Characterization of the structure of the N-terminal portion of yeast Sac1 in complex with the phosphatidylinositol 4-kinase effector Vps74 identifies how Sac1 is directed to dephosphorylate PtdIns4P in the Golgi.
Abstract
Sac1 is a phosphoinositide phosphatase of the endoplasmic reticulum and Golgi apparatus that controls organelle membrane composition principally via regulation of phosphatidylinositol 4-phosphate signaling. We present a characterization of the structure of the N-terminal portion of yeast Sac1, containing the conserved Sac1 homology domain, in complex with Vps74, a phosphatidylinositol 4-kinase effector and the orthologue of human GOLPH3. The interface involves the N-terminal subdomain of the Sac1 homology domain, within which mutations in the related Sac3/Fig4 phosphatase have been linked to Charcot–Marie–Tooth disorder CMT4J and amyotrophic lateral sclerosis. Disruption of the Sac1–Vps74 interface results in a broader distribution of phosphatidylinositol 4-phosphate within the Golgi apparatus and failure to maintain residence of a medial Golgi mannosyltransferase. The analysis prompts a revision of the membrane-docking mechanism for GOLPH3 family proteins and reveals how an effector of phosphoinositide signaling serves a dual function in signal termination.
Introduction
Phosphoinositide signaling modules play fundamental roles in intracellular signaling pathways and in defining organelle identity. They are composed of a phosphatidylinositol kinase, effector proteins that specifically recognize its product, and a phosphatase that terminates signaling. The phosphatidylinositol 4-phosphate (PtdIns4P) signaling modules that function at the Golgi apparatus control the lipid content of Golgi membranes and regulate export of cargo from the trans-Golgi network (TGN; Graham and Burd, 2011; Santiago-Tirado and Bretscher, 2011). PtdIns4P is largely restricted to the TGN and, hence, this lipid is a defining feature of this compartment. In budding yeast, the Golgi phosphatidylinositol 4-kinase (PI4K), called Pik1, localizes to late Golgi compartments and PtdIns4P restriction to the TGN is enforced by a broad specificity phosphatidylinositol phosphatase called Sac1, which localizes to the ER and early Golgi compartments. Sac1 substrates localize to a variety of organelles, including the plasma membrane, endosome, and Golgi apparatus, but the mechanisms by which Sac1 accesses these substrates are poorly understood.
An important structural feature of Sac1 is that the Sac1 homology domain (Sac1-HD), which contains the catalytic subdomain, is linked to the membrane-spanning segments via an ∼70-residue-long sequence that is disordered in the yeast Sac1-HD crystal structure that was determined by x-ray crystallography (Manford et al., 2010). It was proposed that the unstructured segment allows the Sac1 catalytic domain to access substrate in cis (i.e., on the same membrane) and also in trans, at regions where the ER is closely apposed to another organelle membrane (Manford et al., 2010; Stefan et al., 2011), though recent work has challenged the view that Sac1 can act in trans (Mesmin et al., 2013). The Sac1-HD is a composite of a catalytic and an intimately associated N-terminal subdomain (Manford et al., 2010). The function of the N-terminal subdomain is still unknown but is likely physiologically important as mutations in this domain in the Sac1-related protein Sac3/Fig4 are proposed to result in several neuropathies (Chow et al., 2007, 2009).
We recently discovered that a Golgi-localized PI4K effector called Vps74 binds Sac1-HD (Wood et al., 2012). Vps74 is proposed to mediate packaging of medial Golgi glycosyltransferases into coatomer (also called COP1)-coated vesicles that bud from the TGN and ferry cargo to earlier secretory compartments (Schmitz et al., 2008; Tu et al., 2008, 2012), thereby maintaining Golgi residence. In cells lacking Vps74 or Sac1 there is an increase in the amount of PtdIns4P on the medial Golgi compartment (Wood et al., 2012); however, it is unclear if PtdIns4P restriction by Sac1–Vps74 plays a role in the retention of resident glycosyltransferases. Here, we present the crystal structure of the yeast Sac1–Vps74 complex and use this information to determine the consequences of selectively ablating this complex in vivo.
