During kinetochore assembly in budding yeast, the key steps of CENP-A recognition and outer kinetochore recruitment are executed through different yeast CCAN subunits, potentially protecting against inappropriate kinetochore assembly.
Abstract
Kinetochores are megadalton-sized protein complexes that mediate chromosome–microtubule interactions in eukaryotes. How kinetochore assembly is triggered specifically on centromeric chromatin is poorly understood. Here we use biochemical reconstitution experiments alongside genetic and structural analysis to delineate the contributions of centromere-associated proteins to kinetochore assembly in yeast. We show that the conserved kinetochore subunits Ame1CENP-U and Okp1CENP-Q form a DNA-binding complex that associates with the microtubule-binding KMN network via a short Mtw1 recruitment motif in the N terminus of Ame1. Point mutations in the Ame1 motif disrupt kinetochore function by preventing KMN assembly on chromatin. Ame1–Okp1 directly associates with the centromere protein C (CENP-C) homologue Mif2 to form a cooperative binding platform for outer kinetochore assembly. Our results indicate that the key assembly steps, CENP-A recognition and outer kinetochore recruitment, are executed through different yeast constitutive centromere-associated network subunits. This two-step mechanism may protect against inappropriate kinetochore assembly similar to rate-limiting nucleation steps used by cytoskeletal polymers.
Introduction
Kinetochores are key cell division organelles responsible not only for the mechanical connection between chromosomes and microtubules but also for the control of the metaphase–anaphase progression via the spindle assembly checkpoint (Santaguida and Musacchio, 2009). Despite recent advances in the purification of native kinetochore assemblies (Akiyoshi et al., 2010) and a comprehensive parts list of the subunits involved (Santaguida and Musacchio, 2009; Biggins, 2013), kinetochores have generally been more refractory to structural analysis than other macromolecular complexes. As a consequence, a detailed picture of the architecture of a kinetochore, even concerning fundamental aspects such as symmetry, copy number of subunits, and key links within the framework, is still missing. Budding yeast provides an attractive model system to study kinetochores as it offers two important simplifications in the overall configuration: budding yeast kinetochores connect to a single microtubule and their assembly is triggered on a short stretch of DNA, with a strictly positioned centromeric nucleosome over the centromere DNA element II of the yeast point centromere (Krassovsky et al., 2012). Despite a dramatic divergence of the underlying DNA, the complement of proteins that associate with centromeres appears to be largely conserved from yeast to man (Schleiffer et al., 2012; Westermann and Schleiffer, 2013). The structural core of the kinetochore can be subdivided into two supramolecular assemblies each composed of multiple polypeptides (Fig. 1 A). The 10-protein KMN network hosts the two characteristic functions of the kinetochore: first, microtubule binding, achieved by a combination of a structured calponin homology domain and an unstructured tail in the N terminus of the Ndc80 subunit (Ciferri et al., 2008; Alushin et al., 2010), and second, checkpoint signaling, arranged as arrayed binding sites for checkpoint proteins in the extended N terminus of the Spc105KNL-1 subunit (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Primorac et al., 2013; London and Biggins, 2014). These key elements are physically connected and organized by the Mtw1 (Mis12–MIND) complex, which can be regarded as the structural centerpiece of the kinetochore architecture (Cheeseman et al., 2006; Maskell et al., 2010; Petrovic et al., 2010; Hornung et al., 2011). During kinetochore assembly multiple copies of the KMN network need to be positioned on centromeric chromatin, which is one of the key functions of the second supramolecular assembly, the constitutive centromere-associated network (CCAN) complex. As CCAN subunits generally lack enzymatic activities or obvious functional domains, the contributions of the majority of CCAN subunits to kinetochore function has so far remained undefined. A notable exception is centromere protein C (CENP-C; Mif2 in budding yeast), which has been shown to bind to CENP-A (Carroll et al., 2010; Guse et al., 2011; Kato et al., 2013) and to provide a link to the outer kinetochore via a direct interaction with the Mis12 complex in humans and flies (Przewloka et al., 2011; Screpanti et al., 2011). A second link between inner and outer kinetochore is formed by the histone-fold protein CENP-T (Cnn1 in budding yeast), which directly connects to the microtubule-binding Ndc80 complex (Hori et al., 2008a; Bock et al., 2012; Schleiffer et al., 2012; Malvezzi et al., 2013; Nishino et al., 2013). Analysis of kinetochore assembly in budding yeast is facilitated by the clear distinction that can be drawn between essential and nonessential kinetochore subunits. Of the 16 CCAN subunits in budding yeast, only Mif2CENP-C, Ame1CENP-U, and Okp1CENP-Q are essential for viability, implying that they carry out key nonredundant functions. Here we focus our functional analysis on these three subunits and reveal their essential contributions to kinetochore assembly.
Figure 1.
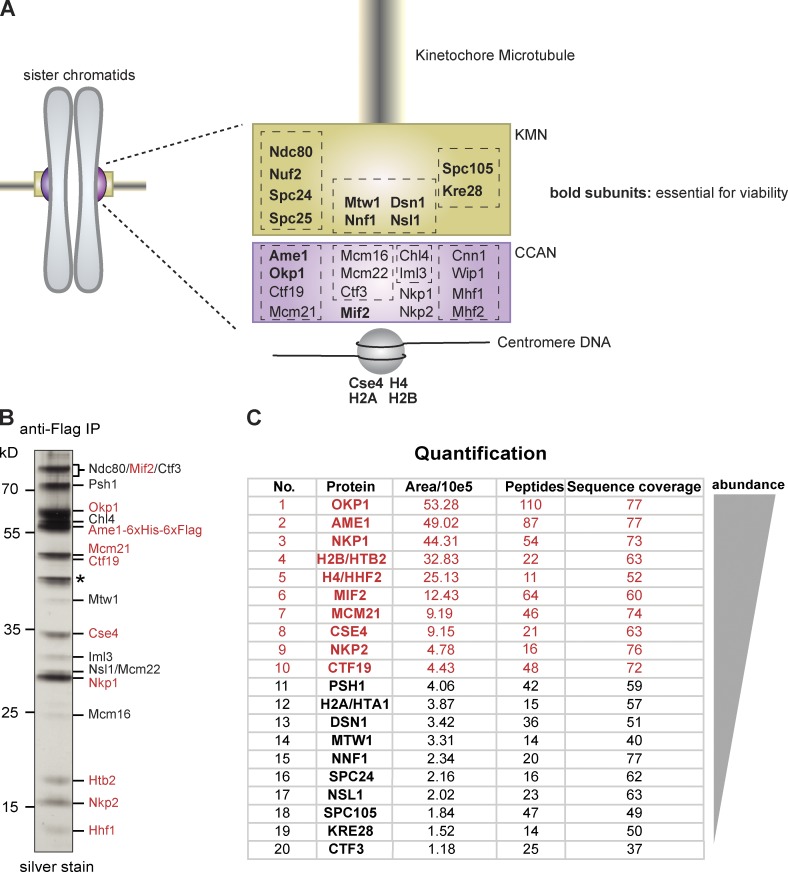
Ame1CENP-U functions at the centromere–kinetochore interface. (A) Schematic representation of yeast kinetochore architecture. Essential subunits are depicted in bold. Dashed boxes indicate subcomplexes within outer and inner kinetochores. (B) Single-step affinity purification of Ame1-6×Flag from logarithmic yeast extracts visualized by silver staining of the gel after Flag peptide elution. Identity of the individual bands was determined by MS analysis of digested peptides. Asterisk denotes background contaminant. (C) Quantification of Ame1 copurifying polypeptides using SWATH-MS. Proteins were ranked according to abundance based on area calculation of three selected peptides for each protein. Red labeling denotes the ten most abundant polypeptides in B and C.
Results
A single-step affinity purification of the budding yeast inner kinetochore
The budding yeast proteins Ame1 and Okp1 associate with Ctf19 and Mcm21 to form the so-called COMA kinetochore complex. The human counterpart of this structure is the CENP-O/P/Q/U/R subcomplex, with the exception that a CENP-R homologue seems to be missing in budding yeast. Although Ame1 and Okp1 are essential proteins in yeast, the phenotypes of CENP-U and CENP-Q depletions in humans cells or deletions in chicken DT40 cells are comparably mild, suggesting that these subunits have supporting rather than essential roles in the kinetochore (Foltz et al., 2006; Hori et al., 2008b). Previous studies have proposed a function in regulating plus end dynamics either through a direct interaction of human CENP-Q with microtubules (Amaro et al., 2010) or through binding of CENP-U to the Ndc80 complex (Hua et al., 2011). In contrast, superresolution imaging of yeast CENP-U, as well as genetic analysis of CENP-Q, seems to indicate a function at the inner kinetochore (Ortiz et al., 1999; Joglekar et al., 2009; Haase et al., 2013). To analyze the functional role of the essential yeast CCAN subunits Ame1CENP-U and Okp1CENP-Q (Fig. 1 A), we first sought to establish their interaction network within the native yeast kinetochore. We integrated a 6×Flag tag at the C terminus of endogenous Ame1 and purified the protein from logarithmically growing yeast under conditions that were previously used for the copurification of large ensembles of kinetochore subunits (Akiyoshi et al., 2010). Silver stain (Fig. 1 B) and mass spectrometry (MS) analysis revealed that Ame1 copurified with a variety of other kinetochore subunits. To establish a ranking of kinetochore subunits that are closely associated with Ame1 we used label-free MS quantification with SWATH acquisition (Gillet et al., 2012; Fig. 1 C). The most abundant polypeptides were inner kinetochore subunits including the components of the centromeric nucleosome, the conserved and essential Mif2CENP-C subunit, the nonessential COMA components Ctf19 and Mcm21, and the poorly characterized subunits Nkp1 and Nkp2. The most abundant KMN component found in the preparation was Mtw1, whereas microtubule-binding components such as Ndc80 or subunits of the Dam1 complex were either absent or present only in trace amounts. These results indicate that yeast Ame1 and Okp1 operate at the inner kinetochore and are closely associated with centromeric chromatin.
