Phosphorylated RPA recruits repair factors to stalled forks, thereby enhancing fork integrity during replication stress.
Abstract
Phosphorylation of replication protein A (RPA) by Cdk2 and the checkpoint kinase ATR (ATM and Rad3 related) during replication fork stalling stabilizes the replisome, but how these modifications safeguard the fork is not understood. To address this question, we used single-molecule fiber analysis in cells expressing a phosphorylation-defective RPA2 subunit or lacking phosphatase activity toward RPA2. Deregulation of RPA phosphorylation reduced synthesis at forks both during replication stress and recovery from stress. The ability of phosphorylated RPA to stimulate fork recovery is mediated through the PALB2 tumor suppressor protein. RPA phosphorylation increased localization of PALB2 and BRCA2 to RPA-bound nuclear foci in cells experiencing replication stress. Phosphorylated RPA also stimulated recruitment of PALB2 to single-strand deoxyribonucleic acid (DNA) in a cell-free system. Expression of mutant RPA2 or loss of PALB2 expression led to significant DNA damage after replication stress, a defect accentuated by poly-ADP (adenosine diphosphate) ribose polymerase inhibitors. These data demonstrate that phosphorylated RPA recruits repair factors to stalled forks, thereby enhancing fork integrity during replication stress.
Introduction
Stalling of the replication machinery during S phase creates a perilous situation for the cell. Such conditions can instigate subsequent replication fork collapse and thereby induce genomic instability such as copy number variation (Arlt et al., 2011), micronuclei formation (Xu et al., 2011), and loss of heterozygosity (Donahue et al., 2006), leading to an increase in tumorigenesis (Kawabata et al., 2011). Although various factors have been recently found to aid the stabilization of stalled replication forks and/or recovery from stress conditions, including SMARCAL1 (Bansbach et al., 2009; Ciccia et al., 2009; Yuan et al., 2009; Yusufzai et al., 2009), the BLM (Bloom syndrome helicase; Davies et al., 2007), Mus81 (Regairaz et al., 2011), and BRCA2 (Schlacher et al., 2011), mechanistic events remain poorly understood.
A key factor in the response to replication stress is replication protein A (RPA), the primary eukaryotic single-stranded DNA (ssDNA)–binding protein (Oakley and Patrick, 2010). Uncoupling of the replicative MCM (minichromosome maintenance) complex helicase and DNA polymerase complexes during stress causes the formation of persistent or exposed ssDNA that is bound by RPA (Byun et al., 2005). The resulting RPA–ssDNA entity causes the recruitment and activation of the ATR (ATM and Rad3 related) and downstream Chk1 checkpoint kinases. The heterotrimeric RPA itself is targeted for modification by ATR and cyclin A–Cdk on the RPA2 subunit, although fork collapse or DNA double-strand breaks (DSBs) lead to additional RPA2 modification by other phosphoinositide 3-kinase–related kinase (PIKK) family members, namely ATM and DNA-PK (DNA-dependent protein kinase; Oakley and Patrick, 2010). Investigation of the functional roles of RPA phosphorylation have demonstrated its importance for homologous recombination (HR; Lee et al., 2010), exit of damaged cells from mitosis (Anantha et al., 2008; Anantha and Borowiec, 2009), and in response to replication stress, DNA synthesis and cell viability (Vassin et al., 2009). It is perhaps not surprising that whole-genome sequencing of lung tumor samples has recently found a mutation of one of the RPA2 PIKK consensus sites (S33Q34 → S33E34; Govindan et al., 2012), suggestive of a causative effect in tumor progression. Even so, phosphorylation does not appreciably affect the affinity of RPA for ssDNA and has relatively modest effects on replication in vitro using an SV40-based reaction (Brush et al., 1994; Henricksen and Wold, 1994; Pan et al., 1995; Oakley et al., 2003; Patrick et al., 2005). Phosphorylation of RPA also does not alter the initial stages of ATR-mediated checkpoint activation (Vassin et al., 2009). RPA modification occurs at the site of damage, with use of RPA phosphorylation mimics indicating that phosphorylated RPA is prevented from being recruited to normal DNA replication forks (Vassin et al., 2004). Phosphorylated RPA therefore marks sites of DNA damage or stress. It has been postulated that the different RPA phosphorylation species, formed in response to replication stress or DSBs, selectively recruit factors important to respond to the insult. However, the critical protein factors whose interaction with RPA is regulated by phosphorylation, and the mechanistic steps affected, are unclear. Because RPA is a central player in DNA repair and the response to DNA replication stress, identification of such factors can reveal key regulated steps in these processes and provide new therapeutic targets for cancer treatment.
PALB2 (partner and localizer of BRCA2), like BRCA2, is a tumor suppressor (Xia et al., 2007) whose defects lead to heightened incidence of both breast and pancreatic cancers (Rahman et al., 2007; Jones et al., 2009). Both PALB2 (Buisson et al., 2010; Dray et al., 2010) and BRCA2 (Jensen et al., 2010; Liu et al., 2010; Thorslund et al., 2010) act as recombination mediators in which they displace RPA from ssDNA and facilitate formation of Rad51 nucleoprotein filaments, an early step in HR. Although BRCA2 and PALB2 have been demonstrated to function in response to HR-mediated repair of DSBs, BRCA2 has also been recently found to function during replication stress, during which it prevents extensive degradation of the nascent DNA and thus inhibits genomic instability (Schlacher et al., 2011). Thus, it is imperative to further elucidate the roles of PALB2 and BRCA2 during replication stress conditions in which DSBs are not a primary lesion.
Using DNA fiber analysis, we find that deregulation of RPA phosphorylation causes defects in fork progression and inhibits recovery of fork movement after relief of replication stress conditions. The effects on recovery are mediated through the PALB2 tumor suppressor. Along with demonstrating that RPA phosphorylation facilitates replisome progression, both during and after replication stress, thereby facilitating replication fork stability, our results reveal a novel mechanism by which PALB2 and BRCA2 are targeted to sites of DNA damage. Although cancers defective in PALB2 and BRCA2 activities are sensitive to agents that inhibit poly-ADP ribose polymerase (PARP; Bryant et al., 2005; Farmer et al., 2005; Lord and Ashworth, 2008; Buisson et al., 2010), such tumors represent only a small fraction of breast cancers (Koboldt et al., 2012). Our work suggests that chemicals that interfere with RPA–PALB2 complex formation can increase the sensitivity of tumors with normal BRCA1/BRCA2/PALB2 activities to PARP inhibitors.
Results
RPA phosphorylation stimulates DNA synthesis at forks during replication stress
We investigated the role of RPA phosphorylation during the response to replication stress using human cells in which endogenous RPA2 was effectively replaced with one of three RPA2 variants (Anantha et al., 2007, 2008; Vassin et al., 2009; Lee et al., 2010). Along with the wild-type (WT) RPA2 control, two RPA2 phosphorylation mutants were tested. The Cdk_A-RPA2 mutant has Ser to Ala substitutions at the two consensus Cdk sites (S23 and S29), whereas the PIKK_A-RPA2 mutant contains Ala substitutions at the two consensus PIKK sites (T21 and S33; Fig. 1 A). Similar to our previous studies (Anantha et al., 2007, 2008; Vassin et al., 2009; Lee et al., 2010), our replacement approach allows expression of the ectopic RPA2 variant at a level similar to that of the natural subunit in the parental U2-OS cell line and efficient silencing of the endogenous RPA (Fig. S1 A). After replacement with either the Cdk_A- or PIKK_A-RPA2 mutant, RPA2 phosphorylation is defective in response to camptothecin (CPT) treatment (Fig. S1 B).
Figure 1.
RPA phosphorylation stimulates DNA synthesis during stress. (A) RPA2 phosphorylation mutants. The seven known human RPA2 phosphorylation sites are indicated by underlining, with the known or putative kinases shown. WT-RPA2, PIKK_A-RPA2 (double T21A/S33A mutation in the two PIKK sites), and Cdk_A-RPA2 (double S23A/S29A mutation in the two Cdk sites) variants were inducibly expressed from U2-OS stable clones. These three RPA2 variants and the PIKK_D (T21D/S33D mutation) and S4A/S8A mutants were transiently expressed in RPE cells. (B) Schematic of IdU/CldU labeling. (C) Representative fibers labeled with IdU (red) and CldU (green) from cells in which the endogenous RPA2 subunit was replaced with ectopic WT-RPA2, PIKK_A-RPA2, or Cdk_A-RPA2, as indicated. Images were resized to normalize IdU lengths, and the images were sorted so that molecules with the shortest CldU tracts were at the top and longest were at the bottom. For each set of tracts, the white line indicates the position of the IdU–CldU transition. These data indicate that cells replaced with WT-RPA2 have longer CldU tracts compared with cells replaced with PIKK_A- or Cdk_A-RPA2. Bar, 20 µm. (D) RPA2 phosphorylation significantly stimulates fork movement under replication stress conditions but not under unperturbed conditions. Quantitation of fork movement, expressed as the replication fork rate. The fork rate for cells replaced with WT-RPA2 was set at 1.5 kb/min. **, P < 0.01, relative to the fork rate of HU-treated cells replaced with WT RPA. Error bars indicate SEMs. (E) Schematic of [3H]TTP labeling involving either an early or late 90-min labeling period in the presence of 1 mM HU. (F) Mutation of RPA2 PIKK or Cdk phosphorylation sites primarily causes a general slowdown in replication fork movement. Endogenous RPA2 was replaced with ectopic WT-, PIKK_A-, or Cdk_A-RPA2. Two parallel batches of the appropriate replaced cell line were either (early) incubated in HU (15 min) and then HU and [3H]TTP for 90 min or (late) incubated in HU (105 min) and then HU and [3H]TTP for 90 min. The experiment was performed in triplicate. The amount of [3H]TTP incorporated into DNA in the early and late labeling periods was quantitated (see Materials and methods), and the value of [3H]TTP incorporated early/[3H]TTP incorporated late was plotted. The data are expressed as means ± SD.