Results and discussion
Structure of the Sac1–Vps74 complex
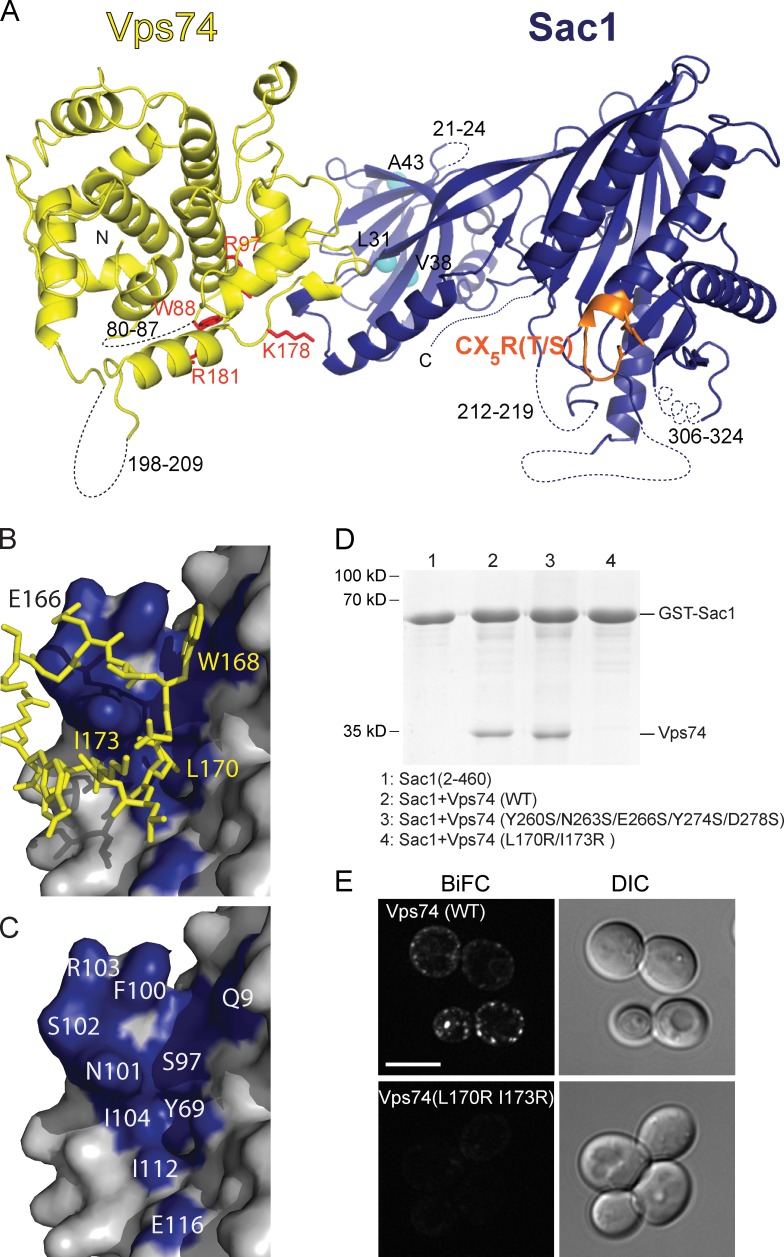
The crystal structure of yeast Sac1-HD (residues 2–449) in a 1:1 complex with Vps74 (residues 64–341) was determined by molecular replacement using individual proteins as search models (Fig. 1, A–C; and Table 1). With the exception of loop regions, Vps74 and Sac1 in the complex adhere closely to the structures of the individual proteins (Protein Data Bank accession numbers 2ZIH and 3LWT; Schmitz et al., 2008; Manford et al., 2010; root-mean-square deviation values for Cα positions are 0.7 and 1.3 Å, respectively [Holm and Park, 2000]). Interestingly, a β hairpin (residues 197–208), which mediates dimerization of Vps74 in the crystal structure of Vps74 alone (Schmitz et al., 2008), is solvent exposed and disordered in the structure of the complex. In Sac1, most of the disordered regions (except for residues 21–24) are in the catalytic domain surrounding the active site, as defined by the “P-loop” containing the CX5R(T/S) catalytic motif. Residues 312–326, which form a helix in the reported structure of Sac1-HD alone (Manford et al., 2010), are among the disordered residues in the Sac1–Vps74 complex, suggesting that this helix may be mobile. Most likely the unstructured regions around the Sac1 active site facilitate its membrane association, as speculated previously (Manford et al., 2010).
Figure 1.
Structure of the Sac1–Vps74 complex. (A) Vps74 (residues 64–341) is yellow with residues in the PtdIns4P binding pocket shown in red. Sac1 (residues 2–449) is blue with the catalytic CX5R(T/S) motif in orange. Unstructured loops are depicted as dotted lines. The homologous positions of mutations that cause CMT4J in the orthologous Sac3/Fig4 are indicated with cyan spheres (Sac1 numbering corresponds to I41T, D48G, and D53Y in Sac3/Fig4; Manford et al., 2010). The complex is tilted back slightly to orient the membrane-binding surface at the bottom toward the reader. (B) Residues 166–176 in Vps74 make extensive contact with Sac1. (C) The surface of Sac1 where Vps74 binds is shown in blue. (D) GST-Sac1 pulldown assays show that Vps74 (L170R and I173R) mutations ablate binding to Sac1, whereas mutations in a second site of contact between Vps74 and Sac1 observed in the crystal do not. (E) Bimolecular fluorescence assay shows that wild-type Vps74 and Sac1 associate in vivo, but Vps74 (L170R and I173R) mutant does not. Bar, 5 µm.
Table 1.
Data collection and refinement statistics
| Data collection | Value |
| Space group | R3 |
| Cell parameters | a = 159.338 Å, c = 143.389 Å α = 90°, β = 90°, γ = 120° |
| Wavelength (Å) | 0.979 |
| Resolution (Å) | 30–3.2 (3.31–3.2) |
| Average redundancy (last shell) | 9.8 (9.7) |
| Rsym (last shell) (%) | 14.5 (81.9) |
| I/σI (last shell) | 18.8 (2.3) |
| Completeness (last shell) (%) | 100 (100) |
| Refinement | |
| Resolution (Å) | 30–3.2 |
| No. of unique reflections (last shell) | 22,586 (2,186) |
| Protein residues | 658 |
| Rwork/Rfree (%) | 22.6/25.5 |
| rmsd bond lengths (Å) | 0.002 |
| rmsd bond angles (°) | 0.453 |
| Average B (Å2) | 128.1 |
| Ramachandran plot | |
| Favored (% residues) | 98.3 |
| Disallowed (% residues) | 0 |
rmsd, root-mean-square deviation.
It was unclear from crystal packing contacts whether the physiologically relevant interaction with Vps74 involves the N-terminal or catalytic subdomains of Sac1. In the crystal, the interface between Vps74 and the N-terminal subdomain of Sac1 occludes ∼1,130 Å2 of surface area (Fig. 1, B and C), whereas the interface between Vps74 and the catalytic Sac1 subdomain is slightly smaller at ∼830 Å2 (Krissinel and Henrick, 2007). Using wild-type GST-tagged Sac1 (residues 2–460) as bait, we were able to pull down wild-type Vps74 as well as a construct with mutations (Y260S, N263S, E266S, Y274S, and D278S) at the smaller interface, indicating that this interface likely is a crystal contact (Fig. 1 D). At the larger interface, residues 166–176 in a Vps74 loop are packed against a hydrophobic surface of the N-terminal subdomain of Sac1 (Fig. 1, B and C). Mutations in Vps74 at this interface (L170R and I173R) abrogate the Sac1–Vps74 interaction in pulldown experiments (Fig. 1 D), indicating that Vps74 interacts with the N-terminal subdomain of Sac1 in solution.