Biochemical reconstitution of the Ame1–Okp1 complex
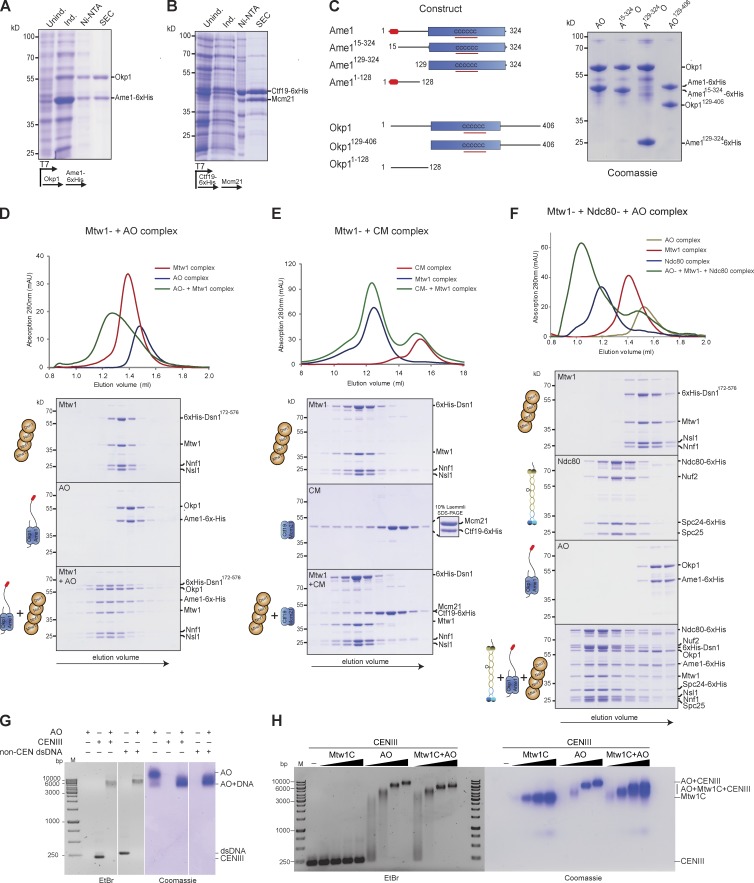
We have previously reported the reconstitution of a four-protein COMA (Ctf19, Okp1, Mcm21, and Ame1) kinetochore complex (Hornung et al., 2011). Given that Ctf19 and Mcm21 are nonessential genes in yeast, we asked whether the essential subunits Ame1CENP-U and Okp1CENP-Q would be sufficient to form a stable subcomplex. Indeed, coexpression of 6×His-tagged Ame1 and Okp1 in bacteria resulted in the purification of a stoichiometric heterodimeric complex (hereafter called the AO complex; Fig. 2 A). In agreement with previous results (Schmitzberger and Harrison, 2012) the nonessential subunits Ctf19 and Mcm21 could also be coexpressed and purified as a complex (Fig. 2 B). During size exclusion chromatography (SEC) the AO complex behaved as an elongated heterodimer, which was extremely stable and resisted salt treatment up to 2.5 M NaCl. Limited proteolysis experiments indicated that the N-terminal parts of Ame1 and Okp1 were sensitive to protease treatment (unpublished data). To determine the molecular requirements for complex formation between Ame1 and Okp1 we coexpressed and purified a variety of truncation mutants: the conserved C-terminal domain of Ame1 containing a predicted coiled-coil domain was necessary for heterodimerization with Okp1, whereas the N-terminal 129 amino acids were dispensable. Similarly, the conserved central domain of Okp1 was required to form a complex with Ame1, whereas the N-terminal extension (residues 1–149) was not (Fig. 2 C).
Figure 2.
A DNA-binding AO complex directly associates with the Mtw1 complex. (A) Coexpression and purification of Ame1-6×His and Okp1 from bacteria. Coomassie-stained gel shows consecutive purification steps. (B) Coexpression and purification of Ctf19–6×His and Mcm21; purification steps as in A. (C) Analysis of Ame1–Okp1 heterodimer formation. Schematic representation of Ame1 and Okp1 domain structure with predicted coiled-coil domains underlined in red. Purification of complexes occurred via a 6×His tag on the Ame1 construct and further separation via gelfiltration. Coomassie-stained gel shows different heterodimeric AO complexes that could be purified. (D) SEC profiles and accompanying SDS-PAGE of the Mtw1 complex (top), the Ame1–Okp1 complex (middle), and a stoichiometric combination of both complexes (5 µM each; bottom). The depicted experiment is representative of more than three similar binding experiments. (E) SEC elution profiles of the Mtw1 complex (top), the Ctf19–Mcm21 complex (middle), and their stoichiometric combination (5 µM each; bottom). The depicted experiment is representative of more than three similar binding experiments. (F) SEC elution profiles and SDS-PAGE gels of the Mtw1 complex (top), recombinant Ndc80 complex (second from top), the Ame1–Okp1 complex (second from bottom), and a stoichiometric combination (5 µM each) of all three complexes (bottom). Note formation of a high molecular mass assembly containing all three complexes. The depicted experiment is representative of more than three similar binding experiments. (G) Electrophoretic mobility shift assay demonstrating DNA-binding activity of the AO complex. Double-stranded 230-bp DNA fragments covering CEN3 or a random noncentromeric fragment were resolved on 1.2% Agarose gels in the presence or absence of the AO complex. Gel was stained with ethidium bromide to visualize DNA and Coomassie to visualize proteins. (H) Binding of AO binding to DNA is concentration dependent and can occur in the presence of Mtw1C. A fixed concentration of CEN3 dsDNA (0.18 µM) was incubated with increasing concentrations of the Mtw1C complex, the AO complex, and both (0.9, 1.9, 3.8, or 6 µM for each complex) and analyzed by electrophoretic mobility shift assay. Note that the Mtw1C does not interact with DNA and does not impede AO binding to DNA.
We next sought to establish the molecular associations between the Ame1–Okp1 complex and other kinetochore subcomplexes. SEC analysis indicated that AO directly engaged with a recombinant four-protein Mtw1 complex—bearing a deletion of the N-terminal 172 residues of Dsn1 to increase stability—into a stoichiometric hexameric assembly (Fig. 2 D). In contrast, the recombinant Ctf19–Mcm21 complex failed to interact with the Mtw1 complex under identical conditions (Fig. 2 E), demonstrating that the association between the COMA and the Mtw1 complex is critically mediated through the essential CCAN subunits Ame1 and Okp1. Addition of the four-protein Ndc80 complex resulted in the formation of a high molecular mass 10-subunit ensemble that contained all proteins in approximately equal stoichiometry (Fig. 2 F). This indicates that binding of AO to the Mtw1 complex is compatible with the further assembly of the outer kinetochore.
Interestingly, during isolation of the recombinant Ame1–Okp1 complex from bacteria, we noticed copurification of bacterial DNA, which could only be removed by repeated high salt washes. This observation prompted us to determine the DNA-binding properties more systematically. Therefore, we incubated DNA-free recombinant AO complex with 230-bp double stranded DNA either encompassing the yeast CEN3 locus or a noncentromeric DNA fragment of similar length and tested their behavior in electrophoretic mobility shift assays. The AO complex displayed DNA-binding activity toward both fragments, as indicated by decreased mobility of the DNA and an altered migration behavior of DNA-bound AO (Fig. 2 G). In contrast, the Mtw1 complex alone did not interact with DNA in this assay (Fig. 2 H). DNA binding by the AO complex occurred in the presence of the Mtw1 complex, and at a DNA concentration of 0.18 µM, a 30-fold excess of AO complex (6 µM) was required to saturate binding (Fig. 2 H). In buffers of comparable ionic strength, no microtubule-binding activity of AO was detected in cosedimentation experiments (Fig. S1). These results indicate that the Ame1–Okp1 complex is a DNA-binding component of the yeast inner kinetochore.
Identification of a conserved Mtw1 receptor motif in Ame1
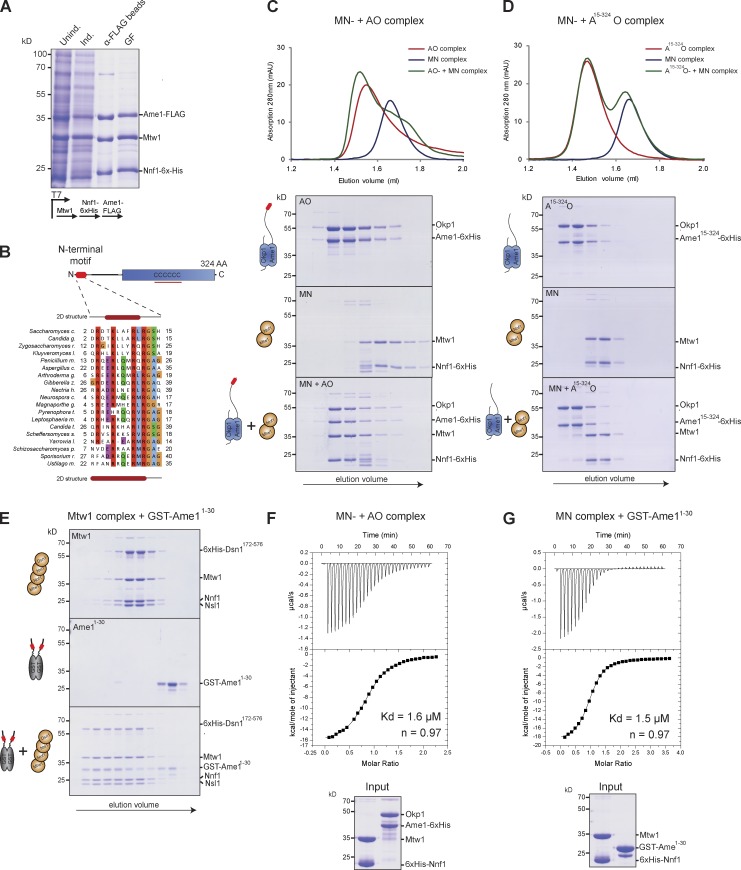
To elucidate the molecular basis for the association between the AO complex and the KMN network, we attempted to coexpress and purify Ame1 or Okp1 individually in different combinations with subunits of the Mtw1 complex. We were able to reconstitute a minimal ternary complex consisting of an Mtw1-Nnf1 heterodimer and Ame1 in the absence of Okp1, demonstrating that Ame1 is the critical subunit mediating the association with the Mtw1 complex (Fig. 3 A).
