Introduction
The somatic mutations in a cancer genome are the aggregate outcome of one or more mutational processes operative through the life of the cancer patient1-3. Each mutational process leaves a characteristic mutational signature determined by the mechanisms of DNA damage and repair that constitute it. A role was recently proposed for the APOBEC family of cytidine deaminases in generating particular genome-wide mutational signatures1,4 and a signature of localized hypermutation called kataegis1,4. A germline copy number polymorphism involving APOBEC3A and APOBEC3B, which effectively deletes APOBEC3B5, has been associated with a modest increased risk of breast cancer6-8. Here, we show that breast cancers in carriers of the deletion show more mutations of the putative APOBEC-dependent genome-wide signatures than cancers in non-carriers. The results suggest that the APOBEC3A/3B germline deletion allele confers cancer susceptibility through increased activity of APOBEC-dependent mutational processes, although the mechanism by which this occurs remains unknown.
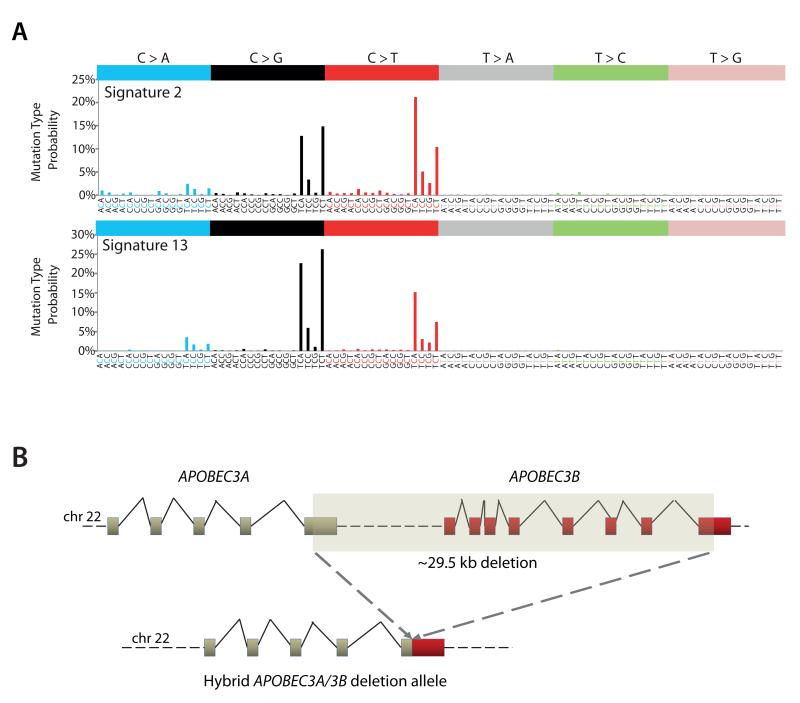
In recent analyses of somatic mutational signatures in 21 whole-genome sequenced primary human breast cancers1, two signatures characterized by C>T and/or C>G mutations at TpCpX trinucleotides were identified (the underlined base is the mutated base and X can be any base). These were subsequently observed in several other cancer types (Fig. 1A, Online methods) and are among the commonest mutational signatures found in human cancer (Supplementary Figures 3A-B)1,9. These signatures have been designated Signatures 2 and 13 (according to the nomenclature of Alexandrov et al 2013)9. Signature 2 is composed predominantly of C>T transitions with fewer C>G transversions at a TpCpX sequence context. In contrast, Signature 13 is dominated by C>G transversions at a TpCpX context1,9. A subset of breast, and other, cancer cases have an extremely large number of mutations of these signatures and we have called these “hypermutators”1,9.
Figure 1. The APOBEC3A/3B germline deletion polymorphism is associated with an increased burden of presumptive apobec-related signatures.
(A) Signatures 2 and 13 extracted by Non-negative Matrix Factorization11 share sequence-specific mutation characteristics to members of the AID/APOBEC family of cytidine deaminases. Both signatures are characterized by C>T transitions and/or C>G transversions at a TpCpX sequence context. Signature 2 is dominated by C>T transitions. Signature 13 is dominated by C>G transversions. (B) The APOBEC3A/3B hybrid deletion allele. The genes are in tandem on chromosome 22. The polymorphism involves a deletion of the APOBEC3B coding sequence fusing the 3′UTR of APOBEC3B to the 3′UTR of APOBEC3A.
