Abstract
Microtubules are cytoskeletal filaments that are dynamically assembled from α/β-tubulin heterodimers. The primary sequence and structure of the tubulin proteins and, consequently, the properties and architecture of microtubules are highly conserved in eukaryotes. Despite this conservation, tubulin is subject to heterogeneity that is generated in two ways: by the expression of different tubulin isotypes and by posttranslational modifications (PTMs). Identifying the mechanisms that generate and control tubulin heterogeneity and how this heterogeneity affects microtubule function are long-standing goals in the field. Recent work on tubulin PTMs has shed light on how these modifications could contribute to a “tubulin code” that coordinates the complex functions of microtubules in cells.
Introduction
Microtubules are key elements of the eukaryotic cytoskeleton that dynamically assemble from heterodimers of α- and β-tubulin. The structure of microtubules, as well as the protein sequences of α- and β-tubulin, is highly conserved in evolution, and consequently, microtubules look alike in almost all species. Despite the high level of conservation, microtubules adapt to a large variety of cellular functions. This adaptation can be mediated by a large panel of microtubule-associated proteins (MAPs), including molecular motors, as well as by mechanisms that directly modify the microtubules, thus either changing their biophysical properties or attracting subsets of MAPs that convey specific functions to the modified microtubules. Two different mechanism can generate microtubule diversity: the expression of different α- and β-tubulin genes, referred to as tubulin isotypes, and the generation of posttranslational modifications (PTMs) on α- and β-tubulin (Figs. 1 and 2). Although known for several decades, deciphering how tubulin heterogeneity controls microtubule functions is still largely unchartered. This review summarizes the current advances in the field and discusses new concepts arising.
Figure 1.
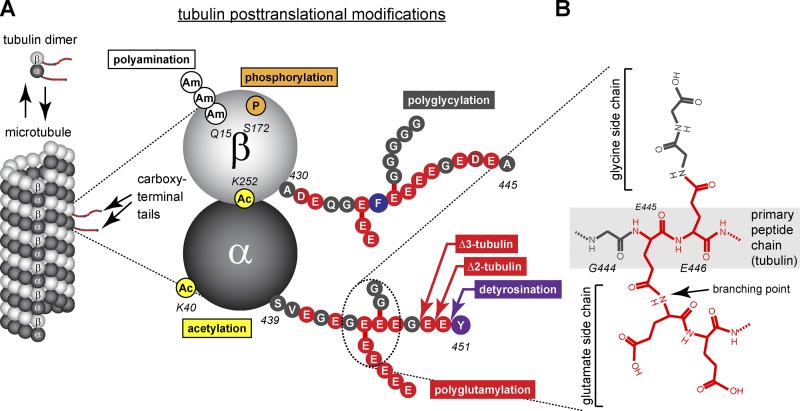
Tubulin heterogeneity generated by PTMs. (A) Schematic representation of the distribution of different PTMs of tubulin on the α/β-tubulin dimer with respect to their position in the microtubule lattice. Acetylation (Ac), phosphorylation (P), and polyamination (Am) are found within the tubulin bodies that assemble into the microtubule lattice, whereas polyglutamylation, polyglycylation, detyrosination, and C-terminal deglutamylation take place within the C-terminal tubulin tails that project away from the lattice surface. The tubulin dimer represents TubA1A and TubB2B (Fig. 2), and modification sites for polyglutamylation and polyglycylation have been randomly chosen. (B) Chemical structure of the branched peptide formed by polyglutamylation and polyglycylation, using the γ-carboxyl groups of the modified glutamate residues as acceptor sites for the isopeptide bonds. Note that in the case of polyglutamylation, the elongation of the side chains generates classical peptide bonds (Redeker et al., 1991).
Figure 2.
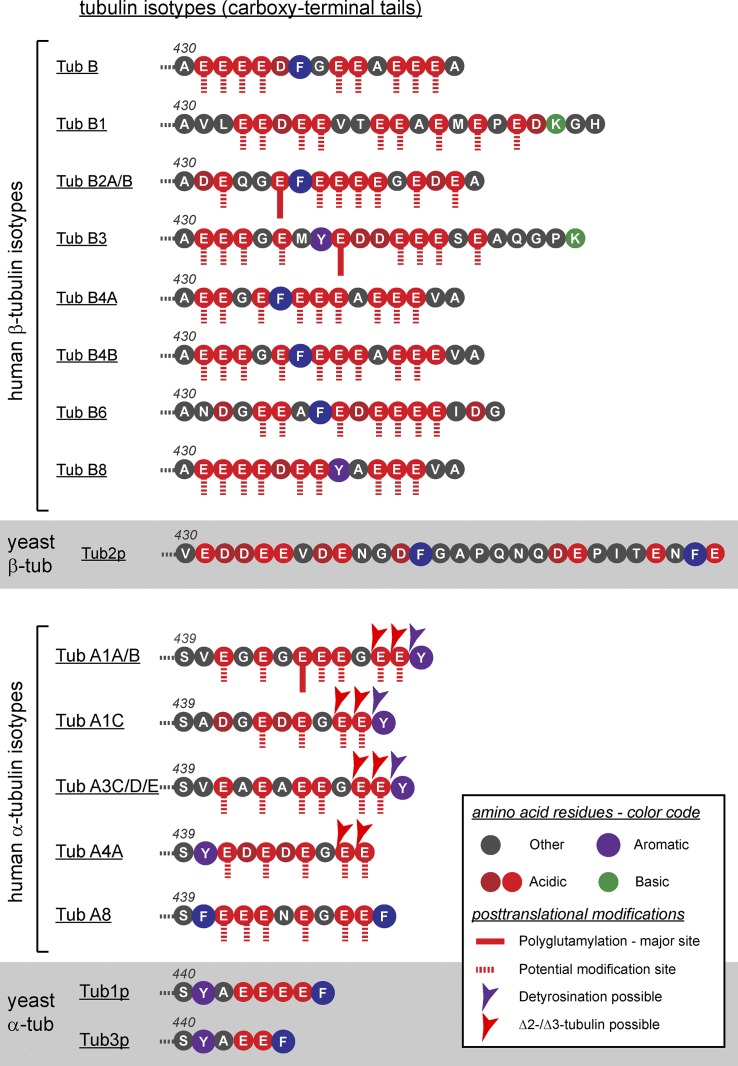
Heterogeneity of C-terminal tails of tubulin isotypes and their PTMs. The amino acid sequences of all tubulin genes found in the human genome are indicated, starting at the last amino acid of the folded tubulin bodies. Amino acids are represented in single-letter codes and color coded according to their biochemical properties. Known sites for polyglutamylation are indicated (Eddé et al., 1990; Alexander et al., 1991; Rüdiger et al., 1992). Potential modification sites (all glutamate residues) are indicated. Known C-terminal truncation reactions of α/β-tubulin (tub) are indicated. The C-terminal tails of the yeast Saccharomyces cerevisiae are shown to illustrate the phylogenetic diversity of these domains.
Tubulin isotypes
The cloning of the first tubulin genes in the late 1970’s (Cleveland et al., 1978) revealed the existence of multiple genes coding for α- or β-tubulin (Ludueña and Banerjee, 2008) that generate subtle differences in their amino acid sequences, particularly in the C-terminal tails (Fig. 2). It was assumed that tubulin isotypes, as they were named, assemble into discrete microtubule species that carry out unique functions. This conclusion was reinforced by the observation that some isotypes are specifically expressed in specialized cells and tissues and that isotype expression changes during development (Lewis et al., 1985; Denoulet et al., 1986). These high expectations were mitigated by a subsequent study showing that all tubulin isotypes freely copolymerize into heterogeneous microtubules (Lewis et al., 1987). To date, only highly specialized microtubules, such as ciliary axonemes (Renthal et al., 1993; Raff et al., 2008), neuronal microtubules (Denoulet et al., 1986; Joshi and Cleveland, 1989), and microtubules of the marginal band of platelets (Wang et al., 1986; Schwer et al., 2001) are known to depend on some specific (β) tubulin isotypes, whereas the function of most other microtubules appears to be independent of their isotype composition.
More recently, a large number of mutations in single tubulin isotypes have been linked to deleterious neurodevelopmental disorders (Keays et al., 2007; Fallet-Bianco et al., 2008; Tischfield et al., 2010; Cederquist et al., 2012; Niwa et al., 2013). Mutations of a single tubulin isotype could lead to an imbalance in the levels of tubulins as a result of a lack of incorporation of mutant isoforms into the microtubule lattice or to incorporation that perturbs the architecture or dynamics of the microtubules. The analysis of tubulin disease mutations is starting to reveal how subtle alterations of the microtubule cytoskeleton can lead to functional aberrations in cells and organisms and might provide novel insights into the roles of tubulin isotypes that have so far been considered redundant.
Tubulin PTMs
Tubulin is subject to a large range of PTMs (Fig. 1), from well-known ones, such as acetylation or phosphorylation, to others that have so far mostly been found on tubulin. Detyrosination/tyrosination, polyglutamylation, and polyglycylation, for instance, might have evolved to specifically regulate tubulin and microtubule functions, in particular in cilia and flagella, as their evolution is closely linked to these organelles. The strong link between those modifications and tubulin evolution has led to the perception that they are tubulin PTMs; however, apart from detyrosination/tyrosination, most of them have other substrates (Regnard et al., 2000; Xie et al., 2007; van Dijk et al., 2008; Rogowski et al., 2009).
Tubulin acetylation.
Tubulin acetylation was discovered on lysine 40 (K40; Fig. 1 A) of flagellar α-tubulin in Chlamydomonas reinhardtii (L’Hernault and Rosenbaum, 1985) and is generally enriched on stable microtubules in cells. Considering that K40 acetylation per se has no effect on the ultrastructure of microtubules (Howes et al., 2014), it is rather unlikely that it directly stabilizes microtubules. As a result of its localization at the inner face of microtubules (Soppina et al., 2012), K40 acetylation might rather affect the binding of microtubule inner proteins, a poorly characterized family of proteins (Nicastro et al., 2011; Linck et al., 2014). Functional experiments in cells have further suggested that K40 acetylation regulates intracellular transport by regulating the traffic of kinesin motors (Reed et al., 2006; Dompierre et al., 2007). These observations could so far not be confirmed by biophysical measurements in vitro (Walter et al., 2012; Kaul et al., 2014), suggesting that in cells, K40 acetylation might affect intracellular traffic by indirect mechanisms.