Figure 3.
A conserved motif in the N terminus of Ame1 is necessary and sufficient for Mtw1 complex binding. (A) Coexpression experiment demonstrating association of Ame1 with a heterodimeric Mtw1–Nnf1 complex. The heterotrimeric complex was purified via a Flag tag on Ame1 and further separated by gel filtration (GF). (B) A conserved motif predicted to form an α helix is present at the N terminus of Ame1-related proteins. Multiple sequence alignment of the N terminus of CENP-U proteins from distantly related yeast species. Conserved or similar residues are shaded according to the ClustalW scheme. Secondary structure predictions are indicated at the top and bottom and a red bar denotes the prediction of an α helix. (C) SEC elution profiles and corresponding SDS-PAGE of the AO complex (top), the Mtw1-Nnf1 heterodimer (MN) of the Mtw1 complex (middle), and a stoichiometric combination of both complexes (5 µM each; bottom). The depicted experiment is representative of more than three similar binding experiments. (D) SEC analysis of a mutant AO complex lacking the N-terminal 15 residues of Ame1 (top), the MN heterodimer, and a stoichiometric combination of both complexes (bottom). Note the abrogation of the interaction upon deletion of the Ame1 motif. The depicted experiment is representative of more than three similar binding experiments. (E) SDS-PAGE showing SEC analysis of the Mtw1 complex (top), GST-Ame11-30 (middle), or a stoichiometric combination of both components (5 µM each; bottom). GST alone was unable to bind the Mtw1 complex (not depicted). (F) ITC analysis of the AO–MN association. SDS-PAGE shows protein complexes used for the experiment. The dissociation constant (Kd) was calculated from n = 3 experiments. (G) ITC analysis of the binding of GST-Ame11-30 to the MN complex. SDS-PAGE shows proteins used for the analysis. The molar concentration of GST-Ame1 was calculated as a monomer, meaning that the dimeric GST-Ame11-30 binds two copies of the MN complex. The dissociation constant was calculated from n = 3 experiments.
A close inspection of sequence alignments between Ame1CENP-U proteins from distantly related fungi revealed, in addition to the highly conserved C-terminal CENP-U domain, the presence of a short peptide motif encoding a predicted α helix at the extreme N terminus of CENP-U proteins (Fig. 3 B). Regular spacing of positively charged residues suggested that they could be positioned on the same side of the predicted α helix and constitute an interaction motif. To further investigate the role of the N-terminal motif we generated an Ame1–Okp1 complex lacking the N-terminal 15 residues of Ame1. Although wild-type Ame1–Okp1 associated stoichiometrically with an Mtw1-Nnf1 heterodimer (Fig. 3 C), the interaction was disrupted in the Ame115–325 mutant (Fig. 3 D). To ask whether the Ame1 binding motif is not only necessary but also sufficient for binding, we fused the N-terminal 30 amino acids of Ame1 to GST and probed the interaction with the Mtw1 complex. SEC demonstrated that GST-Ame11-30 was sufficient to bind to the Mtw1 complex (Fig. 3 E). Quantitative analysis of the interaction by isothermal titration calorimetry (ITC) showed that full-length Ame1–Okp1 bound to an Mtw1-Nnf1 heterodimer with an apparent dissociation constant of 1.6 µM at a 1:1 stoichiometry. When fused to GST the N-terminal 30 residues of Ame1 bound with similarly high affinity (Kd = 1.5 µM) to the Mtw1-Nnf1 heterodimer, suggesting that the motif is the main, if not the sole, site of interaction with MN (Fig. 3 G).
Structural analysis of the Ame1–Okp1 complex
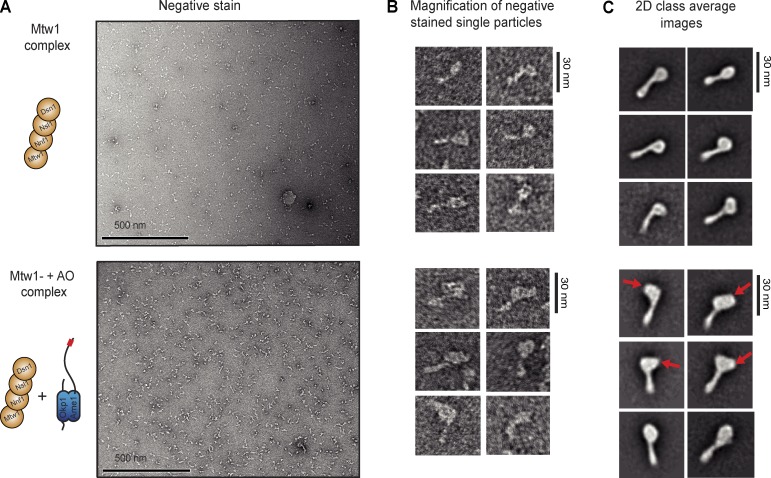
We next structurally characterized the association between the Mtw1 complex and the Ame1–Okp1 complex by single particle negative stain electron microscopy. Visualization of the recombinant Mtw1 complex revealed well-dispersed particles (Fig. 4, A and B) that were suitable for two-dimensional class averaging: the yeast “apo”–Mtw1 complex displayed the previously described characteristic dumbbell shape with a long axis of 25 nm and clearly distinguishable large and small lobes at either end of a stalk (Petrovic et al., 2010; Hornung et al., 2011; Fig. 4 C). The larger lobe showed reduced electron density in its center, hinting at the presence of a cavity (Maskell et al., 2010). In comparison, visualization of the hexameric Mtw1C–AO complex after gel filtration showed an increase in electron density specifically at the larger lobe of the complex, whereas the stalk and smaller lobe appeared relatively unchanged. It seemed that the increase in size of the larger lobe (from 8-nm diameter in the apo–Mtw1 complex to 11–15 nm upon AO binding) did not occur isotropically, but often emerged as extra mass on one side of the Mtw1 complex. In addition, the cavity in the larger lobe was no longer visible, suggesting that the AO complex partially occupies this Mtw1 docking site (Fig. 4 C). The binding mode of the AO complex revealed in these experiments appears different from the previously characterized interaction of a fragment of human CENP-C (residues 1–400) with the Mtw1 complex, which was observed to bind along the entire length of the complex and may rigidify an otherwise flexible conformation (Screpanti et al., 2011).
Figure 4.
Structural analysis of the hexameric Mtw1C–AO complex by single particle electron microscopy. (A) Negative-stained electron microscopy of recombinant full-length Mtw1 complex (top) or a stoichiometric combination of the Mtw1 complex and the Ame1–Okp1 complex (bottom). (B) Close-up views of individual negatively stained particles. (C) 2D class averages derived from electron microscopy images shown in A represent characteristic views of the Mtw1 complex in the absence (top set) and presence (bottom set) of the AO complex. Note the increase in size of the larger lobe of the Mtw1 complex, indicated by red arrows, and the disappearance of the central cavity.