The features of the mutations associated with Signatures 2/13 resemble those of mutations generated by the AID/APOBEC family of cytidine deaminases10,11. Members of this gene family have important physiological roles in antibody diversification (AICDA) and restriction of retroviruses and mobile retro-elements (e.g. APOBEC3A, APOBEC3G) [reviewed11-13]. However, it has been suggested that their DNA editing capabilities could also underlie undesirable mutagenesis leading to cancer4,11,14,15. Indeed, in addition to AICDA, the capacity for editing of nuclear DNA has been demonstrated for APOBEC3A14,16,17, APOBEC1, APOBEC3C and APOBEC3G10.
A common germline copy number deletion polymorphism involving the APOBEC3 gene cluster on chromosome 22 (Fig. 1B) has been associated with an elevated risk of breast cancer. A copy number genome-wide association study (GWAS) of 16,000 cases of eight common diseases highlighted this deletion polymorphism in association with breast cancer in a primary screen although it did not validate in replication6. Subsequently, a GWAS in the Chinese population demonstrated an association with breast cancer (odds ratio (OR) 1.3 one-copy, 1.8 two-copy deletion, p=2.0×10−24)7 that was replicated in a European population (OR 1.2 one-copy, 2.3 two-copy deletion, ptrend = 0.005)8. The deletion allele has a frequency of ~8% in European populations5,6, 37% in East Asians and 93% in Oceania5. The ~29,500bp genomic deletion has delimiting breakpoints in APOBEC3A and APOBEC3B (which are adjacent to each other and in the same orientation on chromosome 22) and results in a chimeric APOBEC3A/3B gene. This hybrid gene is predicted to produce a transcript which is predominantly constituted of APOBEC3A sequence but replaces the APOBEC3A 3′UTR with the APOBEC3B 3′ UTR (Supplementary Note) and encodes a protein which has an identical amino acid sequence to APOBEC3A5 (Fig. 1B). Homozygous carriers of this deletion allele are predicted not to make any APOBEC3B protein. Given its association with breast cancer, we explored the relationship between the deletion allele (Table 1) and the presence of mutational signatures 2/13.
Table 1. In the cohort of 923 breast cancers, the majority of patients had a mutation rate of Signatures 2/13 of less than 1 per Mb.
A subset of patients had mutations comprising mostly (or in some cases entirely) of Signatures 2/13 with a very high mutation rate associated with these signatures (hypermutators). A higher proportion of patients were found to be carriers of at least one copy of the germline deletion allele from amongst patients who had hypermutator breast cancers. A test for trend demonstrates a correlation between the number of copies of the deletion allele in a breast cancer patient and having a hypermutator breast cancer (p=6.251e−5).
| deletion allele status | hypermutators | non-hypermutators | total | hypermutators/total cases |
|---|---|---|---|---|
| homozygous | 4 | 10 | 14 | 0.286 |
| heterozygous | 28 | 100 | 128 | 0.219 |
| non-carrier | 74 | 707 | 781 | 0.095 |
Cochrane-Armitage test for trend p=6.251e-6
Chi-statistic 20.4098
We aggregated a set of 923 breast cancers from multiple different sequencing centres in which normal and neoplastic tissues had been sequenced for somatic mutations, 123 whole-genome and 800 whole-exome (Supplementary Table 1A-B). Using next-generation sequence (NGS) data, we identified 128 patients who were heterozygous and 14 who were homozygous for the APOBEC3A/3B deletion allele (Online methods, Supplementary Note, Supplementary Fig 4A-C, Supplementary Table 2A-C). Applying the non-negative matrix factorization (NNMF) algorithm employed to extract mutational signatures1,9,18 to the somatic mutations, we estimated the numbers and the proportional contribution of mutations attributable to each mutational signature in each cancer case (Supplementary Figure 1C-1O, Supplementary Table 1B). Combining these two sets of results, we observed that cancers with a higher mutational burden of Signatures 2/13 were more likely to be derived from patients who were carriers of at least one copy of the germline APOBEC3A/3B deletion allele (Wilcoxon rank-sum test p=1.7e−3, Online methods, Supplementary Note, Supplementary Figure 3B, E). In particular, the subset of hypermutator cancers19 (Supplementary Table 3A) is associated with the deletion allele. Breast cancers from individuals who are heterozygous or homozygous for the APOBEC3A/3B deletion allele have a relative risk of 2.37 (CI 1.64-3.46) of being hypermutators compared to breast cancers from individuals who do not carry the deletion allele (Table 1, Cochrane-Armitage, p=6.251e−6). By contrast, no association was found between the deletion allele and Signature 1, another mutational signature common in breast and other cancers (p=0.935). The results therefore suggest that the APOBEC3A/3B deletion allele is specifically associated with the burden of Signatures 2/13 mutations in breast cancer (Table 1, Supplementary Note, Supplementary Table 3B addressing population stratification).
