicolas Desse,
Raoulin Soulard,
Elisabeth Cohen-Moyal,
Vincent Lubrano,
Frederic Dhermain,
the POLA Network