Abstract
Motivation and ability both underlie voluntary exercise, each with a potentially unique genetic architecture. Muscle structure and function are one of many morphological and physiological systems acting to simultaneously determine exercise ability. We generated a large (n = 815) advanced intercross line of mice (G4) derived from a line selectively bred for increased wheel running (high runner) and the C57BL/6J inbred strain. We previously mapped quantitative trait loci (QTL) contributing to voluntary exercise, body composition, and changes in body composition as a result of exercise. Using brain tissue in a subset of the G4 (n = 244), we have also previously reported expression QTL (eQTL) colocalizing with the QTL for the higher-level phenotypes. Here, we examined the transcriptional landscape of hind limb muscle tissue via global mRNA expression profiles. Correlations revealed an ∼1,168% increase in significant relationships between muscle transcript expression levels and the same exercise and body composition phenotypes examined previously in the brain. The exercise trait most often significantly correlated with gene expression in the brain was running duration while in the muscle it was maximum running speed. This difference may indicate that time spent engaging in exercise behavior may be more influenced by central (neurobiological) mechanisms, while intensity of exercise may be largely controlled by peripheral mechanisms. Additionally, we used subsets of cis-acting eQTL, colocalizing with QTL, to identify candidate genes based on both positional and functional evidence. We discuss three plausible candidate genes (Insig2, Prcp, Sparc) and their potential regulatory role.
Keywords: adiposity, body weight, eQTL, experimental evolution, wheel running
the predisposition to engage in voluntary activity is variable among humans and rodents and simultaneously influenced by genetics, the environment, and gene-by-environment interactions (27). Although voluntary exercise is exceedingly complex, it is hypothesized that some combination of both ability and motivation play an integral role in regulating the level of activity among individuals, with both of these components having a complex underlying genetic architecture (15).
Neurobiological investigations aimed at uncovering the motivational aspects of voluntary exercise have been discussed previously (see Ref. 26 and references therein). Here we focus on studies chronicling the variation in ability and trainability (broadly characterized as exercise sciences or exercise physiology). One major focus of exercise physiology is uncovering the mechanistic role of gene function and regulation in exercise performance (for a historical perspective see Ref. 5). For example, a total of 214 autosomal genes, seven loci on the X chromosome, and 18 mitochondrial genes were reported as influencing “physical performance” and “health-related fitness” phenotypes in humans (see Ref. 6, 2006–2007 update). This number has almost certainly increased in the subsequent years (e.g., Ref. 34). The performance phenotypes included in Ref. 6's “human gene map” consist of “cardiorespiratory endurance,” “elite endurance athlete status,” “muscle strength,” “other muscle performance traits,” and “exercise intolerance of variable degrees.” The physical fitness traits are grouped into hemodynamic traits including exercise heart rate, blood pressure, and heart morphology; anthropometry and body composition; insulin and glucose metabolism; and blood lipid, lipoprotein, and hemostatic factors (6). Many, if not all, of the traits listed above would be hypothesized to affect the ability to engage in physical activity.
Rodent studies have also demonstrated a genetic basis for individual variation in exercise ability. Importantly, the translational nature of rodent wheel running to human health has been extensively discussed elsewhere (see Refs. 15, 27, and references therein), and we believe that voluntary wheel running appropriately models voluntary exercise in human populations, a complex behavior simultaneously affected by central and peripheral mechanisms. Selective breeding for elevated endurance capacity during forced treadmill running in rats has resulted in greater skeletal muscle capillarity, muscle oxidative enzyme activities, V̇o2max, and peripheral oxygen transport and utilization (see Ref. 21 and references therein). Replicated artificial selection for increased voluntary wheel-running behavior has resulted in an approximate 2.5- to 3.0-fold increase in total revolutions/day (36). Mice bred for high wheel running [high runners (HR) lines] on days 5 and 6 of a 6-day test exhibit a number of constitutive traits (expressed in the absence of wheel access) that clearly or plausibly represent adaptations with respect to wheel-running ability: reduced body mass, less body fat, lower leptin levels, increased levels of adiponectin, resistance to high-fat diet-induced obesity, elevated maximal oxygen consumption during forced treadmill exercise (V̇o2max), greater treadmill endurance, mild cardiac hypertrophy, increased insulin-stimulated glucose uptake in the extensor digitorum longus muscle, a trend toward higher muscle aerobic capacities (via mitochondrial and glycolytic enzyme activities), lower anaerobic capacities, elevated muscle glycogen concentrations, greater muscle (plantaris) capillarity, and altered fiber types in gastrocnemius muscle (see Refs. 3, 16, 18, 37–41 and references therein; not an exhaustive list). The later three phenotypes are unique to selectively bred individuals expressing the minimuscle phenotype, characterized by an ∼50% reduction in mass of the triceps surae muscle complex (gastrocnemius, plantaris, soleus) and in the mass of the entire hindlimb musculature (see Ref. 28 and references therein). This phenotype is caused by an autosomal recessive mutation representing a C-to-T transition located in a 709 bp intron between exons 11 and 12 of the myosin heavy polypeptide 4 (Myh4) skeletal muscle gene (28). It has been observed in two (lab designation line 3 and 6) of the four HR lines and one (lab designation line 5) of the four control lines (28).
In addition to differences observed between HR and control mice in the absence of wheel access (above), we have also observed enhanced plasticity (training effects) in some traits, such as the concentration of the glucose transporter GLUT4 in gastrocnemius muscle. This enhanced plasticity is not explainable by the greater running in HR lines but appears to reflect inherently greater plasticity in the HR lines (i.e., for a given amount of stimulus, such as wheel running/day, individuals in the HR lines show a greater response compared with individuals in the control lines). For complete context and discussion of these traits see Ref. (14).