The effect of phosphorylation site mutation on fork movement during unperturbed and stress conditions was examined using DNA fiber analysis. Cell lines were used in which the ectopic RPA2 variants replaced endogenous RPA2 and synchronized to be primarily in early S phase (Fig. S1 C). Cells were first pulsed with 5′-iodo-2-deoxyuridine (IdU) and then treated with both 5′-chloro-2-deoxyuridine (CldU) and 1 mM hydroxyurea (HU), the latter agent to induce moderate replication stress (Fig. 1 B).
Imaging and quantitation of stretched fibers indicated that the phosphorylation site mutations had no significant effect on the rate of replication fork movement in the absence of HU (Fig. 1, C and D). These data are consistent with past results showing no marked effects of RPA2 phosphorylation site mutations in unperturbed cells (Anantha et al., 2007; Vassin et al., 2009). In contrast, the RPA2 mutations had obvious effects on fork movement during replication stress. In the presence of HU, the fork rate in cells replaced with WT-RPA2 slowed to 0.18 kb/min as compared with that measured in the absence of HU (1.5 kb/min). Fork rates in HU-treated cells replaced with Cdk_A- and PIKK_A-RPA2 were further reduced 3.2-fold (0.057 kb/min) and 3.7-fold (0.049 kb/min), respectively. Although both Cdk and PIKK sites appear to be of similar importance, it has been found previously that mutation of either pair of sites can affect the phosphorylation of the other sites (Fig. S1 B; Anantha et al., 2007; Liu et al., 2012). This phosphorylation cross talk likely contributes to the similar effect of each mutation. Overall, we find that RPA2 phosphorylation at both the Cdk and PIKK sites is necessary to facilitate fork movement during DNA replication stress.
Because past work has demonstrated that RPA phosphorylation is important for the repair of DSBs (Anantha et al., 2007; Lee et al., 2010), we examined whether our HU treatment conditions caused a significant induction of DSBs. We used immunofluorescence microscopy to detect the presence of γ-H2AX, the phosphorylated species of histone variant H2AX that is generated upon DSB formation. Although robust levels of γ-H2AX were formed in the presence of DSB-inducer CPT, HU treatment did not cause a marked increase in γ-H2AX levels above that detected in unperturbed cells, an observation noted for all three RPA2-replaced cell lines (Fig. S2). These data indicate that our HU treatment conditions primarily cause replication stress rather than DSBs.
There are two likely explanations for the shorter tracts in HU-treated cells replaced with either of the two RPA2 phosphorylation site mutants, as compared with cells replaced with WT-RPA2. First, defective RPA phosphorylation could cause forks to stochastically undergo an extended stall as replication stress is prolonged, leading to shortened CldU tracts. Alternatively, defective RPA phosphorylation could cause a general reduction in the overall rate of fork movement under stress conditions. To distinguish between these two possibilities, we performed a [3H]TTP labeling experiment that involved parallel batches of cells being subjected to a 90-min [3H]TTP labeling period at either an “early” or “late” time, in the presence of HU (Fig. 1 E). If the mutant RPA led to an extended stall during replication stress, cells would be expected to incorporate significantly less [3H]TTP in the late labeling period as compared with the early labeling period. In contrast, a general slowing of replication fork movement would be predicted to incorporate similar amounts of [3H]TTP in each of the labeling intervals. We found in fact that cells expressing the three RPA2 variants had similar ratios of late to early incorporation (Fig. 1 F). Similar general results were found using IdU/CldU labeling (Fig. S3, A and B). Although replication forks in the mutant cells likely have a modestly increased probability of undergoing an extended stall during replication stress, it appears that the major effect of RPA2 phosphorylation site mutation is to cause an overall slowing of replication fork movement during stress.
RPA phosphorylation stimulates recovery of fork movement after replication stress
We next examined the role of RPA phosphorylation on the recovery of replication fork movement after release from replication stress. Cells replaced with WT-, PIKK_A-, or Cdk_A-RPA2 were treated with HU for 3 h in the presence of IdU and then allowed to recover for 50 min in fresh media lacking HU but containing CldU (Fig. 2 A). Cells replaced with WT-RPA2 recovered relatively quickly, with the mean fork rate during recovery similar to the fork rate in unperturbed cells (Fig. 2 B). In contrast, both RPA2 mutants were defective in recovering from replication stress. Cells expressing PIKK_A- and Cdk_A-RPA2 had a mean fork rate during recovery that was ∼60 and 50%, respectively, of that seen with the same cells during unperturbed conditions. Similar results were found when cells were examined using [3H]TTP to measure DNA synthesis (Fig. S3, C and D). Both RPA2 mutants also showed a slower recovery from replication stress induced with aphidicolin rather than HU (Fig. 2 C). Overall, these data demonstrate that lack of RPA phosphorylation by either Cdk or ATR causes a defective recovery of replication fork movement after relief of stress conditions.
Figure 2.
Loss of RPA phosphorylation causes defective recovery of DNA replication forks after replication stress. (A) Schematic of fork labeling procedure. Replicating DNA molecules were first labeled with IdU during 3-h incubation under replication stress conditions followed by a 50-min recovery period in which DNA was labeled with CldU. (B) Cells expressing RPA2 mutated at the PIKK or Cdk sites show defective recovery of fork rate after 3-h treatment with 1 mM HU. (C) Defective RPA phosphorylation causes slower fork rate recovery after 3-h treatment with 30 µM aphidicolin (APH). *, P < 0.05; **, P < 0.01, relative to WT-RPA2 values. Error bars indicate SEMs.
Effects of RPA phosphorylation are conserved in a different cell line
The work described in this paper involved U2-OS cells containing an integrated copy of an RPA2 variant. To demonstrate that these results are not an artifact of our cell line, we tested retinal pigment epithelial (RPE) cells using a transient transfection method. Along with testing the WT-, PIKK_A-, and Cdk_A-RPA2 variants, we also tested two other mutants. The first was a PIKK_D mutant in which the two PIKK phosphorylation sites were mutated to phosphomimetic Asp residues (Fig. 1 A), whereas the S4A/S8A-RPA2 variant contained Ala substitutions at the Ser4 and Ser8 positions. Because the S4/S8 sites are primarily phosphorylated under conditions that cause DSBs, mutation of these sites would not be expected to affect fork movement under our replication stress conditions in which DSB formation is low. To better determine the efficiency of replacement, all RPA2 variants were Myc tagged on the C terminus. Past work by the Borowiec laboratory has expressed such RPA2-Myc constructs in human cells, finding that the subunit efficiently forms heterotrimeric RPA and is functional in chromosomal DNA replication, with expression of the WT subunit not having apparent effects on cell cycle progression (Vassin et al., 2004).
Regardless of the mutation, there was efficient replacement of endogenous RPA2 subunit with the ectopic subunit by the transient transfection approach (Fig. 3 A). Testing of each variant using unperturbed conditions (i.e., no HU) found that, with the exception of the PIKK_D mutant, all supported normal rates of fork movement (Fig. 3 B). Cells replaced with the PIKK_D mutant showed a 25% reduction in fork rate in the absence of stress. In the presence of 1 mM HU, cells replaced with the PIKK_A and Cdk_A RPA2 mutants each showed a significant reduction in the rate of fork movement, consistent with the defects in fork progression observed in U2-OS cells (Fig. 1 D). The phosphomimetic PIKK_D mutant also caused a lower rate of rate of fork movement, even though this rate was similar to the other two mutants. In contrast, cells replaced with the S4A/S8A mutant had a modest reduction in fork rate that was not statistically significant. These data indicate that mutation of the PIKK and Cdk sites, but not the N-terminal S4/S8 sites, cause defective replication fork movement during stress. Examining the ability of forks to recover after a 3-h incubation with 1 mM HU, the Cdk variant, and both PIKK mutants showed a ∼50% reduction in fork rates during the 45-min recovery period as compared with WT-RPA2. The S4A/S8A mutant showed an intermediate response. In sum, mutations of the PIKK and Cdk sites, but not the S4/S8 site, caused a reduced ability of forks to progress under replication stress conditions and to recover from this stress, using a different cell line and an alternate replacement method.
Figure 3.