We then examined 1769 cancers of eleven other cancer types in which Signature 2/13 mutations have been found (Table 2, Supplementary Figure 3B-D)11. Of forty patients with acute lymphoblastic leukaemia three were hypermutators (Table 2) and all were carriers of the germline deletion allele, two heterozygous and one homozygous (p=2.51e−5). Enrichment for hypermutators among patients who were heterozygous and homozygous for the deletion allele was also seen in bladder carcinoma, although this did not reach statistical significance (p=0.038, Bonferroni corrected p=0.452, Table 2). Thus, the APOBEC3A/3B deletion allele may be associated with the Signature 2/13 mutation burden in cancers other than breast.
Table 2. Relationship between number of copies of the deletion allele and the burden of Signatures 2/13.
A trend was seen for ALLs but not for other cancers (test for trend).
| hypermutator | non-hypermutator | cancer type | test for trend | Bonferroni | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cancer type | hom | het | non | total | hom | het | non | total | total | chi-statistic | p-value | correction |
| ALL | 1 | 2 | 0 | 3 | 0 | 5 | 32 | 37 | 40 | 17.756 | 2.51E-05 | 3.01E-04 |
| BLCA | 0 | 3 | 6 | 9 | 0 | 13 | 114 | 127 | 136 | 4.3192 | 0.03769 | 4.52E-01 |
| BRCA | 4 | 28 | 74 | 106 | 10 | 100 | 707 | 817 | 923 | 20.4098 | 6.25E-06 | 7.50E-05 |
| CESC | 0 | 1 | 3 | 4 | 0 | 5 | 29 | 34 | 38 | 0.2852 | 0.5933 | - |
| HNSC | 0 | 3 | 33 | 36 | 0 | 33 | 229 | 262 | 298 | 0.5413 | 0.4619 | - |
| KIRP | 0 | 0 | 5 | 5 | 0 | 14 | 81 | 95 | 100 | 0.8568 | 0.3546 | - |
| LUAD | 0 | 3 | 22 | 25 | 0 | 18 | 260 | 278 | 303 | 1.0856 | 0.2975 | - |
| LUSC | 0 | 3 | 12 | 15 | 0 | 17 | 133 | 150 | 165 | 0.9616 | 0.3268 | - |
| MM | 0 | 0 | 2 | 2 | 2 | 10 | 51 | 63 | 65 | 0.4152 | 0.5193 | - |
| STAD | 0 | 1 | 10 | 11 | 0 | 18 | 104 | 122 | 133 | 0.2643 | 0.6072 | - |
| THCA | 0 | 1 | 19 | 20 | 0 | 44 | 220 | 264 | 284 | 1.8977 | 0.1683 | - |
| UCEC | 0 | 0 | 12 | 12 | 4 | 35 | 183 | 222 | 234 | 2.319 | 0.1278 | - |
|
| ||||||||||||
| Total | 5 | 45 | 198 | 248 | 16 | 312 | 2143 | 2471 | 2719 | 10.9215 | 9.51E-04 | - |
In breast and other cancers, several non-carriers of the germline deletion allele had large numbers of Signature 2/13 mutations (Table 2, Supplementary Table 3B). Similarly, several carriers of the deletion allele did not show large numbers of Signature 2/13 mutations in their cancers (Online methods). It thus appears that the germline deletion allele is neither necessary nor sufficient to generate Signature 2/13 mutations. This behaviour is in-keeping with that of a germline susceptibility allele, which has a modest effect on a quantitative trait. Indeed, the marked variation in Signature 2/13 mutation prevalence between different cancers (Supplementary Figure 3C-D) would suggest that multiple factors are likely to influence the burden of Signature 2/13 mutagenesis, such as APOBEC gene inherited variation, APOBEC gene expression, virus/transposon activity and inflammation, and that these may vary in importance in different cancers.
If Signatures 2/13 are due to APOBEC activity, they should bear the known characteristics of the mutations generated by these enzymes. The substitution classes in Signatures 2/13 (C>T transitions and C>G transversions) coupled with the TpC sequence context were responsible for the initial proposition of the role of this enzyme family1. However, APOBEC-induced mutations exhibit other distinctive characteristics, including preferential cytosine to uracil deamination on stretches of single-stranded DNA20-22. Consequently, adjacent APOBEC-induced mutations often arise on the same parental allele (i.e. are in cis with each other) and are on the same DNA strand (i.e. successive mutations may be C>T…C>G…C>T or G>A…G>C…G>A but not C>T…G>A…C>T), a pattern referred to as “strand-coordinated mutagenesis” (Online methods, Supplementary Note).