Previously, we generated an advanced intercross line (AIL; G4) of mice through reciprocal crosses between a line selectively bred for high voluntary wheel running (lab designation line 8) and the inbred strain C57BL/6J (23). The inbred strain was chosen, rather than one of the control lines, in an attempt to maximize the number of fully informative genetic markers. The minimuscle phenotype, discussed above, has never been observed in the HR line utilized to create the AIL. The G4 population has formerly been utilized for investigation of the phenotypic relationships between and identification of QTL for voluntary exercise traits, body composition traits, food consumption, changes in body weight and composition in response to exercise, and skeletal architecture traits (13, 24, 25). Most recently, using whole-brain tissue (26), we reported on the transcriptional landscape relevant to motivational aspects of voluntary exercise, with the presumption that results would be more relevant to motivational aspects than to physical abilities for exercise. We identified genome-wide expression quantitative trait loci (eQTL) and, on the basis of both positional and functional evidence, discussed plausible candidate genes regulating voluntary activity, body composition, and their interactions.
Here, we build upon that initial model of underlying functional genomic architecture by use of hindlimb muscle tissue to capture the transcriptional landscape relevant to certain aspects of the ability to engage in voluntary exercise. Coupled with previous investigations (phenotypic, QTL, eQTL), this study continues to build upon a systems approach toward understanding the predisposition to engage in voluntary exercise. For a detailed discussion of systems approaches aimed at the dissection of complex traits see (see Figs. 1, 4 in Refs. 27 and 33, respectively). Our initial goal in the present paper is to provide a broad comparison of muscle eQTL results to previous eQTL data gathered for brain tissue. As detailed above, we view both motivation and ability as important in the predisposition to engaged in voluntary exercise. However, the extent to which these two factors share a common genetic architecture is unresolved. A long-term goal is to merge all data from this AIL and begin to identify common biological mechanisms influencing exercise, body weight, adiposity, and their interactions (e.g., see Ref. 17).
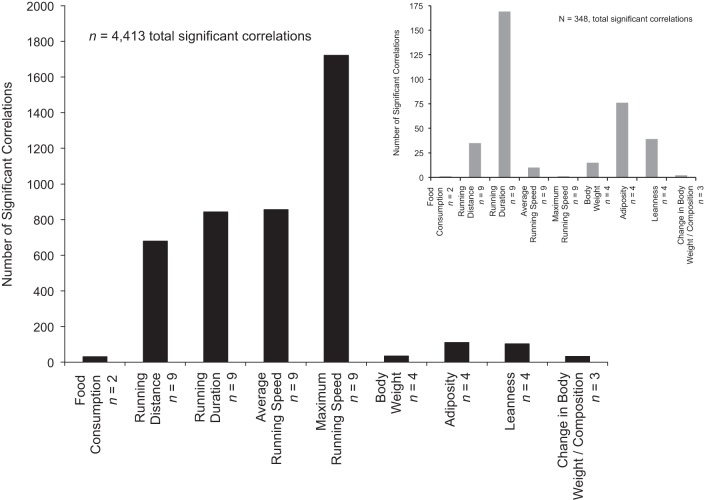
Fig. 1.
The number of statistically significant (P < 0.05, adjusted for multiple comparisons) partial correlations; adjusted for sex and parent of origin, factors with known phenotypic effects (see Ref. 23) between 17,571 significantly expressed transcripts, and exercise (n = 36) and body composition-related phenotypes (n = 17). For comparison, the number of significant partial correlations from expression data in brain tissue is inset (gray bars, Ref. 26).
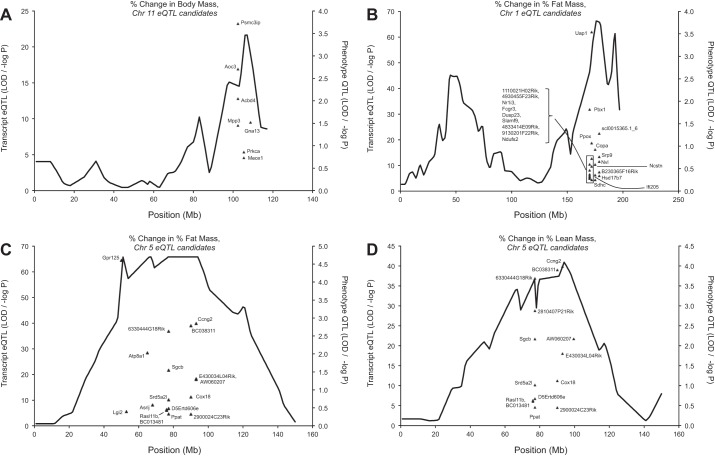
Fig. 4.
Cis-acting eQTL, colocalizing with QTL underlying changes in body weight and composition in response to 6 days of voluntary wheel running. Colocalizing candidate genes that fell within the confidence intervals of the trait QTL are depicted. Gene names in italics were suggestively correlated (partial, controlling for parent and sex) with running distance after correction for multiple comparisons (FDR, P ≤ 0.1, r ≥ 0.20). A: percentage change in body mass (Chr. 11). B: percentage change in percentage fat mass (Chr. 1). Note: eQTL (9130201F22Rik) is 12.9 Mb away from the physical gene midpoint. C: percentage change in percentage fat mass (Chr. 5). Note: eQTL D5Ertd606e, 2900024C23Rik, and AW060207 are 16.4, 18.3, and 15.0 Mb away from their respective physical gene midpoints. D: percentage change in percentage lean mass (Chr. 5). Note: eQTL D5Ertd606e, 2900024C23Rik, and AW060207 are 16.4, 18.3, and 15.0 Mb away from their respective physical gene midpoints.
MATERIALS AND METHODS
Population and phenotyping.