Specific RPA phosphorylation sites are important for replication fork movement during replication stress. (A) Western blot showing efficient replacement of RPA2 by transient transfection. Lysates, prepared 72 h after siRNA transfection, were analyzed by Western blotting for RPA2 and β-actin. For the RPA2 blot, note that both the endogenous (Endo) and Myc-tagged species are seen. Black lines indicate that intervening lanes have been spliced out. (B) An S4A/S8A-RPA2 mutant does not have significant effects on fork movement during replication stress conditions. RPE cells were transiently transfected with the various RPA2 expression constructs and, on the next day, transfected with an siRNA selective for the endogenous RPA2 mRNA. Cells were labeled as shown in the schematic, and fibers were then prepared and imaged. Fork rates are shown in comparison to the rates determined in unperturbed cells replaced with WT-RPA2 (set at 1.5 kb/min). **, P < 0.01, relative to WT-RPA2 value for the same condition. The data are expressed as means ± SEM. (C) The PIKK and Cdk mutants, in contrast to the S4A/S8A mutant, causes defects in fork recovery. RPE cells were replaced with the RPA2 variants and then labeled with CldU and IdU as shown in the schematic. Fork rates were calculated and presented as described for B.
Loss of an RPA phosphatase disrupts fork movement during replication stress
The level of RPA phosphorylation is modulated by the R2–PP4C phosphatase complex (Lee et al., 2010), comprised of the PP4C catalytic and PP4R2 regulatory subunits. Knockdown of the PP4R2 subunit alters the kinetics and pattern of RPA2 phosphorylation, inhibiting HR and increasing sensitivity to DNA damage (Lee et al., 2010). We examined whether the role of R2–PP4C extended to modulating the activity of RPA in response to DNA replication stress. U2-OS cells were treated with an siRNA that significantly reduces PP4R2 protein levels (Fig. 4 A). After treatment with CPT, cells deficient in PP4R2 showed an increase in hyperphosphorylated RPA (Fig. 4 B), demonstrating a loss of PP4 activity toward RPA. Two fiber-labeling experiments were performed to determine the effect of PP4R2 knockdown on fork rates during unperturbed conditions, during HU-induced replication stress, and during recovery from replication stress (Fig. 4 C). Similar to the effects seen with RPA2 phosphorylation site mutation, a reduction in PP4R2 protein levels led to a significant slowing in fork movement both during replication stress (2.5-fold) and the recovery from stress (2.6-fold), relative to control siRNA-treated cells (Fig. 4 D). No effect on fork rates in unperturbed conditions was observed. It should be emphasized that although the RPA2 mutants tested above (i.e., PIKK_A and Cdk_A) reduce the level of RPA phosphorylation, knockdown of PP4R2 increases RPA phosphorylation at early and late times after stress (Lee et al., 2010). These data indicate that both aberrant loss (by RPA2 mutation) and gain (by knockdown of PP4R2; use of the phosphomimetic PIKK_D mutant) of RPA phosphorylation are deleterious to replication fork progression during stress and the recovery from stress.
Figure 4.
Hyperphosphorylation of RPA causes defects in fork movement both during replication stress and the recovery from stress. (A) Western blot analysis showing efficient knockdown of the PP4R2 subunit in U2-OS cells. Cells were transfected with a specific siRNA against PP4R2 or negative (Neg) control siRNA. After transfection (72 h), lysates were prepared and analyzed by Western blotting for PP4R2 or β-actin (loading control). (B) PP4R2 knockdown stimulates RPA phosphorylation. After control or PP4R2 knockdown as in A, cells were treated with 1 µM CPT for various times (as indicated). Cell lysates were then prepared and subjected to Western blot analysis for RPA2. The basal (B; nonphosphorylated) and hyperphosphorylated (H) RPA2 species are indicated. (C) Diagram of the fork labeling procedure used to examine the effect of knockdown of the PP4R2 phosphatase subunit. Two distinct fiber-labeling experiments were performed to examine fork movement during unperturbed conditions and replication stress induced by 1 mM HU treatment (experiment [exp.] 1) and during replication stress and the recovery from stress (experiment 2). (D) Deregulation of RPA phosphorylation causes defects in replisome progression during replication stress and recovery from stress. Fork rates during replication stress (sepia) were the mean rates determined in the first and second experiments (outlined in C). **, P < 0.01, relative to fork rates determined in cells treated with the control siRNA. Error bars indicate SEMs.
RPA phosphorylation facilitates nuclear localization of both PALB2 and BRCA2
PALB2 and BRCA2 are breast cancer tumor suppressors that facilitate repair by HR. Current evidence indicates that a complex of PALB2 and BRCA2 functions as a recombination mediator that catalyzes both the displacement of RPA bound to ssDNA and the coincident loading of the Rad51 recombinase (Buisson et al., 2010; Dray et al., 2010; Jensen et al., 2010; Liu et al., 2010; Thorslund et al., 2010). In addition to its role in HR, BRCA2 has also been found to act during replication stress by protecting nascent DNA from degradation by Mre11 (Schlacher et al., 2011). Although PALB2 and BRCA2 each have recombination mediator activity in the absence of the other, PALB2 also serves to promote the proper intranuclear localization of BRCA2 and thereby the ability of BRCA2 to support HR (Xia et al., 2006). We postulated that the targeting of the PALB2–BRCA2 complex to replication forks is modulated by RPA and in particular the RPA phosphorylation state. We therefore examined the effect of RPA phosphorylation site mutation on nuclear PALB2 foci formation using GFP-PALB2. Control experiments demonstrated nearly complete colocalization of GFP-PALB2 with RPA (Fig. 5 A) and BRCA2 (Fig. S4 A) after treatment with HU, indicating that the GFP tag does not have obvious deleterious effects on PALB2 function. Note that, although a small fraction of the ectopic PALB2 is seen outside of the nucleus, this cytoplasmic localization is dependent on both transfection conditions and cell type and is not always seen (Fig. S4 B). Because the colocalization of PALB2 and RPA could suggest the presence of DSBs at these foci, we performed parallel experiments testing the colocalization of GFP-PALB2 with γ-H2AX. Although colocalization of PALB2 and γ-H2AX was detected after CPT treatment that induces DSBs, only minor γ-H2AX staining was seen under HU treatment or control conditions (Fig. S4 C), again providing evidence that the PALB2 foci formation is not DSB dependent.
Figure 5.
RPA phosphorylation facilitates proper nuclear localization of both PALB2 and BRCA2. (A) RPA and PALB2 show significant colocalization during replication stress. U2-OS cells, transiently transfected with a GFP-PALB2 expression vector, were mock treated or treated with 1 mM HU for 3 h. Cells were then detergent extracted to remove the soluble fraction of the GFP-PALB2 and RPA pools and fixed. After staining for RPA2, cells were imaged for GFP-PALB2, RPA2, and DAPI. (B and C) Mutation of RPA2 phosphorylation sites reduces GFP-PALB2 nuclear retention during replication stress. Cells in which endogenous RPA2 was replaced with ectopic WT-, PIKK_A, or Cdk_A-RPA2 were transfected with a GFP-PALB2 expression vector. Cells were treated with 1 mM HU for 3 h and then imaged as described in A. Images were quantitated using ImageJ (National Institutes of Health), and the relative amount (rel. amt) of PALB2 nuclear staining is shown. Error bars indicate SDs. (D) Mobility of PALB2 under replication stress conditions is significantly higher in cells replaced with either the Cdk_A- or PIKK_A-RPA2 mutant. FRAP analysis of PALB2 mobility is shown, with the data corrected for acquisition bleaching. Prebleach data were used to determine SDs. The calculated t1/2 of recovery is shown below the plot. Each combination of RPA2 variant and condition tested was repeated five to seven times. (E) Mutation of RPA phosphorylation sites causes loss of BRCA2–RPA colocalization during replication stress. U2-OS cells were treated with 1 mM HU for 3 h and then extracted and fixed. Cells were stained for BRCA2, RPA2, and DAPI and imaged. (F) PALB2 and BRCA2 are each in close proximity to RPA in cells, particularly after replication stress. U2-OS cells, either transfected with GFP-PALB2 (two left images) or not transfected (two right images), were either mock treated or incubated with 1 mM HU for 3 h. Cells were analyzed for proximal association of GFP-PALB2 and RPA, or BRCA2 and RPA, using the Duolink immunoassay. In brief, the Duolink assay involves two different DNA-conjugated secondary antibodies that, when adjacent to each other, support formation of a circular DNA molecule that can be amplified by a rolling circle DNA replication. The DNA product is then subsequently detected with a complementary and fluorescently labeled oligonucleotide (Söderberg et al., 2006), giving the observed red dots. Bars: (A) 10 µm; (B, E, and F) 15 µm.
The level of PALB2 nuclear staining was assayed in U2-OS cells replaced with the three RPA2 variants, testing both unperturbed cells (Fig. S5 A) and cells treated with either 1 or 5 mM HU (Fig. 5 B and Fig. S5 B, respectively). Particularly in the presence of HU (Fig. 5 C), cells expressing either the Cdk_A- or PIKK_A-RPA2 mutant were found to have significantly reduced levels of nuclear PALB2 compared with cells replaced with WT-RPA2. Using RPE cells, cells with the PIKK_D mutant also were defective in PALB2 nuclear retention in contrast to the S4A/S8A mutant (Fig. S5 C).