Enzymes involved in K40 acetylation are HDAC6 (histone deacetylase family member 6; Hubbert et al., 2002) and Sirt2 (sirtuin type 2; North et al., 2003). Initial functional studies used overexpression, depletion, or chemical inhibition of these enzymes. These studies should be discussed with care, as both HDAC6 and Sirt2 deacetylate other substrates and have deacetylase-independent functions and chemical inhibition of HDAC6 is not entirely selective for this enzyme (Valenzuela-Fernández et al., 2008). In contrast, acetyl transferase α-Tat1 (or Mec-17; Akella et al., 2010; Shida et al., 2010) specifically acetylates α-tubulin K40 (Fig. 3), thus providing a more specific tool to investigate the functions of K40 acetylation. Knockout mice of α-Tat1 are completely void of K40-acetylated tubulin; however, they show only slight phenotypic aberrations, for instance, in their sperm flagellum (Kalebic et al., 2013). A more detailed analysis of α-Tat1 knockout mice demonstrated that absence of K40 acetylation leads to reduced contact inhibition in proliferating cells (Aguilar et al., 2014). In migrating cells, α-Tat1 is targeted to microtubules at the leading edge by clathrin-coated pits, resulting in locally restricted acetylation of those microtubules (Montagnac et al., 2013). A recent structural study of α-Tat1 demonstrated that the low catalytic rate of this enzyme, together with its localization inside the microtubules, caused acetylation to accumulate selectively in stable, long-lived microtubules (Szyk et al., 2014), thus explaining the link between this PTM and stable microtubules in cells. However, the direct cellular function of K40 acetylation on microtubules is still unclear.
Figure 3.
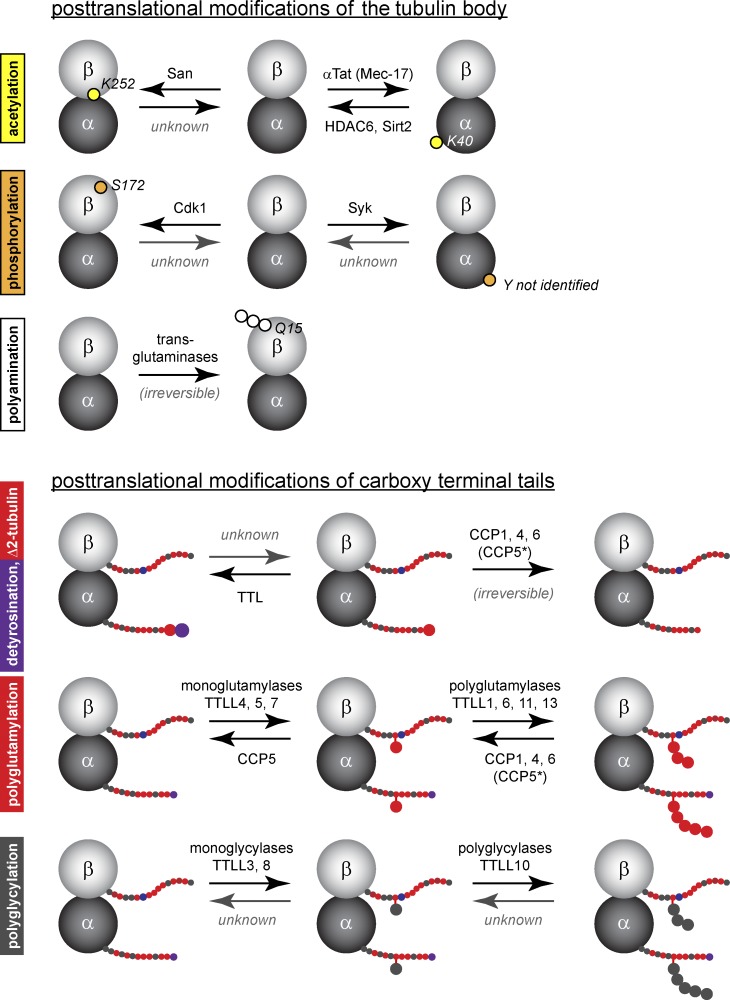
Enzymes involved in PTM of tubulin. Schematic representation of known enzymes (mammalian enzymes are shown) involved in the generation and removal of PTMs shown in Fig. 1. Note that some enzymes still remain unknown, and some modifications are irreversible. (*CCP5 preferentially removes branching points [Rogowski et al., 2010]; however, the enzyme can also hydrolyze linear glutamate chains [Berezniuk et al., 2013]).
Recent discoveries have brought up the possibility that tubulin could be subject to multiple acetylation events. A whole-acetylome study identified >10 novel sites on α- and β-tubulin (Choudhary et al., 2009); however, none of these sites have been confirmed. Another acetylation event has been described at lysine 252 (K252) of β-tubulin. This modification is catalyzed by the acetyltransferase San (Fig. 3) and might regulate the assembly efficiency of microtubules as a result of its localization at the polymerization interface (Chu et al., 2011).
Tubulin detyrosination.
Most α-tubulin genes in different species encode a C-terminal tyrosine residue (Fig. 2; Valenzuela et al., 1981). This tyrosine can be enzymatically removed (Hallak et al., 1977) and religated (Fig. 3; Arce et al., 1975). Mapping of tyrosinated and detyrosinated microtubules in cells using specific antibodies (Gundersen et al., 1984; Geuens et al., 1986; Cambray-Deakin and Burgoyne, 1987a) revealed that subsets of interphase and mitotic spindle microtubules are detyrosinated (Gundersen and Bulinski, 1986). As detyrosination was mostly found on stable and long-lived microtubules, especially in neurons (Cambray-Deakin and Burgoyne, 1987b; Robson and Burgoyne, 1989; Brown et al., 1993), it was assumed that this modification promotes microtubule stability (Gundersen et al., 1987; Sherwin et al., 1987). Although a direct stabilization of the microtubule lattice was considered unlikely (Khawaja et al., 1988), it was found more recently that detyrosination protects cellular microtubules from the depolymerizing activity of kinesin-13–type motor proteins, such as KIF2 or MCAK, thus increasing their longevity (Peris et al., 2009; Sirajuddin et al., 2014).
Besides kinesin-13 motors, plus end–tracking proteins with cytoskeleton-associated protein glycine-rich (CAP-Gly) domains, such as CLIP170 or p150/glued, specifically interact with tyrosinated microtubules (Peris et al., 2006; Bieling et al., 2008) via this domain (Honnappa et al., 2006). In contrast, kinesin-1 moves preferentially on detyrosinated microtubules tracks in cells (Liao and Gundersen, 1998; Kreitzer et al., 1999; Konishi and Setou, 2009). The effect of detyrosination on kinesin-1 motor behavior was recently measured in vitro, and a small but significant increase in the landing rate and processivity of the motor has been found (Kaul et al., 2014). Such subtle changes in the motor behavior could, in conjunction with other factors, such as regulatory MAPs associated with cargo transport complexes (Barlan et al., 2013), lead to a preferential use of detyrosinated microtubules by kinesin-1 in cells.
Despite the early biochemical characterization of a detyrosinating activity, the carboxypeptidase catalyzing detyrosination of α-tubulin has yet to be identified (Hallak et al., 1977; Argaraña et al., 1978, 1980). In contrast, the reverse enzyme, tubulin tyrosine ligase (TTL; Fig. 3; Raybin and Flavin, 1975; Deanin and Gordon, 1976; Argaraña et al., 1980), has been purified (Schröder et al., 1985) and cloned (Ersfeld et al., 1993). TTL modifies nonpolymerized tubulin dimers exclusively. This selectivity is determined by the binding interface between the TTL and tubulin dimers (Szyk et al., 2011, 2013; Prota et al., 2013). In contrast, the so far unidentified detyrosinase acts preferentially on polymerized microtubules (Kumar and Flavin, 1981; Arce and Barra, 1983), thus modifying a select population of microtubules within cells (Gundersen et al., 1987).
In most organisms, only one unique gene for TTL exists. Consequently, TTL knockout mice show a huge accumulation of detyrosinated and particularly Δ2-tubulin (see next section). TTL knockout mice die before birth (Erck et al., 2005) with major developmental defects in the nervous system that might be related to aberrant neuronal differentiation (Marcos et al., 2009). TTL is strictly tubulin specific (Prota et al., 2013), indicating that all observed defects in TTL knockout mice are directly related to the deregulation of the microtubule cytoskeleton.
Δ2-tubulin and further C-terminal modification.
A biochemical study of brain tubulin revealed that ∼35% of α-tubulin cannot be retyrosinated (Paturle et al., 1989) because of the lack of the penultimate C-terminal glutamate residue of the primary protein sequence (Fig. 2; Paturle-Lafanechère et al., 1991). This so-called Δ2-tubulin (for two C-terminal amino acids missing) cannot undergo retyrosination as a result of structural constraints within TTL (Prota et al., 2013) and thus is considered an irreversible PTM.
Δ2-tubulin accumulates in long-lived microtubules of differentiated neurons, axonemes of cilia and flagella, and also in cellular microtubules that have been artificially stabilized, for instance, with taxol (Paturle-Lafanechère et al., 1994). The generation of Δ2-tubulin requires previous detyrosination of α-tubulin; thus, the levels of this PTM are indirectly regulated by the detyrosination/retyrosination cycle. This mechanistic link is particularly apparent in the TTL knockout mice, which show massive accumulation of Δ2-tubulin in all tested tissues (Erck et al., 2005). Loss of TTL and the subsequent increase of Δ2-tubulin levels were also linked to tumor growth and might contribute to the aggressiveness of the tumors by an as-yet-unknown mechanism (Lafanechère et al., 1998; Mialhe et al., 2001). To date, no specific biochemical role of Δ2-tubulin has been determined; thus, one possibility is that the modification simply locks tubulin in the detyrosinated state.
The enzymes responsible for Δ2-tubulin generation are members of a family of cytosolic carboxypeptidases (CCPs; Fig. 3; Kalinina et al., 2007; Rodriguez de la Vega et al., 2007), and most of them also remove polyglutamylation from tubulin (see next section; Rogowski et al., 2010). These enzymes are also able to generate Δ3-tubulin (Fig. 1 A; Berezniuk et al., 2012), indicating that further degradation of the tubulin C-terminal tails are possible; however, the functional significance of this event is unknown.
Polyglutamylation.
Polyglutamylation is a PTM that occurs when secondary glutamate side chains are formed on γ-carboxyl groups of glutamate residues in a protein (Fig. 1, A and B). The modification was first discovered on α- and β-tubulin from the brain (Eddé et al., 1990; Alexander et al., 1991; Rüdiger et al., 1992; Mary et al., 1994) as well as on axonemal tubulin from different species (Mary et al., 1996, 1997); however, it is not restricted to tubulin (Regnard et al., 2000; van Dijk et al., 2008). Using a glutamylation-specific antibody, GT335 (Wolff et al., 1992), it was observed that tubulin glutamylation increases during neuronal differentiation (Audebert et al., 1993, 1994) and that axonemes of cilia and flagella (Fouquet et al., 1994), as well as centrioles of mammalian centrosomes (Bobinnec et al., 1998), are extensively glutamylated.
Enzymes catalyzing polyglutamylation belong to the TTL-like (TTLL) family (Regnard et al., 2003; Janke et al., 2005). In mammals, nine glutamylases exist, each of them showing intrinsic preferences for modifying either α- or β-tubulin as well as for initiating or elongating glutamate chains (Fig. 3; van Dijk et al., 2007). Two of the six well-characterized TTLL glutamylases also modify nontubulin substrates (van Dijk et al., 2008).
