Abstract
Background
Prostate cancer is a common malignancy in men, and inevitably some patients experience biochemical recurrence after radical prostatectomy. To date, there are no reliable predictors for prostate cancer recurrence, and novel predictors are urgently needed. PCDH10 (protocadherin-10) is a novel tumor suppressor gene, which is down-regulated by promoter methylation in prostate cancer. The aim of this study was to evaluate the feasibility of using PCDH10 methylation to predict the biochemical recurrence (BCR) of prostate cancer after radical prostatectomy.
Material/Methods
Fresh tissue samples were obtained from 151 patients with primary prostate cancer, and from 34 patients with benign prostatic hyperplasia (BPH) as control. The methylation status of PCDH10 in prostate cancer tissues and controls were examined using methylation-specific PCR (MSP), and then associated with clinicopathological features and BCR-free survival of patients with prostate cancer.
Results
We found that PCDH10 methylation was detected in 79 (52.3%) patients with prostate cancer, but no methylation was found in controls (P<0.0001). Moreover, PCDH10 methylation was significantly associated with higher preoperative prostate-specific antigen (PSA) level (P <0.0001), higher Gleason Score (P<0.0001), advanced clinical stage (P=0.0002), lymph node metastasis (P=0.0389), angiolymphatic invasion (P=0.0303), and biochemical recurrence (P=0.0068). Moreover, PCDH10 methylation was associated with poor BCR-free survival (P<0.0001), and may be used as an independent predictor of BCR-free survival (P=0.0046).
Conclusions
Our results indicate that PCDH10 methylation in prostate cancer tissue is an independent prognostic biomarker of worse BCR-free survival of patients with prostate cancer after radical prostatectomy.
MeSH Keywords: Biological Markers, DNA Methylation, Prostatic Neoplasms
Background
Prostate cancer is one of the most common malignancies in men and the second leading cause of cancer-related death in the United States, with an estimated 233 000 new cases and 29 480 deaths in 2014 [1]. For patients with localized disease, radical prostatectomy is the criterion standard treatment, including laparoscopic, robotic and open retropubic approaches [2]. Despite recent advances in the treatment of localized prostate cancer, some men will experience a recurrence of disease, typically detected by a rise in serum PSA level [3]. The use of PSA screening has lead to the early detection of clinical localized prostate cancer, but to date there are no reliable predictors of prostate cancer recurrence [4]. Accurate prediction of tumor recurrence after radical prostatectomy remains an important problem. The ability to predict disease recurrence at diagnosis is essential to improve therapeutic strategies. Thus, accurate predictive biomarkers are needed.
Prostate cancer is one of the most complicated human malignancies, and the initiation and progression of the disease is associated with genetic and epigenetic alterations [5]. DNA methylation, the most common and best-characterized epigenetic change in human tumors, is an enzyme-induced chemical modification that principally concerns the cytosine molecule, on which a methyl residue is transferred at the 5′ position. This modification mainly occurs in cytosine-guanine dinucleotide-rich areas, known as CpG islands, in the gene promoter regions. DNA methylation of the promoter regions is a major mechanism of inactivation of tumor suppressor genes and can act as a substitute to gene deletion or mutation. The methylation of promoter CPG islands of tumor suppressor genes has been identified in human cancers, including prostate cancer [5]. Aberrant promoter methylation and the associated down-regulation of gene expression are believed to play crucial roles in tumor development and progression [6]. Because aberrant promoter methylation of candidate genes can be measured in tumor tissue or body fluids, this may identify novel biomarkers for early diagnosis or prognosis [7]. In recent years, the association of PCDH10 with human malignancies has been proposed. PCDH10 is a member of the cadherin superfamily. Recent studies have demonstrated that PCDH10 is a putative tumor suppressor gene in several human cancer types, including lymphoid malignancies, colorectal cancer, lung cancer, bladder cancer, and prostate cancer. The down-regulation of PCDH10 expression promotes tumor cell proliferation, migration and invasion [8–14]. Moreover, a recent study demonstrated that PCDH10 is down-regulated in prostate cancer mainly due to promoter methylation, but PCDH10 methylation is not found in normal prostate tissues [14]. These findings led us to investigate the role of PCDH10 methylation in predicting the biochemical recurrence of prostate cancer after radical prostatectomy.