These data suggest that RPA phosphorylation increases the stability of PALB2 binding to chromatin during DNA damage. To test this, we examined the effect of RPA phosphorylation on PALB2 mobility using FRAP. Simply, the rate of recovery of GFP-PALB2 fluorescence in a nuclear region of interest, after photobleaching, is an indicator of PALB2 mobility and, hence, PALB2 binding to other nuclear entities. In the absence of HU, no significant differences in the half-time of GFP-PALB2 fluorescence recovery (t1/2) were found between the three RPA2 variants (Fig. 5 D). Remarkably, the t1/2 of recovery for GFP-PALB2 in cells replaced with PIKK_A- and Cdk_A-RPA2 each decreased ∼50% in the presence of HU compared with unperturbed cells. Only a statistically insignificant difference in GFP-PALB2 fluorescence recovery was noted in cells replaced with WT-RPA2, comparing unperturbed and HU-treated cells. To explain the lack of stress-increased PALB2 mobility in these cells, we speculate that the PALB2 in unstressed cells is present at nuclear storage sites. In the presence of stress, this PALB2 can be released to associate with stalled forks. Unlike cells expressing the mutant RPA2, cells replaced with WT-RPA2 coincidentally have a similar strength of association to the storage sites and stalled forks, meaning a similar degree of PALB2 mobility in unperturbed and stressed cells. Overall, these data demonstrate that defective RPA phosphorylation causes an increase in the mobility of PALB2 selectively under replication stress conditions.
Because PALB2 regulates BRCA2 localization (Xia et al., 2006), we also examined the effect of RPA2 phosphorylation site mutation on BRCA2 localization. Although HU-treated cells replaced with WT-RPA2 showed relatively strong colocalization of BRCA2 and RPA, BRCA2 did not show significant localization with either of the two mutant RPA2 variants (Fig. 5 E). This effect was primarily seen in cells with numerous small RPA foci, indicative of early to mid–S-phase cells (Dimitrova and Gilbert, 2000), as opposed to cells with a low number of large RPA foci that are in late S phase or G2 (unpublished data). To further demonstrate a close (<40 nm) association of RPA with GFP-PALB2 and BRCA2 in cells, we used a proximity ligation assay (Söderberg et al., 2006). Complexes of RPA with GFP-PALB2 were seen in the presence or absence of HU (Fig. 5 F), as suggested by the GFP-PALB2–RPA immunofluorescence analysis (Fig. 5 A). For BRCA2–RPA association, the number of complexes showed a clear increase under replication stress conditions (Fig. 5 F). Although a majority of the detected GFP-PALB2–RPA complexes were apparently nuclear, a subset was detected in the cytoplasm as also seen by immunofluorescence imaging of the two factors (Fig. 5 A). In sum, RPA phosphorylation stimulates the proper nuclear localization of both PALB2 and BRCA2, leading to each of these factors being in close proximity to RPA.
RPA phosphorylation stimulates recruitment of the tumor suppressor PALB2 to ssDNA
We examined the ability of RPA to recruit PALB2 to ssDNA in a cell-free system. A bead-bound dT90 substrate was preloaded with unphosphorylated RPA or RPA phosphorylated on the two RPA2 Cdk sites with cyclin B–Cdc2 (Fig. 6 A). RPA–ssDNA complexes were incubated with extracts of unperturbed U2-OS cells, and the bead-bound material was then subjected to stringent wash conditions (e.g., 200 mM NaCl). PALB2 from unperturbed extracts was observed to be associated with the nonphosphorylated RPA (Fig. 6 B, lane 1). When the same extracts were incubated with phosphorylated RPA, a 4.6-fold increase in PALB2 binding was observed (Fig. 6 B, lane 2). We also tested the effect of HU treatment on PALB2 recruitment by RPA–ssDNA complexes, using extracts from either unperturbed or HU-treated cells (Fig. 6 C). Control extracts demonstrated that PALB2 association to the RPA–ssDNA complexes was again stimulated by RPA phosphorylation (Fig. 6, C [compare lanes 1 and 3] and D). Although a similar 4.2-fold enhancement of PALB2 binding was seen using extracts from HU-treated cells (Fig. 6 C, lanes 2 and 4), these extracts also showed a ∼50% loss of PALB2 binding to the RPA–ssDNA complex compared with extracts of unperturbed cells. Because the data are normalized to the amount of RPA on the beads (Fig. 6 C, bottom), this effect was apparently not caused by components within the HU-treated extracts causing a severe degradation of the ssDNA substrate. The level of PALB2 in each extract was also similar (unpublished data). We speculate that the reduced PALB2 binding seen in extracts from HU-treated cells is caused by factors that modulate the PALB2–RPA interaction or by the stress conditions altering the PALB2 modification state, thereby affecting the association of PALB2 with RPA. Overall, these data indicate that RPA recruits PALB2 to ssDNA, and this recruitment is stimulated by RPA phosphorylation.
Figure 6.
RPA phosphorylation stimulates recruitment of PALB2 to ssDNA. (A) Phosphorylation of RPA by Cdk. RPA bound to a dT90 substrate was either mock treated (lane 1) or incubated (lane 2) with cyclin B–Cdk1. (B) PALB2 and RPA interact in vitro. (left) The relative amounts of PALB2 recovered, corrected for RPA2 levels, are shown under the PALB2 blot. (right) The amount of pS29 phosphorylation declines modestly during incubation with extract. NE, nuclear extract. Black lines indicate that intervening lanes have been spliced out. (C) HU affects PALB2 binding to RPA. The approach was similar to that used in B, with the exception that both nonperturbed and HU-treated extracts were tested. (D) Quantitation of PALB2 binding to nonphosphorylated (RPA) or phosphorylated (pRPA) RPA. rel. amt, relative amount. The data are expressed as means ± SD.
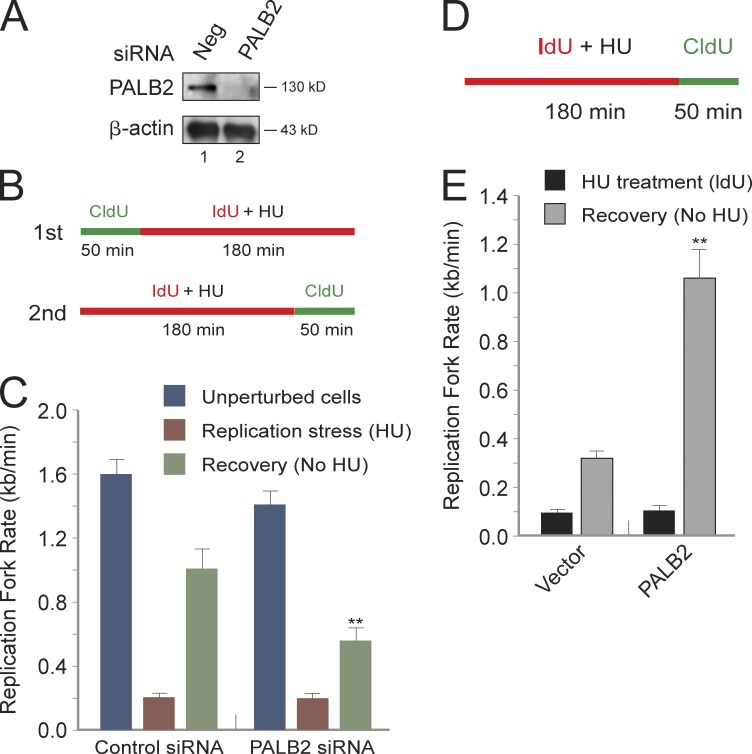
PALB2 selectively stimulates recovery of the DNA replication fork after stress
The findings described in this paper indicate that mutation of RPA phosphorylation sites affects PALB2 chromatin assembly, nuclear localization and dynamics, and association with RPA2 in vitro. These data suggest that PALB2 also affects replication fork movement during stress. Similar to the test of the PP4R2 regulatory subunit, two separate fiber-labeling experiments were performed to examine the effect of siRNA PALB2 knockdown on fork movement in unperturbed U2-OS cells, during HU-induced replication stress and the recovery period after this stress (Fig. 7, A and B). Although knockdown of PALB2 did not cause significant effects on unperturbed fork movement or the rate of fork progression during replication stress, cells deficient in PALB2 had an apparent twofold reduction in fork rate during the 50-min stress recovery period (Fig. 7 C). To provide additional evidence for this effect, we tested the effect of introducing PALB2 into the PALB2-null cell line EUFA1341 on fork progression (Xia et al., 2007). Expression of PALB2 selectively stimulated fork movement approximately threefold during recovery from HU-mediated replication stress (Fig. 7, D and E).
Figure 7.
The loss of PALB2 impedes fork recovery after replication stress. (A) Western blot showing efficient knockdown of PALB2 in U2-OS cells. Cells were transfected with a specific siRNA against PALB2 or negative (Neg) control siRNA. (B) Diagram showing the fork labeling procedure used to examine the effect of knockdown of PALB2 on fork movement. (C) PALB2 knockdown causes significant defects in the recovery from stress. (D) Diagram of the fork labeling procedure used to examine the effect of PALB2 on HU-induced replication stress and recovery in WT- and PALB2-rescued PALB2-null EUFA1341 cells. (E) Recovery of DNA synthesis after stress is stimulated by PALB2. Note that the presence of PALB2 did not affect replication fork rates during replication stress. **, P < 0.01, relative to fork rates determined in control cells. Error bars indicate SEMs.