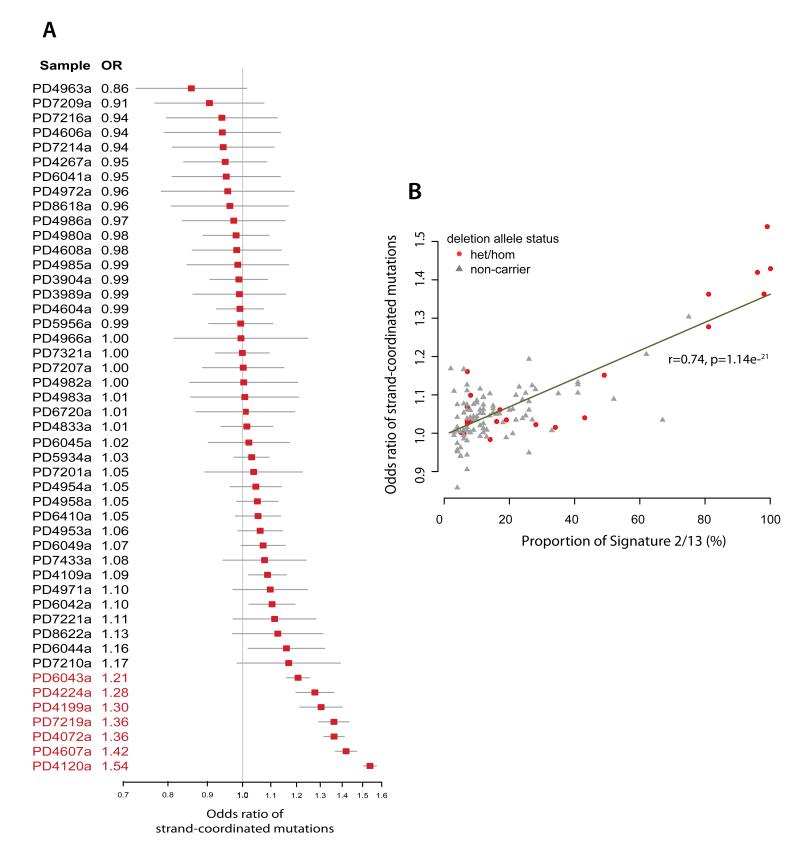
To investigate the presence of strand-coordinated mutagenesis in Signatures 2/13 (Supplementary Figure 4A-C), we examined the frequency of two successive mutations arising on the same strand and on different strands in the 123 breast cancers which had been whole-genome sequenced. Several cancers demonstrate more strand-coordinated pairs of mutations than expected by chance (corrected for mutation spectrum and mutation burden, Fig. 2A) and this is directly correlated to the proportion of Signature 2/13 mutations in these cancers (Fig. 2B, r=0.74, p=1.1e−21). Furthermore, examination of NGS reads in these cancers show that strand-coordinated mutations are usually in cis (Supplementary Note, Supplementary Table 4, Supplementary Figure 4D, p<0.0001), confirming that they are linked to each other on the same parental haplotype. Together, these findings are compatible with the model that Signature 2/13 mutations often arise on stretches of single-stranded DNA similar to mutations induced by APOBEC enzymes4,20.
Figure 2. Additional features of Signatures 2/13 that are similar to mutagenic patterns of APOBECs.
(A) Several cancers showed an excess of mutations arising on the same strand or strand-coordinated mutagenesis. For reasons of space only a subset of cancers is depicted here. Odds ratio (OR) of observed strand-coordinated mutations over expected is presented (as a red box) with 95% confidence intervals (grey line). The OR was calculated from the observed number of same strand/different strand mutations divided by the number of expected same strand/different strand mutations (where the expected numbers were corrected for the overall mutation rate of the cancer and the mutation spectra). A higher OR indicates more same strand mutations than expected. Patients highlighted in red have hypermutator breast cancers. (B) A direct correlation is seen between the OR of strand-coordinated mutagenesis and the fractional burden of Signatures 2/13. Patients who are homozygous/heterozygous for the deletion allele are highlighted (red dots) to show the enrichment of deletion carriers amongst breast cancers with a high burden of Signatures 2/13.
The association between the germline APOBEC3A/3B deletion allele and Signature 2/13 mutation burden (OR 2.68 one-copy, 3.82 two-copy deletion, ptrend 6.251e−6; combined OR 2.78 CI 1.75-4.41) is in-keeping with the reported modest increased risk of breast cancer conferred by the deletion allele based on GWAS. However, the mechanism by which the germline APOBEC3A/3B fusion confers elevated APOBEC mutagenic activity is unclear. The amino acid sequence of the predicted fusion protein is identical to APOBEC3A, although the transcript is a chimaera of APOBEC3A and a segment of the APOBEC3B UTR and this could confer altered transcriptional or translational regulation of APOBEC3A. The other consequence of the APOBEC3A/3B germline deletion allele is deletion of the APOBEC3B coding sequence and thus absence of APOBEC3B in homozygous patients (Supplementary Figure 5A). It is not immediately clear, however, how this would directly increase APOBEC-related mutagenesis.