An AIL (G4, n = 815) was created by reciprocally crossing mice selectively bred for high voluntary wheel running (HR line) and the inbred strain C57BL/6J (B6). Complete methods regarding the creation and phenotyping of the G4 population, single nucleotide polymorphisms utilized for QTL analyses (n = 530), and RNA isolation and microarray analysis procedures may be found elsewhere (23–26). Only a brief methodological description will be provided here. All procedures were approved by and are in accordance with guidelines set forth by the Institutional Animal Care and Use Committee at the University of North Carolina (UNC) at Chapel Hill.
G4 mice (∼8 wk of age) were weighed, body composition assessed (% fat tissue and % lean tissue; EchoMRI-100, Echo Medical Systems, Houston, TX), and individually housed with access to running wheels (circumference = 1.1 m, model 80850; Lafayette Instruments, Lafayette, IN) for 6 days. Distance (total revolutions), time spent running (cumulative 1 min intervals in which at least one revolution was recorded), average speed (total revolutions/time spent running), and maximum speed were calculated daily, as were the mean values on days 5 and 6 (the criterion for which the HR line was selectively bred; see Ref. 36). Mice were removed from the wheels following the completion of the 6th day of wheel access (i.e., the morning of day 7) and killed the same day in the order in which they were given wheel access (which was randomly chosen across both sex and parent-of-origin types). Following postwheel access weight and body composition measures, mice were decapitated, and hindlimb (triceps surae complex, including lateral and medial heads of the gastrocnemius, plantaris, and soleus) muscles were harvested, flash-frozen in liquid nitrogen, and stored at −80°C.
RNA isolation and microarray analysis.
Isolation and purification of total RNA with TRIzol (Invitrogen, Carlsbad, CA) was performed from a homogenate of the right triceps surae complex. A subset (n = 243, 4 individuals were removed from the final analyses because of a lack of genotype information) of the total G4 population (n = 815) was utilized and represented the population-wide variation in running distance, each of two parent-of-origin types [whether a G4 individual was descended from a progenitor (F0) cross of HR♀ × B6♂ or B6♀ × HR♂], and both sexes. These 243 individuals overlapped with those previously used in Ref. 26. We used the MouseWG-6 v2.0 Beadchip (Illumina, San Diego, CA) to profile 45,281 transcripts and processed them with the Illumina Microarray Services at Expression Analysis, (Durham, NC). Profiles were normalized by Loess-Quantile normalization methods with R v. 2.8.1 statistical software (R Development Core Team; http://www.r-project.org, lumi package), and detection scores ≥ 0.95 were utilized for correlation and eQTL analyses (8, 19, 32).
Correlation analysis.
Genes significantly expressed above background (detection scores ≥ 0.95) were tested for correlation with exercise (n = 36) and body composition (n = 17) phenotypes previously measured in the G4 population by the PROC CORR procedure in SAS (version 9.1, SAS Institute, Cary, NC). Correlations were adjusted for sex and parent-of-origin type, factors with known phenotypic effects (23). P values were adjusted for multiple comparisons in SAS (PROC MULTTEST procedure) by the false discovery rate (FDR) procedure controlling the overall type I error rate at 5% (10).
eQTL analysis.
We identified eQTL by the multiple imputation method within R/qtl for the R environment (7, 35). Statistical models included sex and parent-of-origin type. Following Ref. 26, a significance threshold [logarithm of odds (LOD) > 3.8] was calculated via permutation tests (n = 1,000) of 100 randomly selected transcripts (an approach also similar to Ref. 43). Cis-acting (or local) eQTL were defined as being 10 Mb or less away from the midpoint of the physical location of the gene each represented, while trans-acting eQTL were >10 Mb away (following Ref. 12).
RESULTS
Correlation analysis.
Transcripts were normalized (Loess-Quantile normalization), and 12,794 (of 45,281) were identified with a detection score (calculated across all 243 mice) ≥ 0.95. After adjustment for multiple testing, 4,413 (0.66% of total possible) partial correlations were found to be statistically significant (P < 0.05), indicating potential functional relevance (Fig. 1). Relationships between exercise-related traits and transcript levels accounted for the largest proportion (92.9%) of observed significant correlations (Fig. 1). Among the exercise traits, maximum running speed represented the largest percentage of significant relationships with transcript levels (39.0%). Collectively, body weight and composition-related traits accounted for 5.6% of significant correlations (Fig. 1). Changes in body weight and composition, as a result of 6 days of exercise, represented 0.7% of significant correlations with transcript levels. Correlations with the greatest magnitude between exercise/body composition traits and transcript levels are presented in supporting information (Supplemental Table S1).1
eQTL analysis.