Knockout or depletion of glutamylating enzymes in different model organisms revealed an evolutionarily conserved role of glutamylation in cilia and flagella. In motile cilia, glutamylation regulates beating behavior (Janke et al., 2005; Pathak et al., 2007; Ikegami et al., 2010) via the regulation of flagellar dynein motors (Kubo et al., 2010; Suryavanshi et al., 2010). Despite the expression of multiple glutamylases in ciliated cells and tissues, depletion or knockout of single enzymes often lead to ciliary defects, particularly in motile cilia (Ikegami et al., 2010; Vogel et al., 2010; Bosch Grau et al., 2013; Lee et al., 2013), suggesting essential and nonredundant regulatory functions of these enzymes in cilia.
Despite the enrichment of polyglutamylation in neuronal microtubules (Audebert et al., 1993, 1994), knockout of TTLL1, the major polyglutamylase in brain (Janke et al., 2005), did not show obvious neuronal defects in mice (Ikegami et al., 2010; Vogel et al., 2010). This suggests a tolerance of neuronal microtubules to variations in polyglutamylation.
Deglutamylases, the enzymes that reverse polyglutamylation, were identified within a novel family of CCPs (Kimura et al., 2010; Rogowski et al., 2010). So far, three out of six mammalian CCPs have been shown to cleave C-terminal glutamate residues, thus catalyzing both the reversal of polyglutamylation and the removal of gene-encoded glutamates from the C termini of proteins (Fig. 3). The hydrolysis of gene-encoded glutamate residues is not restricted to tubulin, in which it generates Δ2- and Δ3-tubulin, but has also been reported for other proteins such as myosin light chain kinase (Rusconi et al., 1997; Rogowski et al., 2010). One enzyme of the CCP family, CCP5, preferentially removes branching points generated by glutamylation, thus allowing the complete reversal of the polyglutamylation modification (Kimura et al., 2010; Rogowski et al., 2010). However, CCP5 can also hydrolyze C-terminal glutamate residues from linear peptide chains similar to other members of the CCP family (Berezniuk et al., 2013).
CCP1 is mutated in a well-established mouse model for neurodegeneration, the pcd (Purkinje cell degeneration) mouse (Mullen et al., 1976; Greer and Shepherd, 1982; Fernandez-Gonzalez et al., 2002). The absence of a key deglutamylase leads to strong hyperglutamylation in brain regions that undergo degeneration, such as the cerebellum and the olfactory bulb (Rogowski et al., 2010). When glutamylation levels were rebalanced by depletion or knockout of the major brain polyglutamylase TTLL1 (Rogowski et al., 2010; Berezniuk et al., 2012), Purkinje cells survived. Although the molecular mechanisms of hyperglutamylation-induced degeneration remain to be elucidated, perturbation of neuronal transport, as well as changes in the dynamics and stability of microtubules, is expected to be induced by hyperglutamylation. Increased polyglutamylation levels have been shown to affect kinesin-1–mediated transport in cultured neurons (Maas et al., 2009), and the turnover of microtubules can also be regulated by polyglutamylation via the activation of microtubule-severing enzymes such as spastin (Lacroix et al., 2010).
Subtle differences in polyglutamylation can be seen on diverse microtubules in different cell types. The functions of these modifications remain to be studied; however, its wide distribution strengthens the idea that it could be involved in fine-tuning a range of microtubule functions.
Polyglycylation.
Tubulin polyglycylation or glycylation, like polyglutamylation, generates side chains of glycine residues within the C-terminal tails of α- and β-tubulin (Fig. 1, A and B). The modification sites of glycylation are considered to be principally the same as for glutamylation, and indeed, both PTMs have been shown to be interdependent in cells (Rogowski et al., 2009; Wloga et al., 2009). Initially discovered on Paramecium tetraurelia tubulin (Redeker et al., 1994), glycylation has been extensively studied using two antibodies, TAP952 and AXO49 (Bressac et al., 1995; Levilliers et al., 1995; Bré et al., 1996). In contrast to polyglutamylation, glycylation is restricted to cilia and flagella in most organisms analyzed so far.
Glycylating enzymes are also members of the TTLL family, and homologues of these enzymes have so far been found in all organisms with proven glycylation of ciliary axonemes (Rogowski et al., 2009; Wloga et al., 2009). In mammals, initiating (TTLL3 and TTLL8) and elongating (TTLL10) glycylases work together to generate polyglycylation (Fig. 3). In contrast, the two TTLL3 orthologues from Drosophila melanogaster can both initiate and elongate glycine side chains (Rogowski et al., 2009).
In mice, motile ependymal cilia in brain ventricles acquire monoglycylation upon maturation, whereas polyglycylation is observed only after several weeks (Bosch Grau et al., 2013). Sperm flagella, in contrast, acquire long glycine chains much faster, suggesting that the extent of polyglycylation could correlate with the length of the axonemes (Rogowski et al., 2009). Depletion of glycylases in mice (ependymal cilia; Bosch Grau et al., 2013), zebrafish (Wloga et al., 2009; Pathak et al., 2011), Tetrahymena thermophila (Wloga et al., 2009), and D. melanogaster (Rogowski et al., 2009) consistently led to ciliary disassembly or severe ciliary defects. How glycylation regulates microtubule functions remains unknown; however, the observation that glycylation-depleted axonemes disassemble after initial assembly (Rogowski et al., 2009; Bosch Grau et al., 2013) suggests a role of this PTM in stabilizing axonemal microtubules. Strikingly, human TTLL10 is enzymatically inactive; thus, humans have lost the ability to elongate glycine side chains (Rogowski et al., 2009). This suggests that the elongation of the glycine side chains is not an essential aspect of the function of this otherwise critical tubulin PTM.
Other tubulin PTMs.
Several other PTMs have been found on tubulin. Early studies identified tubulin phosphorylation (Eipper, 1974; Gard and Kirschner, 1985; Díaz-Nido et al., 1990); however, no specific functions were found. The perhaps best-studied phosphorylation event on tubulin takes place at serine S172 of β-tubulin (Fig. 1 A), is catalyzed by the Cdk1 (Fig. 3), and might regulate microtubule dynamics during cell division (Fourest-Lieuvin et al., 2006; Caudron et al., 2010). Tubulin can be also modified by the spleen tyrosine kinase Syk (Fig. 3; Peters et al., 1996), which might play a role in immune cells (Faruki et al., 2000; Sulimenko et al., 2006) and cell division (Zyss et al., 2005; Sulimenko et al., 2006).
Polyamination has recently been discovered on brain tubulin (Song et al., 2013), after having been overlooked for many years as a result of the low solubility of polyaminated tubulin. Among several glutamine residues of α- and β-tubulin that can be polyaminated, Q15 of β-tubulin is considered the primary modification site (Fig. 1 A). Polyamination is catalyzed by transglutaminases (Fig. 3), which modify free tubulin as well as microtubules in an irreversible manner, and most likely contribute to the stabilization of microtubules (Song et al., 2013).
Tubulin was also reported to be palmitoylated (Caron, 1997; Ozols and Caron, 1997; Caron et al., 2001), ubiquitinated (Ren et al., 2003; Huang et al., 2009; Xu et al., 2010), glycosylated (Walgren et al., 2003; Ji et al., 2011), arginylated (Wong et al., 2007), methylated (Xiao et al., 2010), and sumoylated (Rosas-Acosta et al., 2005). These PTMs have mostly been reported without follow-up studies, and some of them are only found in specific cell types or organisms and/or under specific metabolic conditions. Further studies will be necessary to gain insights into their potential roles for the regulation of the microtubule cytoskeleton.
Current advances and future perspectives
The molecular heterogeneity of microtubules, generated by the expression of different tubulin isotypes and by the PTM of tubulin has fascinated the scientific community for ∼40 years. Although many important advances have been made in the past decade, the dissection of the molecular mechanisms and a comprehensive understanding of the biological functions of tubulin isotypes and PTMs will be a challenging field of research in the near future.
Direct measurements of the impact of tubulin heterogeneity.
The most direct and reliable type of experiments to determine the impact of tubulin heterogeneity on microtubule behavior are in vitro measurements with purified proteins. However, most biophysical work on microtubules has been performed with tubulin purified from bovine, ovine, or porcine brains, which can be obtained in large quantities and with a high degree of purity and activity (Vallee, 1986; Castoldi and Popov, 2003). Brain tubulin is a mixture of different tubulin isotypes and is heavily posttranslationally modified and thus inept for investigating the functions of tubulin heterogeneity (Denoulet et al., 1986; Cambray-Deakin and Burgoyne, 1987b; Paturle et al., 1989; Eddé et al., 1990). Thus, pure, recombinant tubulin will be essential to dissect the roles of different tubulin isoforms and PTMs.
Attempts to produce recombinant, functional α- and β-tubulin in bacteria have failed so far (Yaffe et al., 1988), most likely because of the absence of the extensive tubulin-specific folding machinery (Yaffe et al., 1992; Gao et al., 1993; Tian et al., 1996; Vainberg et al., 1998) in prokaryotes. An alternative source of tubulin with less isotype heterogeneity and with almost no PTMs is endogenous tubulin from cell lines such as HeLa, which in the past has been purified using a range of biochemical procedures (Bulinski and Borisy, 1979; Weatherbee et al., 1980; Farrell, 1982; Newton et al., 2002; Fourest-Lieuvin, 2006). Such tubulin can be further modified with tubulin-modifying enzymes, such as polyglutamylases, either by expressing those enzymes in the cells before tubulin purification (Lacroix and Janke, 2011) or in vitro with purified enzymes (Vemu et al., 2014). Despite some technical limitations of these methods, HeLa tubulin modified in cells has been successfully used in an in vitro study on the role of polyglutamylation in microtubule severing (Lacroix et al., 2010).
Naturally occurring variants of tubulin isotypes and PTMs can be purified from different organisms, organs, or cell types, but obviously, only some combinations of tubulin isotypes and PTMs can be obtained by this approach. The recent development of an affinity purification method using the microtubule-binding TOG (tumor overexpressed gene) domain of yeast Stu2p has brought a new twist to this approach, as it allows purifying small amounts of tubulin from any cell type or tissue (Widlund et al., 2012).
The absence of tubulin heterogeneity in yeast has made budding and fission yeast potential expression systems for recombinant, PTM-free tubulin (Katsuki et al., 2009; Drummond et al., 2011; Johnson et al., 2011). However, the expression of mammalian tubulin in this system has remained impossible. This problem was then partially circumvented by expressing tubulin chimeras that consist of a yeast tubulin body fused to mammalian C-terminal tubulin tails, thus mimicking different tubulin isotypes (Sirajuddin et al., 2014). Moreover, detyrosination can be generated by deleting the key C-terminal residue from endogenous or chimeric α-tubulin (Badin-Larçon et al., 2004), and polyglutamylation is generated by chemically coupling glutamate side chains to specifically engineered tubulin chimeras (Sirajuddin et al., 2014). These approaches allowed the first direct measurements of the impact of tubulin isotypes and PTMs on the behavior of molecular motors in vitro (Sirajuddin et al., 2014) and the analysis of the effects of tubulin heterogeneity on microtubule behavior and interactions inside the yeast cell (Badin-Larçon et al., 2004; Aiken et al., 2014).