The TpC sequence context of mutations generated by APOBEC123,24, APOBEC3A22 and APOBEC3B22,25,26 closely mirrors the sequence context of Signature 2/13 mutations in human cancers indicating that these particular members of the APOBEC enzyme family are likely candidates for generating these mutational signatures4,22. Thus far, there have been no recurrent somatic mutations identified within the APOBEC gene family that can be associated with Signature 2/13. Based on gene expression studies, recent reports have suggested that APOBEC3B is responsible27,28,29. However, the existence of Signature 2/13 hypermutator breast cancers in individuals with germline homozygosity for the APOBEC3A/3B deletion allele, which completely removes APOBEC3B coding sequences (Fig. 1B) and in which APOBEC3B expression is absent (Online methods, Supplementary Note, Supplementary Fig 5A-B)30, indicates that over-activity of APOBEC3B is unlikely to be exclusively responsible for Signatures 2/13 mutations.
The burden of somatic mutations due to Signatures 2/13 is one of the highest attributable to any mutational signature across the spectrum of human cancer11. Thus, elucidation of the mechanisms underlying Signatures 2/13 will advance understanding of carcinogenesis in several cancer types and potentially influence strategies for cancer prevention and treatment. The effect of the APOBEC3A/3B germline deletion allele on the Signature 2/13 mutation burden reported here provides independent evidence for the underlying role of members of the APOBEC gene family in generating these mutations. Furthermore, it provides a plausible biological mechanism by which this breast cancer predisposition allele could confer its effect. The geographic variation in population frequency of the APOBEC3A/3B germline deletion allele5 suggests that there may be selection in favour of it (Online methods). Since some APOBECs are involved in innate immunity4,11 to infection it may be that protection to infection is conferred by the deletion allele. This may be balanced, to some extent, by predisposition to cancer. If true, this would be remarkable since both effects would be mediated by the same underlying mechanism; the double-edged sword of the mutagenic activity of the APOBEC proteins.
Online methods
1. Background information
Next-generation whole-genome and exome-sequenced cancer samples were previously sequenced by members of the International Cancer Genome Consortium, The Cancer Genome Atlas and other centres9,18. High-confidence somatic substitutions were obtained from these consortia or other peer-reviewed publications not related to these consortia, filtered further for potential false positive calls using dbSNP, 1000 genomes, the NHLBI GO Exome Sequencing Project, the 69 Complete Genomics panel as well as a bespoke panel of BAM files of unmatched normal tissues containing more than 120 normal genomes and 500 exomes9. These data were then parsed through an algorithm previously developed to extract mutational signatures in human cancers18 called Non-negative Matrix Factorisation (NNMF)9,18.
Six main substitution classes (C>A:G>T, C>G:G>C, C>T:G>A, T>A:A>T, T>C:A>G and T>G:A>C) were subdivided according to the 5 prime and 3 prime flanking sequence context. Since there are six classes of base substitution and 16 possible sequence contexts for each mutated base (A, C, G or T at the 5′ base and A, C, G or T at the 3′ base), there are 96 possible mutated trinucleotides for each cancer. Herewith, the convention for describing a mutated trinucleotide will be XpCpX, where X can be any base and the mutated base is underlined1,2,9,18.
A total of 7,042 samples were analyzed from 30 types of cancer. 21 distinct mutational signatures were extracted. The commonest signatures were Signature 1A and Signature 1B, both characterized by C>T mutations at a XpCpG trinucleotide (Supplementary Figure 1A), and Signature 2 and Signature 13, characterized by dominant C>T transitions at a TpCpX sequence context in Signature 2 and C>G transversions at a TpCpX sequence context in Signature 13 (Supplementary Figure 1A-B)1,2,9,18. Signatures 1A/1B are likely to be caused by deamination at methylated CpGs whereas Signatures 2/13 are thought to be due to the APOBEC family of cytidine deaminases. Therefore, for the purposes of this analysis, Signatures 2 and 13 are considered together. NNMF is able to estimate the number of mutations associated with extracted mutation signatures for individual cancers in a given set of samples (summarized in Supplementary Table 1B, Figure 1C-1O).