To date, there has been no investigation of the association between PCDH10 methylation and prostate cancer recurrence. In the current study, we investigated the methylation status of PCDH10 in prostate cancer tissues by MSP, and associated our findings with clinicopathological features known to be important for the recurrence of prostate cancer and BCR-free survival of prostate cancer patients after radical prostatectomy.
Material and Methods
Patients and tissue samples
A total of 151 tumor samples were obtained from patients who had been diagnosed pathologically between 2003 and 2006 during open retropubic radical prostatectomy at the Third Hospital of Hebei Medical University. These tissue samples were reviewed on HE-stained tissue sections to confirm diagnosis and tumor content of at least 70% of tumor cells in the tissue samples. The criteria for the enrollment of prostate cancer cases (age range 55–88 years, median 69 years) were histopathologic diagnosis of prostate cancer for the first time, no history of other malignancies, did not receive any anti-cancer therapy, and availability of follow-up information. Patients who received adjuvant or neoadjuvant chemotherapy or hormonal or radiation treatment before tumor recurrence were excluded. The Gleason score were determined by 2 senior pathologists who were experienced in the diagnosis of prostate cancer; they looked at the samples and then discussed the cases. The detailed clinical features of prostate cancer patients are shown in Table 1. The end-point of this trial was the 5-year rate of biochemical recurrence and the time to recurrence for those in whom cancer recurred. BCR-free survival was defined as from the date of surgery to the date of recurrence, and we defined biochemical recurrence after radical prostatectomy as serum PSA >0.2 ng/ml at any stage of postoperative follow-up, as reported previously [2]. Normal prostate tissues were collected as control from 34 patients (age range 52–87 years, median 68 years) with benign prostatic hyperplasia (BPH) during transurethral resection of prostate; these tissues were examined pathologically to exclude the possibility of incidental tumors. The controls were matched with prostate cancer patients by age. All tissue samples were immediately snap-frozen in liquid nitrogen and stored at –80°C for DNA extraction. Written informed consent was obtained from all the patients, and this study was approved by the Ethics Committee of the Third Hospital of Hebei Medical University (No. HMU20030079). All of the tissue samples were handled and made anonymous according to legal and ethics standards.
Table 1.
The main characteristics of patients with prostate cancer, and the relationship between PCDH10 methylation and clinicopathologic parameters (n=151).
| Features | Variables | No. | M (%) | U (%) | P |
|---|---|---|---|---|---|
| Age | <65 | 66 | 31 (47.0) | 35 (53.0) | 0.2463 |
| ≥65 | 85 | 48 (56.5) | 37 (43.5) | ||
| Clinical stage | T1 | 77 | 29 (37.7) | 48 (62.3) | 0.0002 |
| T2/T3 | 74 | 50 (67.6) | 24 (32.4) | ||
| Preoperative PSA | <4 | 5 | 1 (20.0) | 4 (80.0) | <0.0001 |
| 4–10 | 54 | 13 (24.1) | 41 (75.9) | ||
| >10 | 92 | 65 (70.7) | 27 (29.3) | ||
| Gleason score | <7 | 83 | 30 (36.1) | 53 (63.9) | <0.0001 |
| 7 | 29 | 19 (65.5) | 10 (34.5) | ||
| >7 | 39 | 30 (76.9) | 9 (23.1) | ||
| Surgical margin status | Presence | 11 | 6 (54.6) | 5 (45.4) | 0.8779 |
| Absence | 140 | 73 (52.1) | 67 (47.9) | ||
| Seminal vesicle invasion | Presence | 29 | 18 (62.1) | 11 (37.9) | 0.2421 |
| Absence | 122 | 61 (50.0) | 61 (50.0) | ||
| Lymph node metastasis | Presence | 14 | 11 (78.6) | 3 (21.4) | 0.0389 |
| Absence | 137 | 68 (49.6) | 69 (50.4) | ||
| Angiolymphatic invasion | Presence | 30 | 21 (70.0) | 9 (30.0) | 0.0303 |
| Absence | 121 | 58 (47.9) | 63 (52.1) | ||
| Biochemical recurrence | Presence | 43 | 30 (69.8) | 13 (30.2) | 0.0068 |
| Absence | 108 | 49 (45.4) | 59 (54.6) |
M – methylation; U – unmethylation.