We further confirmed the role of Vps74 residues 166–176 in recognition of Sac1 using a YFP bimolecular fluorescence complementation assay. As previously reported (Wood et al., 2012), when the N-terminal segment of YFP is fused to the N terminus of Vps74 (N•YFP-Vps74) and the C-terminal segment of YFP is fused to the C terminus of Sac1 (C•YFP-Sac1), fluorescence complementation is observed (Fig. 1 E). However, when the Vps74 (L170R and I173R) mutant was assayed, a fluorescence complementation signal was not detectable. The L170R and I173R mutations do not interfere with Golgi targeting of either GFP-tagged protein in vivo (Fig. 2 A) or binding of pure recombinant protein to PtdIns4P-containing liposomes in vitro (Fig. 2 B), indicating that these mutations do not perturb the structure of Vps74. Thus, these data establish a function of the N-terminal subdomain of Sac1 in mediating interactions with Vps74. We speculate that other proteins may similarly recognize this subdomain of Sac1 and the precedent established by the Sac1–Vps74 complex may be especially relevant for investigating the contributions of mutations located in the N-terminal subdomain of a related Sac1 family member, Sac3 (also known as Fig4), which are proposed to contribute to Charcot–Marie–Tooth disorder CMT4J and amyotrophic lateral sclerosis (Chow et al., 2007, 2009; Fig. 1 A).
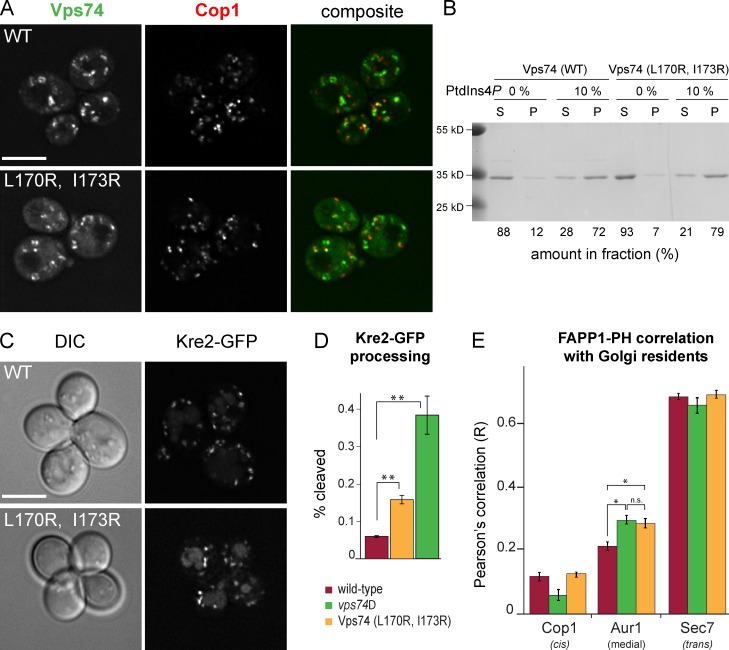
Figure 2.
Recognition of Sac1 by Vps74 is required to maintain medial PtdIns4P level on medial Golgi compartments. (A) GFP-Vps74 and Vps74 (L170R and I173R) localize to early Golgi compartments decorated by Cop1-mKate2. Maximum projections of a z stack are shown. Bar, 5 µm. (B) Vps74 and Vps74 (L170R and I173R) possess similar PtdIns4P binding properties in vitro. Purified Vps74 proteins were incubated with liposomes containing 0% or 10% PtdIns4P and the proportion of Vps74 that sedimented with liposomes was determined from Coomassie blue–stained gels. S, supernatant; P, pellet. Molecular mass standards are indicated to the left. (C) Golgi residence of Kre2-GFP is perturbed in cells expressing Vps74 (L170R and I173R). Maximum projections of a z stack are shown. Bar, 5 µm. (D) The proportion of GFP released by vacuolar proteolysis of Kre2-GFP in the indicated strains was determined by anti-GFP immunoblotting of cell lysates. The mean (n = 4) and standard error of the mean are plotted. **, P < 0.01. (E) The distribution of PtdIns4P across Golgi compartments was inferred by calculating Pearson’s correlation coefficients between the PtdIns4P biosensor, GFP-FAPP1•PH and mKate2-tagged Cop1 (cis/medial), Aur1 (medial), and Sec7 (trans) in at least 25 cells for each condition. Bars are color coded as in D. The standard error of each determination is indicated. *, P < 0.05; n.s., not significant (P = 0.45).
Vps74 functions in Golgi PI4K signaling via interactions with PtdIns4P, Golgi residents, coatomer, and Sac1. All functions of Vps74 require PtdIns4P binding, and the identification of mutations in Vps74 that ablate recognition of Sac1 but do not affect PtdIns4P binding provided us with the ability to selectively address the function of the Sac1–Vps74 complex. As shown previously, a GFP-tagged form of Kre2, an α1,2-mannosyltransferase, resides in punctate medial Golgi compartments but is delivered to the vacuole in vps74Δ cells, resulting in GFP accumulation in the vacuole lumen (Schmitz et al., 2008; Tu et al., 2008). In cells expressing Vps74 (L170R and I173R) mutant, Kre2-GFP localizes to the vacuole lumen as well as punctate compartments (Fig. 2 C). Quantitation of Kre2-GFP missorting by determining the amounts of GFP released from full-length Kre2-GFP by vacuolar proteolysis shows that ∼16% of Kre2-GFP is cleaved in Vps74 (L170R and I173R) cells as compared with 6% in wild-type cells and 38% in vps74Δ cells (Fig. 2 D). These data indicate that recognition of Sac1 by Vps74 is key for the role of Vps74 in maintaining localization of Golgi residents.