RPA phosphorylation and PALB2 protect against micronuclei formation after stress
Formation of micronuclei in mammalian cells after stress is an established indicator of genomic instability (Heddle et al., 1991; Fenech et al., 2011). These micronuclei contain chromosome fragments, and the presence of this broken DNA causes them to be detectable by TUNEL staining. We therefore tested the effect of mutation of RPA phosphorylation sites and absence of PALB2 on micronuclei formation after stress. U2-OS cells replaced with WT or Cdk_A-RPA2 were treated with HU for 4 h and allowed to recover for 5 h, and then, micronuclei formation was analyzed (Fig. 8 A). Note that these experiments involve longer treatment conditions compared with those used for DNA fiber analysis. As expected, cells expressing the phosphorylation-defective RPA2 had approximately threefold higher level of cells with TUNEL-positive micronuclei compared with WT-RPA2 (Fig. 8, B and C).
Figure 8.
Cells with defective RPA phosphorylation or lacking PALB2 show increased micronuclei formation as a result of stress. (A) Scheme used to assess DNA damage in U2-OS cells expressing an RPA phosphorylation mutant on recovery from HU treatment. Cells were either exposed to 5 mM HU for 4 h and then allowed to recover for 5 h (i), treated with the PARP inhibitor veliparib (PARPi) for 25 h (ii), or treated overnight with veliparib followed by 4-h HU treatment and 5-h recovery phases (iii), also in the presence of veliparib. (B) TUNEL staining in U2-OS cells in which endogenous RPA2 was replaced with WT-RPA2 or Cdk_A-RPA2. Cells are shown after recovery from HU treatment or after recovery from HU treatment in the presence of veliparib (as indicated). Cells were stained with DAPI (blue) and processed using a TUNEL assay involving fluorescein-12-dUTP incorporation (green). Micronuclei are evident as green bodies found at the outer edges of nuclei. To increase the number of cells shown, the image of cells expressing WT-RPA2 and recovering from HU/PARP inhibitor treatment is a composite. Below the main images is an enlarged image showing micronuclei formation observed in HU/PARP inhibitor–treated cells replaced with Cdk_A-RPA2. Bars: (main images) 40 µm; (enlarged images) 15 µm. (C) Mutation of RPA2 phosphorylation sites increases micronuclei formation after replication stress. The TUNEL-positive micronuclei signal of U2-OS cells replaced with WT-, Cdk_A-, or PIKK_A-RPA2 showing TUNEL-positive micronuclei formation in unperturbed cells or cells treated with veliparib alone for 25 h, 5 h after a 4-h HU treatment, and 5 h after 4-h HU treatment in the presence of veliparib are shown. Mutation of Cdk sites, and to a lesser extent PIKK sites, on RPA2 causes an increase in micronuclei formation after HU/PARP inhibitor treatment. **, P < 0.01, relative to micronuclei levels determined in WT-RPA cells for the similar condition. (D) PALB2 protects against micronuclei formation during recovery from replication stress. The TUNEL-positive micronuclei signals of U2-OS cells treated with siRNA to knockdown PALB2 or control siRNA (Neg., negative) and treated with HU and veliparib, as indicated, are shown. The data are expressed as means ± SD.
Deleterious mutations in BRCA1, BRCA2, or PALB2 cause defective HR with BRCA1 also functioning in DNA interstrand cross-link repair (Bunting et al., 2012). Cells expressing mutants in any of these three genes can be ∼1,000-fold more sensitive to inhibition of PARP, an enzyme that acts in an alternate DNA repair pathway (Bryant et al., 2005; Farmer et al., 2005; Lord and Ashworth, 2008; Buisson et al., 2010). We examined the effect of the PARP inhibitor veliparib in cells replaced with the RPA2 variants (Fig. 8 A, condition iii). Cells replaced with WT-RPA2 and treated with veliparib in addition to HU showed only a minor amount of micronuclei formation during the recovery from HU treatment (Fig. 8, B and C). In contrast, veliparib treatment of cells replaced with Cdk_A-RPA2 showed a dramatic increase in micronuclei formation, both in terms of the fraction of cells with micronuclei and the intensity of TUNEL staining in these bodies, indicative of a high level of genomic instability during recovery from HU treatment (Fig. 8 B). Similar results were observed when testing the PIKK_A-RPA2 mutant (Fig. 8 C).
We next tested cells in which PALB2 was knocked down by siRNA treatment. Similar to the effect of mutating RPA2 phosphorylation sites, knockdown of PALB2 caused an increase in micronuclei formation after stress (Fig. 8 D). Combined with the observed association between RPA and PALB2 (Fig. 6, B and C), the similar effects of PALB2 knockdown and defective RPA2 phosphorylation on micronuclei formation demonstrate that PALB2 and RPA cooperate to protect genomic DNA from instability in response to DNA replication stress.
Discussion
The functional significance of RPA phosphorylation has only recently become apparent. RPA phosphorylation has been found to facilitate HR after induction of DSBs (Lee et al., 2010), to facilitate exit from a damaged mitosis (Anantha et al., 2008), and to increase cell viability and DNA synthesis during replication stress (Vassin et al., 2009). Even so, the mechanistic events and molecular players through which RPA phosphorylation mediates its effects have remained elusive. We make the novel finding that phosphorylated RPA recruits DNA repair factors (PALB2 and BRCA2) to sites of DNA damage or stress. RPA phosphorylation stimulates replication fork movement during stress and recovery from stress, with both hypophosphorylation and hyperphosphorylation causing defects in fork progression. Our data demonstrate that these effects of RPA phosphorylation on replisome recovery are mediated by recruitment of the PALB2 and BRCA2 tumor suppressors to sites of DNA replication stress, linking these three factors in protecting the cell against genomic instability.
PALB2 and BRCA2 act as recombination mediators that displace RPA bound to ssDNA and simultaneously support binding of Rad51 (Buisson et al., 2010; Dray et al., 2010; Jensen et al., 2010; Liu et al., 2010; Thorslund et al., 2010). Our findings elucidate why PALB2 and BRCA2 are targeted to sites of replication stress but not unperturbed replication forks. RPA is normally unphosphorylated or hypophosphorylated in unperturbed interphase cells, and phosphorylated RPA is poorly recruited to unperturbed DNA replication forks (Vassin et al., 2004). Upon fork stalling, the action of ATR and Cdk on the bound RPA marks the replisome as being anomalous, and our data indicate that a major effect of RPA phosphorylation is to stimulate recruitment of PALB2 and BRCA2 to sites of replication stress. We are currently characterizing RPA–PALB2 complex formation in vitro to understand the conditions causing PALB2 to act as a recombination mediator that displaces RPA and loads Rad51 onto DNA (Buisson et al., 2010; Dray et al., 2010).
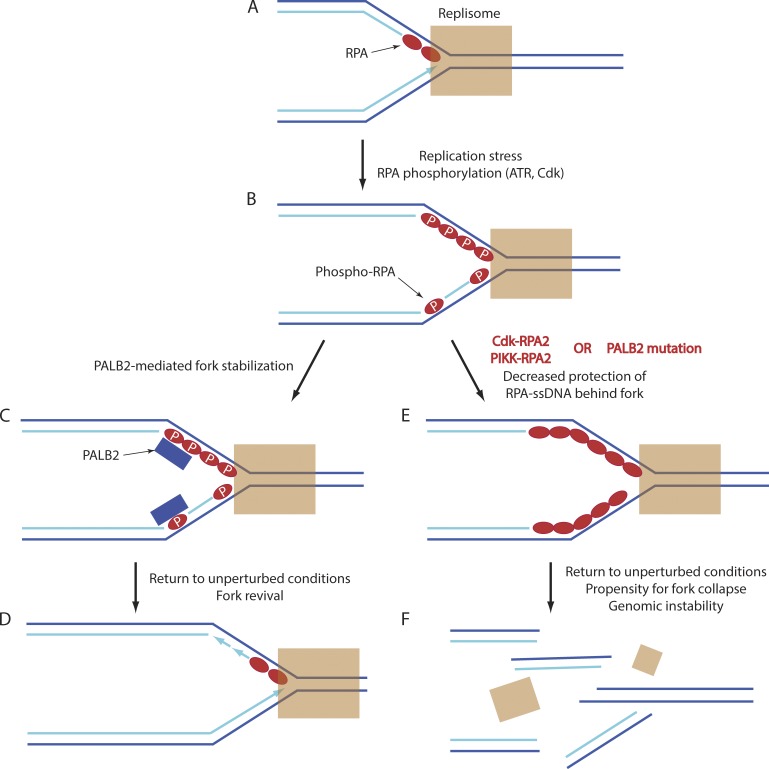
Our data suggest a model connecting PALB2 with phosphorylated RPA (Fig. 9). Replication fork stalling leads to uncoupling of the replicative helicase and DNA polymerase complexes, the generation of persistent or exposed ssDNA bound by RPA (Nam and Cortez, 2011), and RPA phosphorylation by ATR and Cdk (Anantha et al., 2007; Vassin et al., 2009). The phosphorylated RPA recruits the PALB2–BRCA2 complex for fork stabilization. Upon alleviation of stress conditions, the loss of RPA phosphorylation by the R2–PP4C phosphatase complex (Lee et al., 2010) and other phosphatases facilitates the loss of PALB2–BRCA2 binding and the revival of replication fork movement. If RPA phosphorylation is deregulated by RPA2 mutation or by the absence of R2–PP4C activity, the reduction in DNA synthesis during stress (this work; Vassin et al., 2009) leads to the generation of abnormally long lengths of ssDNA and reduced PALB2 association with stalled forks. With the mitigation of DNA stress, these defects cause an increase in fork collapse and the formation of micronuclei.