Methylation-specific PCR (MSP)
The MSP was performed as in our previous study; genomic DNA was extracted using DNeasy Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instruction. The extracted DNA was treated with bisulfite before MSP using EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s instruction [13]. The following PCDH10 MSP primers were used: methylated: forward 5′-TCGTTAAATAGATACGTTACGC-3′, and reverse 5′-TAAAAACTAAAAACT TTCCGCG-3′; unmethylated: forward 5′-GTTGTTAAATAGATATGTTATGT-3′, and reverse 5′-CTA AAA ACTAAA AACTTTCCACA-3′ (12). In vitro methylated DNA and unmethylated DNA (New England Biolabs, Beverly, MA, USA) were used as methylation and non-methylation positive control. The PCR products were separated in 2% agarose gel, stained with ethidium bromide and visualized under ultraviolet illumination. Samples were scored as methylation-positive when methylated alleles were visualized as bands in the methylated DNA lane or both in methylated and unmethylated DNA lanes, and scored as methylation-negative when bands were seen only in the unmethylated DNA lane [12].
Statistical analysis
Statistical analysis was performed using SAS version 8.0 (SAS Institute, Cary, NC, USA). The data was analyzed systematically. Fisher’s exact test was used to assess the difference of PCDH10 methylation between prostate cancer patients and controls. Chi-square test or Fisher’s exact test was used to assess the relationship between PCDH10 methylation and clinical features. Kaplan-Meier survival analysis and log-rank test were used to assess the difference in BCR-free survival between patients with PCDH10 methylated and unmethylated. Univariate and multivariate Cox proportional hazard model analysis was used to assess the prognostic effect of PCDH10 methylation. A two-sided p value <0.05 was considered statistically significant.
Results
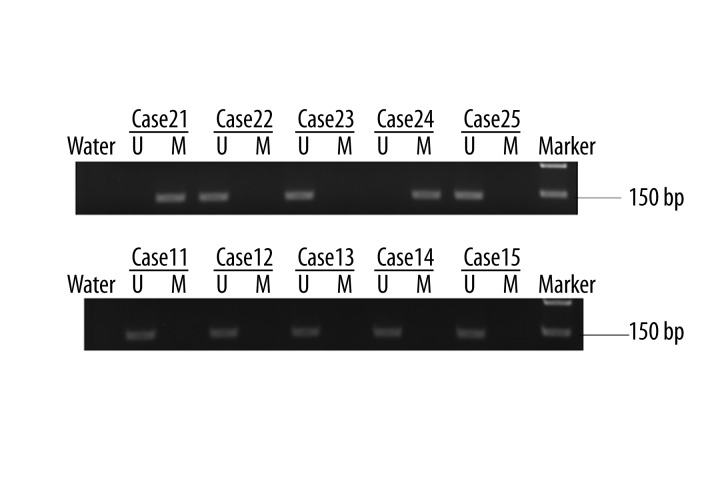
A total of 151 prostate cancer patients and 34 patients with BPH were included in this study. MSP was used to detect the methylation status of PCDH10 in prostate cancer tissues and controls. The promoter methylation of PCDH10 was detected in 79 (52.3%) patients with prostate cancer, but no methylation was found in patients with BPH, and the difference was statistically significant (P<0.0001). The representative MSP results are shown in Figure 1. Subsequently, we investigated the association between PCDH10 methylation and clinicopathological features, in prostate cancer patients only, and the results are shown in Table 1. We found that PCDH10 methylation was significantly associated with the higher preoperative PSA level (P<0.0001), higher Gleason Score (P<0.0001), advanced clinical stage (P=0.0002), lymph node metastasis (P=0.0389), angiolymphatic invasion (P=0.0303), and biochemical recurrence (P=0.0068). Moreover, PCDH10 methylation was associated with poor BCR-free survival (P<0.0001), and could be used as an independent predictor of BCR-free survival (P=0.0046). However, the methylation of PCDH10 was not associated with age, surgical margin status, or seminal vesicle invasion.
Figure 1.
Representative MSP results. U indicates unmethylated gene; M indicates methylated gene. (A) MSP results in patients with prostate cancer, cases 21 and 24 show methylated PCDH10, whereas cases 22, 23, and 25 show unmethylated PCDH10. (B) MSP results in patients with BPH; all cases show unmethylated PCDH10.