We previously reported that the level of PtdIns4P on medial Golgi compartments is increased in vps74Δ and sac1Δ cells, leading us to propose that binding of Sac1 by Vps74 promotes dephosphorylation of PtdIns4P on the medial Golgi compartment (Wood et al., 2012). The elucidation of the Sac1–Vps74 interface allowed us to probe the role of the Sac1–Vps74 complex in controlling the distribution of PtdIns4P across Golgi compartments. To do so, we determined Pearson’s correlation coefficients (R) for a PtdIns4P biosensor, GFP-tagged PH domain from human FAPP1 (Godi et al., 2004), and COP1-mKate2, Aur1-mKate2, and Sec7-mKate2—markers of early, medial, and late Golgi compartments, respectively—in >25 cells of each strain. The results are summarized in Fig. 2 E. In wild-type cells, GFP-FAPP1•PH localizes predominantly to Golgi compartments decorated with Sec7 (R = 0.70) and to a lesser extent to earlier compartments decorated by Aur1-mKate2 (R = 0.22) or Cop1-mKate2 (R = 0.12). In cells expressing Vps74 (L170R and I173R), we observed a 1.39-fold increase in the correlation with Aur1, essentially equivalent to the 1.33-fold increase observed for vps74Δ cells. These results confirm our earlier proposal (Wood et al., 2012) that Vps74 directs Sac1-mediated dephosphorylation of PtdIns4P on early Golgi compartments.
Role of the Vps74–GOLPH3 β hairpin in membrane binding
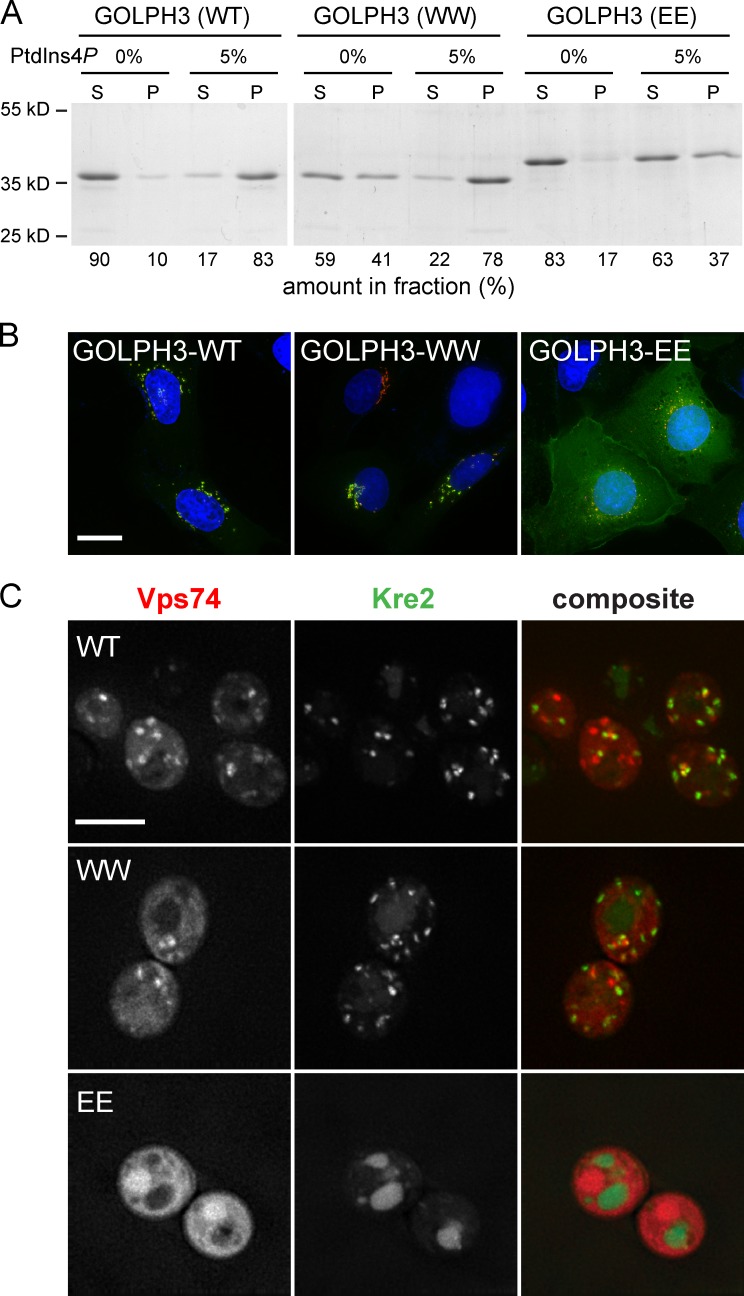
In the crystal structures of a Vps74 tetramer and a GOLPH3 dimer, residues 197–208 in Vps74 and 189–200 in GOLPH3 form a β hairpin (Schmitz et al., 2008). Hydrophobic residues at the tip of the hairpin, F201FLF in Vps74 and F193LLF in GOLPH3, pack into pockets in an adjacent molecule to form dimers, and we reported previously that deletion or shortening of the hairpin ablates formation of the Vps74 tetramer and results in a substantial loss of Golgi targeting and function of Vps74 and GOLPH3 in vivo (Schmitz et al., 2008). In the structure of the Sac1–Vps74 complex presented here, Vps74 occurs as a monomer and residues 198–200, corresponding to the hairpin loop, are disordered. Importantly, these residues are solvent exposed and on the same face of the complex as the PtdIns4P binding pocket of Vps74 and the putative membrane binding surface of Sac1 (Fig. 1 A). We therefore considered the possibility that the hairpin loop might also contribute to membrane binding via hydrophobic interactions with the membrane. To test this, we made substitutions in the two residues at the tip of the hairpin whose side chains would be expected to interact with a membrane if our hypothesis is correct and assayed several functional attributes of the mutant proteins. We used GOLPH3 for in vitro membrane binding studies because it binds PtdIns4P with approximately threefold higher affinity than Vps74, so that lipid binding can be assayed more reliably (Wood et al., 2009). In one mutant, L194 and L195 were changed to tryptophan (L194W and L195W), with the rationale that these substitutions would preserve or enhance the hydrophobicity of the hairpin and, hence, membrane association. In a second mutant, these residues were substituted with glutamate (L194E and L195E) to introduce negative charge that would disfavor membrane association. In liposome binding assays (Fig. 3 A), we found that wild-type and the Trp-substituted mutant protein associate avidly with PtdIns4P-containing liposomes (83% and 78% bound, respectively), whereas the association of the Glu-substituted mutant is reduced by approximately one half (37% bound). The Trp-substituted mutant protein also shows increased association with liposomes without PtdIns4P (41% bound compared with 10% for wild-type GOLPH3), indicating that these substitutions enhance nonspecific membrane binding. Hence, these results are consistent with our hypothesis that the β hairpin of GOLPH3 contributes to membrane binding.