In this analysis, a total of 2,719 samples were previously characterized by NNMF and also had BAM files available for inspection (Supplementary Table 1A-B). BAM files were downloaded from CGhub (https://cghub.ucsc.edu/) between 9 May 2013 and 26 June 2013. For the ease of tracking samples through this analysis, we have kept the naming convention attached to the cancer sample for tables and figures, even if the germline deletion polymorphism was sourced from a matched normal because the signatures of somatic mutagenesis will have been identified in the tumor samples in the first place. This is also for the purpose of continuity between publications. For samples originating from the Sanger Institute, PDXXXX denotes a specific individual, with suffix “a”,”c” or “d” denoting tumor samples and suffix “b” for the matched normal sample.
2. Detection of germline APOBEC3A/3B deletion polymorphism
In order to detect this deletion polymorphism from next-generation sequencing data, multiple loci within and flanking the coordinates of the deletion were sampled (Supplementary Figure 2A, Supplementary Table 2A) from BAM files (overall workflow and directions to processing data in Supplementary Figure 2B). Raw short read data had been aligned back to the reference genome (NCBI build 37) with duplicates and unmapped reads removed. Externally sourced BAM files were sourced from the TCGA data hub https://browser.cghub.ucsc.edu/.
Matched normal BAM files were sought for calling this deletion allele. However, a tumor BAM file was used if a normal BAM was not available (Supplementary Table 2B). Samples that did not have BAM files available for examination were excluded from analysis. In total, the APOBEC3A/3B polymorphism detection was sourced from 561 tumors (99 BLCA, 117 BRCA, 1 CESC, 19 HNSC, 2 KIRP, 303 LUAD, 12 STAD, 2 THCA and 6 UCEC) and 2158 normals.
For samples with whole genome data, the expected sequencing depth in the absence of the deletion polymorphism, i.e. wild-type copy number (CN) of 2, d2, was calculated as the average depth of the 60 loci in the flanking regions. The expected depth in the presence of a heterozygous deletion allele, d1, is then given by d1 = d2 / 2 and the expected depth in the presence of a homozygous deletion allele, d0, was set to the expected number of misreads, estimated as d2/20. A maximum likelihood test was performed to identify the most likely CN from the set 31, with corresponding expected depths represented as Poisson distributions {Pois(d0), Pois(d1), Pois(d2)}, given the observed sequencing depths within the region of the deletion polymorphism (Supplementary Dataset 1).
For exome-sequenced samples, an expectation-maximisation algorithm was utilized, with the copy number (CN) of each sample and the ratio of sequencing depth within and outside the deletion polymorphism used as latent variables. The CN of each sample was initialized as 2 and the depth ratio of loci within the deletion polymorphism region to those outside, r, was modelled non-parametrically by bootstrap resampling (n=1000). For samples with CN of 2, 1 and 0, respectively, the expected depths within the deletion polymorphism region are then given by
At each maximisation step, the copy number of each sample was assigned as that whose distribution showed most overlap with din for that sample, after bootstrap resampling. At each expectation step, r was recalculated using bootstrap resampling of loci within just those samples classified as CN=2 in the previous maximisation step. The EM algorithm was continued until no samples were reclassified from one iteration to the next, or for a maximum of 100 iterations (Supplementary Figure 2B, Supplementary Dataset 2).
The results of the calling of the polymorphism status in all the samples is provided in Supplementary Table 2B. The reproducibility of the calling method was sought by examining the concordance between calling on the tumor and normal BAM files from the same patient (Supplementary Note, Supplementary Table 2C, Supplementary Figure 2C) as well as concordance between genome- and exome-sequenced samples in the same patient.
3. The relationship between the APOBEC3A/3B germline deletion allele and somatic mutational signatures in cancer
The dataset comprised genome-sequenced (123) as well as exome-sequenced (800) cancers. In order to perform the analyses, the rate of mutation was calculated for each cancer (rate of Signature 2/13 per Mb), correcting for whether the samples had been genome- or exome-sequenced.
Because the rates of Signatures 2/13 were not normally distributed (Supplementary 3A-D), a one-sided Wilcoxon rank-sum test was performed to see whether carrying one copy of the deletion allele had an overall effect on the mutation rate of the signatures.
We sought to include more cancer samples in order to increase the power of the analyses. There were no further available breast cancer samples with BAM files ready for download, hence we sought inclusion of other cancer types that had previously been analyzed9,18. However, it was noted that the distribution of rates of Signatures 2/13 varied considerably between cancer-types (Supplementary Figure 3B-D) and clear outliers were present in all the cancer types skewing the distribution of mutation rates (Supplementary Figure 3A,D-E).
Some cancers were observed to have a strikingly high proportion of total mutations associated with Signatures 2/13 and/or have higher rates of mutagenesis associated with this signature (Supplementary Figure 1C-1O, 5B). Using the rate of Signatures 2/13 mutagenesis, outliers were identified as patients with cancers that had a mutation rate exceeding 1.5 times the length of the interquartile range from the 75th percentile for each type of cancer19. These outliers will hitherto be referred to as “hypermutators” although we do not suggest that there is an on-going biological process attached to this name. Given the considerable variation of the mutation rates for different cancer tissue-types (Supplementary Figure 1A,1B), each cancer type was analyzed separately. A summary of the hypermutators versus non-hypermutators is provided in the Supplementary Table 3A.