To investigate the prognostic value of PCDH10 promoter methylation in prostate cancer, we assessed the association between PCDH10 methylation and BCR-free survival using Kaplan-Meier survival analysis and log-rank test. We found that patients with PCDH10 methylated had shorter BCR-free survival than prostate cancer patients with PCDH10 unmethylated (P<0.0001) (Figure 2). To determine whether PCDH10 methylation has independent prognostic value, univariate and multivariate analysis was performed. The detailed findings are shown in Table 2. The results suggest that PCDH10 methylation is inversely associated with BCR-free survival independently (P=0.0046). In addition, the clinical stage (P=0.0315) and Gleason score (P=0.0132) was also independently associated with BCR-free survival of patients with prostate cancer after radical prostatectomy.
Figure 2.
Kaplan-Meier survival curves for 151 patients with prostate cancer, according to PCDH10 methylation status in tumor tissues. Patients with PCDH10 methylated (n=79) versus PCDH10 unmethylated (n=72), P<0.0001 log-rank test, demonstrating the effect of PCDH10 on the biochemical recurrence-free survival of the patients.
Table 2.
Prognostic value of PCDH10 methylation for the BCR-free survival in univariate and multivariate analysis.
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| PCDH10 methylation | 3.116 | 1.372–6.721 | 0.0003 | 3.044 | 1.132–6.533 | 0.0046 |
| Gleason score | 2.035 | 1.265–7.135 | 0.0017 | 1.933 | 1.087–5.211 | 0.0132 |
| Clinical stage | 1.957 | 1.089–4.536 | 0.0158 | 1.427 | 1.013–3.879 | 0.0315 |
| Surgical margin status | 1.563 | 0.913–2.016 | 0.1325 | |||
| Seminal vesicle invasion | 1.477 | 0.828–1.789 | 0.3527 | |||
| Lymph node metastasis | 1.441 | 0.915–2.036 | 0.3894 | |||
| Angiolymphatic invasion | 1.233 | 0.759–1.632 | 0.4682 | |||
| Preoperative PSA | 1.173 | 0.863–2.945 | 0.5874 | |||
| Age | 1.033 | 0.752–1.874 | 0.677 | |||
HR – hazard ratio.
Discussion
Prostate cancer is a common malignancy in men in the Western world, and the incidence is gradually increasing in Asian countries [15]. Currently, radical prostatectomy is used as potential curative therapy in patients with clinically localized disease. Unfortunately, some men inevitably will experience biochemical recurrence after surgery [16]. Now, a combination of clinical and pathological parameters – especially Gleason score, preoperative PSA, and staging – is used to predicate the recurrence of the disease in general, but the prediction is insufficiently accurate for use with individual patients [17] despite a report by Bryniarski suggesting that preoperative PSA mass is a better predictor of biochemical recurrence after radical prostatectomy than PSA concentration [18]. Given this background, there is an urgent need for biomarkers to predict the recurrence of disease after surgery, and thereby to identify patients at high risk for recurrence, and then determining which patients need early adjuvant therapy after radical prostatectomy.
Aberrant DNA methylation is a characteristic of prostate cancer, and plays an important role in tumor initiation and progression [19]. Although the exact mechanism by which these epigenetic alterations arise in prostate cancer is unknown, this alteration occurs frequently and may be an attractive potential biomarker for prostate cancer [20–23]. To aid in prostate cancer decision-making, biomarkers are needed to distinguish patients with aggressive tumors from patients with clinically insignificant tumors [24,25]. In recent years, some DNA methylation biomarkers have been investigated, such as APC, RASSF1, GSTP1, and PITX2. Especially, the promoter methylation of PITX2 gene is a potential predictor of biochemical recurrence in prostate cancer patients after radical prostatectomy. However, these findings need to be confirmed by future studies, and new biomarkers are needed [26]. In the current study, using MSP, we investigated the methylation status of PCDH10 in prostate cancer tissues and normal prostate tissues and found that PCDH10 methylation occurred frequently in prostate cancer, but the methylation of PCDH10 was not detected in normal prostate tissues. Our findings are in accordance with previous studies on the methylation status of PCDH10 in other genitourinary tumors [12,13]. These findings indicate that PCDH10 methylation may be a useful biomarker in prostate cancer.