Figure 3.
The β hairpin of GOLPH3 and Vps74 is required for membrane binding. (A) GOLPH3 liposome binding assays. Pure GOLPH3 proteins (wild type, L194W and L195W, and L194E and L195E) were incubated with liposomes containing 0% or 5% PtdIns4P and the liposomes were then collected by centrifugation. The proportions of GOLPH3 proteins in each supernatant (S) and pellet (P) fraction were determined from Coomassie blue–stained gels. Molecular mass standards are indicated to the left. (B) Localization of mutant GOLPH3 proteins in vivo. GFP-tagged GOLPH3 proteins were expressed in U2-OS cells that stably express mKate2-tagged β1,4-galactosyltransferase I (amino acids 1–81). Cell nuclei are blue. Bar, 20 µm. (C) Yeast Vps74 and Kre2-GFP protein localization. mCherry-tagged wild-type, L202W/L203W, and L202E/L203E Vps74 proteins were expressed in vps74Δ cells that also express GFP-tagged Kre2. Maximum projections are shown. Bar, 5 µm.
The functional consequences of the substitutions in the β hairpin were addressed by expressing wild-type and mutant forms of GOLPH3 as C-terminal fusions to GFP (Fig. 3 B). Both the wild-type and Trp-substituted mutant proteins localize to the Golgi, whereas the Glu-substituted mutant mislocalizes to the cytosol. To extend this analysis to Vps74, we constructed analogous mutations in Vps74 (F202W and L203W and F202E and L203E) and expressed them as C-terminal fusions to mCherry in vps74Δ cells (Fig. 3 C). Similar to GOLPH3, the Trp-substituted protein localizes to punctate Golgi compartments (Fig. 3 C). In contrast, Golgi localization of the Glu-substituted mutant was abolished. Consistent with these localizations, Kre2-GFP resides predominantly in the Golgi of Vps74 (F202W and L203W) cells, but accumulates in the vacuole of cells with the Vps74 (F202E and L203E) cells. Altogether, the results demonstrate that membrane binding, Golgi targeting, and the function of Vps74–GOLPH3 tolerate hydrophobic substitutions of the residues at the tip of the β hairpin, but not changes to Glu. We therefore suggest that Vps74 and GOLPH3 dock to Golgi membranes with the tip of the β hairpin inserted into the interfacial region, augmenting PtdIns4P recognition and stabilizing membrane association.
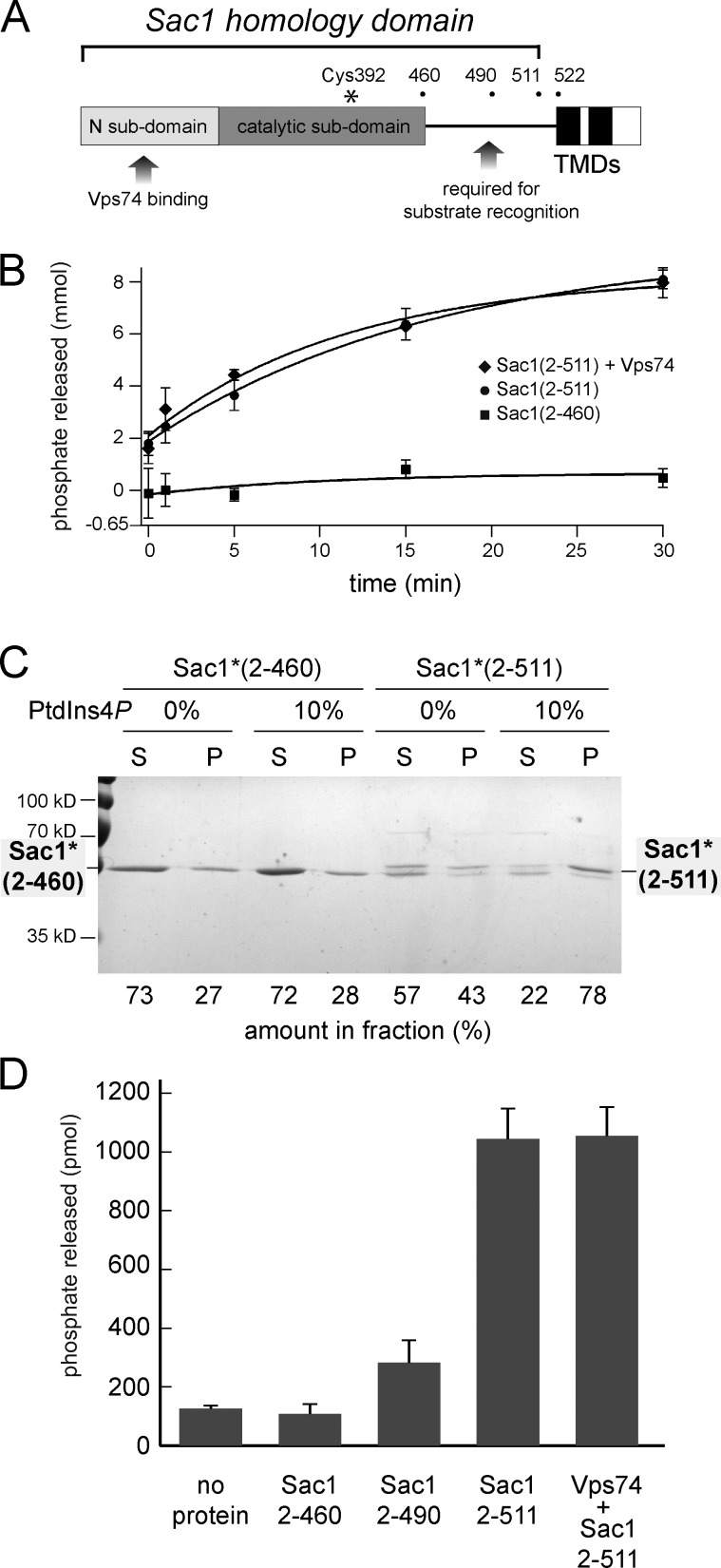
Linker region of Sac1 is required for catalytic activity
We previously reported that Vps74 does not influence Sac1(2–521) PtdIns4P phosphatase activity when both proteins were added separately to the reaction (Wood et al., 2012) and we find that this is also the case for the Sac1(2–511)–Vps74 complex prepared by coexpression (Fig. 4, A and B). While preparing for these experiments, we discovered that a shorter version of Sac1, Sac1(2–460), truncated at the end of the structured region, exhibits essentially no PtdIns4P phosphatase activity (Fig. 4 B). In the first description of the Sac1-HD crystal structure, residues 450–522 were surmised to be unstructured in the full-length, membrane-associated protein, and it was proposed that this segment serves as a flexible linker that allows the catalytic domain to access substrate on the same (in cis) or a different (in trans) membrane (Manford et al., 2010; Fig. 4 A). As Sac1(2–460) and Sac1(2–511) exhibit identical expression and purification properties, the data raise the possibility that residues 461–511 actively promote phosphatase activity. To test if the lack of activity of Sac1(2–460) is a result of impaired PtdIns4P-dependent membrane binding, we assayed liposome binding by Sac1(2–460) and Sac1(2–511). An inactivating mutation (C392S) was introduced into the active sites of Sac1(2–460) and Sac1(2–511) and cosedimentation with liposomes containing 0% or 10% PtdIns4P was determined (Fig. 4 C). Indeed, the presence of PtdIns4P in liposomes does not elicit substantial membrane binding by Sac1(2–460), but Sac1(2–511) does, suggesting a role for residues 461–511 in membrane binding and/or substrate recognition. To discriminate these possibilities, we measured the activities of Sac1(2–460), a