4: Strand-coordinated mutagenesis
In theory, neighboring mutations could arise on either of two strands of a double-helix (Supplementary Figure 4A) particularly if they had arisen as independent events during different cycles of cell division. If more mutations are observed to occur on the same strand than expected by chance (Supplementary Figure 4B), this would imply one of two scenarios: Either those neighboring mutations arose over different rounds of cell division with preferential targeting of one strand over another or they arose during a single round of cell division and potentially occurred in the same instance.
We therefore sought to formally document that neighboring mutations are occurring on the same strand more often than expected or “strand-coordinated mutagenesis”. In order to demonstrate genome-wide strand coordination, analysis was carried out on all whole-genome sequence data for which BAM files were available (Supplementary Table 4A). Given a set of mutations, each occurring at a base of type (A, C, G, T) on the + strand, we identify all pairs of mutations and classify them as ‘same’ if both mutations are of the same originating base and ‘diff’ if not (i.e. first and second mutations of each pair respectively: A>X and A>X; G>X and G>X; C>X and C>X; and T>X and T>X, with no prior selection for mutations at a TpC context and where X can be any base). The distance between successive pairs or intermutation distance is also calculated (Supplementary Figure 4C).
The proportion of pairs of mutations that are expected to occur on the same base assuming randomly ordered mutations is given by , where pX is the fraction of mutations that occur at nucleotide X. To depict the deviation of the observed pairs of mutations found on the same strand from that of the expected pairs of mutations on the same strand, a standard Forest plot was constructed (data of expected and observed same strand mutations for all 124 samples are provided in Supplementary Table 4A, columns B-E). For the reason of space, only a subset of samples were presented in Figure 2A.
Because same-strand mutations were ascertained in an unbiased way from any mutation type (not restricted to just cytosine mutations at TpCs), to see whether strand-coordinated mutations were a particular feature of Signatures 2/13, we sought a relationship between the degree of “strand-coordination”, given by the OR of strand-coordination, and the fractional burden of Signatures 2/13 in each cancer (Supplementary Figure 4D).
We sought additional characteristics of the mutations in the whole-genome sequenced breast cancers that support the suggestion that mutations associated with Signatures 2/13 have arisen due to the APOBEC family of enzymes (Supplementary Note, Supplementary Figure 4D, Supplementary Table 4).
5: Relationship between expression of APOBEC family members and rates of mutation of Signatures 2/13
RNA-seq derived expression data was obtained from the https://browser.cghub.ucsc.edu/ for relevant samples. In total, there were 1691 patients for whom comparable data were obtainable. Expression levels for each APOBEC family member were standardized relative to the levels of TBP (TATA-binding protein) and the relationship between the APOBEC3B expression levels and germline deletion allele status in these cancers (Supplementary Figure 5A-B, Supplementary Table 5A).
6. Selection for the APOBEC3A/3B deletion
The germline APOBEC3A/3B deletion polymorphism highlighted in this analysis was reported to display a strikingly differentiated worldwide distribution of allele frequencies5. The FST value (measuring population differentiation) was re-examined using the reported deletion allele frequencies and additional SNP genotype data from the CEPH-HGDP panel published after the CNP study32. This value will depend on the way the populations are grouped, and needs to be compared with other variants of similar frequency to measure how unusual it is. We used two published grouping schemes, into five continental geographical/genetic groups (1. Sub-Saharan Africans, 2. Middle Easterners plus Europeans, 3. East Asians, 4. Native Americans and 5. Oceanians) or into 32 population groups33, and matched SNP frequencies measured as minor allele frequency to ± 0.1%. For the five continental groups, FST was 0.330 (97.4th percentile compared with 2,716 frequency-matched SNPs), and for the 32 population groups it was 0.285 (96.6th percentile compared to 2,059 frequency-matched SNPs). The level of population differentiation was thus higher than expected by chance, which can result from positive selection34, and we therefore examined other statistics sensitive to positive selection. Cross-population extended haplotype homozygosity (XP-EHH) and integrated haplotype score (iHS) values were obtained from the HGDP Selection Browser (http://hgdp.uchicago.edu/). These haplotype-based tests for positive selection35 utilise information 500kb upstream and downstream of the deletion, and thus the two sides can be examined separately. Neither side showed any significantly high XP-EHH (>2.5) or iHS value (∣iHS∣>2.0) in any continental group or individual population. Finally, we looked at allele frequency spectrum-based tests (Tajima’s D, Fay & Wu’s H and Nielsen et al.’s Composite Likelihood Ratio test) using the 1000 Genomes Phase 1 re-sequenced data in the East Asian populations (CHB, CHS and JPT) (1000G Phase 1)36 in the regions surrounding the deletion, as described (1000G Pilot)31. There was no evidence for positive selection in these populations, although in this case, the power of these tests is limited because the frequency of the deletion in these populations is not high enough.