Currently, the most promising development has been the successful purification of fully functional recombinant tubulin from the baculovirus expression system (Minoura et al., 2013). Using this system, defined α/β-tubulin dimers can be obtained using two different epitope tags on α- and β-tubulin, respectively. Although these epitope tags are essential for separating recombinant from the endogenous tubulin, they could also affect tubulin assembly or microtubule–MAP interactions. Thus, future developments should focus on eliminating these tags.
Current efforts have brought the possibility of producing recombinant tubulin into reach. Further improvement and standardization of these methods will certainly provide a breakthrough in understanding the mechanisms by which tubulin heterogeneity contributes to microtubule functions.
Complexity of tubulin—understanding the regulatory principles.
The diversity of tubulin genes (isotypes) and the complexity of tubulin PTMs have led to the proposal of the term “tubulin code” (Verhey and Gaertig, 2007; Wehenkel and Janke, 2014), in analogy to the previously coined histone code (Jenuwein and Allis, 2001). Tubulin molecules consist of a highly structured and thus evolutionarily conserved tubulin body and the unstructured and less conserved C-terminal tails (Nogales et al., 1998). As PTMs and sequence variations within the tubulin body are expected to affect the conserved tubulin fold and therefore the properties of the microtubule lattice, they are not likely to be involved in generating the tubulin code. In contrast, modulations of the C-terminal tails could encode signals on the microtubule surface without perturbing basic microtubule functions and properties (Figs. 1 A and 4). Indeed, the highest degree of gene-encoded diversity (Fig. 2) and the highest density and complexity of PTMs (Fig. 1) are found within these tail domains.
Figure 4.
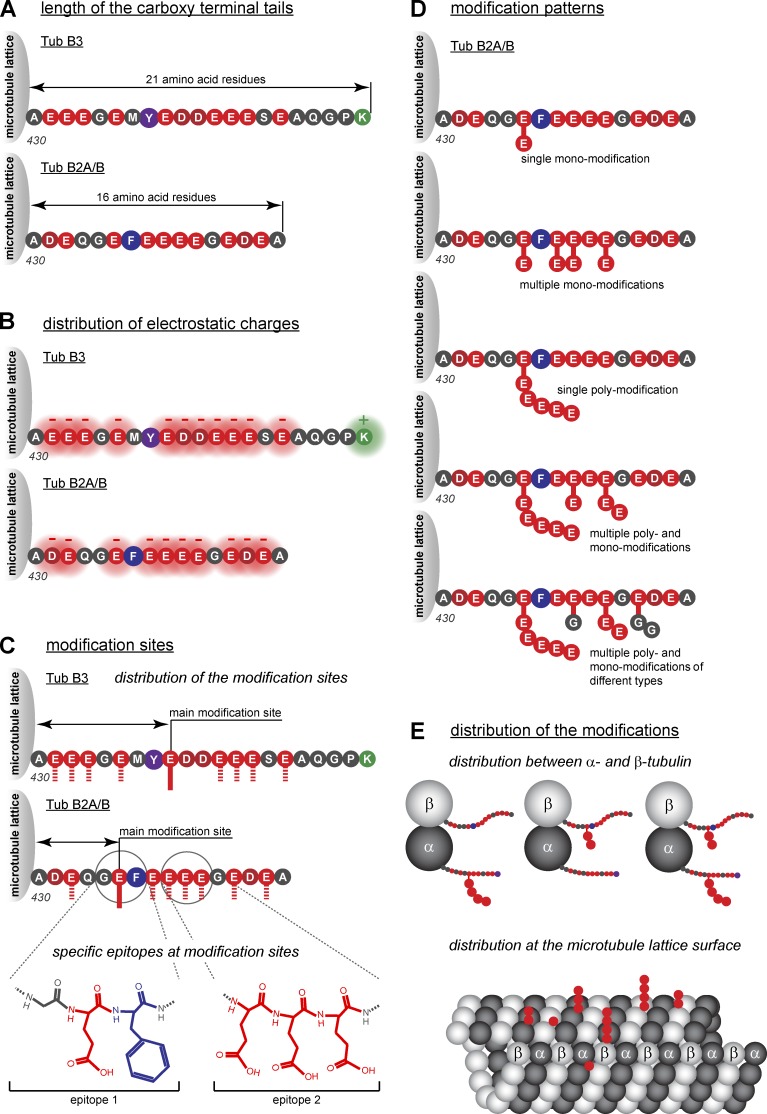
Molecular components of the tubulin code. Schematic representation of potential coding elements that could generate specific signals for the tubulin code. (A) The length of the C-terminal tails of different tubulin isotypes differ significantly (Fig. 2) and could have an impact on the interactions between microtubules and MAPs. (B) Tubulin C-terminal tails are rich in charged amino acid residues. The distribution of these residues and local densities of charges could influence the electrostatic interactions with the tails and the readers. (C) Although each glutamate residue within the C-terminal tails could be considered a potential modification site, only some sites have been found highly occupied in tubulin purifications from native sources. This indicates selectivity of the modification reactions, which can participate in the generation of specific modification patterns (see D). Modification sites might be distinguished by their neighboring amino acid residues, which could create specific modification epitopes. (D) As a result of the large number of modification sites and the variability of side chains, a large variety of modification patterns could be generated within a single C-terminal tail of tubulin. (E) Modification patterns as shown in D can be distinct between α- and β-tubulin. These modification patterns could be differentially distributed at the surface of the microtubule lattice, thus generating a higher-order patterning. Tub, tubulin. For color coding, see Fig. 2.
Considering the number of tubulin isotypes plus all potential combinations of PTMs (e.g., each glutamate residue within the C-terminal tubulin tail could be modified by either polyglutamylation or polyglycylation, each of them generating side chains of different lengths; Fig. 4), the number of distinct signals generated by the potential tubulin code would be huge. However, as many of these potential signals represent chemical structures that are similar and might not be reliably distinguished by readout mechanisms, it is possible that the tubulin code generates probabilistic signals. In this scenario, biochemically similar modifications would have similar functional readouts, and marginal differences between those signals would only bias biological processes but not determine them. This stands in contrast to the concept of the histone code, in which precise patterns of different PTMs on the histone proteins encode distinct biological signals.
The concept of probabilistic signaling is already inscribed in the machinery that generates the tubulin code. Polyglutamylases and polyglycylases from the TTLL family have preferential activities for either α- or β-tubulin and for generating different lengths of the branched glutamate or glycine chains. Although under conditions of low enzyme concentrations, as found in most cells and tissues, the enzymes seem to selectively generate their preferential type of PTM, higher enzyme concentrations induce a more promiscuous behavior, leading, for instance, to a loss of selectivity for α- or β-tubulin (van Dijk et al., 2007). Similarly, the modifying enzymes might prefer certain modification sites within the C-terminal tails of tubulin but might be equally able to modify other sites, which could be locally regulated in cells. For example, β-tubulin isotypes isolated from mammalian brain were initially found to be glutamylated on single residues (Alexander et al., 1991; Rüdiger et al., 1992), which in the light of the comparably low sensitivity of mass spectrometry at the time might rather indicate a preferential than a unique modification of these sites. Nevertheless, the neuron-specific polyglutamylase for β-tubulin TTLL7 (Ikegami et al., 2006) can incorporate glutamate onto many more modification sites of β-tubulin in vitro (Mukai et al., 2009), which clearly indicates that not all of the possible modification events take place under physiological conditions.
Several examples supporting a probabilistic signaling mode of the tubulin code are found in the recent literature. In T. thermophila, a ciliate without tubulin isotype diversity (Gaertig et al., 1993) but with a huge repertoire of tubulin PTMs and tubulin-modifying enzymes (Janke et al., 2005), tubulin can be easily mutagenized to experimentally eliminate sites for PTMs. Mutagenesis of the most commonly occupied glutamylation/glycylation sites within the β-tubulin tails did not generate a clear decrease of glycylation levels nor did it cause obvious phenotypic alterations. This indicates that the modifying enzymes can deviate toward alternative modification sites and that similar PTMs on different sites can compensate the functions of the mutated site. However, when all of the key modification sites were mutated, glycylation became prominently decreased, which led to severe phenotypes, including lethality (Xia et al., 2000). Most strikingly, these phenotypes could be recovered by replacing the C-terminal tail of α-tubulin with the nonmutated β-tubulin tail. This α–β-tubulin chimera became overglycylated and functionally compensated for the absence of modification sites on β-tubulin. The conclusion of this study is that PTM- and isotype-generated signals can fulfill a biological function within a certain range of tolerance.
But how efficient is such compensation? The answer can be found in a variety of already described deletion mutants for tubulin-modifying enzymes in different model organisms. Most single-gene knockouts for TTLL genes (glutamylases or glycylases) did not result in prominent phenotypic alterations in mice, even for enzymes that are ubiquitously expressed. Only some highly specialized microtubule structures show functional aberrations upon the deletion of a single enzyme. These “tips of the iceberg” are usually the motile cilia and sperm flagella, which carry very high levels of polyglutamylation and polyglycylation (Bré et al., 1996; Kann et al., 1998; Rogowski et al., 2009). It thus appears that some microtubules are essentially dependent on the generation of specific PTM patterns, whereas others can tolerate changes and appear to function normally. How “normal” these functions are remains to be investigated in future studies. It is possible that defects are subtle and thus overlooked but could become functionally important under specific conditions.
A tubulin code also requires readout mechanisms. The most likely “readers” of the tubulin code are MAPs and molecular motors. Considering the probabilistic signaling hypothesis, the expected effects of the signals would be in most cases rather gradual changes, for instance, to fine-tune molecular motor traffic and/or to bias motors toward defined microtubule tracks but not to obliterate motor activity or MAP binding to microtubules. An in vitro study using recombinant tubulin chimeras purified from yeast confirmed this notion (Sirajuddin et al., 2014). By analyzing which elements of the tubulin code can regulate the velocity and processivity of the molecular motors kinesin and dynein, these researchers found that the C-terminal tails of α- and β-tubulin differentially influence the kinetic parameters of the tested motors; however, the modulation was rather modest. One of their striking observations was that a single lysine residue, present in the C-terminal tails of two β-tubulin isotypes (Figs. 2 and 4), significantly affected motor traffic and that this effect can be counterbalanced by polyglutamylation. These observations are the first in vitro evidence for the interdependence of different elements of the tubulin code and provide another indication for its probabilistic mode of signaling.
Future directions.
One of the greatest technological challenges to understanding the function of the tubulin code is to detect and interpret subtle and complex regulatory events generated by this code. It will thus be instrumental to further develop tools to better distinguish graded changes in PTM levels on microtubules in cells and tissues (Magiera and Janke, 2013) and to reliably measure subtle modulations of microtubule behavior in reconstituted systems.
The current advances in the field and especially the availability of whole-organism models, as well as first insights into the pathological role of tubulin mutations (Tischfield et al., 2011), are about to transform our way of thinking about the regulation of microtubule cytoskeleton. Tubulin heterogeneity generates complex probabilistic signals that cannot be clearly attributed to single biological functions in most cases and that are not essential for most cellular processes. Nevertheless, it has been conserved throughout evolution of eukaryotes and can hardly be dismissed as not important. To understand the functional implications of these processes, we might be forced to reconsider how we define biologically important events and how we measure events that might encode probabilistic signals. The answers to these questions could provide novel insights into how complex systems, such as cells and organisms, are sustained throughout difficult and challenging life cycles, resist to environmental stress and diseases, and have the flexibility needed to succeed in evolution.