The conserved Ame1 motif is essential for kinetochore assembly in vivo
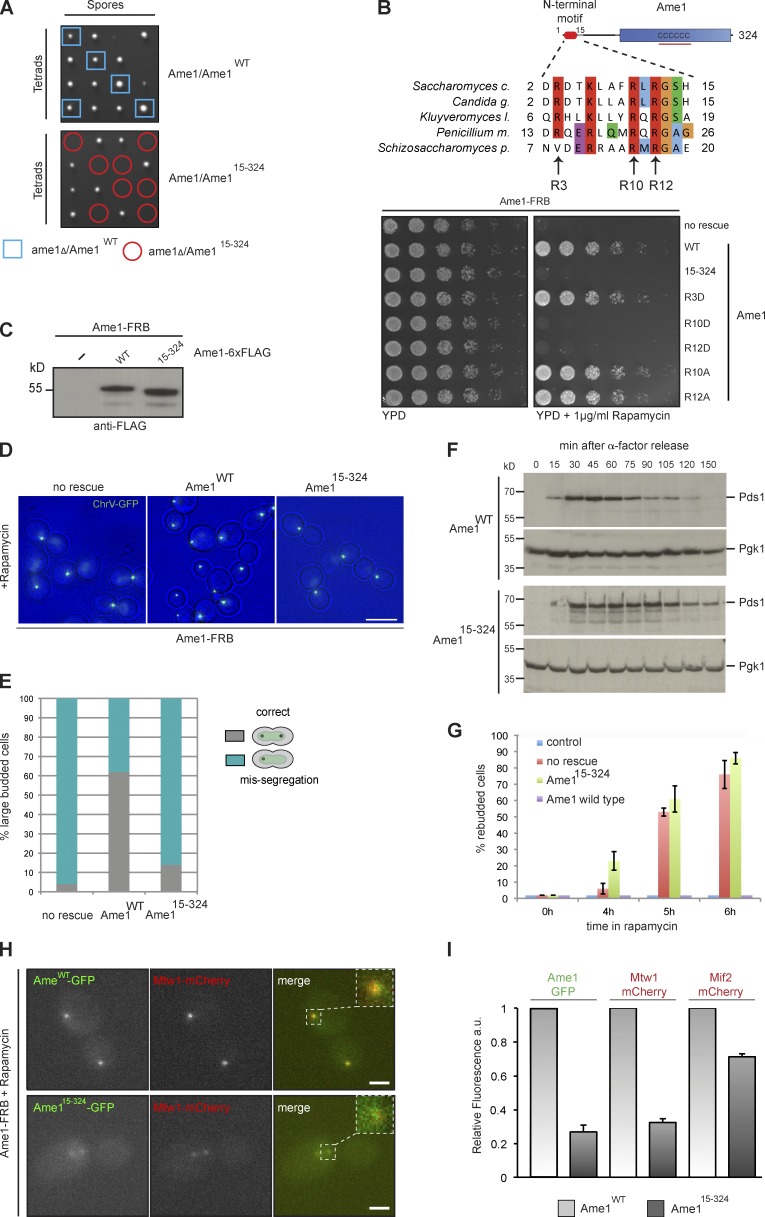
Binding of AO to the Mtw1 complex might be one of many redundant connections within the kinetochore architecture, or alternatively play a unique and crucial role during assembly. To analyze the contribution of the Ame1–Mtw1 complex interaction to kinetochore function in vivo, we generated yeast strains that express either Ame1 wild type or mutant protein lacking the Mtw1 receptor motif as the sole source of the protein in the cell. We failed to recover viable haploid spores that expressed only Ame115–324, suggesting that deletion of the Mtw1 binding motif in Ame1 is lethal (Fig. 5 A). To confirm this and create a conditional system for the analysis of terminal phenotypes we used the anchor-away technique to remove the endogenous copy of Ame1 from the yeast nucleus by addition of rapamycin, while expressing wild-type or mutant Ame1 rescue alleles under their endogenous promoter. Serial dilution assays indicated that Ame115–324 was unable to rescue the lethality of Ame1 depletion on rapamycin-containing plates, despite being expressed at levels comparable to a wild-type control (Fig. 5, B and C). Point mutations in the Ame1 motif had differential effects on viability: charge-reversing mutations in the highly conserved arginine residues Arg10 and Arg12 were lethal, whereas mutations of the same residues to alanine or replacement of the less conserved Arg3 residue with aspartate were tolerated (Fig. 5 B).
Figure 5.
The conserved Ame1 motif is essential for outer kinetochore assembly in vivo. (A) Tetrad analysis of the Ame115–324 mutant. The inferred genotype of individual haploid spores is indicated by blue squares and red circles. (B) Analysis of Ame1 mutants using the anchor-away technique. Ame1-FRB strains containing RPL13-FKBP12 for ribosome anchoring and additionally harboring the indicated rescue alleles were plated in serial dilution on YPD or YPD + rapamycin plates and incubated at 30°C. (C) Western blot analysis confirming expression of 6×Flag-tagged Ame1 wild type or Ame115–324 mutant. (D) Segregation of fluorescently labeled chromosome V in Ame1-FRB strains with different rescue alleles. Bar, 5 µm. (E) Quantification of chromosome missegregation in Ame1-FRB strains. Segregation was scored in large-budded cells 3 h after treatment with 1 µg/ml rapamycin. (F) Pds1/Securin levels analyzed by Western blot in Ame1 wild-type and mutant strains after release from an α factor arrest into rapamycin to anchor-away Ame1-FRB. Note stabilization of Pds1 levels in the mutant. (G) Quantification of cellular morphology upon nuclear depletion of Ame1-FRB in the absence or presence of Ame1 rescue alleles. The percentage of cells in the population containing more than one bud is indicated. Error bars denote SEM; n = 3. (H) Localization of Ame1 and Mtw1 analyzed by live cell microscopy after anchoring-away endogenous Ame1. Bar, 2 µm. Inset represents twofold magnification of indicated area. (I) Quantification of Ame1, Mtw1, and Mif2 fluorescence in kinetochore clusters in Ame1 wild type and Ame115–324 mutant. Fluorescence intensity in Ame1 wild type was normalized to 1. Error bars denote SEM; n = 3 for each strain.
To analyze the phenotype of Ame1 motif mutants we followed the segregation of fluorescently labeled chromosome V after treating strains with rapamycin. Ame1-FRB strains lacking a rescue allele accumulated large budded cells that failed to segregate chromosome V to the daughter cell (Fig. 5 D). Although cells with a wild-type rescue allele successfully segregated chromosome V in 60% of the cases, cells with the Ame115–324 allele resembled cells lacking a rescue allele (Fig. 5 E). We additionally followed the levels of Pds1/Securin after release of Ame1-FRB strains from an α factor arrest into rapamycin-containing medium. Cells with a wild-type Ame1 allele displayed normal Pds1 degradation kinetics, whereas Ame115–324 cells delayed Pds1 degradation, consistent with the accumulation of large-budded cells (Fig. 5 F). These results indicate that immediately after rapamycin treatment, Ame1 mutant strains display checkpoint activity. After prolonged treatment with rapamycin (>3 h) we noticed, however, a rapid accumulation of multi-budded cells in Ame1-FRB strains, indicating that the cells were not able to maintain the mitotic arrest—a phenotype typically seen in checkpoint-defective mutants (Li and Murray, 1991). Presence of a wild-type allele prevented accumulation of large and multi-budded cells, whereas Ame115–325 cells behaved very similar to cells lacking any rescue allele (Fig. 5 G). To determine the underlying reason for the severe phenotype, we examined the localization of GFP-tagged wild-type and mutant Ame1 rescue alleles as well as the Mtw1 complex and Mif2 1 h after treating a logarithmic culture with rapamycin. Although wild-type Ame1-GFP showed the typical kinetochore localization to two clusters in large-budded cells, localization of Ame115–325 to kinetochores was severely reduced (Fig. 5, F and G). Importantly, the reduction in Ame1 localization was accompanied by a proportional decrease of the Mtw1-mCherry signal in kinetochore clusters. The drastic reduction in localization of the Mtw1 complex, which serves as an assembly hub for the outer kinetochore (Petrovic et al., 2014), can explain the severe phenotype of the Ame115–324 mutant. In addition, localization of the CENP-C homologue Mif2 was reduced in the Ame1 mutant, although to a lesser extent compared with the Mtw1 complex. We conclude that the interaction between Ame1 and the Mtw1 complex mediated by the conserved N-terminal motif is essential for their codependent localization to the kinetochore.
Mif2CENP-C provides CENP-A recognition but is dispensable for direct KMN binding
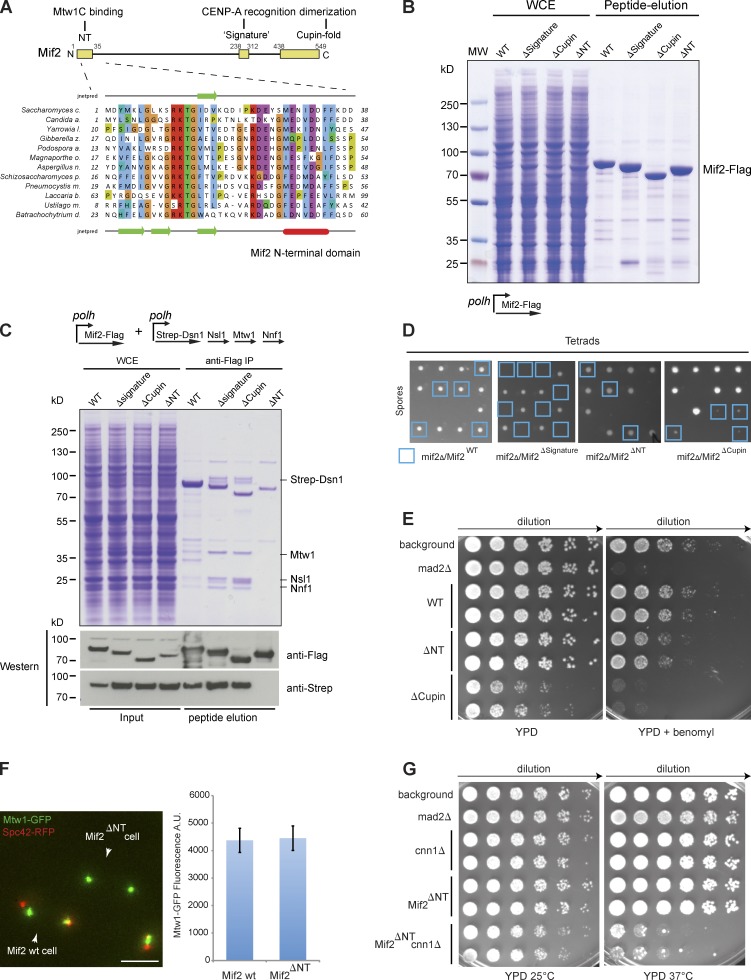
Preventing an interaction between Ame1 and the Mtw1 complex led to a loss of both proteins from the kinetochore, implying that their recruitment to centromeres depends on additional CCAN subunits. The CENP-C homologue Mif2 is a prime candidate for such a factor because it is an essential CCAN component in all systems. Previous work has defined three important domains that are common to CENP-C proteins: a C-terminal cupin fold, which mediates the homodimerization of the molecule (Cohen et al., 2008); a so-called CENP-C signature motif that has recently been shown to provide a direct contact to the hydrophobic C terminus of CENP-A (Kato et al., 2013), thus discriminating between CENP-A and H3 nucleosomes; and an N-terminal conserved domain, which confers a direct interaction between human CENP-C and the Mis12 complex (Screpanti et al., 2011; Fig. 6 A). To dissect the contributions of these domains to kinetochore assembly, we generated recombinant baculoviruses and purified full-length Mif2 as well as mutant versions lacking the N-terminal motif (ΔN), the signature sequence (ΔSignature), or the cupin fold (ΔCupin) from insect cells (Fig. 6 B). As predicted, deletion of the cupin domain prevented dimerization, resulting in an increased elution volume from the gel filtration column (Fig. S2 A). After coexpression, the four-protein Mtw1 complex copurified with Mif2-Flag from insect cells and formed a defined complex. Copurification of the Mtw1 complex with Mif2-Flag was maintained in ΔSignature and ΔCupin mutants, but abrogated in the ΔN mutant (Fig. 6 C). Thus, similar to human CENP-C, yeast Mif2 contains a binding domain for the Mtw1 complex located in the extreme N terminus of the molecule. We next analyzed the phenotypes of these Mif2 mutants in vivo. Tetrad analysis indicated that deletion of the CENP-C signature motif was lethal, whereas deletion of the cupin fold produced slowly growing but viable spores (Fig. 6 D). Surprisingly, deletion of the N-terminal Mtw1 complex binding domain was well tolerated, causing only a minor growth defect in the presence of the microtubule-destabilizing drug benomyl (Fig. 6 E). Consistent with the lack of a pronounced growth phenotype, the level of Mtw1-GFP at kinetochore clusters appeared unchanged as judged by live cell microscopy (Fig. 6 F). Nevertheless, the N terminus of Mif2 contributes to outer kinetochore function as indicated by a synthetic growth phenotype with a deletion of the yeast CENP-T homologue Cnn1 (Fig. 6 G). We conclude that CENP-A recognition via the signature motif is essential for Mif2 kinetochore function in vivo, whereas binding to the Mtw1 complex via the N terminus or homodimerization via the cupin fold plays a functionally less important role.