slightly longer construct, Sac1(2–490), and Sac1(2–511) toward a soluble PtdIns4P analogue (diC8-PtdIns4P), which allows probing of Sac1 activity in the absence of a membrane bilayer. Only Sac1(2–511) exhibits substantial PtdIns4P phosphatase activity (Fig. 4 D), as observed for PtdIns4P liposomes as substrate. Together, these results indicate that Sac1 residues 451–511 are required for PtdIns4P recognition, either by participating in recognition directly or indirectly by influencing the structure of the catalytic domain in a manner not revealed by crystallographic analyses. Importantly, these results suggest that the portion of Sac1 available to act as a flexible linker is shorter than the ∼70 amino acids that were proposed (Manford et al., 2010) and it may not, therefore, provide sufficient freedom to allow Sac1 to function in trans at organelle membrane contact sites. Accordingly, we propose that as Sac1 transits the Golgi (Faulhammer et al., 2005, 2007) it encounters Vps74, which directs local dephosphorylation of PtdIns4P. Vps74 localizes predominantly to early Golgi compartments (Wood et al., 2012), so this mechanism serves to restrict PI4K signaling events to the TGN. More generally, these findings indicate that the recognition of the N-terminal subdomain of Sac1, and likely other proteins of the Sac1 family, by receptors such as Vps74 direct phosphoinositide phosphatase activity at a molecular scale, modifying the local membrane environment. In the yeast Golgi apparatus, this ensures that the TGN-specific lipid PtdIns4P does not accompany early Golgi residents in the retrograde pathway and, hence, maintains cisternal identity.
Figure 4.
The proposed tether of Sac1 is required for recognition and hydrolysis of PtdIns4P. (A) A schematic diagram of pertinent structural features of Sac1 is shown. The positions of the last amino acid of truncated Sac1 proteins are indicated. The proposed tether (Manford et al., 2010) spans approximately residues 450–522. The trans-membrane–spanning domains (TMDs) are colored black. (B) diC16-PtdIns4P phosphatase assays. The amounts of phosphate released from liposomes containing PtdIns4P are plotted. The means (n = 4) and standard deviations for each time point are indicated. (C) Vps74 liposome binding assays. The indicated catalytically inactive Sac1 (C392S; Sac1*) proteins were incubated with liposomes containing 0% or 10% PtdIns4P and the liposomes were collected by centrifugation. The proportions of each protein in the supernatant (S) and pellet (P) fractions were determined from Coomassie blue–stained gels. A substantial portion of the unstructured region of Sac1*(2–511) is cleaved during purification and the bound portion is indicated only by the full-length protein. (D) diC8-PtdIns4P phosphatase assays. The amounts of phosphate released from diC8-PtdIns4P after incubation (20 min at RT). The means (n = 3) and standard deviations are indicated.
Materials and methods
Recombinant protein expression
Coding sequences for Sac1 (residues 2–460 or 2–511) and Vps74 (residues 46–345) of Saccharomyces cerevisiae were PCR amplified from yeast genomic DNA and cloned into Duet vectors (EMD Millipore) for complex formation. Sac1 was cloned into pCDFDuet-1, and Vps74 was cloned into pCOLADuet-1 vector. For purification purposes, Vps74 has an N-terminal hexa-histidine-SUMO tag, which was removed by cleavage with SUMO protease. For the phosphatase activity assay, Sac1 constructs were cloned into pET28a vector with an N-terminal hexa-histidine-SUMO tag. For the protein pulldown assay, Sac1 constructs were cloned into pGEX-4T-3 vector. The constructs of Sac1 (C392S) used for PtdIns4P binding assay and Vps74 mutant used in the pulldown assay (L170R and I173R and Y260S, N263S, E266S, Y274S, and D278S) were generated by site-directed mutagenesis from the wild-type constructs (Quickchange system; Agilent Technologies). The wild-type GOLPH3 construct for the liposome binding assay is described in Wood et al. (2009), and mutants were generated by PCR.