Acknowledgments
I would like to thank M.M. Magiera, A. Wehenkel (Institut Curie), M. Steinmetz (Paul Scherrer Institute, Villigen, Switzerland), and the anonymous reviewers for insightful discussions and critical reading of the manuscript.
This work was supported by the Institut Curie, the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Agence Nationale de la Recherche awards ANR-12-BSV2-0007 and ANR-10-LBX-0038, part of the Initiatives d’Excellence Paris Sciences et Lettres ANR-10-IDEX-0001-02 PSL, the INCA_6517 grant, the Association pour la Recherche sur le Cancer Programme Labellisé SL220120605303, and European Molecular Biology Organization Young Investigator Programme.
Footnotes
Abbreviations used in this paper:
- CCP
- cytosolic carboxypeptidase
- MAP
- microtubule-associated protein
- PTM
- posttranslational modification
- TTL
- tubulin tyrosine ligase
- TTLL
- TTL-like
References
- Aguilar, A., Becker L., Tedeschi T., Heller S., Iomini C., and Nachury M.V.. 2014. α-tubulin K40 acetylation is required for contact inhibition of proliferation and cell-substrate adhesion. Mol. Biol. Cell. 25:1854–1866 10.1091/mbc.E13-10-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken, J., Sept D., Costanzo M., Boone C., Cooper J.A., and Moore J.K.. 2014. Genome-wide analysis reveals novel and discrete functions for tubulin carboxy-terminal tails. Curr. Biol. 24:1295–1303 10.1016/j.cub.2014.03.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella, J.S., Wloga D., Kim J., Starostina N.G., Lyons-Abbott S., Morrissette N.S., Dougan S.T., Kipreos E.T., and Gaertig J.. 2010. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 467:218–222 10.1038/nature09324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, J.E., Hunt D.F., Lee M.K., Shabanowitz J., Michel H., Berlin S.C., MacDonald T.L., Sundberg R.J., Rebhun L.I., and Frankfurter A.. 1991. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc. Natl. Acad. Sci. USA. 88:4685–4689 10.1073/pnas.88.11.4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce, C.A., and Barra H.S.. 1983. Association of tubulinyl-tyrosine carboxypeptidase with microtubules. FEBS Lett. 157:75–78 10.1016/0014-5793(83)81119-3 [DOI] [PubMed] [Google Scholar]
- Arce, C.A., Rodriguez J.A., Barra H.S., and Caputo R.. 1975. Incorporation of L-tyrosine, L-phenylalanine and L-3,4-dihydroxyphenylalanine as single units into rat brain tubulin. Eur. J. Biochem. 59:145–149 10.1111/j.1432-1033.1975.tb02435.x [DOI] [PubMed] [Google Scholar]
- Argaraña, C.E., Barra H.S., and Caputto R.. 1978. Release of [14C]tyrosine from tubulinyl-[14C]tyrosine by brain extract. Separation of a carboxypeptidase from tubulin-tyrosine ligase. Mol. Cell. Biochem. 19:17–21 10.1007/BF00231230 [DOI] [PubMed] [Google Scholar]
- Argaraña, C.E., Barra H.S., and Caputto R.. 1980. Tubulinyl-tyrosine carboxypeptidase from chicken brain: properties and partial purification. J. Neurochem. 34:114–118 10.1111/j.1471-4159.1980.tb04628.x [DOI] [PubMed] [Google Scholar]
- Audebert, S., Desbruyères E., Gruszczynski C., Koulakoff A., Gros F., Denoulet P., and Eddé B.. 1993. Reversible polyglutamylation of alpha- and beta-tubulin and microtubule dynamics in mouse brain neurons. Mol. Biol. Cell. 4:615–626 10.1091/mbc.4.6.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert, S., Koulakoff A., Berwald-Netter Y., Gros F., Denoulet P., and Eddé B.. 1994. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J. Cell Sci. 107:2313–2322 [DOI] [PubMed] [Google Scholar]
- Badin-Larçon, A.C., Boscheron C., Soleilhac J.M., Piel M., Mann C., Denarier E., Fourest-Lieuvin A., Lafanechère L., Bornens M., and Job D.. 2004. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc. Natl. Acad. Sci. USA. 101:5577–5582 10.1073/pnas.0307917101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan, K., Rossow M.J., and Gelfand V.I.. 2013. The journey of the organelle: teamwork and regulation in intracellular transport. Curr. Opin. Cell Biol. 25:483–488 10.1016/j.ceb.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezniuk, I., Vu H.T., Lyons P.J., Sironi J.J., Xiao H., Burd B., Setou M., Angeletti R.H., Ikegami K., and Fricker L.D.. 2012. Cytosolic carboxypeptidase 1 is involved in processing α- and β-tubulin. J. Biol. Chem. 287:6503–6517 10.1074/jbc.M111.309138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezniuk, I., Lyons P.J., Sironi J.J., Xiao H., Setou M., Angeletti R.H., Ikegami K., and Fricker L.D.. 2013. Cytosolic carboxypeptidase 5 removes α- and γ-linked glutamates from tubulin. J. Biol. Chem. 288:30445–30453 10.1074/jbc.M113.497917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling, P., Kandels-Lewis S., Telley I.A., van Dijk J., Janke C., and Surrey T.. 2008. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183:1223–1233 10.1083/jcb.200809190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec, Y., Khodjakov A., Mir L.M., Rieder C.L., Eddé B., and Bornens M.. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch Grau, M., Gonzalez Curto G., Rocha C., Magiera M.M., Marques Sousa P., Giordano T., Spassky N., and Janke C.. 2013. Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J. Cell Biol. 202:441–451 10.1083/jcb.201305041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bré, M.H., Redeker V., Quibell M., Darmanaden-Delorme J., Bressac C., Cosson J., Huitorel P., Schmitter J.M., Rossler J., Johnson T., et al. 1996. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J. Cell Sci. 109:727–738 [DOI] [PubMed] [Google Scholar]
- Bressac, C., Bré M.H., Darmanaden-Delorme J., Laurent M., Levilliers N., and Fleury A.. 1995. A massive new posttranslational modification occurs on axonemal tubulin at the final step of spermatogenesis in Drosophila. Eur. J. Cell Biol. 67:346–355 [PubMed] [Google Scholar]
- Brown, A., Li Y., Slaughter T., and Black M.M.. 1993. Composite microtubules of the axon: quantitative analysis of tyrosinated and acetylated tubulin along individual axonal microtubules. J. Cell Sci. 104:339–352 [DOI] [PubMed] [Google Scholar]
- Bulinski, J.C., and Borisy G.G.. 1979. Self-assembly of microtubules in extracts of cultured HeLa cells and the identification of HeLa microtubule-associated proteins. Proc. Natl. Acad. Sci. USA. 76:293–297 10.1073/pnas.76.1.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray-Deakin, M.A., and Burgoyne R.D.. 1987a. Acetylated and detyrosinated alpha-tubulins are co-localized in stable microtubules in rat meningeal fibroblasts. Cell Motil. Cytoskeleton. 8:284–291 10.1002/cm.970080309 [DOI] [PubMed] [Google Scholar]
- Cambray-Deakin, M.A., and Burgoyne R.D.. 1987b. Posttranslational modifications of α-tubulin: acetylated and detyrosinated forms in axons of rat cerebellum. J. Cell Biol. 104:1569–1574 10.1083/jcb.104.6.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, J.M.1997. Posttranslational modification of tubulin by palmitoylation: I. In vivo and cell-free studies. Mol. Biol. Cell. 8:621–636 10.1091/mbc.8.4.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, J.M., Vega L.R., Fleming J., Bishop R., and Solomon F.. 2001. Single site alpha-tubulin mutation affects astral microtubules and nuclear positioning during anaphase in Saccharomyces cerevisiae: possible role for palmitoylation of alpha-tubulin. Mol. Biol. Cell. 12:2672–2687 10.1091/mbc.12.9.2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi, M., and Popov A.V.. 2003. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88 10.1016/S1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Caudron, F., Denarier E., Thibout-Quintana J.-C., Brocard J., Andrieux A., and Fourest-Lieuvin A.. 2010. Mutation of Ser172 in yeast β tubulin induces defects in microtubule dynamics and cell division. PLoS ONE. 5:e13553 10.1371/journal.pone.0013553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist, G.Y., Luchniak A., Tischfield M.A., Peeva M., Song Y., Menezes M.P., Chan W.-M., Andrews C., Chew S., Jamieson R.V., et al. 2012. An inherited TUBB2B mutation alters a kinesin-binding site and causes polymicrogyria, CFEOM and axon dysinnervation. Hum. Mol. Genet. 21:5484–5499 10.1093/hmg/dds393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., and Mann M.. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 325:834–840 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Chu, C.-W., Hou F., Zhang J., Phu L., Loktev A.V., Kirkpatrick D.S., Jackson P.K., Zhao Y., and Zou H.. 2011. A novel acetylation of β-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol. Biol. Cell. 22:448–456 10.1091/mbc.E10-03-0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., Kirschner M.W., and Cowan N.J.. 1978. Isolation of separate mRNAs for alpha- and beta-tubulin and characterization of the corresponding in vitro translation products. Cell. 15:1021–1031 10.1016/0092-8674(78)90286-6 [DOI] [PubMed] [Google Scholar]
- Deanin, G.G., and Gordon M.W.. 1976. The distribution of tyrosyltubulin ligase in brain and other tissues. Biochem. Biophys. Res. Commun. 71:676–683 10.1016/0006-291X(76)90841-X [DOI] [PubMed] [Google Scholar]
- Denoulet, P., Eddé B., and Gros F.. 1986. Differential expression of several neurospecific beta-tubulin mRNAs in the mouse brain during development. Gene. 50:289–297 10.1016/0378-1119(86)90333-1 [DOI] [PubMed] [Google Scholar]
- Díaz-Nido, J., Serrano L., López-Otín C., Vandekerckhove J., and Avila J.. 1990. Phosphorylation of a neuronal-specific beta-tubulin isotype. J. Biol. Chem. 265:13949–13954 [PubMed] [Google Scholar]
- Dompierre, J.P., Godin J.D., Charrin B.C., Cordelières F.P., King S.J., Humbert S., and Saudou F.. 2007. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J. Neurosci. 27:3571–3583 10.1523/JNEUROSCI.0037-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, D.R., Kain S., Newcombe A., Hoey C., Katsuki M., and Cross R.A.. 2011. Purification of tubulin from the fission yeast Schizosaccharomyces pombe. Methods Mol. Biol. 777:29–55 10.1007/978-1-61779-252-6_3 [DOI] [PubMed] [Google Scholar]
- Eddé, B., Rossier J., Le Caer J.P., Desbruyères E., Gros F., and Denoulet P.. 1990. Posttranslational glutamylation of alpha-tubulin. Science. 247:83–85 10.1126/science.1967194 [DOI] [PubMed] [Google Scholar]
- Eipper, B.A.1974. Properties of rat brain tubulin. J. Biol. Chem. 249:1407–1416 [PubMed] [Google Scholar]