Figure 6.
Molecular dissection of Mif2CENP-C function. (A) Domain organization of Mif2 and presumptive functions of individual domains. Close-up view shows multiple sequence alignments of a conserved N-terminal domain in CENP-C proteins form divergent yeasts, with secondary structure predictions on top and bottom. Green arrow denotes β fold and red bar denotes α helix. (B) Expression and purification of Mif2 variants in SF9 insect cells. Coomassie-stained gel shows whole cell extracts and purified proteins after Flag peptide elution. (C) A Mif2 N-terminal deletion mutant is defective in interaction with the Mtw1 complex. Mif2-Flag wild type and mutants were coinfected with a virus containing Strep-tagged Mtw1 complex. Coomassie-stained gel and Western blots of input and flag peptide elution are shown. (D) Tetrad analysis of different Mif2 mutants. Note that haploid Mif2ΔSignature spores were not recovered and that haploid Mif2ΔCupin mutants have a small spore size. (E) Serial dilution growth assay of Mif2 mutants on YPD or YPD + 20 µg/ml benomyl. (F) Analysis of Mtw1-GFP fluorescence at kinetochore clusters in Mif2 wild-type and ΔNT cells. Bar, 5 µm. Quantification of fluorescence intensity; n = 30 clusters. (G) Serial dilution assay showing temperature-sensitive synthetic growth phenotype of Mif2ΔNT mutant in combination with a deletion of the yeast CENP-T homologue Cnn1.
Mif2 and Ame1–Okp1 interact to form a binding scaffold for Mtw1 complexes at the inner kinetochore
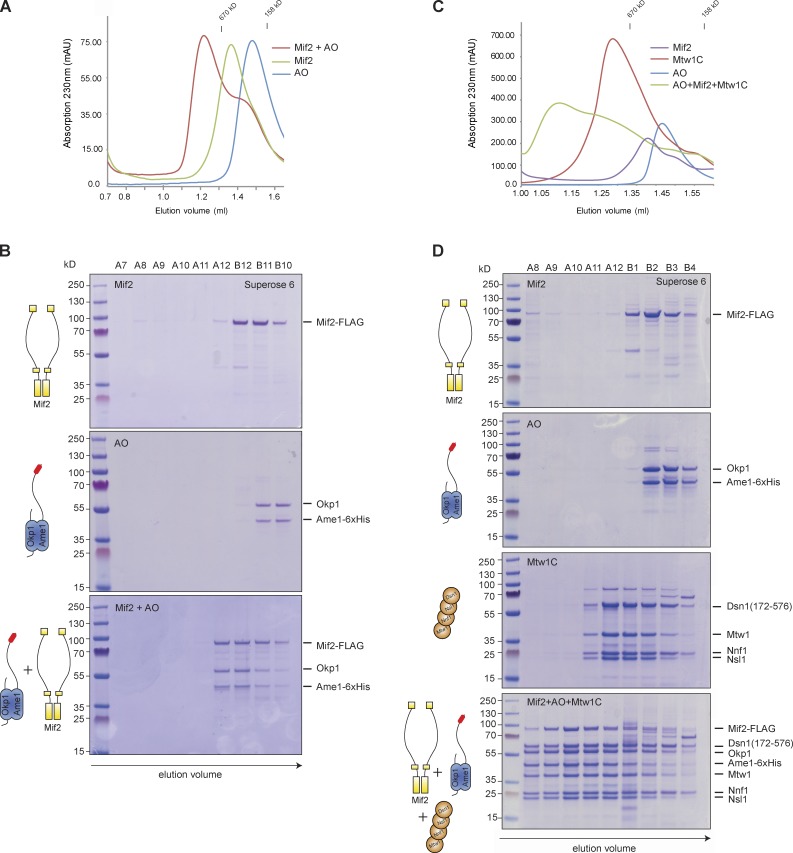
Besides recognition of the CENP-A nucleosome Mif2 may additionally contribute to inner kinetochore formation through direct binding of AO. Indeed, SEC indicated the formation of a stable Mif2–AO complex (Fig. 7, A and B) that did not require dimerization of Mif2 (Fig. S2 B). We next asked whether Mif2 and the AO complex compete for Mtw1 complex interaction, or whether their association is compatible with the binding of multiple Mtw1 complexes. In comparison to the individual components, a combination of Mif2, AO, and the Mtw1 complex eluted early from SEC with the peak fractions containing stoichiometric amounts of all seven components (Fig. 7, E and D). Comparison to the elution position of the Mtw1 complex alone indicates that the main molecular species formed under these conditions contains at least two copies of the Mtw1 complex, suggesting a 2:2:2 stoichiometry of the Mif2–AO–Mtw1C complex. The elution profile remained relatively broad, however; therefore we cannot exclude the possibility of higher-order associations. Removing Mtw1 binding sites through the Ame115–325 mutation eliminated the fast eluting species (Fig. S3). Thus, Mif2, the AO complex, and the Mtw1 complex can associate into a stable complex whose formation depends on the conserved motif in the Ame1 N terminus.
Figure 7.
Mif2 and the AO complex interact to form a stable binding platform for Mtw1 complexes. (A) SEC analysis of Mif2–AO–Mtw1C assembly. SEC elution profiles (A) and Coomassie-stained gels (B) showing elution behavior of Mif2-Flag alone (top), Ame–Okp1 alone (middle), and a stoichiometric combination (5 µM) of both (bottom). Elution position of marker proteins Thyroglobulin (670 kD) and γ-globulin (158 kD) are indicated at the top. The depicted experiment is representative of n > 3 binding assays under identical conditions. The same Ame1–Okp1 data are also presented in Fig. S2 B. (C) SEC elution profiles and Coomassie-stained gels (D) showing elution behavior of Mif2-Flag alone (5 µM), the AO complex (5 µM), the Mtw1 complex (10 µM), and a combination of Mif2-Flag, Mtw1 complex, and AO complex (bottom). The depicted experiment is representative of n > 3 binding assays under identical conditions. The same Mif2 + AO + Mtw1C data are also presented in Fig. S3.