To evaluate the prognostic value of PCDH10 methylation for biochemical recurrence of prostate cancer, we analyzed the association between PCDH10 methylation and traditional clinico-pathological characteristics in prostate cancer. We observed that PCDH10 methylation was significantly associated with a higher level of preoperative PAS, higher Gleason score, advanced clinical stage, lymph node metastasis, angiolymphatic invasion, and biochemical recurrence. These factors are commonly considered to be associated with malignant behaviors and poor outcomes of prostate cancer [27–30]. Our results indicate that PCDH10 methylation in tumor specimens may add significant prognostic value to currently used routine predictors. To further clarify the relationship between PCDH10 methylation and biochemical recurrence, we performed Kaplan-Meier survival analysis and log-rank testing. Interestingly, the BCR-free survival of patients with PCDH10 methylated was significantly shorter than that of patients with PCDH10 unmethylated. Moreover, multivariate analysis demonstrated that PCDH10 methylation independently predicts biochemical recurrence in patients with prostate cancer after radical prostatectomy. Overall, the findings of our study suggest that PCDH10 is an independent predictor of biochemical recurrence after radical prostatectomy. Detection of PCDH10 methylation in tumor samples after surgery may help identify patients with high risk of biochemical recurrence. Additional early-stage radiotherapy, chemotherapy, or anti-androgen therapy may be needed to save more lives. Although, future studies are needed before clinical implementation, PCDH10 methylation may add significant value to existing models in predicting the biochemical recurrence of prostate cancer.
Conclusions
Our data indicate that PCDH10 methylation in prostate cancer tissues is a frequent event, and is associated with higher preoperative PAS level, Gleason score, advanced clinical stage, lymph node metastasis, angiolymphatic invasion, and biochemical recurrence. Moreover, PCDH10 methylation independently predicts worse BCR-free survival in patients with prostate cancer after radical prostatectomy
Footnotes
Source of support: Departmental sources
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Mir MC, Joseph B, Zhao R, et al. Effectiveness of epidural versus alternate analgesia for pain relief after radical prostatectomy and correlation with biochemical recurrence in men with prostate cancer. Res Rep Urol. 2013;5:139–45. doi: 10.2147/RRU.S49219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Punnen S, Cooperberg MR, D’Amico AV, et al. Management of biochemical recurrence after primary treatment of prostate cancer: a systematic review of the literature. Eur Urol. 2013;64(6):905–15. doi: 10.1016/j.eururo.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 4.Qin GQ, He HC, Han ZD, et al. Combined overexpression of HIVEP3 and SOX9 predicts unfavorable biochemical recurrence-free survival in patients with prostate cancer. Onco Targets Ther. 2014;7:137–46. doi: 10.2147/OTT.S55432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majumdar S, Buckles E, Estrada J, et al. Aberrant DNA methylation and prostate cancer. Curr Genomics. 2011;12(7):486–505. doi: 10.2174/138920211797904061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasiljević N, Wu K, Brentnall AR, et al. Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. Dis Markers. 2011;30(4):151–61. doi: 10.3233/DMA-2011-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goering W, Kloth M, Schulz WA. DNA methylation changes in prostate cancer. Methods Mol Biol. 2012;863:47–66. doi: 10.1007/978-1-61779-612-8_4. [DOI] [PubMed] [Google Scholar]
- 8.Narayan G, Xie D, Freddy AJ, et al. PCDH10 promoter hypermethylation is frequent in most histologic subtypes of mature lymphoid malignancies and occurs early in lymphomagenesis. Genes Chromosomes Cancer. 2013;52(11):1030–41. doi: 10.1002/gcc.22098. [DOI] [PubMed] [Google Scholar]
- 9.Danese E, Minicozzi AM, et al. Epigenetic alteration: new insights moving from tissue to plasma – the example of PCDH10 promoter methylation in colorectal cancer. Br J Cancer. 2013;109(3):807–13. doi: 10.1038/bjc.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma JG, He ZK, Ma JH, et al. Downregulation of protocadherin-10 expression correlates with malignant behaviour and poor prognosis in human bladder cancer. J Int Med Res. 2013;41(1):38–47. doi: 10.1177/0300060513476989. [DOI] [PubMed] [Google Scholar]