- Erck, C., Peris L., Andrieux A., Meissirel C., Gruber A.D., Vernet M., Schweitzer A., Saoudi Y., Pointu H., Bosc C., et al. 2005. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl. Acad. Sci. USA. 102:7853–7858 10.1073/pnas.0409626102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld, K., Wehland J., Plessmann U., Dodemont H., Gerke V., and Weber K.. 1993. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 120:725–732 10.1083/jcb.120.3.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallet-Bianco, C., Loeuillet L., Poirier K., Loget P., Chapon F., Pasquier L., Saillour Y., Beldjord C., Chelly J., and Francis F.. 2008. Neuropathological phenotype of a distinct form of lissencephaly associated with mutations in TUBA1A. Brain. 131:2304–2320 10.1093/brain/awn155 [DOI] [PubMed] [Google Scholar]
- Farrell, K.W.1982. Isolation of tubulin from nonneural sources. Methods Enzymol. 85(part B):385–393 10.1016/0076-6879(82)85039-8 [DOI] [PubMed] [Google Scholar]
- Faruki, S., Geahlen R.L., and Asai D.J.. 2000. Syk-dependent phosphorylation of microtubules in activated B-lymphocytes. J. Cell Sci. 113:2557–2565 [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez, A., La Spada A.R., Treadaway J., Higdon J.C., Harris B.S., Sidman R.L., Morgan J.I., and Zuo J.. 2002. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 295:1904–1906 10.1126/science.1068912 [DOI] [PubMed] [Google Scholar]
- Fouquet, J.P., Eddé B., Kann M.L., Wolff A., Desbruyeres E., and Denoulet P.. 1994. Differential distribution of glutamylated tubulin during spermatogenesis in mammalian testis. Cell Motil. Cytoskeleton. 27:49–58 10.1002/cm.970270106 [DOI] [PubMed] [Google Scholar]
- Fourest-Lieuvin, A.2006. Purification of tubulin from limited volumes of cultured cells. Protein Expr. Purif. 45:183–190 10.1016/j.pep.2005.05.011 [DOI] [PubMed] [Google Scholar]
- Fourest-Lieuvin, A., Peris L., Gache V., Garcia-Saez I., Juillan-Binard C., Lantez V., and Job D.. 2006. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol. Biol. Cell. 17:1041–1050 10.1091/mbc.E05-07-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig, J., Thatcher T.H., McGrath K.E., Callahan R.C., and Gorovsky M.A.. 1993. Perspectives on tubulin isotype function and evolution based on the observation that Tetrahymena thermophila microtubules contain a single alpha- and beta-tubulin. Cell Motil. Cytoskeleton. 25:243–253 10.1002/cm.970250305 [DOI] [PubMed] [Google Scholar]
- Gao, Y., Vainberg I.E., Chow R.L., and Cowan N.J.. 1993. Two cofactors and cytoplasmic chaperonin are required for the folding of alpha- and beta-tubulin. Mol. Cell. Biol. 13:2478–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard, D.L., and Kirschner M.W.. 1985. A polymer-dependent increase in phosphorylation of β-tubulin accompanies differentiation of a mouse neuroblastoma cell line. J. Cell Biol. 100:764–774 10.1083/jcb.100.3.764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens, G., Gundersen G.G., Nuydens R., Cornelissen F., Bulinski J.C., and DeBrabander M.. 1986. Ultrastructural colocalization of tyrosinated and detyrosinated α-tubulin in interphase and mitotic cells. J. Cell Biol. 103:1883–1893 10.1083/jcb.103.5.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer, C.A., and Shepherd G.M.. 1982. Mitral cell degeneration and sensory function in the neurological mutant mouse Purkinje cell degeneration (PCD). Brain Res. 235:156–161 10.1016/0006-8993(82)90206-2 [DOI] [PubMed] [Google Scholar]
- Gundersen, G.G., and Bulinski J.C.. 1986. Distribution of tyrosinated and nontyrosinated α-tubulin during mitosis. J. Cell Biol. 102:1118–1126 10.1083/jcb.102.3.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen, G.G., Kalnoski M.H., and Bulinski J.C.. 1984. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 38:779–789 10.1016/0092-8674(84)90273-3 [DOI] [PubMed] [Google Scholar]
- Gundersen, G.G., Khawaja S., and Bulinski J.C.. 1987. Postpolymerization detyrosination of α-tubulin: a mechanism for subcellular differentiation of microtubules. J. Cell Biol. 105:251–264 10.1083/jcb.105.1.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak, M.E., Rodriguez J.A., Barra H.S., and Caputto R.. 1977. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 73:147–150 10.1016/0014-5793(77)80968-X [DOI] [PubMed] [Google Scholar]
- Honnappa, S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F.K., and Steinmetz M.O.. 2006. Key interaction modes of dynamic +TIP networks. Mol. Cell. 23:663–671 10.1016/j.molcel.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Howes, S.C., Alushin G.M., Shida T., Nachury M.V., and Nogales E.. 2014. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol. Biol. Cell. 25:257–266 10.1091/mbc.E13-07-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K., Diener D.R., and Rosenbaum J.L.. 2009. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 186:601–613 10.1083/jcb.200903066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert, C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.-F., and Yao T.-P.. 2002. HDAC6 is a microtubule-associated deacetylase. Nature. 417:455–458 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- Ikegami, K., Mukai M., Tsuchida J., Heier R.L., Macgregor G.R., and Setou M.. 2006. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J. Biol. Chem. 281:30707–30716 10.1074/jbc.M603984200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami, K., Sato S., Nakamura K., Ostrowski L.E., and Setou M.. 2010. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc. Natl. Acad. Sci. USA. 107:10490–10495 10.1073/pnas.1002128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Rogowski K., Wloga D., Regnard C., Kajava A.V., Strub J.-M., Temurak N., van Dijk J., Boucher D., van Dorsselaer A., et al. 2005. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 308:1758–1762 10.1126/science.1113010 [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis C.D.. 2001. Translating the histone code. Science. 293:1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Ji, S., Kang J.G., Park S.Y., Lee J., Oh Y.J., and Cho J.W.. 2011. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids. 40:809–818 10.1007/s00726-010-0698-9 [DOI] [PubMed] [Google Scholar]
- Johnson, V., Ayaz P., Huddleston P., and Rice L.M.. 2011. Design, overexpression, and purification of polymerization-blocked yeast αβ-tubulin mutants. Biochemistry. 50:8636–8644 10.1021/bi2005174 [DOI] [PubMed] [Google Scholar]
- Joshi, H.C., and Cleveland D.W.. 1989. Differential utilization of β-tubulin isotypes in differentiating neurites. J. Cell Biol. 109:663–673 10.1083/jcb.109.2.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic, N., Sorrentino S., Perlas E., Bolasco G., Martinez C., and Heppenstall P.A.. 2013. αTAT1 is the major α-tubulin acetyltransferase in mice. Nat. Commun. 4:1962 10.1038/ncomms2962 [DOI] [PubMed] [Google Scholar]
- Kalinina, E., Biswas R., Berezniuk I., Hermoso A., Aviles F.X., and Fricker L.D.. 2007. A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 21:836–850 10.1096/fj.06-7329com [DOI] [PubMed] [Google Scholar]
- Kann, M.L., Prigent Y., Levilliers N., Bré M.H., and Fouquet J.P.. 1998. Expression of glycylated tubulin during the differentiation of spermatozoa in mammals. Cell Motil. Cytoskeleton. 41:341–352 [DOI] [PubMed] [Google Scholar]
- Katsuki, M., Drummond D.R., Osei M., and Cross R.A.. 2009. Mal3 masks catastrophe events in Schizosaccharomyces pombe microtubules by inhibiting shrinkage and promoting rescue. J. Biol. Chem. 284:29246–29250 10.1074/jbc.C109.052159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul, N., Soppina V., and Verhey K.J.. 2014. Effects of α-tubulin K40 acetylation and detyrosination on kinesin-1 motility in a purified system. Biophys. J. 106:2636–2643 10.1016/j.bpj.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays, D.A., Tian G., Poirier K., Huang G.-J., Siebold C., Cleak J., Oliver P.L., Fray M., Harvey R.J., Molnár Z., et al. 2007. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 128:45–57 10.1016/j.cell.2006.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawaja, S., Gundersen G.G., and Bulinski J.C.. 1988. Enhanced stability of microtubules enriched in detyrosinated tubulin is not a direct function of detyrosination level. J. Cell Biol. 106:141–149 10.1083/jcb.106.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., Kurabe N., Ikegami K., Tsutsumi K., Konishi Y., Kaplan O.I., Kunitomo H., Iino Y., Blacque O.E., and Setou M.. 2010. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs). J. Biol. Chem. 285:22936–22941 10.1074/jbc.C110.128280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, Y., and Setou M.. 2009. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 12:559–567 10.1038/nn.2314 [DOI] [PubMed] [Google Scholar]
- Kreitzer, G., Liao G., and Gundersen G.G.. 1999. Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Mol. Biol. Cell. 10:1105–1118 10.1091/mbc.10.4.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, T., Yanagisawa H.A., Yagi T., Hirono M., and Kamiya R.. 2010. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 20:441–445 10.1016/j.cub.2009.12.058 [DOI] [PubMed] [Google Scholar]
- Kumar, N., and Flavin M.. 1981. Preferential action of a brain detyrosinolating carboxypeptidase on polymerized tubulin. J. Biol. Chem. 256:7678–7686 [PubMed] [Google Scholar]
- Lacroix, B., and Janke C.. 2011. Generation of differentially polyglutamylated microtubules. Methods Mol. Biol. 777:57–69 10.1007/978-1-61779-252-6_4 [DOI] [PubMed] [Google Scholar]
- Lacroix, B., van Dijk J., Gold N.D., Guizetti J., Aldrian-Herrada G., Rogowski K., Gerlich D.W., and Janke C.. 2010. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J. Cell Biol. 189:945–954 10.1083/jcb.201001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafanechère, L., Courtay-Cahen C., Kawakami T., Jacrot M., Rüdiger M., Wehland J., Job D., and Margolis R.L.. 1998. Suppression of tubulin tyrosine ligase during tumor growth. J. Cell Sci. 111:171–181 [DOI] [PubMed] [Google Scholar]
- Lee, G.-S., He Y., Dougherty E.J., Jimenez-Movilla M., Avella M., Grullon S., Sharlin D.S., Guo C., Blackford J.A. Jr, Awasthi S., et al. 2013. Disruption of Ttll5/stamp gene (tubulin tyrosine ligase-like protein 5/SRC-1 and TIF2-associated modulatory protein gene) in male mice causes sperm malformation and infertility. J. Biol. Chem. 288:15167–15180 10.1074/jbc.M113.453936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levilliers, N., Fleury A., and Hill A.M.. 1995. Monoclonal and polyclonal antibodies detect a new type of post-translational modification of axonemal tubulin. J. Cell Sci. 108:3013–3028 [DOI] [PubMed] [Google Scholar]