For the overexpression of Sac1 or the Sac1–Vps74 complex, the single plasmid or two Duet plasmids were cotransformed into Escherichia coli BL21(DE3) cells. The cells were grown at 37°C to an OD600 of ∼0.6–0.8, and then shifted to 20°C before induction with 0.5 mM IPTG. Cells were harvested 18 h after induction. Both Sac1 and Sac1–Vps74 complex were purified by Ni-NTA chromatography (QIAGEN) and the Sac1–Vps74 complex was purified as a 1:1 complex, as assessed by size exclusion chromatography. SUMO tags were removed by treatment with SUMO protease. Protein samples were further purified by gel filtration on a Superdex200 column (GE Healthcare). The gel filtration buffer contained 20 mM Hepes, pH 8.0, 150 mM NaCl, and 0.5 mM TCEP.
GOLPH3 proteins were expressed in E. coli with an N-terminal His tag from pET28a-derived vectors. Cells were mechanically lysed in lysis buffer (20 mM Na2HPO4/NaH2PO4, pH 7.4, 500 mM NaCl, and 25 mM imidazole supplemented with Complete mini EDTA-free protease inhibitor [Roche]) using a cell disruptor (Avenstin). Cleared cell extracts were applied to a HisTrap column using an AKTA Prime chromatography system (GE Healthcare) and bound GOLPH3 was eluted with an imidazole gradient. Fractions containing GOLPH3 were combined and the buffer was exchanged to 25 mM Hepes, pH 7.5, and 150 mM NaCl using a Zebra desalting column (Thermo Fisher Scientific).
Crystallization and data collection
Crystals of the Sac1–Vps74 complex were grown at 4°C by the hanging-drop vapor diffusion method. Drops consisted of 1.5 µl of protein solution (8–15 mg/ml) and 1.5 µl of reservoir solution containing 200 mM Hepes, pH 7.0, 35% (vol/vol) MPD, and 0.3 M NDSB-195. Crystals were cryoprotected by rapidly passing them through a solution of mother liquor, mounting in a nylon loop, and plunging the mounted crystal into liquid nitrogen. Data were collected at beamline 24-ID-C at the Advanced Photon Source, Argonne National Laboratory. Crystals belong to space group R3 (a = 159.338 Å, b =159.338 Å, and c =143.389 Å; α = 90°, β = 90°, and γ = 120°) and diffract to 3.2 Å. Datasets were processed and data from two crystals were scaled together with HKL2000 (Otwinowski and Minor, 1997; Table 1).
Structure determination and refinement
The Sac1–Vps74 structure was determined by molecular replacement using the coordinates of Sac1 (Protein Data Bank [PDB] accession number 3LWT) and Vps74 (PDB accession number 2ZIH) in PHENIX (Adams et al., 2002). Coordinates and structure factors have been deposited in the PDB (accession number 4TU3). PDB accession number 3LWT was rerefined to improve its geometry before it was used as a search model. The model was built in Coot (Emsley and Cowtan, 2004) and refined in PHENIX. One Sac1–Vps74 complex was modeled in the asymmetric unit (Fig. 1, A–C; and Table 1), except for several regions that are disordered. An initial round of torsion angle dynamics was followed by cycles of manual rebuilding in Coot. We used XYZ coordinate, TLS refinement, and individual B factor refinement options and allowed for twinning. Refinement statistics are in Table 1.
In vitro protein–protein interaction assay
GST-fused Sac1 was purified and immobilized on glutathione beads. Both wild-type and mutant Vps74 (50 nM) were purified and incubated overnight at 4°C with GST-Sac1 (10 nM) in binding buffer (20 mM Hepes, pH 7.5, 0.5 mM TCEP, and 0. 2% Triton X-100). The beads were washed and eluted in SDS-PAGE sample buffer. Bound protein was visualized by Coomassie staining a 13% SDS polyacrylamide gel.
Sac1 activity assay
Phosphate release from PtdIns4P by Sac1 proteins and the Sac1(2–511)–Vps74 complex was measured using a published malachite green absorbance assay (Maehama et al., 2000). PtdIns4P liposomes were produced from pure synthetic lipids (Avanti Polar Lipids, Inc.) by mixing 75 mol percent dioleoyl-phosphatidylcholine, 15 mol percent dioleoyl-phosphatidylserine, and 10 mol percent l-α-PtdIns4P in buffer (25 mM Hepes and 150 mM NaCl, pH 7.5) by extrusion through 1-µm-pore filters using a mini-extruder (Avanti Polar Lipids, Inc.). Proteins (0.5 mM each or Sac1–Vps74 complex) were incubated with 2 mM lipids. For activity measurements toward soluble PtdIns4P, proteins (0.5 µM) were incubated with diC8-PtdIns4P (50 µM) in buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, and 0.5 mM TCEP) for 20 min at RT.
Liposome binding experiments
Liposomes used for Vps74 protein sedimentation assays were produced from pure synthetic lipids (Avanti Polar Lipids, Inc., or Echelon Biosciences) by mixing 75 mol percent (85% for liposomes not containing PtdIns4P) dioleoyl-phosphatidylcholine, 15 mol percent dioleoyl-phosphatidylserine, and 10 mol percent diC16-PtdIns4P in buffer (25 mM Hepes and 150 mM NaCl, pH 7.5) by extrusion through 1-µm-pore filters using a mini-extruder. Liposomes used for GOLPH3 protein sedimentation assays contained 5% mol percent diC16-PtdIns4P and 80% mol percent dioleoyl-phosphatidylcholine. For binding assays, liposomes (2 mM lipid) were incubated with purified proteins (1 µM) for 60 min at 4°C. Liposomes were collected by centrifugation at 200,000 g for 20 min at 4°C. Supernatants were collected and pellets were suspended in an equal volume of sample buffer. The protein band densities were determined using Image Lab software (Bio-Rad Laboratories).