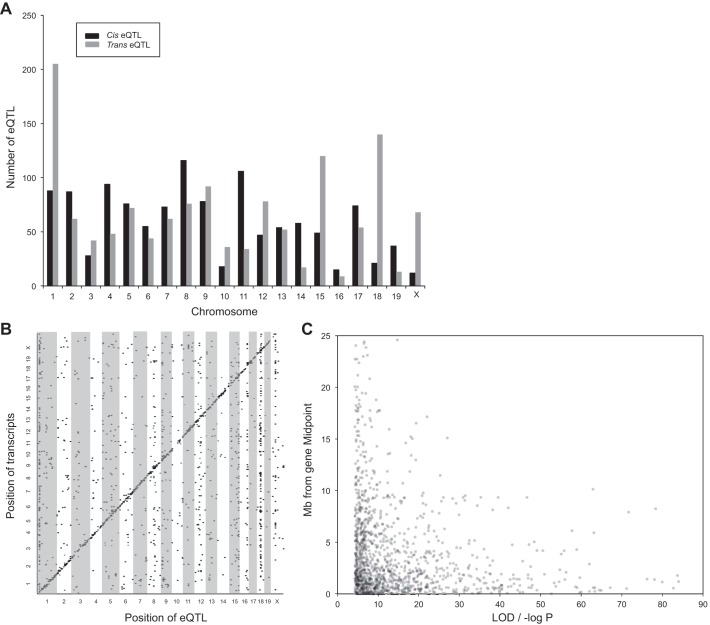
In total, 1,186 cis-acting and 1,330 trans-acting statistically significant eQTL were observed (Fig. 2, A and B). The average LOD score for cis-acting eQTL was 14.9 (range = 4.3–84.0), while for trans-acting eQTL the mean LOD score was 5.5 with a range of 4.3–95.9. Among cis-acting eQTL, the median distance of the mapped location to the midpoint of the physical location of the gene was 2.00 Mb, and the distance was generally negatively correlated with the significance level (Fig. 2C). For comparison, our prior work using brain tissue in the same population yielded a median distance of 1.94 Mb (26). Moreover, in a recombinant inbred mouse strain panel, the pre-Collaborative Cross, the median liver eQTL-gene distance was 0.92 Mb (4). Trans-acting eQTL were identified on all chromosomes, with a potential master regulatory region observed on the proximal end Chr. 1 at ∼16.7–20.1 Mb harboring 126 eQTL (Fig. 2B). An additional potential master regulatory region was observed on Chr. 18 at ∼56.7–59.8 Mb with 106 eQTL (Fig. 2B). A potential master regulatory region on Chr. 1 was also previously identified from brain expression data. However, this region was found distally at ∼170–180 Mb and contained 332 trans-acting eQTL.
Fig. 2.
A: the number of cis-acting (black bars) and trans-acting (gray bars) expression quantitative trait loci (eQTL) across all chromosomes with a logarithm of odds (LOD) ≥ 4.3. B: physical gene location as a function of mapped position of each QTL. Potential master regulatory regions are observed on chromosomes 1 and 18. C: distance (Mb) from gene midpoint of mapped location of Cis-acting eQTL. Cis eQTL were defined as falling within 10 Mb of the gene's physical midpoint location.
Using the G4 population we previously identified 39 significant and 18 suggestive QTL representing various exercise traits (21). Here, as in Ref. 26 we compared the locations of cis-acting eQTL within the confidence intervals (CI, defined by 1 LOD drop) of QTL observed for subsets of the mean exercise traits (distance, duration, average speed, and maximum speed; Fig. 3).
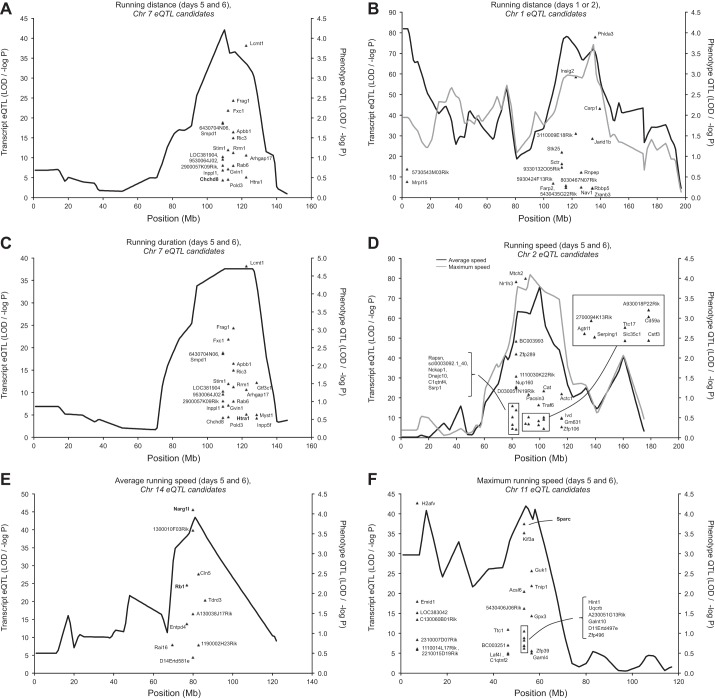
Fig. 3.
Cis-acting eQTL colocalizing with exercise QTL. Colocalizing candidate genes that fell within the confidence intervals of the trait QTL are depicted. Gene names in boldface were significantly correlated (partial, controlling for parent and sex) with running distance after correction for multiple comparisons false discovery rate (FDR), P ≤ 0.05, r ≥ 0.25]. Gene names in italics were suggestively correlated (partial, controlling for parent and sex) with running distance after correction for multiple comparisons (FDR, P ≤ 0.1, r ≥ 0.20). A: mean running distance (Chr. 7) on days 5 and 6 of a 6-day test. Note: eQTL Htra1 is 15.7 Mb away from the physical gene midpoint. B: running distance (Chr. 1) on each of days 1 (black line) and 2 (gray line). Note: the following eQTL are 13.8 Mb (5930424F13Rik), 17.1 Mb (Stk25), 26.4 (9330132O05Rik), 18.0 Mb (5430435G22Rik), and 20.0 Mb (Farp2) away from their physical gene midpoints. C: mean running duration (Chr. 7) on days 5 and 6 of a 6-day test. Note: eQTL Htra1 is 15.7 Mb away from the physical gene midpoint. D: average (black line) and maximum (gray line) running speed (Chr. 2) on days 5 and 6 of a 6-day test. E: average running speed (Chr. 14) on days 5 and 6 of a 6-day test. Note: eQTL D14Ertd581e is 19.8 Mb away and Cln5 is 20.8 Mb away from their physical gene midpoint. F: maximum running speed (Chr. 11) on days 5 and 6 of a 6-day test. Note: eQTL Garn14 is 17.7 Mb away and eQTL BC003251 is 11.9 Mb away from the physical gene midpoints.