- Lewis, S.A., Lee M.G., and Cowan N.J.. 1985. Five mouse tubulin isotypes and their regulated expression during development. J. Cell Biol. 101:852–861 10.1083/jcb.101.3.852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S.A., Gu W., and Cowan N.J.. 1987. Free intermingling of mammalian beta-tubulin isotypes among functionally distinct microtubules. Cell. 49:539–548 10.1016/0092-8674(87)90456-9 [DOI] [PubMed] [Google Scholar]
- L’Hernault, S.W., and Rosenbaum J.L.. 1985. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 24:473–478 10.1021/bi00323a034 [DOI] [PubMed] [Google Scholar]
- Liao, G., and Gundersen G.G.. 1998. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J. Biol. Chem. 273:9797–9803 10.1074/jbc.273.16.9797 [DOI] [PubMed] [Google Scholar]
- Linck, R., Fu X., Lin J., Ouch C., Schefter A., Steffen W., Warren P., and Nicastro D.. 2014. Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2+-binding proteins, and stable protofilaments. J. Biol. Chem. 289:17427–17444 10.1074/jbc.M114.568949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludueña, R.F., and Banerjee A.. 2008. The isotypes of tubulin: distribution and functional significance. Cancer Drug Discovery and Development: The Role of Microtubules in Cell Biology, Neurobiology, and Oncology. Fojo T., editor Humana Press, Totowa, NJ: 123–175 [Google Scholar]
- Maas, C., Belgardt D., Lee H.K., Heisler F.F., Lappe-Siefke C., Magiera M.M., van Dijk J., Hausrat T.J., Janke C., and Kneussel M.. 2009. Synaptic activation modifies microtubules underlying transport of postsynaptic cargo. Proc. Natl. Acad. Sci. USA. 106:8731–8736 10.1073/pnas.0812391106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiera, M.M., and Janke C.. 2013. Investigating tubulin posttranslational modifications with specific antibodies. Methods Cell Biol. 115:247–267 10.1016/B978-0-12-407757-7.00016-5 [DOI] [PubMed] [Google Scholar]
- Marcos, S., Moreau J., Backer S., Job D., Andrieux A., and Bloch-Gallego E.. 2009. Tubulin tyrosination is required for the proper organization and pathfinding of the growth cone. PLoS ONE. 4:e5405 10.1371/journal.pone.0005405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary, J., Redeker V., Le Caer J.P., Promé J.C., and Rossier J.. 1994. Class I and IVa beta-tubulin isotypes expressed in adult mouse brain are glutamylated. FEBS Lett. 353:89–94 10.1016/0014-5793(94)01018-8 [DOI] [PubMed] [Google Scholar]
- Mary, J., Redeker V., Le Caer J.P., Rossier J., and Schmitter J.M.. 1996. Posttranslational modifications in the C-terminal tail of axonemal tubulin from sea urchin sperm. J. Biol. Chem. 271:9928–9933 10.1074/jbc.271.17.9928 [DOI] [PubMed] [Google Scholar]
- Mary, J., Redeker V., Le Caer J.P., Rossier J., and Schmitter J.M.. 1997. Posttranslational modifications of axonemal tubulin. J. Protein Chem. 16:403–407 10.1023/A:1026336722124 [DOI] [PubMed] [Google Scholar]
- Mialhe, A., Lafanechère L., Treilleux I., Peloux N., Dumontet C., Brémond A., Panh M.H., Payan R., Wehland J., Margolis R.L., and Job D.. 2001. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 61:5024–5027 [PubMed] [Google Scholar]
- Minoura, I., Hachikubo Y., Yamakita Y., Takazaki H., Ayukawa R., Uchimura S., and Muto E.. 2013. Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 587:3450–3455 10.1016/j.febslet.2013.08.032 [DOI] [PubMed] [Google Scholar]
- Montagnac, G., Meas-Yedid V., Irondelle M., Castro-Castro A., Franco M., Shida T., Nachury M.V., Benmerah A., Olivo-Marin J.-C., and Chavrier P.. 2013. αTAT1 catalyses microtubule acetylation at clathrin-coated pits. Nature. 502:567–570 10.1038/nature12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai, M., Ikegami K., Sugiura Y., Takeshita K., Nakagawa A., and Setou M.. 2009. Recombinant mammalian tubulin polyglutamylase TTLL7 performs both initiation and elongation of polyglutamylation on β-tubulin through a random sequential pathway. Biochemistry. 48:1084–1093 10.1021/bi802047y [DOI] [PubMed] [Google Scholar]
- Mullen, R.J., Eicher E.M., and Sidman R.L.. 1976. Purkinje cell degeneration, a new neurological mutation in the mouse. Proc. Natl. Acad. Sci. USA. 73:208–212 10.1073/pnas.73.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, C.N., DeLuca J.G., Himes R.H., Miller H.P., Jordan M.A., and Wilson L.. 2002. Intrinsically slow dynamic instability of HeLa cell microtubules in vitro. J. Biol. Chem. 277:42456–42462 10.1074/jbc.M207134200 [DOI] [PubMed] [Google Scholar]
- Nicastro, D., Fu X., Heuser T., Tso A., Porter M.E., and Linck R.W.. 2011. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci. USA. 108:E845–E853 10.1073/pnas.1106178108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, S., Takahashi H., and Hirokawa N.. 2013. β-Tubulin mutations that cause severe neuropathies disrupt axonal transport. EMBO J. 32:1352–1364 10.1038/emboj.2013.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales, E., Wolf S.G., and Downing K.H.. 1998. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 391:199–203 10.1038/34465 [DOI] [PubMed] [Google Scholar]
- North, B.J., Marshall B.L., Borra M.T., Denu J.M., and Verdin E.. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 11:437–444 10.1016/S1097-2765(03)00038-8 [DOI] [PubMed] [Google Scholar]
- Ozols, J., and Caron J.M.. 1997. Posttranslational modification of tubulin by palmitoylation: II. Identification of sites of palmitoylation. Mol. Biol. Cell. 8:637–645 10.1091/mbc.8.4.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, N., Obara T., Mangos S., Liu Y., and Drummond I.A.. 2007. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell. 18:4353–4364 10.1091/mbc.E07-06-0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, N., Austin C.A., and Drummond I.A.. 2011. Tubulin tyrosine ligase-like genes ttll3 and ttll6 maintain zebrafish cilia structure and motility. J. Biol. Chem. 286:11685–11695 10.1074/jbc.M110.209817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paturle, L., Wehland J., Margolis R.L., and Job D.. 1989. Complete separation of tyrosinated, detyrosinated, and nontyrosinatable brain tubulin subpopulations using affinity chromatography. Biochemistry. 28:2698–2704 10.1021/bi00432a050 [DOI] [PubMed] [Google Scholar]
- Paturle-Lafanechère, L., Eddé B., Denoulet P., Van Dorsselaer A., Mazarguil H., Le Caer J.P., Wehland J., and Job D.. 1991. Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry. 30:10523–10528 10.1021/bi00107a022 [DOI] [PubMed] [Google Scholar]
- Paturle-Lafanechère, L., Manier M., Trigault N., Pirollet F., Mazarguil H., and Job D.. 1994. Accumulation of delta 2-tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. J. Cell Sci. 107:1529–1543 [DOI] [PubMed] [Google Scholar]
- Peris, L., Thery M., Fauré J., Saoudi Y., Lafanechère L., Chilton J.K., Gordon-Weeks P., Galjart N., Bornens M., Wordeman L., et al. 2006. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 174:839–849 10.1083/jcb.200512058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris, L., Wagenbach M., Lafanechère L., Brocard J., Moore A.T., Kozielski F., Job D., Wordeman L., and Andrieux A.. 2009. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 185:1159–1166 10.1083/jcb.200902142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J.D., Furlong M.T., Asai D.J., Harrison M.L., and Geahlen R.L.. 1996. Syk, activated by cross-linking the B-cell antigen receptor, localizes to the cytosol where it interacts with and phosphorylates alpha-tubulin on tyrosine. J. Biol. Chem. 271:4755–4762 10.1074/jbc.271.9.4755 [DOI] [PubMed] [Google Scholar]
- Prota, A.E., Magiera M.M., Kuijpers M., Bargsten K., Frey D., Wieser M., Jaussi R., Hoogenraad C.C., Kammerer R.A., Janke C., and Steinmetz M.O.. 2013. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 200:259–270 10.1083/jcb.201211017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, E.C., Hoyle H.D., Popodi E.M., and Turner F.R.. 2008. Axoneme beta-tubulin sequence determines attachment of outer dynein arms. Curr. Biol. 18:911–914 10.1016/j.cub.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybin, D., and Flavin M.. 1975. An enzyme tyrosylating alpha-tubulin and its role in microtubule assembly. Biochem. Biophys. Res. Commun. 65:1088–1095 10.1016/S0006-291X(75)80497-9 [DOI] [PubMed] [Google Scholar]
- Redeker, V., Le Caer J.P., Rossier J., and Promé J.C.. 1991. Structure of the polyglutamyl side chain posttranslationally added to alpha-tubulin. J. Biol. Chem. 266:23461–23466 [PubMed] [Google Scholar]
- Redeker, V., Levilliers N., Schmitter J.M., Le Caer J.P., Rossier J., Adoutte A., and Bré M.H.. 1994. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science. 266:1688–1691 10.1126/science.7992051 [DOI] [PubMed] [Google Scholar]
- Reed, N.A., Cai D., Blasius T.L., Jih G.T., Meyhofer E., Gaertig J., and Verhey K.J.. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16:2166–2172 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Regnard, C., Desbruyères E., Huet J.C., Beauvallet C., Pernollet J.C., and Eddé B.. 2000. Polyglutamylation of nucleosome assembly proteins. J. Biol. Chem. 275:15969–15976 10.1074/jbc.M000045200 [DOI] [PubMed] [Google Scholar]
- Regnard, C., Fesquet D., Janke C., Boucher D., Desbruyéres E., Koulakoff A., Insina C., Travo P., and Eddé B.. 2003. Characterisation of PGs1, a subunit of a protein complex co-purifying with tubulin polyglutamylase. J. Cell Sci. 116:4181–4190 10.1242/jcs.00743 [DOI] [PubMed] [Google Scholar]
- Ren, Y., Zhao J., and Feng J.. 2003. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J. Neurosci. 23:3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal, R., Schneider B.G., Miller M.M., and Ludueña R.F.. 1993. Beta IV is the major beta-tubulin isotype in bovine cilia. Cell Motil. Cytoskeleton. 25:19–29 10.1002/cm.970250104 [DOI] [PubMed] [Google Scholar]
- Robson, S.J., and Burgoyne R.D.. 1989. Differential localisation of tyrosinated, detyrosinated, and acetylated alpha-tubulins in neurites and growth cones of dorsal root ganglion neurons. Cell Motil. Cytoskeleton. 12:273–282 10.1002/cm.970120408 [DOI] [PubMed] [Google Scholar]
- Rodriguez de la Vega, M., Sevilla R.G., Hermoso A., Lorenzo J., Tanco S., Diez A., Fricker L.D., Bautista J.M., and Avilés F.X.. 2007. Nna1-like proteins are active metallocarboxypeptidases of a new and diverse M14 subfamily. FASEB J. 21:851–865 10.1096/fj.06-7330com [DOI] [PubMed] [Google Scholar]
- Rogowski, K., Juge F., van Dijk J., Wloga D., Strub J.-M., Levilliers N., Thomas D., Bré M.H., Van Dorsselaer A., Gaertig J., and Janke C.. 2009. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 137:1076–1087 10.1016/j.cell.2009.05.020 [DOI] [PubMed] [Google Scholar]