Overall, this locus shows unusually high differentiation among continents and populations. However, there remains a lack of other evidence for positive selection and so we cannot convincingly conclude that this deletion has been positively selected in human populations.
Supplementary Material
Acknowledgements
We would like to thank the Wellcome Trust for support (grant reference 098051). SN-Z is a Wellcome-Beit Prize Fellow and is supported through a Wellcome Trust Intermediate Fellowship (grant reference WT100183MA). PJC is personally funded through a Wellcome Trust Senior Clinical Research Fellowship (grant reference WT088340MA). NB is an EHA fellow and is supported by a Lady Tata Memorial Trust award. The H.L. Holmes Award from the National Research Council Canada and an EMBO Fellowship supports AS. We would like to thank Matt Hurles and Carl Anderson of the WTSI for their input. We would also like to acknowledge funding from the Breakthrough Breast Cancer Research (ICGC 08/09) and the BASIS project, funded by the European Community’s Seventh Framework Programme (FP7/2010-2014) under the grant agreement number 242006. We would like to thank The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) for access to mutation catalogues used in the Alexandrov et al 2013 and for access to BAM files. This study was performed within the Research Ethics Approval of 09/h0306/36.
Footnotes
Competing interests: There are no competing interests to declare.
References
- 1.Nik-Zainal S, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowarski R, Kotler M. APOBEC3 Cytidine Deaminases in Double-Strand DNA Break Repair and Cancer Promotion. Cancer Res. 2013;73:3494–8. doi: 10.1158/0008-5472.CAN-13-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craddock N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–20. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long J, et al. A common deletion in the APOBEC3 genes and breast cancer risk. J Natl Cancer Inst. 2013;105:573–9. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xuan D, et al. APOBEC3 deletion polymorphism is Associated with Breast Cancer Risk among women of European Ancestry. Carcinogenesis. 2013;34:2240–2243. doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov LB, N. ZS, Wedge DC. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–53. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 11.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–74. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Koito A, Ikeda T. Intrinsic restriction activity by AID/APOBEC family of enzymes against the mobility of retroelements. Mob Genet Elements. 2011;1:197–202. doi: 10.4161/mge.1.3.17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suspene R, et al. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A. 2011;108:4858–63. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petit V, Vartanian JP, Wain-Hobson S. Powerful mutators lurking in the genome. Philos Trans R Soc Lond B Biol Sci. 2009;364:705–15. doi: 10.1098/rstb.2008.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–50. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suspene R, Aynaud MM, Vartanian JP, Wain-Hobson S. Efficient Deamination of 5-Methylcytidine and 5-Substituted Cytidine Residues in DNA by Human APOBEC3A Cytidine Deaminase. PLoS One. 2013;8:e63461. doi: 10.1371/journal.pone.0063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–59. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodge VJ, Austin J. A survey of outlier detection methodologies. Artificial Intelligence Review. 2004;22:85–126. [Google Scholar]
- 20.Nowarski R, et al. APOBEC3G enhances lymphoma cell radioresistance by promoting cytidine deaminase-dependent DNA repair. Blood. 2012;120:366–75. doi: 10.1182/blood-2012-01-402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtz CM, Sadler HA, Mansky LM. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 2013;41:6139–48. doi: 10.1093/nar/gkt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byeon IJ, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petit V, et al. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol. 2009;385:65–78. doi: 10.1016/j.jmb.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 24.Beale RC, et al. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–96. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 25.Taylor BJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinohara M, et al. APOBEC3B can impair genomic stability by inducing base substitutions in genomic DNA in human cells. Sci Rep. 2012;2:806. doi: 10.1038/srep00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013 doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rousseeuw PJ, Ruts I, Tukey JW. The bagplot: A bivariate boxplot. American Statistician. 1999;53:382–387. [Google Scholar]
- 31.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 33.Perry GH, et al. Evolutionary genetics of the human Rh blood group system. Hum Genet. 2012;131:1205–16. doi: 10.1007/s00439-012-1147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y, et al. Population differentiation as an indicator of recent positive selection in humans: an empirical evaluation. Genetics. 2009;183:1065–77. doi: 10.1534/genetics.109.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–37. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.