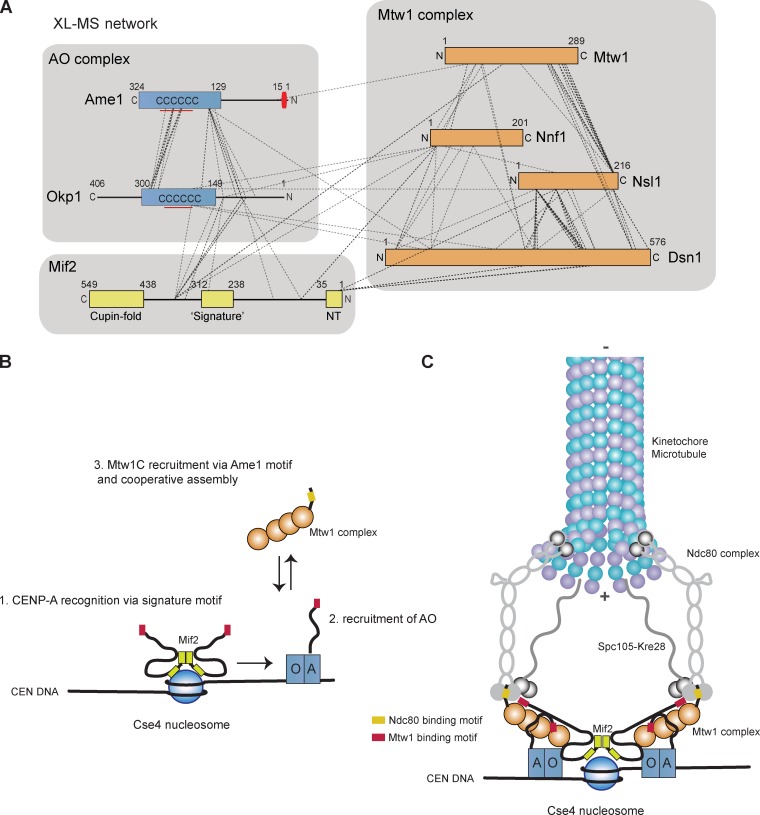
To gain further insights into the topology of this network of inner kinetochore proteins, we performed chemical cross-linking of the Mif2–AO–Mtw1C assembly followed by detection of cross-linked peptides by MS (Fig. 8 A). The method exploits the ability of Bis[sulfosuccinimidyl] suberate to cross-link primary amines of lysine residues within a distance compatible with the cross-linker (11 Å). In total, 154 intersubunit cross-links between the seven proteins were mapped (Table S3). The detection of extensive cross-links between the conserved CENP-U and -Q domains confirmed the biochemical characterization of the requirements for AO heterodimerization. Furthermore, in accordance with our biochemical analysis, the N-terminal Mif2 residues Lys9 and Lys12 were involved in cross-links to the Mtw1 complex subunits Dsn1 and Nsl1. Importantly, cross-links between the N terminus of Ame1 (Lys30) and the N-terminal domain of Mtw1 (Lys49 and Lys51) were detected, further illustrating the key role of the conserved Ame1 motif. We failed to identify cross-links involving lysine residues in the Ame1 motif itself, possibly because they are buried in the interaction interface and are therefore not accessible for the cross-linker. A key role for Okp1 in establishing the contact to Mif2 is suggested by the detection of multiple cross-links between these subunits. In particular the Mif2 residues Lys342, Lys343, and Lys363, located between the cupin fold and the Signature motif, were found to be physically close to Okp1. In summary, the cross-linking map provides a comprehensive overview of the connectivity between these essential kinetochore subunits.
Figure 8.
Network of protein interactions at the inner kinetochore and critical steps in yeast kinetochore assembly. (A) Cross-link (XL) MS analysis of the Mif2–AO–Mtw1 complex using the cross-linker Bis[sulfosuccinimidyl] suberate. Topological map of the Mif2–AO–Mtw1 complex assembly based on the identified intersubunit cross-links. See Table S3 for a full list of identified cross-links. (B) CENP-A recognition and initiation of outer KT assembly are distinct steps performed by different CCAN subunits. CENP-A recognition depends on the signature motif in Mif2. Although Mif2 contains a binding site for the Mtw1 complex, this association is insufficient to trigger outer kinetochore assembly. Only after AO recruitment and the localization of the Mtw1 complex via the conserved Ame1 motif, yeast centromeres become competent to assemble the outer kinetochore. Arrows indicate localization dependencies. (C) Modular architecture of the yeast kinetochore illustrating the central role of the AO complex and cooperative binding of Mtw1 complexes mediated by Mif2 and AO.
Discussion
An essential role for the AO complex in the budding yeast kinetochore
Our biochemical and genetic experiments have revealed key molecular requirements for the assembly of the yeast kinetochore. Fundamentally, the goal of kinetochore formation is to trigger the specific association of multiple copies of the KMN network in the immediate vicinity of a nucleosome in which the histone H3 is replaced by Cse4/CENP-A. The severe kinetochore phenotype of biochemically defined mutants demonstrates that in budding yeast the critical connection between inner and outer kinetochore hinges on the Ame1CENP-U subunit and a short binding motif that establishes a link to the Mtw1 complex as the central KMN component (Fig. 8, B and C).
What is the underlying reason for this key role of Ame1CENP-U in yeast? Other organisms such as Caenorhabditis elegans and Drosophila melanogaster seem to lack an extended CCAN (including CENP-U) entirely and solely rely on CENP-C to connect the KMN network to CENP-A chromatin. In budding yeast the interaction of a Mif2 homodimer with a single centromeric nucleosome may not configure a sufficient number of high-affinity binding sites for the assembly of multiple copies of the KMN network, which could necessitate the usage of an additional Mtw1 recruitment factor that can enhance affinity through cooperative binding. This requirement may be more relaxed in systems where multiple CENP-A nucleosomes are brought together to provide a binding platform for the CCAN. In human cells it has been shown that overexpression of the N terminus of CENP-C disrupts kinetochore function by a dominant-negative effect (Screpanti et al., 2011). As the loss of Mis12 complex binding by CENP-C has not been directly tested, this leaves open the possibility of alternative routes of Mtw1 complex recruitment. Although a clear conservation of the Ame1 motif can only be detected with confidence for fungal CENP-U proteins, secondary structure predictions indicate the presence of short structured domains also at the N terminus of human CENP-U, hinting that a Mis12 complex binding function of the OPQUR subcomplex might be conserved.
Implications for kinetochore architecture
Our study contributes to a deeper understanding of the design principles of kinetochores. Extending an architectural theme already found in CENP-T and CENP-C proteins, yeast CENP-U uses a linear, peptide-like motif in an otherwise flexible extension of the molecule to establish the connection to the Mtw1 complex. Given the emergence of the Mtw1 complex as a binding scaffold for proteins containing RWD domains (Petrovic et al., 2014), it may be somewhat surprising that the RWD domain–containing proteins Ctf19–Mcm21 do not seem to be able to bind to the Mtw1 complex in isolation. We cannot exclude, however, that in the context of the full COMA complex, Ctf19–Mcm21 might be contacting the Mtw1 complex with their RWD domains and therefore contribute to the overall affinity and structure of the assembly. A further characterization of the DNA-binding activity of the AO complex will be necessary to evaluate the extent of its contribution to kinetochore function. For a variety of fungal CENP-U proteins, “AT-hook” domains similar to the DNA binding motifs found in CENP-C are predicted (unpublished data), but for Ame1 itself this assignment is uncertain. It seems plausible that both Mif2 and Ame1–Okp1 associate with the AT-rich DNA that wraps around the Cse4 nucleosome. Cooperative binding of multiple AO complexes could then provide several binding sites for the Mtw1 complex. Our structural analysis gives insights into the topology of the Mtw1 complex within the kinetochore organization: the AO complex interacts specifically with the larger lobe of the Mtw1 complex. This conclusion is strengthened by our cross-linking analysis, which detected physical proximity between the N terminus of Ame1 and the N terminus of Mtw1. Maskell et al. (2010) have shown that MBP fusions to the N terminus of Mtw1 are localized at the larger lobe of the Mtw1 complex, resembling the AO binding detected in our study. Based on recent electron microscopy results (Petrovic et al., 2014), this places the DNA-binding components AO juxtaposed to the two outer members of the KMN network, Ndc80 and Spc105, which are at the other lobe of the Mtw1 complex, thus rationalizing the central role of AO within the kinetochore architecture. A key goal for future structural analysis will be to visualize higher order assemblies that reveal how multiple copies of the Mtw1 complex are organized. The biochemical insights provided in our study allow a good starting point for such an analysis.
A multistep assembly pathway of the budding yeast kinetochore
In the process of kinetochore assembly, cells have to balance conflicting requirements. On the one hand, assembly should be rapid, such that after replication yeast centromeres quickly become competent to bind to microtubules and initiate the process of sister chromatid biorientation on the spindle. On the other hand, kinetochore assembly needs to be highly specific to avoid the deleterious effects of assembling multiple kinetochores on the same chromosome. This conflict is especially evident in budding yeast, where kinetochore assembly is initiated around a single CENP-A nucleosome, which is overall very similar to its H3 counterpart (Furuyama and Biggins, 2007; Tachiwana et al., 2011). Moreover, CENP-A nucleosomes frequently become deposited in noncentromeric regions, yet this usually does not lead to the assembly of a functional kinetochore, even under conditions of CENP-A overexpression (Lefrançois et al., 2013). Our results provide indications for how specific and cooperative assembly of a kinetochore is achieved. Although Mif2CENP-C is required for the specificity of the assembly via its CENP-A binding motif, it is only upon association of the AO complex that yeast centromeres become competent to assemble the outer kinetochore. The additional requirement of a key molecular factor that initiates outer kinetochore assembly may provide a form of kinetic proofreading that protects against inappropriate assembly. Such assembly intrinsic mechanisms may work together with additional factors that control the level of CENP-A nucleosomes at chromosome arms (Hewawasam et al., 2010; Ranjitkar et al., 2010). The essential kinetochore components, Mif2, Mtw1 complex, and Ame1–Okp1, may also be important points of regulation during assembly as indicated by the presence of critical phosphorylation sites on these subunits (Westermann et al., 2003; Akiyoshi et al., 2013). This might be especially important immediately before and after replication of centromeric DNA. Our experiments provide a biochemical framework in which the regulation of the kinetochore assembly process, for example through cell cycle kinases, can be tested in the future.