- 11.Tang X, Yin X, Xiang T, et al. Protocadherin 10 is frequently downregulated by promoter methylation and functions as a tumor suppressor gene in non-small cell lung cancer. Cancer Biomark. 2013;12(1):11–19. doi: 10.3233/CBM-2012-00280. [DOI] [PubMed] [Google Scholar]
- 12.Lin YL, Li ZG, He ZK, et al. Clinical and prognostic significance of protocadherin-10 (PCDH10) promoter methylation in bladder cancer. J Int Med Res. 2012;40(6):2117–23. doi: 10.1177/030006051204000609. [DOI] [PubMed] [Google Scholar]
- 13.Lin YL, Li ZG, Guan TY. The clinical significance of PCDH10 promoter methylation in patients with bladder transitional cell carcinoma. Urol Int. 2013;90(2):219–24. doi: 10.1159/000345053. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Li W, Xie J, et al. Epigenetic inactivation of PCDH10 in human prostate cancer cell lines. Cell Biol Int. 2011;35(7):671–76. doi: 10.1042/CBI20100568. [DOI] [PubMed] [Google Scholar]
- 15.Ito K. Prostate cancer in Asian men. Nat Rev Urol. 2014;11(4):197–212. doi: 10.1038/nrurol.2014.42. [DOI] [PubMed] [Google Scholar]
- 16.Cottrell S, Jung K, Kristiansen G, et al. Discovery and validation of 3 novel DNA methylation markers of prostate cancer prognosis. J Urol. 2007;177(5):1753–58. doi: 10.1016/j.juro.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Shariat SF, Scherr DS, Gupta A, et al. Emerging biomarkers for prostate cancer diagnosis, staging, and prognosis. Arch Esp Urol. 2011;64(8):681–94. [PubMed] [Google Scholar]
- 18.Bryniarski P, Paradysz A, Fryczkowski M. PSA mass as a marker of prostate cancer progression after radical prostatectomy. Med Sci Monit. 2011;17(2):CR104–9. doi: 10.12659/MSM.881395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiam K, Ricciardelli C, Bianco-Miotto T. Epigenetic biomarkers in prostate cancer: Current and future uses. Cancer Lett. 2014;342(2):248–56. doi: 10.1016/j.canlet.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Haldrup C, Mundbjerg K, Vestergaard EM, et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J Clin Oncol. 2013;31(26):3250–58. doi: 10.1200/JCO.2012.47.1847. [DOI] [PubMed] [Google Scholar]
- 21.Mahapatra S, Klee EW, Young CY, et al. Global methylation profiling for risk prediction of prostate cancer. Clin Cancer Res. 2012;18(10):2882–95. doi: 10.1158/1078-0432.CCR-11-2090. [DOI] [PubMed] [Google Scholar]
- 22.Chao C, Chi M, Preciado M, et al. Methylation markers for prostate cancer prognosis: a systematic review. Cancer Causes Control. 2013;24(9):1615–41. doi: 10.1007/s10552-013-0249-2. [DOI] [PubMed] [Google Scholar]
- 23.Yu YP, Ding Y, Chen R, et al. Whole-genome methylation sequencing reveals distinct impact of differential methylations on gene transcription in prostate cancer. Am J Pathol. 2013;183(6):1960–70. doi: 10.1016/j.ajpath.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain S, Haidar A, Bloom RE, et al. Bicalutamide-induced hepatotoxicity: A rare adverse effect. Am J Case Rep. 2014;15:266–70. doi: 10.12659/AJCR.890679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho A, Gabriel A, Bhatnagar A, et al. Seasonality pattern of breast, colorectal, and prostate cancer is dependent on latitude. Med Sci Monit. 2014;20:818–24. doi: 10.12659/MSM.890062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietrich D, Meller S, Uhl B, et al. Nucleic acid-based tissue biomarkers of urologic malignancies. Crit Rev Clin Lab Sci. 2014;30:1–27. doi: 10.3109/10408363.2014.906130. [DOI] [PubMed] [Google Scholar]
- 27.Mearini L, Costantini E, Bellezza G, et al. Is there any clinical parameter able to predict prostate cancer after initial diagnosis of atypical small acinar proliferation? Urol Int. 2008;81(1):29–35. doi: 10.1159/000137637. [DOI] [PubMed] [Google Scholar]
- 28.Briganti A, Suardi N, Gallina A, et al. Predicting the risk of bone metastasis in prostate cancer. Cancer Treat Rev. 2014;40(1):3–11. doi: 10.1016/j.ctrv.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 29.DeLancey JO, Wood DP, Jr, He C, et al. Evidence of perineural invasion on prostate biopsy specimen and survival after radical prostatectomy. Urology. 2013;81(2):354–57. doi: 10.1016/j.urology.2012.09.034. [DOI] [PubMed] [Google Scholar]
- 30.Schnoeller T, Jentzmik F, Rinnab L, et al. Circulating free testosterone is an independent predictor of advanced disease in patients with clinically localized prostate cancer. World J Urol. 2013;31(2):253–59. doi: 10.1007/s00345-012-0902-5. [DOI] [PubMed] [Google Scholar]