Yeast strains and culture conditions
Yeast strains were constructed in BY4742 (MATα his3-1, leu2-0, met15-0, and ura3-0). Cells were grown in standard synthetic complete medium lacking nutrients required to maintain selection for auxotrophic markers and/or plasmids (Sherman et al., 1979). Cell extracts were prepared for immunoblotting by incubating cells with 10% trichloroacetic acid on ice for 10 min, washing twice with acetone, and then performing mechanical lysis with glass beads in SDS-PAGE sample buffer. Student’s unpaired t test was used to determine statistical analyses of Kre2-GFP processing.
Cell culture and transfection
U2-OS cells were cultured in DMEM supplemented with 10% fetal bovine serum. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 24 h, the cells were fixed and stained with Hoechst 33342.
Light microscopy and image analysis
Yeast cells expressing GFP- or mCherry-tagged proteins harvested from cultures grown to OD600 ≈ 0.5 were mounted in growth medium, and 3D image stacks were collected at 0.3-µm z increments on a DeltaVision Elite workstation (Applied Precision) based on an inverted microscope (IX-70; Olympus) using a 100×, 1.4 NA oil immersion lens. Images were captured at 24°C with a 12-bit charge-coupled device camera (CoolSnap HQ; Photometrics) and deconvolved with softWoRx (v.6.0) software using the iterative-constrained algorithm and the measured point spread function. All image analysis and preparation was done using ImageJ (v.1.48; Rasband, 2007). Student’s unpaired t test was used to determine statistical analyses of colocalization data.
Acknowledgments
We are grateful to the staff at the Northeastern Collaborative Access Team at the Advanced Photon Source for assistance with data collection and to colleagues for discussions and critical reading of the manuscript.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under awards R01GM095766 and R01GM080616.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- HD
- homology domain
- PI4K
- phosphatidylinositol 4-kinase
- PtdIns4P
- phosphatidylinositol 4-phosphate
- TGN
- trans-Golgi network
References
- Adams, P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., and Terwilliger T.C.. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- Chow, C.Y., Zhang Y., Dowling J.J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M.E., Li J., Zhang X., et al. 2007. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 448:68–72 10.1038/nature05876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, C.Y., Landers J.E., Bergren S.K., Sapp P.C., Grant A.E., Jones J.M., Everett L., Lenk G.M., McKenna-Yasek D.M., Weisman L.S., et al. 2009. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am. J. Hum. Genet. 84:85–88 10.1016/j.ajhg.2008.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Faulhammer, F., Konrad G., Brankatschk B., Tahirovic S., Knödler A., and Mayinger P.. 2005. Cell growth–dependent coordination of lipid signaling and glycosylation is mediated by interactions between Sac1p and Dpm1p. J. Cell Biol. 168:185–191 10.1083/jcb.200407118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulhammer, F., Kanjilal-Kolar S., Knödler A., Lo J., Lee Y., Konrad G., and Mayinger P.. 2007. Growth control of Golgi phosphoinositides by reciprocal localization of sac1 lipid phosphatase and pik1 4-kinase. Traffic. 8:1554–1567 10.1111/j.1600-0854.2007.00632.x [DOI] [PubMed] [Google Scholar]
- Godi, A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D.R., Kular G.S., Daniele T., Marra P., Lucocq J.M., and De Matteis M.A.. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6:393–404 10.1038/ncb1119 [DOI] [PubMed] [Google Scholar]
- Graham, T.R., and Burd C.G.. 2011. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 21:113–121 10.1016/j.tcb.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L., and Park J.. 2000. DaliLite workbench for protein structure comparison. Bioinformatics. 16:566–567 10.1093/bioinformatics/16.6.566 [DOI] [PubMed] [Google Scholar]
- Krissinel, E., and Henrick K.. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Maehama, T., Taylor G.S., Slama J.T., and Dixon J.E.. 2000. A sensitive assay for phosphoinositide phosphatases. Anal. Biochem. 279:248–250 10.1006/abio.2000.4497 [DOI] [PubMed] [Google Scholar]
- Manford, A., Xia T., Saxena A.K., Stefan C., Hu F., Emr S.D., and Mao Y.. 2010. Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 29:1489–1498 10.1038/emboj.2010.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin, B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B.. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 155:830–843 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor W.. 1997. Processing of x-ray diffraction data collected in oscillation mode. Macromolecular Crystallography. Carter C.W. Jr, and Sweet R.M., Academic Press, New York: 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Rasband, W.S.2007. ImageJ. National Institutes of Health, Bethesda, MD: http://rsb.info.nih.gov/ij/ (accessed July 22, 2013) [Google Scholar]
- Santiago-Tirado, F.H., and Bretscher A.. 2011. Membrane-trafficking sorting hubs: cooperation between PI4P and small GTPases at the trans-Golgi network. Trends Cell Biol. 21:515–525 10.1016/j.tcb.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, K.R., Liu J., Li S., Setty T.G., Wood C.S., Burd C.G., and Ferguson K.M.. 2008. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell. 14:523–534 10.1016/j.devcel.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink G.R., and Lawrence L.W.. 1979. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 98 pp [Google Scholar]
- Stefan, C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., and Emr S.D.. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144:389–401 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Tu, L., Tai W.C., Chen L., and Banfield D.K.. 2008. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 321:404–407 10.1126/science.1159411 [DOI] [PubMed] [Google Scholar]
- Tu, L., Chen L., and Banfield D.K.. 2012. A conserved N-terminal arginine-motif in GOLPH3-family proteins mediates binding to coatomer. Traffic. 13:1496–1507 10.1111/j.1600-0854.2012.01403.x [DOI] [PubMed] [Google Scholar]
- Wood, C.S., Schmitz K.R., Bessman N.J., Setty T.G., Ferguson K.M., and Burd C.G.. 2009. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187:967–975 10.1083/jcb.200909063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, C.S., Hung C.S., Huoh Y.S., Mousley C.J., Stefan C.J., Bankaitis V., Ferguson K.M., and Burd C.G.. 2012. Local control of phosphatidylinositol 4-phosphate signaling in the Golgi apparatus by Vps74 and Sac1 phosphoinositide phosphatase. Mol. Biol. Cell. 23:2527–2536 10.1091/mbc.E12-01-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]