Materials and methods
Protein expression and purification
Expression constructs for kinetochore proteins were created by amplifying the respective genes from yeast genomic DNA and cloning them using appropriate restriction sites into pST39, pET28, pETDuett, pACYCDuet, pGEX6P, or pCOLA plasmids (bacterial expression) or pFL plasmids (insect cell expression). For list of constructs please see Table S1. Unless indicated otherwise the following conditions are valid for all protein expressions performed in this work. Cells were grown at 37°C until OD600 of 0.6 and subsequently induced with 0.5 mM IPTG. Expressions were conducted overnight at 18°C with the exception of the MNc (0.5 mM IPTG; 4 h at 37°C). Expression of Mtw1c and MNc was performed in BL21 (DE3; Novagen). AOc and CMc were expressed in Rosetta 2(pLys) (DE3) cells (EMD Millipore). Lysis, wash, and elution buffers as well as chromatography steps varied for the different protein complexes and are described in the individual sections. Sonication of bacteria was performed in the presence of protease inhibitors (Roche) and PMSF. The 6×Histidine fusion proteins were isolated with Ni-NTA agarose beads (QIAGEN), whereas M2 affinity agarose (Sigma-Aldrich) was used for FLAG-tagged proteins. Incubation with affinity beads was performed in batch format in a 50-ml Falcon tube for 1 h at 4°C.
Ndc80 complex
Expression and purification of yeast Ndc80 complex was performed as described previously (Lampert et al., 2010). In brief, the two subcomplexes of the Ndc80 complex (Ndc80–6×His–Nuf2-EGFP and Spc24–6×His–Spc25p) were separately expressed from the pETDuett or pACYCDuet-1 vectors (Novagen). Both plasmids were cotransfected into BL21 DE3 (EMD Millipore). Bacteria were grown to OD600 = 0.6 at 37°C, induced with 0.2 mM IPTG, and grown for 12–15 h at 18°C. The two subcomplexes were eluted with 150 mM NaCl, 10 mM Hepes, pH 7.0, and 250 mM imidazol from the Ni-NTA beads, and then subjected to in vitro reconstitution of the full-length Ndc80 complex and further purified on the Superdex 200 HiLoad 16/60 (GE Healthcare). Gel filtration was conducted in 150 mM NaCl and 10 mM Hepes, pH 7.
Mtw1 complex
Lysis buffer for Mtw1 complex (N-terminal 6×His tag on the Dsn1 subunit) was 50 mM Tris-HCL, pH 7.5, 500 mM NaCl, 30 mM imidazole, 10% glycerol, and 0.5% Tween 20 as described previously (Maskell et al., 2010). Elution was performed in batch with 30 mM Tris, pH 8.5, 80 mM NaCl, and 250 mM imidazole. Subsequently, proteins were directly loaded onto anion exchange chromatography (MonoQ5/50 GL column; GE Healthcare). The column was developed with 2–column volume (CV) gradient wash from 0–20% buffer B, followed by 15-CV wash of 20% buffer B. The conductivity is at 25 mS/cm. The chromatography was performed with a step gradient consisting of buffer (30 mM Tris HCl, pH 8.5, and 5% glycerol from 80 mM [A] to 1 M NaCl [B]) using a flow rate of 1 ml/min. The protein is eluted within 2 CV by increasing buffer B to 50%.
MN complex purification
Purification was performed as for Mtw1 complex. Additionally, after the ion exchange chromatography a SEC was conducted with buffer (30 mM Hepes, pH 7.5, 150 mM NaCl, and 5% glycerol).
AO complex purification
AO15–324 as well as the CM complex purification pellets were resuspended in lysis buffer (30 mM Na/K phosphate, pH 7.0, 30 mM imidazole, 1 M NaCl, and 1 mM DTT). To remove the E. Coli DNA that was bound to the AO complex, beads were washed six times for 10 min at 4°C while rotating with wash buffer (30 mM Hepes, pH 7.5, 30 mM imidazole, 5 mM CaCl2, 1 mM DTT, and 600 mM NaCl). AO-6×His was eluted from a Ni-NTA column using a step gradient consisting of a 50-mM imdazole step followed by 300 mM imidazole for elution. Subsequent purification over gel filtration was performed in 30 mM Na/K phosphate, pH 7.0, and 50 mM ammonium sulfate.
Mif2 and Mtw1 complex purification from insect cells
Open reading frames encoding kinetochore subunits were amplified from yeast genomic DNA and cloned into the pFL vector for insect cell expression. Generation of recombinant viruses expressing multiple subunits was performed according to the multi-Bac system (Trowitzsch et al., 2010). For expression and purification, insect cells were opened with a Dounce homogenizer in Lysis buffer (20 mM Na2HPO4/NaH2PO4, pH 7.0, 300 mM NaCl, and 1 mM EDTA) with Complete EDTA-free protease inhibitors (Roche). Cleared extracts were incubated with M2 anti-Flag agarose (Sigma-Aldrich) for 2 h, washed 3× with buffer, and eluted with 2 mg/ml 3×Flag peptide in Lysis buffer.
For purification of 6×His-tagged proteins, insect cells were lysed in 20 mM Na2HPO4/NaH2PO4, pH 7.0, 300 mM NaCl, 20 mM imidazole, 1 mM PMSF, and Complete protease inhibitors. Cleared extracts were incubated with Ni-NTA agarose (QIAGEN) for 2 h and washed three times, and proteins were eluted with 200 mM imidazole in Lysis buffer.
Coexpression and purification of MNc with Ame1
Ame1-FLAG was cloned into cassette 4 of the pST39 plasmid that already contained the subunits Mtw1, Nsl1, and Nnf1-6×His. Bacterial expression was performed at 37°C for 4 h. Lysis buffer was PBS with 5% glycerol. After binding to the M2 affinity gel (Sigma-Aldrich) beads were washed with PBS, 5% glycerol, and either 150 or 600 mM NaCl. Elution was performed with 2 mg/ml 3×FLAG peptide. Complex formation was detectable even under conditions of 600 mM NaCl.
Interaction studies using SEC
Analytical SEC experiments were performed on Superose 6 10/30 (CMc + Mtw1c) or on Superose 6 3.2/30 columns. All SEC interaction studies were conducted under isocratic elution conditions at 4°C. The protein elution was monitored by absorbance at a wavelength of 280 nm. For analytical SEC, proteins were mixed in equimolar ratios between 5 and 15 µM and incubated for 10–30 min on ice. The eluates were fractioned, loaded onto SDS-PAGE, and stained with Coomassie.
ITC
ITC experiments were performed in 30 mM Hepes, pH 7.5, 150 mM NaCl, and 2% glycerol at 28°C using a VP-ITC calorimeter (Microcal Inc.). In the individual experiments, the following protein concentrations were titrated into the device cell that contained the protein of lower molarity: GST-Ame 1–30 (400 mM) into MN (25 mM); MN (243 mM) into AO (24 mM); and, as a control experiment, MN (271 mM) into GST (18 mM). The calculated molarities were based on the assumption that GST is present as a monomer in solution. Thermodynamic values and stoichiometries were obtained using Origin 7 (OriginLab Corp.) by fitting the binding isotherms with the nonlinear least squares method, assuming one set of binding sites.
Electron microscopy
Mtw1 and Mtw1AO complexes were further prepared for electron microscopy analysis using the GraFIX method (Kastner et al., 2008). 200 µg of the complex was layered onto a 10–20% glycerol gradient in 25 mM Hepes, pH 7.5, and 100 mM NaCl, which also contained 0.02–0.15% glutaraldehyde gradient, and ultracentrifuged at 56,000 rpm for 14 h at 4°C. Fractions were analyzed by SDS-PAGE and fractions containing monomeric cross-linked complex were deposited onto a freshly glow-discharged, carbon-coated copper grid, blotted after 1 min, and stained with 2% uranyl formate for 1 min. Images were collected on an 11-megapixel charge coupled device camera (Morada; Olympus) using a Morgani 268D electron microscope (FEI) operated at 80 kV and 71,000 magnification. Approximately 2,000 particles that appeared to correspond to full Mtw1 and Mtw1AO complexes were picked in EMAN2 boxer, and class averages were calculated by XMIPP ML2D. Other measurements were performed using ImageJ (National Institutes of Health).
Ame1-Flag purification and MS
Ame1-associated proteins were obtained by a single-step antibody purification with a 6×Flag tag fused to the Ame1 C terminus according to a previously published protocol with a few exceptions (Akiyoshi et. al., 2010). The logarithmic yeast culture was collected at OD600 = 1, and the cells were harvested and frozen as droplets in liquid N2. Yeast cells were subsequently lysed with a freezer mill (Biospec). The cleared lysate was incubated for 2 h with 50 µl of Dynabeads coupled with M2 flag antibody. The bound protein complex was predigested overnight at 37°C by addition of 500 ng LysC (sequencing grade; Wako Pure Chemical Industries). Cysteines were reduced (6.5 mM TCEP; 30 min at 56°C) and alkylated (40 mM MMTS; 30 min at RT in dark) followed by overnight digestion at 37°C by addition of 500 ng trypsin (sequencing grade). Before the MS analysis the samples were acidified with 10% TFA. To check the immunopurification, 15 µl of beads were used to elute with 3×Flag peptide and SDS-PAGE gel was run.
MS analysis was performed on a 5600 TripleTOF (AB Sciex) coupled with an Ultramate 3000 RCLC nanosystem (Thermo Fisher Scientific). The samples were measured with a 120-min linear gradient. For data-dependent analysis experiments (protein identification and spectral library generation), the mass spectrometer was operated in a manner where a TOF-MS scan was collected, from which the top 20 ions were selected, in a total cycle time of 2.3 s. For SWATH MS-based experiments (protein quantification), the method was set in a similar manner to a previously establish method (Gillet et al., 2012). The isolation window was set to 35 D and the total cycle time was 1.29 s. All data-dependent analysis MS files were searched using ProteinPilot software v. 4.5 (AB Sciex) with the Paragon algorithm with the following parameters: MMTS cysteine alkylation, digestion by trypsin, and search against yeast database (http://www.yeastgenome.org). Spectral alignment and targeted data extraction of SWATH samples was performed using PeakView v.1.2 (AB Sciex). For the protein quantification, three manually selected peptides with three transitions per peptide were used in Marker View v.1.2 (AB Sciex).