Cis-acting eQTL localizing with running distance QTL (mean on days 5 and 6) revealed 19 positional candidate genes on Chr. 7 (Fig. 3A). Among these 19 candidate eQTL, cytochrome c oxidase assembly factor 4 (Coa4; Chchd8, old name) was significantly (FDR, P = 0.0248, r = 0.2737) correlated with running distance. For running distance on days 1 or 2, 18 potential candidate genes were identified under 2 QTL on Chr. 1 (Fig. 3B). For mean running duration on days 5 and 6, we identified 22 candidate genes on Chr. 7 (Fig. 3C). The candidate genes unique to running duration resulted from an expansion of the CI for running duration loci (91–129 Mb) compared with the CI for the running distance QTL (99–124 Mb). A statistically significant partial correlation (FDR, P = 0.0014, r = 0.3439) was observed between running duration and HtrA serine peptidase 1 (Htra1) (Fig. 3C). We observed 28 significant cis-acting candidate eQTL that mapped under the previously identified QTL (Chr. 2) for running speed (average and maximum) on days 5 and 6 of the 6-day wheel exposure (Fig. 3D). We identified 10 cis-acting candidate eQTL for average running speed on Chr. 14 (Fig. 3E). Of these, partial correlations indicated that N (alpha)-acetyl transferase 16 (Naa16; Narg1l, old name) and retinoblastoma 1 (Rb1) were statistically significantly correlated with average running speed on days 5 and 6 (FDR, P = 0.0328, r = −0.2652; P = 0.0455, r = 0.2536, respectively). In addition to those identified on Chr. 2, 31 cis-acting candidate eQTL were identified on Chr. 11 for maximum running speed (Fig. 3F). Of these correlation analysis revealed that secreted acidic cysteine rich glycoprotein (Sparc) was significantly correlated with maximal running speed (FDR, P = 0.0033, r = −0.3253).
We also examined colocalizing cis-acting candidate eQTL and loci previously identified for change in body weight and body composition in response to 6 days of exercise. Comparisons between cis-acting eQTL and loci observed for percentage change in body mass, as a result of 6 days of exercise, revealed only seven candidate genes on Chr. 11 (Fig. 4A). For percentage change in percentage fat mass we identified 21 candidate genes on Chr. 1 and 17 on Chr. 5 (Fig. 4, B and C). In addition, we observed 15 candidate genes on Chr. 5 for percentage change in percentage lean mass (Fig. 4D). No colocalizing cis-acting candidate eQTL was significantly correlated with any change in body weight or body composition variable in response to 6 days of exercise.
In comparison with previously mapped candidate eQTL in brain tissue (see Ref. 26), muscle tissue revealed fewer cis-acting candidate eQTL colocalizing with QTL for exercise and body composition phenotypes. Brain and muscle tissue shared the following number of cis-acting candidate eQTL colocalizing with exercise and body composition phenotypes: running distance on days 5 and 6 (Chr. 7), 11 out of a possible 19; running distance (days 1 or 2, Chr. 1), 11 out of a possible 18; running duration on days 5 and 6 (Chr. 7), 13 out of a possible 22; running speed (average and maximum) on days 5 and 6 (Chr. 2), 13 out of a possible 28; average running speed on days 5 and 6 (Chr. 14), 7 out of a possible 10; maximum running speed on days 5 and 6 (Chr. 11), 18 out of a possible 29; percent change in body mass following exercise (Chr. 11), 4 out of a possible 7; percent change in percent fat mass following exercise, 10 out of a possible 21 (Chr. 1) and 11 out of a possible 17 (Chr. 5); and percent change in percent lean mass following exercise, 6 out of possible 19 (Chr. 5).
DISCUSSION
Motivation vs. ability.
We previously utilized brain tissue to examine the transcriptional landscape relevant to the motivation to engage in voluntary exercise (26), although we recognize that the brain also plays an obvious role in neuromuscular stimulation and control, so could also be involved in running ability. We revealed hundreds of cis-acting candidate genes underlying previously identified genomic regions (QTL) that account for some of the variation in phenotypes associated with exercise, body composition, and the change in body composition in response to 6 days of voluntary wheel running. In addition to describing the general mode of gene action and reporting on correlational analyses, we specifically discussed six plausible candidate genes (Insig2, Socs2, DBY, Arrdc4, Prcp, IL15) and their potential role in the regulation of voluntary activity, body composition, and their interactions in a neurophysiological framework. Here, we have extended that initial model of underlying functional genomic architecture using hindlimb muscle tissue to capture the transcriptional landscape relevant to the ability to engage in voluntary exercise. The primary purpose of this article is to compare the current results to those in Ref. 26; however, we do acknowledge that eQTL that are closely linked may cluster and yet be different. Therefore, overlapping eQTL may not necessarily encompass the same underlying genetic loci that govern both motivation and ability. In cases of brain and muscle eQTL being closely associated, an important question for further study would be how such genetic architecture may have evolved by correlated natural or artificial selection acting on the complex phenotype of voluntary exercise.
Although we previously observed greater transcript expression levels and diversity in brain tissue (26), we detected far fewer significant relationships between gene expression and phenotypes compared with muscle tissue (Fig. 1). We observed an increase of ∼1,168% in statistically significant relationships between muscle transcript expression levels and the same exercise and body composition phenotypes examined by Ref. 26. This increase is most notably reflected in the elevated number of significant partial correlations (controlling for sex and parent-of-origin type) between muscle transcript expression and exercise-related traits. Of note, the exercise trait most often significantly correlated with gene expression in the brain was running duration, while in the muscle it was maximum running speed. The differences in the distribution of proportions of correlations potentially indicate that total time spent engaging in exercise behavior may be more influenced by central (neurobiological) mechanisms, while intensity of exercise may be largely controlled by peripheral mechanisms (in the current case muscle morphology or physiology). Lending support to the above hypothesis, Ref. 31 observed that noninvasive brain stimulation over the temporal cortex in trained cyclists regulates activity of autonomic nervous system and the perception of effort during exercise.