Figure 9.
Model indicating the protective effect of RPA phosphorylation and PALB2 on fork stability during replication stress. (A) An unperturbed replication fork has nonphosphorylated RPA bound to the lagging strand template, with this RPA turned over rapidly during fork movement. (B) Replication fork stalling and consequent helicase-DNA polymerase uncoupling (not depicted) cause the generation of persistent ssDNA on both the leading and lagging strand templates that is stably bound by RPA. This RPA becomes phosphorylated on RPA2 by ATR and Cdk. (C) Phosphorylated RPA recruits PALB2 (and likely BRCA2) to the fork, stabilizing the stalled fork complex. (D) Alleviation of stress conditions leads to RPA dephosphorylation, reducing PALB2 binding to the RPA–-ssDNA complex. The previous binding of PALB2 to the replisome facilitates a rapid revival of fork movement. (E) Deregulated RPA phosphorylation or loss of PALB2 causes reduced protection of the RPA–ssDNA complex. (F) This defective protection is evinced upon the return to nonstress conditions because recovery of fork movement is diminished, and forks are more prone to collapse. P, phosphorylation.
Although RPA phosphorylation has effects on fork movement during replication stress and on recovery from stress, we only observe involvement of PALB2 in the recovery phase. These data indicate that phosphorylated RPA stimulates the recruitment of factors other than PALB2 to facilitate an increase in the fork rate during replication stress. RPA phosphorylation has been shown to modulate the association with various factors including Rad51, Rad52, ATM, DNA-PK, and DNA-dependent protein kinase, catalytic subunit (Oakley et al., 2003; Patrick et al., 2005; Wu et al., 2005; Lee et al., 2010). A regulatory role for phosphorylation on the Rad51–RPA interaction has been shown, with expression of a hyperphosphorylated RPA mimic inappropriately sequestering soluble RPA–Rad51 from chromatin (Lee et al., 2010). Nevertheless, the functional impact of many of these interactions has remained elusive.
As shown previously for HR (Lee et al., 2010), phosphorylation of RPA does not appear to be merely a signal to increase the activity of RPA under cell stress conditions. Instead, we find that both aberrant loss (by mutation of RPA2 phosphorylation sites) and gain (by knockdown of the R2 subunit of the PP4 RPA phosphatase; use of an RPA2 phosphomimetic) of RPA phosphorylation cause defects in replication fork movement during stress and during recovery from stress. These data suggest that an active cycle of RPA phosphorylation and dephosphorylation is needed for proper maintenance of replication fork movement during stress. More generally, our findings have clinical significance. Cancers with defective RPA phosphorylation (e.g., Govindan et al., 2012) would be expected to be sensitive to PARP inhibitors. Furthermore, although PARP inhibitors have shown promise against breast cancers with BRCA1/2 mutations (Tutt et al., 2010), a recent test of a large number of primary breast tumors by a comprehensive genomic analysis found that only 2–3% of luminal/ER+ cancers, the most numerous tumor type, had mutations in BRCA1 or BRCA2 genes (Koboldt et al., 2012). Our study suggests that therapeutic targeting of the RPA–PALB2 interaction may provide a route to sensitize tumors lacking BRCA1, BRCA2, or PALB2 mutations to PARP inhibitors.
Materials and methods
Cell culture, antibodies, and reagents
U2-OS cells were grown in McCoy’s medium supplemented with 10% (vol/vol) FBS. Clonal U2-OS cell lines allowing inducible expression of WT-, PIKK_A-, and Cdk_A-RPA2 were generated by infection with the pRetro-Off retroviral vector (Takara Bio Inc.) containing an integrated copy of the appropriate RPA2 variant (Anantha et al., 2007; Vassin et al., 2009). Cells were maintained in complete McCoy’s medium containing 1 µg/ml doxycycline. PALB2-null EUFA1341 fibroblasts, derived from an individual with Fanconi anemia (Xia et al., 2007), were made available for our use by H. Joenje (VU University Medical Center, Amsterdam, Netherlands) and provided by P. Andreassen (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH). The EUFA1341 cells were grown in a mixture containing 50% DMEM and 50% F10 media (vol/vol) and supplemented with 10% (vol/vol) FBS.
Primary antibodies used were rabbit polyclonal anti-PALB2 (Bethyl Laboratories, Inc./Sigma-Aldrich), rabbit polyclonal (Bethyl Laboratories, Inc./Thermo Fisher Scientific), mouse monoclonal (Thermo Fisher Scientific) anti-RPA2, rabbit polyclonal anti–pS4/pS8- and anti–pS33-RPA2 (Bethyl Laboratories, Inc.), rabbit polyclonal anti–β-actin (Abcam), mouse monoclonal anti–γ-H2AX (EMD Millipore/Abcam), rabbit polyclonal anti-PP4R2 (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-GFP (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-BRCA2 (Santa Cruz Biotechnology, Inc.), anti-CldU (rat anti-BrdU obtained from Accurate Chemical & Scientific Corporation and Abcam), and mouse monoclonal anti-IdU (BD and Sigma-Aldrich). The rabbit polyclonal anti–pS29-RPA2 antibody was custom synthesized by Bethyl Laboratories, Inc. using a CSPGGFGpSPAPSQ phosphopeptide (Anantha et al., 2007). Secondary antibodies were Alexa Fluor 488 goat anti–mouse, Alexa Fluor 546 goat anti–rabbit, Texas red goat anti–mouse, Alexa Fluor 594 goat anti–rat (Invitrogen), FITC donkey anti–rat, FITC donkey anti–rabbit, and Cy5 donkey anti–rabbit (Jackson ImmunoResearch Laboratories, Inc.). HU, aphidicolin, and CPT were purchased from Sigma-Aldrich. CldU and IdU were purchased from MP Biomedicals, and veliparib was obtained from Selleck Chemicals.
Microscopy
For fiber experiments, images were acquired at ambient temperature with epifluorescent illumination using a microscope (Axiophot; Carl Zeiss; 63× Plan Neofluar, 1.25 NA oil immersion objective) and a camera (AxioCam HR; Carl Zeiss), using AxioVision (v4.8) acquisition software (Carl Zeiss). For cell immunofluorescence and the proximity ligation assay, images were similarly captured using a microscope (Axiophot; 20× Plan Neofluar, 0.50 NA; 40× Plan Neofluar, 0.75 NA; 63× Plan Neofluar, 1.25 NA oil immersion; and 100× Plan Apochromat, 1.4 NA oil immersion) at room temperature. Images were analyzed using Illustrator CS4 (Adobe) or Photoshop CS4 (Adobe). For FRAP experiments, bleaching and acquisition was performed using a confocal microscope (LSM 510; Carl Zeiss) with a 40× Plan Neofluar, 1.3 NA oil immersion objective, using LSM (v4.2) acquisition software (Carl Zeiss). The dimensions of the nuclear bleach region were identical for all imaged cells. Five images were taken before bleaching, after which images were acquired every ∼200 ms for 300 frames. Data were analyzed using Volocity (PerkinElmer), with acquisition bleaching corrected using Volocity tools.
RPA2 replacement and protein knockdown
Endogenous RPA2 was replaced with ectopic variants and silenced as previously described (Anantha et al., 2007). In brief, the desired U2-OS clone was grown for 24 h in medium lacking doxycycline to allow expression of the ectopic RPA2. Knockdown of endogenous RPA2 was accomplished by siRNA transfection using HiPerFect (QIAGEN), with cells tested 72 h after transfection. Custom-synthesized siRNAs (GE Healthcare) used were RPA2, sense 5′-AACCUAGUUUCACAAUCUGUU-3′ and antisense 5′-CAGAUUGUGAAACUAGGUUUU-3′ (targeting the 3′-untranslated region); and PP4R2, sense 5′-UAUACUGAGAGGUCUAAUAUU-3′ and antisense 5′-UAUUAGACCUCUCAGUAUAUU-3′. For RPE cells, cells were first transfected with the siRNA targeting endogenous RPA2 and, 24 h later, transiently transfected with the RPA2 expression plasmids. The RPA2 variants, synthesized by GenScript, were expressed with a C-terminal Myc tag from the pEF6/Myc expression plasmid (Vassin et al., 2004). Cells were analyzed 48 h after DNA transfection. PALB2-null EUFA1341 cells were rescued for PALB2 by transfecting with a PALB2 expression plasmid. Commercial reagents for the siRNA against PALB2 (catalog no. sc-93396; Santa Cruz Biotechnology, Inc.) and control siRNA (catalog no. 1027280; AllStars Negative Control siRNA; QIAGEN) were used. DNA plasmids were transfected using Effectene (QIAGEN), whereas siRNA transfection used HiPerFect.