Chemical cross-linking and MS of the Mtw1–Mif2–Ame1–Okp1 complex
Recombinant protein complex at a concentration of ∼0.2 mg/ml was cross-linked with 100 µM isotopically labeled (d0/d12) Bis[sulfosuccinimidyl] suberate (Creative Molecules) at 35°C for 30 min. Reaction was quenched with 50 mM ammonium bicarbonate and supplemented with 8 M urea to a final concentration of 6 M. After reduction and alkylation cross-linked proteins were digested with Lys-C (1:50 wt/wt; Wako Pure Chemical Industries) for 3 h, diluted with 50 mM ammonium bicarbonate to 1 M urea, and digested with trypsin (1:50 wt/wt; Promega) overnight. Cross-linked peptides were purified by reversed phase chromatography using C18 cartridges (Sep-Pak; Waters). Cross-linked peptides were enriched by peptide SEC and analyzed by tandem MS (Orbitrap Elite; Thermo Fisher Scientific; Herzog et al., 2012). Fragment ion spectra were searched and cross-links were identified by the dedicated software program xQuest (Walzthoeni et al., 2012).
Yeast genetics
Yeast strains were constructed in the S288C background. Yeast strain generation and methods were performed by standard procedures. The anchor-away approach for characterization of Ame1 mutants was performed as described (Haruki et al., 2008), using the ribosomal RPL13-FKBP12 anchor. Final rapamycin concentration in plates or liquid media was 1 µg/ml. For a list of strains see Table S2.
For live cell microscopy, cells were grown in synthetic medium and imaged on concanavalin A–coated coverslips by live cell Deltavision deconvolution microscopy (Applied Precision) on an IX-71 microscope (Olympus) controlled by Softworx and equipped with a Plan Apochromat 100× 1.4 NA objective (Olympus) and a Coolsnap HQ camera (Photometrics) at 25°C. Z stacks (8 × 0.35 µm apart) were acquired and projected into 2D images. Images were processed and analyzed using ImageJ, Photoshop (Adobe), and Metamorph (Molecular Devices). Quantification of kinetochore fluorescence intensity was performed on maximum intensity projections of equally scaled images in ImageJ. A rectangular region of interest was defined around a kinetochore cluster, and the integrated kinetochore fluorescence intensity was measured after background subtraction of an equally sized nonkinetochore area. Kinetochore fluorescence was quantified for at least 30 clusters in three independent biological experiments.
Online supplemental material
Fig. S1 shows microtubule cosedimentation assay of the AO complex. Fig. S2 shows gel filtration analysis of Mif2 mutants and additional interaction studies of Mif2 mutants with the AO complex. Fig. S3 provides additional gel filtration analysis of the binding between a mutant AO complex lacking the conserved Ame1 motif and the Mif2 and Mtw1 complexes. Table S1 lists plasmids used in this study and Table S2 lists yeast strains. Table S3 provides a comprehensive list of all interprotein cross-links found by the MS analysis of the Mif2–AO–Mtw1 complex. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201403081/DC1.
Supplementary Material
Acknowledgments
The authors thank all members of the Westermann laboratory for discussions and critical reading of the manuscript.
This work received funding from the European Research Council under the European Community’s Seventh Framework Program (S. Westermann; FP7/2007-2013)/ERC grant agreement no. 203499) and from the Austrian Science Fund (S. Westermann; SFB F34-B03). The Research Institute of Molecular Pathology is funded by Boehringer Ingelheim.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere-associated network
- CENP
- centromere protein
- ITC
- isothermal titration calorimetry
- MS
- mass spectrometry
- SEC
- size exclusion chromatography
References
- Akiyoshi, B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., and Biggins S.. 2010. Tension directly stabilizes reconstituted kinetochore–microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi, B., Nelson C.R., and Biggins S.. 2013. The aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics. 194:785–789 10.1534/genetics.113.150839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin, G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., and Nogales E.. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro, A.C., Samora C.P., Holtackers R., Wang E., Kingston I.J., Alonso M., Lampson M., McAinsh A.D., and Meraldi P.. 2010. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12:319–329 10.1038/ncb2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S.2013. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 194:817–846 10.1534/genetics.112.145276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, L.J., Pagliuca C., Kobayashi N., Grove R.A., Oku Y., Shrestha K., Alfieri C., Golfieri C., Oldani A., Dal Maschio M., et al. 2012. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat. Cell Biol. 14:614–624 10.1038/ncb2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, C.W., Milks K.J., and Straight A.F.. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Ciferri, C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., and Simons K.T.. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491 10.1091/mbc.E08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz, D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Furuyama, S., and Biggins S.. 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 104:14706–14711 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet, L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., and Aebersold R.. 2012. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. O11:016717 10.1074/mcp.O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse, A., Carroll C.W., Moree B., Fuller C.J., and Straight A.F.. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 477:354–358 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, J., Mishra P.K., Stephens A., Haggerty R., Quammen C., Taylor R.M. II, Yeh E., Basrai M.A., and Bloom K.. 2013. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr. Biol. 23:1939–1944 10.1016/j.cub.2013.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki, H., Nishikawa J., and Laemmli U.K.. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31:925–932 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Herzog, F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F.U., Ban N., et al. 2012. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 337:1348–1352 10.1126/science.1221483 [DOI] [PubMed] [Google Scholar]
- Hewawasam, G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., Florens L., Workman J.L., and Gerton J.L.. 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell. 40:444–454 10.1016/j.molcel.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008a. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Hori, T., Okada M., Maenaka K., and Fukagawa T.. 2008b. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell. 19:843–854 10.1091/mbc.E07-06-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung, P., Maier M., Alushin G.M., Lander G.C., Nogales E., and Westermann S.. 2011. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J. Mol. Biol. 405:548–559 10.1016/j.jmb.2010.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, S., Wang Z., Jiang K., Huang Y., Ward T., Zhao L., Dou Z., and Yao X.. 2011. CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment. J. Biol. Chem. 286:1627–1638 10.1074/jbc.M110.174946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A.P., Bloom K., and Salmon E.D.. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner, B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., et al. 2008. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 5:53–55 10.1038/nmeth1139 [DOI] [PubMed] [Google Scholar]
- Kato, H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., and Bai Y.. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 340:1110–1113 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassovsky, K., Henikoff J.G., and Henikoff S.. 2012. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 109:243–248 10.1073/pnas.1118898109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert, F., Hornung P., and Westermann S.. 2010. The Dam1 complex confers microtubule plus end–tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois, P., Auerbach R.K., Yellman C.M., Roeder G.S., and Snyder M.. 2013. Centromere-like regions in the budding yeast genome. PLoS Genet. 9:e1003209 10.1371/journal.pgen.1003209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., and Murray A.W.. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531 10.1016/0092-8674(81)90015-5 [DOI] [PubMed] [Google Scholar]
- London, N., and Biggins S.. 2014. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 28:140–152 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London, N., Ceto S., Ranish J.A., and Biggins S.. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi, F., Litos G., Schleiffer A., Heuck A., Mechtler K., Clausen T., and Westermann S.. 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32:409–423 10.1038/emboj.2012.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell, D.P., Hu X.W., and Singleton M.R.. 2010. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190:823–834 10.1083/jcb.201002059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino, T., Rago F., Hori T., Tomii K., Cheeseman I.M., and Fukagawa T.. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J., Stemmann O., Rank S., and Lechner J.. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140–1155 10.1101/gad.13.9.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic, A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852 10.1083/jcb.201002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic, A., Mosalaganti S., Keller J., Mattiuzzo M., Overlack K., Krenn V., De Antoni A., Wohlgemuth S., Cecatiello V., Pasqualato S., et al. 2014. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell. 53:591–605 10.1016/j.molcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Primorac, I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., and Musacchio A.. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2:e01030 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka, M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., and Glover D.M.. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Ranjitkar, P., Press M.O., Yi X., Baker R., MacCoss M.J., and Biggins S.. 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell. 40:455–464 10.1016/j.molcel.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida, S., and Musacchio A.. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer, A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., and Westermann S.. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14:604–613 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- Schmitzberger, F., and Harrison S.C.. 2012. RWD domain: a recurring module in kinetochore architecture shown by a Ctf19–Mcm21 complex structure. EMBO Rep. 13:216–222 10.1038/embor.2012.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti, E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., and Musacchio A.. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd, L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., and Millar J.B.. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana, H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.Y., et al. 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 476:232–235 10.1038/nature10258 [DOI] [PubMed] [Google Scholar]
- Trowitzsch, S., Bieniossek C., Nie Y., Garzoni F., and Berger I.. 2010. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 172:45–54 10.1016/j.jsb.2010.02.010 [DOI] [PubMed] [Google Scholar]
- Walzthoeni, T., Claassen M., Leitner A., Herzog F., Bohn S., Förster F., Beck M., and Aebersold R.. 2012. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods. 9:901–903 10.1038/nmeth.2103 [DOI] [PubMed] [Google Scholar]
- Westermann, S., and Schleiffer A.. 2013. Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol. 23:260–269 10.1016/j.tcb.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Westermann, S., Cheeseman I.M., Anderson S., Yates J.R. III, Drubin D.G., and Barnes G.. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215–222 10.1083/jcb.200305100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi, Y., Yang C.H., Tanno Y., and Watanabe Y.. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.