We do acknowledge that a limitation to the current approach is the choice of a nonspecific brain region as our measure of comparison. This choice may have produced a significant signal dilution relative to skeletal muscle and may contribute, or account for, the large differences in the number of statistically significant relationships between muscle and brain transcript levels and relevant phenotypes. As we stated in Ref. 26, in our opinion, there is no one particular brain (or muscle) region sufficient to account for the diversity of behavioral and physiological traits measured in the G4 population (e.g., running distance, weight regulation, food consumption). Our compromise was to bisect the hemispheres, run our initial expression assays on one hemisphere, and reserve the remaining hemisphere for potential follow-up studies in a more focused/targeted fashion.
Identification of potential candidate genes.
As was done previously (see Ref. 26), we will only discuss a fraction of the potential candidate genes depicted in Figs. 3 and 4. We have chosen the genes discussed below based on some combination of their cis-acting nature, colocalization with previously identified phenotypic QTL, and significant correlation with phenotypes of interest.
Insulin induced gene 2 (Insig2) was found to be a highly significant cis-acting eQTL (LOD = 58.6), colocalizing with loci previously identified for exercise and body composition-related traits on Chr. 1. Insig2 has been previously associated with human obesity, total plasma cholesterol levels, cholesterol biosynthesis, and lipid and cholesterol metabolism (2, 9, 11, 20, 30). Similar findings for Insig2 were previously reported in brain tissue (LOD = 100.0 in Ref. 26), potentially indicating a dual role in underlying both motivation and ability for exercise behavior. Perhaps more importantly, these findings taken together reinforce the implication of Insig2 in the regulation of the relationship between exercise and body weight.
Prolyl carboxypeptidase (angiotensinase C) (Prcp; 2510048K03Rik, old name) was found to be a highly significant (LOD = 62.9) cis-acting eQTL colocalizing with previously identified QTL for running distance and duration on Chr. 7. Prcp-null mice showed elevated levels of brain α-MSH and reduced food intake, were leaner and shorter than wild-type controls, and were resistant to high-fat diet-induced obesity (22, 43). Similar phenotypes have been observed in the HR strain of mice utilized here (37, 41). Prcp was also a focal candidate gene in brain tissue (LOD = 99.5, in Ref. 26), colocalizing with the same phenotypes and as described may play a pivotal nontissue specific role in the regulation of the relationship between exercise and body weight.
Secreted acidic cysteine rich glycoprotein (Sparc) was found to be a highly significant (LOD = 37.5) cis-acting eQTL mapped to Chr. 11, a region contained within the confidence intervals of previously identified QTL for running maximum running on days 5 and 6. Sparc muscle secretion has been previously reported to increase following exercise and has been shown to inhibit colon tumorigenesis by increasing apoptosis (1). Partial correlations also revealed that Sparc was significantly correlated with maximal running speed, further increasing the likelihood of its functional relevance. Although not specifically highlighted in our previous work on brain tissue, Sparc was a significant cis-acting eQTL (LOD = 63.1) colocalizing with same phenotypes described above, however, not significantly correlating with any focal phenotype.
The validation of the results presented above will necessitate future studies and additional lines of evidence as described elsewhere (27, 29). A high priority should be to validate the functional role of these candidate genes, in the context of both motivation and ability to engage in voluntary exercise behavior. Regardless, these results coupled with those of Ref. 26 further develop an initial model of the underlying functional genomic architecture of predisposition to engage in voluntary exercise and its effects on body weight and composition in the context of a neurobiological and muscular physiological framework.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant DK-076050 to D. Pomp. S. A. Kelly was partially supported through an NIH-funded (5T32MH-075854-04) Interdisciplinary Obesity Training program. Phenotypes were collected using the Animal Metabolism Phenotyping core facility within UNC's Clinical Nutrition Research Center (funded by NIH Grant DK-056350).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: S.A.K., D.L.N., K.H., T.G., and D.P. conception and design of research; S.A.K., D.L.N., and K.H. performed experiments; S.A.K. and D.L.N. analyzed data; S.A.K., T.G., and D.P. interpreted results of experiments; S.A.K. prepared figures; S.A.K. drafted manuscript; S.A.K., D.L.N., K.H., T.G., and D.P. edited and revised manuscript; S.A.K., D.L.N., K.H., T.G., and D.P. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Z. Yun for assistance with animal care and data collection. We thank Chris Wiesen at UNC's Odum Institute for Research in Social Science statistical consultation.