- Rogowski, K., van Dijk J., Magiera M.M., Bosc C., Deloulme J.-C., Bosson A., Peris L., Gold N.D., Lacroix B., Bosch Grau M., et al. 2010. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 143:564–578 10.1016/j.cell.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Rosas-Acosta, G., Russell W.K., Deyrieux A., Russell D.H., and Wilson V.G.. 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell. Proteomics. 4:56–72 10.1074/mcp.M400149-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger, M., Plessman U., Klöppel K.D., Wehland J., and Weber K.. 1992. Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 308:101–105 10.1016/0014-5793(92)81061-P [DOI] [PubMed] [Google Scholar]
- Rusconi, F., Potier M.C., Le Caer J.P., Schmitter J.M., and Rossier J.. 1997. Characterization of the chicken telokin heterogeneity by time-of-flight mass spectrometry. Biochemistry. 36:11021–11026 10.1021/bi970752e [DOI] [PubMed] [Google Scholar]
- Schröder, H.C., Wehland J., and Weber K.. 1985. Purification of brain tubulin-tyrosine ligase by biochemical and immunological methods. J. Cell Biol. 100:276–281 10.1083/jcb.100.1.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, H.D., Lecine P., Tiwari S., Italiano J.E. Jr, Hartwig J.H., and Shivdasani R.A.. 2001. A lineage-restricted and divergent beta-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr. Biol. 11:579–586 10.1016/S0960-9822(01)00153-1 [DOI] [PubMed] [Google Scholar]
- Sherwin, T., Schneider A., Sasse R., Seebeck T., and Gull K.. 1987. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated α-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol. 104:439–446 10.1083/jcb.104.3.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida, T., Cueva J.G., Xu Z., Goodman M.B., and Nachury M.V.. 2010. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA. 107:21517–21522 10.1073/pnas.1013728107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin, M., Rice L.M., and Vale R.D.. 2014. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16:335–344 10.1038/ncb2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., Kirkpatrick L.L., Schilling A.B., Helseth D.L., Chabot N., Keillor J.W., Johnson G.V.W., and Brady S.T.. 2013. Transglutaminase and polyamination of tubulin: posttranslational modification for stabilizing axonal microtubules. Neuron. 78:109–123 10.1016/j.neuron.2013.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina, V., Herbstman J.F., Skiniotis G., and Verhey K.J.. 2012. Luminal localization of α-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PLoS ONE. 7:e48204 10.1371/journal.pone.0048204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulimenko, V., Dráberová E., Sulimenko T., Macurek L., Richterová V., Dráber P., and Dráber P.. 2006. Regulation of microtubule formation in activated mast cells by complexes of gamma-tubulin with Fyn and Syk kinases. J. Immunol. 176:7243–7253 10.4049/jimmunol.176.12.7243 [DOI] [PubMed] [Google Scholar]
- Suryavanshi, S., Eddé B., Fox L.A., Guerrero S., Hard R., Hennessey T., Kabi A., Malison D., Pennock D., Sale W.S., et al. 2010. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. 20:435–440 10.1016/j.cub.2009.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk, A., Deaconescu A.M., Piszczek G., and Roll-Mecak A.. 2011. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 18:1250–1258 10.1038/nsmb.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk, A., Piszczek G., and Roll-Mecak A.. 2013. Tubulin tyrosine ligase and stathmin compete for tubulin binding in vitro. J. Mol. Biol. 425:2412–2414 10.1016/j.jmb.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk, A., Deaconescu A.M., Spector J., Goodman B., Valenstein M.L., Ziolkowska N.E., Kormendi V., Grigorieff N., and Roll-Mecak A.. 2014. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell. 157:1405–1415 10.1016/j.cell.2014.03.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, G., Huang Y., Rommelaere H., Vandekerckhove J., Ampe C., and Cowan N.J.. 1996. Pathway leading to correctly folded beta-tubulin. Cell. 86:287–296 10.1016/S0092-8674(00)80100-2 [DOI] [PubMed] [Google Scholar]
- Tischfield, M.A., Baris H.N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W.-M., Andrews C., Demer J.L., Robertson R.L., et al. 2010. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 140:74–87 10.1016/j.cell.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield, M.A., Cederquist G.Y., Gupta M.L. Jr, and Engle E.C.. 2011. Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr. Opin. Genet. Dev. 21:286–294 10.1016/j.gde.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg, I.E., Lewis S.A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H.L., and Cowan N.J.. 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 93:863–873 10.1016/S0092-8674(00)81446-4 [DOI] [PubMed] [Google Scholar]
- Valenzuela, P., Quiroga M., Zaldivar J., Rutter W.J., Kirschner M.W., and Cleveland D.W.. 1981. Nucleotide and corresponding amino acid sequences encoded by alpha and beta tubulin mRNAs. Nature. 289:650–655 10.1038/289650a0 [DOI] [PubMed] [Google Scholar]
- Valenzuela-Fernández, A., Cabrero J.R., Serrador J.M., and Sánchez-Madrid F.. 2008. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 18:291–297 10.1016/j.tcb.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Vallee, R.B.1986. Reversible assembly purification of microtubules without assembly-promoting agents and further purification of tubulin, microtubule-associated proteins, and MAP fragments. Methods Enzymol. 134:89–104 10.1016/0076-6879(86)34078-3 [DOI] [PubMed] [Google Scholar]
- van Dijk, J., Rogowski K., Miro J., Lacroix B., Eddé B., and Janke C.. 2007. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 26:437–448 10.1016/j.molcel.2007.04.012 [DOI] [PubMed] [Google Scholar]
- van Dijk, J., Miro J., Strub J.-M., Lacroix B., van Dorsselaer A., Eddé B., and Janke C.. 2008. Polyglutamylation is a post-translational modification with a broad range of substrates. J. Biol. Chem. 283:3915–3922 10.1074/jbc.M705813200 [DOI] [PubMed] [Google Scholar]
- Vemu, A., Garnham C.P., Lee D.-Y., and Roll-Mecak A.. 2014. Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods Enzymol. 540:149–166 10.1016/B978-0-12-397924-7.00009-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey, K.J., and Gaertig J.. 2007. The tubulin code. Cell Cycle. 6:2152–2160 10.4161/cc.6.17.4633 [DOI] [PubMed] [Google Scholar]
- Vogel, P., Hansen G., Fontenot G., and Read R.. 2010. Tubulin tyrosine ligase-like 1 deficiency results in chronic rhinosinusitis and abnormal development of spermatid flagella in mice. Vet. Pathol. 47:703–712 10.1177/0300985810363485 [DOI] [PubMed] [Google Scholar]
- Walgren, J.L.E., Vincent T.S., Schey K.L., and Buse M.G.. 2003. High glucose and insulin promote O-GlcNAc modification of proteins, including alpha-tubulin. Am. J. Physiol. Endocrinol. Metab. 284:E424–E434 [DOI] [PubMed] [Google Scholar]
- Walter, W.J., Beránek V., Fischermeier E., and Diez S.. 2012. Tubulin acetylation alone does not affect kinesin-1 velocity and run length in vitro. PLoS ONE. 7:e42218 10.1371/journal.pone.0042218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Villasante A., Lewis S.A., and Cowan N.J.. 1986. The mammalian β-tubulin repertoire: hematopoietic expression of a novel, heterologous β-tubulin isotype. J. Cell Biol. 103:1903–1910 10.1083/jcb.103.5.1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee, J.A., Luftig R.B., and Weihing R.R.. 1980. Purification and reconstitution of HeLa cell microtubules. Biochemistry. 19:4116–4123 10.1021/bi00558a033 [DOI] [PubMed] [Google Scholar]
- Wehenkel, A., and Janke C.. 2014. Towards elucidating the tubulin code. Nat. Cell Biol. 16:303–305 10.1038/ncb2938 [DOI] [PubMed] [Google Scholar]
- Widlund, P.O., Podolski M., Reber S., Alper J., Storch M., Hyman A.A., Howard J., and Drechsel D.N.. 2012. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol. Biol. Cell. 23:4393–4401 10.1091/mbc.E12-06-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga, D., Webster D.M., Rogowski K., Bré M.H., Levilliers N., Jerka-Dziadosz M., Janke C., Dougan S.T., and Gaertig J.. 2009. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell. 16:867–876 10.1016/j.devcel.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Wolff, A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., and Denoulet P.. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432 [PubMed] [Google Scholar]
- Wong, C.C.L., Xu T., Rai R., Bailey A.O., Yates J.R. III, Wolf Y.I., Zebroski H., and Kashina A.. 2007. Global analysis of posttranslational protein arginylation. PLoS Biol. 5:e258 10.1371/journal.pbio.0050258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, L., Hai B., Gao Y., Burnette D., Thazhath R., Duan J., Bré M.H., Levilliers N., Gorovsky M.A., and Gaertig J.. 2000. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 149:1097–1106 10.1083/jcb.149.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H., El Bissati K., Verdier-Pinard P., Burd B., Zhang H., Kim K., Fiser A., Angeletti R.H., and Weiss L.M.. 2010. Post-translational modifications to Toxoplasma gondii alpha- and beta-tubulins include novel C-terminal methylation. J. Proteome Res. 9:359–372 10.1021/pr900699a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, R., Clark K.M., and Gorovsky M.A.. 2007. Endoplasmic reticulum retention signal-dependent glycylation of the Hsp70/Grp170-related Pgp1p in Tetrahymena. Eukaryot. Cell. 6:388–397 10.1128/EC.00366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G., Paige J.S., and Jaffrey S.R.. 2010. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28:868–873 10.1038/nbt.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.B., Levison B.S., Szasz J., and Sternlicht H.. 1988. Expression of a human alpha-tubulin: properties of the isolated subunit. Biochemistry. 27:1869–1880 10.1021/bi00406a012 [DOI] [PubMed] [Google Scholar]
- Yaffe, M.B., Farr G.W., Miklos D., Horwich A.L., Sternlicht M.L., and Sternlicht H.. 1992. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 358:245–248 10.1038/358245a0 [DOI] [PubMed] [Google Scholar]
- Zyss, D., Montcourrier P., Vidal B., Anguille C., Mérezègue F., Sahuquet A., Mangeat P.H., and Coopman P.J.. 2005. The Syk tyrosine kinase localizes to the centrosomes and negatively affects mitotic progression. Cancer Res. 65:10872–10880 10.1158/0008-5472.CAN-05-1270 [DOI] [PubMed] [Google Scholar]