DNA fiber analysis
DNA single molecule experiments for U2-OS cells were performed essentially as previously described (Norio and Schildkraut, 2001; Guan et al., 2009). In brief, cells were synchronized with nocodazole for 12 h, and mitotic cells were collected by mechanical shaking and then grown in fresh medium for an additional 12 h to allow cells to reach S phase. Synchronization of cells for DNA fiber experiments was verified by flow cytometry performed on a flow cytometer (FACSCalibur; BD). Cells were pulsed with 30 µM IdU and 30 µM CldU for the times indicated. As needed, 1 mM HU was added to the medium to induce replication stress. To detect incorporated IdU and CldU, DNA was stretched on silanized slides, blocked with 3% (wt/vol) BSA, and immunostained with primary and secondary antibodies. For RPE cells, cells were labeled using 50 µM IdU and 50 µM CldU for the indicated times, and the fiber spreading technique described by Terret et al. (2009) was used. In brief, aliquots of the cells on slides were lysed with 200 mM Tris-HCl, pH 7.4, 0.5% (wt/vol) SDS, and 50 mM EDTA, and the slides were tilted to a 15° angle for 30–60 s to stretch the DNA fibers. Fibers were then stained as described for U2-OS cells, and images were captured.
Immunofluorescence
To detect PALB2, GFP-PALB2, RPA2, γ-H2AX, and BRCA2 foci, cells were first extracted with cytoskeletal buffer (10 mM Hepes-KOH, pH 7.4, 300 mM sucrose, 100 mM NaCl, and 3 mM MgCl2) containing 0.5% Triton X-100 for 2 min on ice. Cells were then fixed with 4% (wt/vol) paraformaldehyde in PBS for 45 min and permeabilized with PBS containing 0.5% NP-40 for 2 min. After blocking in 1% (wt/vol) nonfat dry milk and 0.1% Tween 20 in PBS, cells were stained as needed with primary and secondary antibodies in blocking solution. For quantitation of nuclear PALB2 staining, images of DAPI-stained cells were first analyzed by Volocity software (v5.5) to identify nuclei. The GFP intensities in identified nuclei were then determined. For PALB2 staining in RPE cells, we added the additional criteria of using only RPA-positive cells (i.e., cells in S phase) for measurement of GFP signals.
Proximity ligation assay
Detection of closely associated RPA and PALB2, or RPA and BRCA2, used the Duolink In Situ kit (Sigma-Aldrich). For RPA–PALB2 detection, U2-OS cells were transfected with GFP-PALB2 using the Effectene Transfection Reagent (QIAGEN). At 24 h after transfection, cells were treated to cause replication stress, extracted with cytoskeletal buffer, and fixed as described in the previous paragraph. Cells were then stained with primary antibodies against GFP and RPA2 for 1 h at 37°C. For RPA/BRCA2 detection, untransfected U2-OS cells were tested, using primary antibodies against RPA2 and BRCA2. The cells were then treated with proximity ligation assay probe MINUS and PLUS secondary antibodies (Duolink) as described using the manufacturer’s instructions. Cells were stained with DAPI and visualized by epifluorescence microscopy.
FRAP
Inducible RPA2 U2-OS clones were cultured in McCoy’s 5A Iwakata & Grace Modification (Cellgro) supplemented with 10% FBS. Clones were silenced for endogenous RPA2 after ectopic RPA2 induction (24 h) as described in the RPA2 replacement and protein knockdown paragraph. After silencing (24 h), cells were transfected with PALB2-GFP using Effectene Transfection kit (24 h) and then treated with 100 ng/ml of nocodazole for 12 h. Mitotic cells were shaken off and replated on 35-mm uncoated glass-bottom cell culture dishes (MatTek Corporation). The fluorescence bleaching experiments commenced 14 h later, when the cell population reached S phase (Fig. S1).
Tritiated thymidine incorporation
For cell labeling, 2 µCi [3H]TTP (80 Ci/mmol; PerkinElmer) was added to asynchronous cells in 2 ml of medium in the presence or absence of HU. After labeling, cells were washed with ice-cold PBS, subjected to trypsin-EDTA treatment for 30 min on ice, and then incubated with 20% TCA on ice for 30 min. Precipitates were collected, and radioactivity was quantitated. Each labeling condition was repeated in triplicate.
PALB2–RPA interaction in vitro
Human RPA was expressed in Escherichia coli strain C43(DE3) (Lucigen Corporation) from the p11d-tRPA plasmid (a gift of M. Wold, University of Iowa, Iowa City, IA) and purified as previously described using Affi-Gel blue and Mono-Q resins (Henricksen et al., 1994). For interaction experiments, 200 nM RPA was incubated for 1 h at 37°C with 150 pM 5′-biotin–tagged dT90 ssDNA substrate (Gene Link) in 50 µl of 40-mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, and 50 µM ATP. The high ratio of RPA to ssDNA substrate allows complete saturation of the DNA template under these binding conditions. To produce phosphorylated RPA, 230 pg Cdk1–cyclin B (a gift of J. Gautier, Columbia University Medical Center, New York, NY) was added to the reaction. After incubation, the reaction mixture was combined with 50 µl streptavidin–agarose bead slurry (Sigma-Aldrich), and the excess RPA and Cdk1–cyclin B complex was removed by washing beads with 20× slurry volume of RPA binding buffer (10 mM Tris-HCl, pH 7.5, 200 mM NaCl, 10% [vol/vol] glycerol, 0.2 mM EDTA, and 0.1 mM DTT). For extract preparation, either unperturbed U2-OS cells or HU-treated cells (5 mM HU for 2 h) were used. Cells were harvested, and nuclear extracts were prepared as previously described (Dignam et al., 1983). In brief, cells in hypotonic buffer were lysed with a Dounce homogenizer, and the nuclei were pelleted and then extracted with a moderate salt buffer. Nuclear extract (16 µg) was added to the bead-bound RPA–ssDNA complexes in 1 ml RPA binding buffer, and the reaction was rocked at 4°C for 1 h. After washing the beads three times with 1 ml RPA binding buffer, the bead-bound material was released by the addition of SDS-loading buffer and then analyzed by 12% PAGE and Western blotting.
Micronuclei formation assay
U2-OS cells were either (1) treated with 10 µM veliparib (ABT-888) for 25 h, (2) treated with 5 mM HU for 4 h and allowed to recover in media lacking HU for 5 h, or (3) treated with 10 µM veliparib (ABT-888) for 16 h and then incubated with 5 mM HU for 4 h and allowed to recover for 5 h. The latter two steps of condition 3 also took place in the presence of veliparib. Cells were then immediately fixed with 4% (wt/vol) formaldehyde for 30 min on ice and subjected to TUNEL staining using a TUNEL assay (DeadEnd Fluorometric TUNEL System; Promega), as per the manufacturer’s instructions.
Flow cytometry
To analyze cell cycle distribution, cells were trypsinized and fixed with 70% ethanol. Cells were then stained with PBS containing 0.02% (wt/vol) propidium iodide (Sigma-Aldrich), 0.1% (vol/vol) Triton X-100, and 200 µg/ml RNase A. FACS was performed on a flow cytometer (FACSCalibur) by using CellQuest software (Flow Cytometry and Cell Sorting Center, New York University Langone Medical Center; BD). Cell cycle analysis was performed using the FlowJo software (Tree Star).
Online supplemental material
Fig. S1 shows the efficient replacement of RPA2 using U2-OS clones, demonstrating that cells replaced with either of the two RPA2 mutants have defective RPA2 phosphorylation. Fig. S2 demonstrates that the mild replication stress conditions used in our study do not induce significant DSB formation in replaced U2-OS cells. Fig. S3 provides evidence that reduction of RPA phosphorylation slows rather than stops replication fork movement during replication stress and that cells replaced with mutant RPA2 are defective in recovery from HU treatment. Fig. S4 shows that PALB2 colocalizes with BRCA2 and RPA but not γ-H2AX during replication stress. Fig. S5 demonstrates that specific RPA2 phosphorylation sites are more important for nuclear PALB2 retention. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201404111/DC1.
Supplementary Material
Acknowledgments
We thank M. Wold, H. Joenje, P. Andreassen, and J. Gautier for reagents and J. Chen and T. Huang for assistance with the single DNA molecule experiments involving the tilt method.