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Aoi W, Naito Y, Takagi T, Tanimura Y, Takanami Y, Kawai Y, Sakuma K, Hang LP, Mizushima K, Hirai Y, Koyama R, Wada S, Higashi A, Kokura S, Ichikawa H, Yoshikawa T. A novel myokine, secreted protein acidic and rich in cysteine (SPARC), suppresses colon tumorigenesis via regular exercise. Gut 62: 882–889, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Asselbergs FW, Guo Y, van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B, Appelman YE, Barnard J, Baumert J, Beitelshees AL, Bhangale TR, Chen YD, Gaunt TR, Gong Y, Hopewell JC, Johnson T, Kleber ME, Langaee TY, Li M, Li YR, Liu K, McDonough CW, Meijs MF, Middelberg RP, Musunuru K, Nelson CP, O'Connell JR, Padmanabhan S, Pankow JS, Pankratz N, Rafelt S, Rajagopalan R, Romaine SP, Schork NJ, Shaffer J, Shen H, Smith EN, Tischfield SE, van der Most PJ, van Vliet-Ostaptchouk JV, Verweij N, Volcik KA, Zhang L, Bailey KR, Bailey KM, Bauer F, Boer JM, Braund PS, Burt A, Burton PR, Buxbaum SG, Chen W, Cooper-Dehoff RM, Cupples LA, deJong JS, Delles C, Duggan D, Fornage M, Furlong CE, Glazer N, Gums JG, Hastie C, Holmes MV, Illig T, Kirkland SA, Kivimaki M, Klein R, Klein BE, Kooperberg C, Kottke-Marchant K, Kumari M, LaCroix AZ, Mallela L, Murugesan G, Ordovas J, Ouwehand WH, Post WS, Saxena R, Scharnagl H, Schreiner PJ, Shah T, Shields DC, Shimbo D, Srinivasan SR, Stolk RP, Swerdlow DI, Taylor HA, Jr, Topol EJ, Toskala E, van Pelt JL, van Setten J, Yusuf S, Whittaker JC, Zwinderman AH, LifeLines Cohort Study, Anand SS, Balmforth AJ, Berenson GS, Bezzina CR, Boehm BO, Boerwinkle E, Casas JP, Caulfield MJ, Clarke R, Connell JM, Cruickshanks KJ, Davidson KW, Day IN, de Bakker PI, Doevendans PA, Dominiczak AF, Hall AS, Hartman CA, Hengstenberg C, Hillege HL, Hofker MH, Humphries SE, Jarvik GP, Johnson JA, Kaess BM, Kathiresan S, Koenig W, Lawlor DA, März W, Melander O, Mitchell BD, Montgomery GW, Munroe PB, Murray SS, Newhouse SJ, Onland-Moret NC, Poulter N, Psaty B, Redline S, Rich SS, Rotter JI, Schunkert H, Sever P, Shuldiner AR, Silverstein RL, Stanton A, Thorand B, Trip MD, Tsai MY, van der Harst P, van der Schoot E, van der Schouw YT, Verschuren WM, Watkins H, Wilde AA, Wolffenbuttel BH, Whitfield JB, Hovingh GK, Ballantyne CM, Wijmenga C, Reilly MP, Martin NG, Wilson JG, Rader DJ, Samani NJ, Reiner AP, Hegele RA, Kastelein JJ, Hingorani AD, Talmud PJ, Hakonarson H, Elbers CC, Keating BJ, Drenos F. Large-scale gene-centric meta-analyses across 32 studies identifies multiple lipid loci. Am J Hum Genet 91: 823–838, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audet GN, Meek TH, Garland T, Jr, Olfert IM. Expression of angiogenic regulators and skeletal muscle capillarity in selectively bred high aerobic capacity mice. Exp Physiol 96: 1138–1150, 2011. [DOI] [PubMed] [Google Scholar]
- 4.Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, de Villena FP, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res 21: 1213–1222, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin KM, Haddad F. Research in the exercise sciences: where are we and where do we go from here - part II. Sport Sci Rev 38: 42–50, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray MS, Hagberg JM, Pérusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc 41: 35–73, 2009. [DOI] [PubMed] [Google Scholar]
- 7.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890, 2003. [DOI] [PubMed] [Google Scholar]
- 8.Burns B, Schmidt K, Williams SR, Kim S, Giriajan S, Elsea SH. Rai1 haploinsufficiency causes reduced Bdnf expression resulting in hyperphagia, obesity and altered fat distribution in mice and humans with no evidence of metabolic syndrome. Hum Mol Genet 19: 4026–4042, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cervino AC, Li G, Edwards S, Zhu J, Laurie C, Tokiwa G, Lum PY, Wang S, Castellani LW, Lusis AJ, Carlson S, Sachs AB, Schadt EE. Integrating QTL and high-density SNP analyses in mice to identify Insig2 as a susceptibility gene for plasma cholesterol levels. Genomics 86: 505–517, 2005. [DOI] [PubMed] [Google Scholar]
- 10.Curran-Everett D. Multiple comparisons: philosophies and illustrations. Am J Physiol Regul Integr Comp Physiol 279: R1–R8, 2000. [DOI] [PubMed] [Google Scholar]
- 11.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, Charles MA, Balkau B, Froguel P. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science 315: 187, 2007. [DOI] [PubMed] [Google Scholar]
- 12.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res 15: 681–691, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farber CR, Kelly SA, Baruch E, Yu D, Hua K, Nehrenberg DL, de Villena FP, Buus RJ, Garland T, Jr, Pomp D. Identification of quantitative trait loci influencing skeletal architecture in mice: emergence of Cdh11 as a primary candidate gene regulating femoral morphology. J Bone Miner Res 26: 2174–2183, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garland T, Jr, Kelly SA. Phenotypic plasticity and experimental evolution. J Exp Biol 209: 2344–2361, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Garland T, Jr, Schutz H, Chappell MA, Keeney BK, Meek TH, Copes LE, Acosta W, Drenowatz C, Maciel RC, van Dijk G, Kotz CM, Eisenmann JC. The biological control of voluntary exercise, spontaneous physical activity, and daily energy expenditure in relation to obesity: human and rodent perspectives. J Exp Biol 214: 206–229, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garland T, Jr, Kelly SA, Malisch JL, Kolb EM, Hannon RM, Keeney BK, Van Cleave SL, Middleton KM. How to run far: multiple solutions and sex-specific responses to selective breeding for high voluntary activity levels. Proc Biol Soc 278: 574–581, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh S, Vivar JC, Sarzynski MA, Sung YJ, Timmons JA, Bouchard C, Rankinen T. Integrative pathway analysis of genome-wide association study of V̇o2max response to exercise training. J Appl Physiol 115: 1343–1359, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard I, Rezende EL, Garland T., Jr Leptin levels and body composition of mice selectively bred for high voluntary activity. Physiol Biochem Zool 80: 568–579, 2007. [DOI] [PubMed] [Google Scholar]