This work was supported by National Institutes of Health grant R01 GM083185 (J.A. Borowiec), an Exceptional Project Award Grant from the Breast Cancer Alliance (J.A. Borowiec), National Cancer Institute grant P30CA16087 (J.A. Borowiec), National Institutes of Health grant R01CA142698 (D. Chowdhury), a Basic Scholar Grant from the American Cancer Society (D. Chowdhury), National Institutes of Health grant 5R01 GM045751 (C.L. Schildkraut), and the Empire State Stem Cell Fund through New York State contract C024348 (C.L. Schildkraut).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CldU
- 5′-chloro-2-deoxyuridine
- CPT
- camptothecin
- DSB
- DNA double-strand break
- HR
- homologous recombination
- HU
- hydroxyurea
- IdU
- 5′-iodo-2-deoxyuridine
- PARP
- poly-ADP ribose polymerase
- PIKK
- phosphoinositide 3-kinase–related kinase
- RPA
- replication protein A
- RPE
- retinal pigment epithelial
- ssDNA
- single-stranded DNA
- WT
- wild type
References
- Anantha, R.W., and Borowiec J.A.. 2009. Mitotic crisis: the unmasking of a novel role for RPA. Cell Cycle. 8:357–361 10.4161/cc.8.3.7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantha, R.W., Vassin V.M., and Borowiec J.A.. 2007. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 282:35910–35923 10.1074/jbc.M704645200 [DOI] [PubMed] [Google Scholar]
- Anantha, R.W., Sokolova E., and Borowiec J.A.. 2008. RPA phosphorylation facilitates mitotic exit in response to mitotic DNA damage. Proc. Natl. Acad. Sci. USA. 105:12903–12908 10.1073/pnas.0803001105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt, M.F., Ozdemir A.C., Birkeland S.R., Wilson T.E., and Glover T.W.. 2011. Hydroxyurea induces de novo copy number variants in human cells. Proc. Natl. Acad. Sci. USA. 108:17360–17365 10.1073/pnas.1109272108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach, C.E., Bétous R., Lovejoy C.A., Glick G.G., and Cortez D.. 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 23:2405–2414 10.1101/gad.1839909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush, G.S., Anderson C.W., and Kelly T.J.. 1994. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc. Natl. Acad. Sci. USA. 91:12520–12524 10.1073/pnas.91.26.12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., and Helleday T.. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 434:913–917 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Buisson, R., Dion-Côté A.M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., and Masson J.Y.. 2010. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17:1247–1254 10.1038/nsmb.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting, S.F., Callén E., Kozak M.L., Kim J.M., Wong N., López-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A., et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 46:125–135 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun, T.S., Pacek M., Yee M.C., Walter J.C., and Cimprich K.A.. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–1052 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia, A., Bredemeyer A.L., Sowa M.E., Terret M.E., Jallepalli P.V., Harper J.W., and Elledge S.J.. 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23:2415–2425 10.1101/gad.1832309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, S.L., North P.S., and Hickson I.D.. 2007. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 14:677–679 10.1038/nsmb1267 [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz R.M., and Roeder R.G.. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova, D.S., and Gilbert D.M.. 2000. Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex. Exp. Cell Res. 254:321–327 10.1006/excr.1999.4770 [DOI] [PubMed] [Google Scholar]
- Donahue, S.L., Lin Q., Cao S., and Ruley H.E.. 2006. Carcinogens induce genome-wide loss of heterozygosity in normal stem cells without persistent chromosomal instability. Proc. Natl. Acad. Sci. USA. 103:11642–11646 10.1073/pnas.0510741103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray, E., Etchin J., Wiese C., Saro D., Williams G.J., Hammel M., Yu X., Galkin V.E., Liu D., Tsai M.S., et al. 2010. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 17:1255–1259 10.1038/nsmb.1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434:917–921 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Fenech, M., Kirsch-Volders M., Natarajan A.T., Surralles J., Crott J.W., Parry J., Norppa H., Eastmond D.A., Tucker J.D., and Thomas P.. 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 26:125–132 10.1093/mutage/geq052 [DOI] [PubMed] [Google Scholar]
- Govindan, R., Ding L., Griffith M., Subramanian J., Dees N.D., Kanchi K.L., Maher C.A., Fulton R., Fulton L., Wallis J., et al. 2012. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 150:1121–1134 10.1016/j.cell.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Z., Hughes C.M., Kosiyatrakul S., Norio P., Sen R., Fiering S., Allis C.D., Bouhassira E.E., and Schildkraut C.L.. 2009. Decreased replication origin activity in temporal transition regions. J. Cell Biol. 187:623–635 10.1083/jcb.200905144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddle, J.A., Cimino M.C., Hayashi M., Romagna F., Shelby M.D., Tucker J.D., Vanparys P., and MacGregor J.T.. 1991. Micronuclei as an index of cytogenetic damage: past, present, and future. Environ. Mol. Mutagen. 18:277–291 10.1002/em.2850180414 [DOI] [PubMed] [Google Scholar]
- Henricksen, L.A., and Wold M.S.. 1994. Replication protein A mutants lacking phosphorylation sites for p34cdc2 kinase support DNA replication. J. Biol. Chem. 269:24203–24208 [PubMed] [Google Scholar]
- Henricksen, L.A., Umbricht C.B., and Wold M.S.. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121–11132 [PubMed] [Google Scholar]
- Jensen, R.B., Carreira A., and Kowalczykowski S.C.. 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 467:678–683 10.1038/nature09399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S., Hruban R.H., Kamiyama M., Borges M., Zhang X., Parsons D.W., Lin J.C., Palmisano E., Brune K., Jaffee E.M., et al. 2009. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 324:217 10.1126/science.1171202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata, T., Luebben S.W., Yamaguchi S., Ilves I., Matise I., Buske T., Botchan M.R., and Shima N.. 2011. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell. 41:543–553 10.1016/j.molcel.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt, D.C., Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F., Fulton L.L., Dooling D.J., Ding L., Mardis E.R., et al. ; Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490:61–70 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.H., Pan Y., Kanner S., Sung P., Borowiec J.A., and Chowdhury D.. 2010. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 17:365–372 10.1038/nsmb.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Doty T., Gibson B., and Heyer W.D.. 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17:1260–1262 10.1038/nsmb.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Opiyo S.O., Manthey K., Glanzer J.G., Ashley A.K., Amerin C., Troksa K., Shrivastav M., Nickoloff J.A., and Oakley G.G.. 2012. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 40:10780–10794 10.1093/nar/gks849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, C.J., and Ashworth A.. 2008. Targeted therapy for cancer using PARP inhibitors. Curr. Opin. Pharmacol. 8:363–369 10.1016/j.coph.2008.06.016 [DOI] [PubMed] [Google Scholar]
- Nam, E.A., and Cortez D.. 2011. ATR signalling: more than meeting at the fork. Biochem. J. 436:527–536 10.1042/BJ20102162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio, P., and Schildkraut C.L.. 2001. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 294:2361–2364 10.1126/science.1064603 [DOI] [PubMed] [Google Scholar]
- Oakley, G.G., and Patrick S.M.. 2010. Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci (Landmark Ed). 15:883–900 10.2741/3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, G.G., Patrick S.M., Yao J., Carty M.P., Turchi J.J., and Dixon K.. 2003. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry. 42:3255–3264 10.1021/bi026377u [DOI] [PubMed] [Google Scholar]
- Pan, Z.Q., Park C.H., Amin A.A., Hurwitz J., and Sancar A.. 1995. Phosphorylated and unphosphorylated forms of human single-stranded DNA-binding protein are equally active in simian virus 40 DNA replication and in nucleotide excision repair. Proc. Natl. Acad. Sci. USA. 92:4636–4640 10.1073/pnas.92.10.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, S.M., Oakley G.G., Dixon K., and Turchi J.J.. 2005. DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry. 44:8438–8448 10.1021/bi048057b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, N., Seal S., Thompson D., Kelly P., Renwick A., Elliott A., Reid S., Spanova K., Barfoot R., Chagtai T., et al. ; Breast Cancer Susceptibility Collaboration (UK). 2007. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 39:165–167 10.1038/ng1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regairaz, M., Zhang Y.W., Fu H., Agama K.K., Tata N., Agrawal S., Aladjem M.I., and Pommier Y.. 2011. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I–DNA complexes. J. Cell Biol. 195:739–749 10.1083/jcb.201104003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher, K., Christ N., Siaud N., Egashira A., Wu H., and Jasin M.. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 145:529–542 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg, O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Terret, M.E., Sherwood R., Rahman S., Qin J., and Jallepalli P.V.. 2009. Cohesin acetylation speeds the replication fork. Nature. 462:231–234 10.1038/nature08550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund, T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., and West S.C.. 2010. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17:1263–1265 10.1038/nsmb.1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt, A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., Friedlander M., Arun B., Loman N., Schmutzler R.K., et al. 2010. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 376:235–244 10.1016/S0140-6736(10)60892-6 [DOI] [PubMed] [Google Scholar]
- Vassin, V.M., Wold M.S., and Borowiec J.A.. 2004. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 24:1930–1943 10.1128/MCB.24.5.1930-1943.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin, V.M., Anantha R.W., Sokolova E., Kanner S., and Borowiec J.A.. 2009. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 122:4070–4080 10.1242/jcs.053702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Yang Z., Liu Y., and Zou Y.. 2005. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem. J. 391:473–480 10.1042/BJ20050379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., and Livingston D.M.. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 22:719–729 10.1016/j.molcel.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Xia, B., Dorsman J.C., Ameziane N., de Vries Y., Rooimans M.A., Sheng Q., Pals G., Errami A., Gluckman E., Llera J., et al. 2007. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 39:159–161 10.1038/ng1942 [DOI] [PubMed] [Google Scholar]
- Xu, B., Sun Z., Liu Z., Guo H., Liu Q., Jiang H., Zou Y., Gong Y., Tischfield J.A., and Shao C.. 2011. Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS ONE. 6:e18618 10.1371/journal.pone.0018618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J., Ghosal G., and Chen J.. 2009. The annealing helicase HARP protects stalled replication forks. Genes Dev. 23:2394–2399 10.1101/gad.1836409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai, T., Kong X., Yokomori K., and Kadonaga J.T.. 2009. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 23:2400–2404 10.1101/gad.1831509 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.