- 19.Gordon RR, La Merrill M, Hunter KW, Sørensen P, Threadgill DW, Pomp D. Dietary fat-dependent transcriptional architecture and copy number alterations associated with modifiers of mammary cancer metastasis. Clin Exp Metastasis 27: 279–293, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science 312: 279–283, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Howlett RA, Kirkton SD, Gonzalez NC, Wagner HE, Britton SL, Koch LG, Wagner PD. Peripheral oxygen transport and utilization in rats following continued selective breeding for endurance running capacity. J Appl Physiol 106: 1819–1825, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong JK, Szabo G, Raso GM, Meli R, Diano S. Deletion of prolyl carboxypeptidase attenuates the metabolic effects of diet-induced obesity. Am J Physiol Endocrinol Metab 302: E1502–E1510, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly SA, Nehrenberg DL, Hua K, Gordon RR, Garland T, Jr, Pomp D. Parent-of-origin effects on voluntary exercise levels and body composition in mice. Physiol Genomics 40: 111–120, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly SA, Nehrenberg DL, Peirce JL, Hua K, Steffy BM, Wiltshire T, Pardo-Manuel de Villena F, Garland T, Jr, Pomp D. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol Genomics 42: 190–200, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly SA, Nehrenberg DL, Hua K, Garland T, Jr, Pomp D. Exercise, weight loss, and changes in body composition in mice: phenotypic relationships and genetic architecture Physiol Genomics 43: 199–212, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly SA, Nehrenberg DL, Hua K, Garland T, Jr, Pomp D. Functional genomic architecture of predisposition to voluntary exercise in mice: expression QTL in the brain. Genetics 191: 266–284, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly SA, Pomp D. Genetic determinants of voluntary exercise. Trends Genet 29: 348–357, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly SA, Bell TA, Selitsky SR, Buus RJ, Hua K, Weinstock GM, Garland T, Jr, Pardo-Manuel de Villena F, Pomp D. A novel intronic SNP in the Myosin heavy polypeptide 4 gene is responsible for the Mini-Muscle phenotype characterized by major reduction in hindlimb muscle mass in mice. Genetics 195: 1385–1395, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lightfoot JT. Current understanding of the genetic basis for physical activity. J Nutr 141: 526–530, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, Zhu X, Thorleifsson G, Gunnarsdottir S, Walters GB, Thorsteinsdottir U, Kong A, Gulcher J, Nguyen TT, Scherag A, Pfeufer A, Meitinger T, Brönner G, Rief W, Soto-Quiros ME, Avila L, Klanderman B, Raby BA, Silverman EK, Weiss ST, Laird N, Ding X, Groop L, Tuomi T, Isomaa B, Bengtsson K, Butler JL, Cooper RS, Fox CS, O'Donnell CJ, Vollmert C, Celedón JC, Wichmann HE, Hebebrand J, Stefansson K, Lange C, Hirschhorn JN. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet 3: e61, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okano AH, Fontes EB, Montenegro RA, Farinatti PD, Cyrino ES, Li LM, Bikson M, Noakes TD. Brain stimulation modulates the autonomic nervous system, rating of perceived exertion and performance during maximal exercise. Br J Sports Med [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 32.O'Leary J, Osborne LR. Global analysis of gene expression in the developing brain of Gtf2ird1 knockout mice. PLoS One 6: e23868, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomp D, Nehrenberg D, Estrada-Smith D. Complex genetics of obesity in mouse models. Annu Rev Nutr 28: 331–345, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth SM, Rankinen T, Hagberg JM, Loos RJ, Pérusse L, Sarzynski MA, Wolfarth B, Bouchard C. Advances in exercise, fitness, and performance genomics in 2011. Med Sci Sports Exerc 44: 809–817, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics 159: 371–387, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swallow JG, Carter PA, Garland T., Jr Artificial selection for increased wheel-running behavior in house mice. Behav Genet 28: 227–237, 1998. [DOI] [PubMed] [Google Scholar]
- 37.Swallow JG, Koteja P, Carter PA, Garland T., Jr Artificial selection for increased wheel-running activity in house mice results in decreased body mass at maturity. J Exp Biol 202: 2513–2520, 1999. [DOI] [PubMed] [Google Scholar]
- 38.Swallow JG, Koteja P, Carter PA, Garland T., Jr Food consumption and body composition in mice selected for high wheel-running activity. J Comp Physiol B 171: 651–659, 2001. [DOI] [PubMed] [Google Scholar]
- 39.Swallow JG, Hayes JP, Koteja P, Garland T., Jr Selection experiments and experimental evolution of performance and physiology, in: Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments, edited by Garland T, Jr, Rose MR. Berkeley, CA: Univ. of California Press, 2009. [Google Scholar]
- 40.Vaanholt LM, Meerlo P, Garland T, Jr, Visser GH, van Dijk G. Plasma adiponectin is increased in mice selectively bred for high wheel-running activity, but not by wheel running per se. Horm Metab Res 39: 377–383, 2007. [DOI] [PubMed] [Google Scholar]
- 41.Vaanholt LM, Jonas I, Doornbos M, Schubert KA, Nyakas C, Garland T, Jr, Visser GH, van Dijk G. Responses in energy balance to high-fat feeding in mice selectively bred for high wheel-running activity. Int J Obes 32: 1566–1575, 2008. [DOI] [PubMed] [Google Scholar]
- 42.van Nas A, Ingram-Drake L, Sinsheimer JS, Wang SS, Schadt EE, Drake T, Lusis AJ. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics 185: 1059–1068, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallingford N, Perroud B, Gao Q, Coppola A, Gyengesi E, Liu ZW, Gao XB, Diament A, Haus KA, Shariat-Madar Z, Mahdi F, Wardlaw SL, Schmaier AH, Warden CH, Diano S. Prolycarboxpeptidase regulates food intake by inactivating alpha-MSH in rodents. J Clin Invest 119: 2291–2303, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.