Abstract
Objective
Substantial clinical, pathological and genetic overlap exists between amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). TDP-43 inclusions have been found in both ALS and FTD cases (FTD-TDP). Recently, a repeat expansion in C9orf72 was identified as the causal variant in a proportion of ALS and FTD cases. We sought to identify additional evidence for a common genetic basis for the spectrum of ALS-FTD.
Methods
We used published GWAS data of 4,377 ALS patients and 13,017 controls and 435 pathology-proven FTD-TDP cases and 1,414 controls for genotype imputation. Data were analyzed in a joint meta-analysis, by replicating topmost associated hits of one disease in the other, and by using a conservative rank products analysis, allocating equal weight to ALS and FTD-TDP sample sizes.
Results
Meta-analysis identified 19 genome-wide significant single nucleotide polymorphisms (SNPs) at C9orf72 on chromosome 9p21.2 (lowest p=2.6×10−12) and one SNP in UNC13A on chromosome 19p13.11 (p=1.0×10−11) as shared susceptibility loci for ALS and FTD-TDP. Conditioning on the 9p21.2 genotype increased statistical significance at UNC13A. A third signal, on chromosome 8q24.13 at the SPG8 locus coding for strumpellin, (p=3.91×10−7) was replicated in an independent cohort of 4,056 ALS patients and 3,958 controls (p=0.026; combined analysis p=1.01×10−7).
Interpretation
We identified common genetic variants at C9orf72, but in addition in UNC13A that are shared between ALS and FTD. UNC13A provides a novel link between ALS and FTD-TDP, and identifies changes in neurotransmitter release and synaptic function as a converging mechanism in the pathogenesis of ALS and FTD-TDP.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by progressive muscle weakness due to the loss of motor neurons in both brain and spinal cord. No cure exists and disease etiology has not yet been fully elucidated. Important overlap exists with frontotemporal dementia (FTD), which is characterized by changes in cognition, behavior and language. Clinically, approximately 5–15% of patients with ALS have FTD, while about 3–14% of FTD patients also fulfill the criteria for ALS.1,2 Neuropathologically, the majority of FTD cases can be divided in two subtypes, characterized by cellular inclusions of either tau (FTD-tau) or TDP-43 (FTD-TDP). TDP-43 inclusions have been found in neurons of both ALS and FTD-TDP patients.3 Lastly, substantial genetic overlap between ALS and FTD has been reported. Linkage studies identified a locus of several megabases on chromosome 9p21 in families of patients with both ALS and FTD.4–6 Previous genome-wide association studies (GWAS) of non-familial ALS helped fine-map this region, and recently, a hexanucleotide repeat expansion in C9orf72 was discovered in this region.7–11 The C9orf72 repeat expansion is present in approximately 6% of sporadic ALS and sporadic FTD patients, and in up to 37% and 25% of familial ALS and FTD cases, respectively.12 Additionally, mutations in VCP have been implicated in both ALS patients and in FTD.13 Furthermore, mutations in the gene for TDP-43 (TARDBP) have been found in ALS and FTD with motor neuron degeneration (FTD-MND), but are rarely present in FTD-TDP cases without motor neuron symptoms.14,15
The majority of gene mutations have been found in familial cases of ALS and FTD, but these mutations are less frequent in cases without a positive family history.11,12,16 Meta-analysis of GWAS data is a powerful tool to discover new susceptibility loci for non-familial disease.17 The association signals from a GWAS may represent common variants acting as ‘genetic risk factors’, but may also form a proxy for rare, moderately penetrant genetic variants, such as the repeat expansion in C9orf72.7,9 The discovery of the C9orf72 repeat expansion has, additionally, shown that intronic, non-coding variation may be causal to disease. Previously, the most recent and largest GWAS of sporadic ALS identified the locus on chromosome 9p21.2 (comprising C9orf72) and UNC13A as susceptibility loci.10,11 Recently, the first GWAS of FTD-TDP patients has been published, identifying three common variants in TMEM106B associated with susceptibility to sporadic FTD.16 The association with TMEM106B variants has now been replicated in independent cohorts including FTD-TDP patients.18,19
Both ALS and FTD may form parts of a spectrum of neurodegenerative disease. This spectrum ranges from pure motor ALS, to ALS with mild cognitive impairment, to FTD-MND, and ultimately, to pure FTD without motor neuron symptoms.20 In the present study, we sought to identify a common genetic basis for this spectrum of neurodegenerative disease. Therefore, we conducted a meta-analysis of all available GWAS data in ALS and TDP-43 positive FTD aimed at the discovery of additional common variants that would affect susceptibility to both neurodegenerative diseases.
Methods
Subjects
ALS cohorts were derived from all available previously published GWAS of ALS patients.10,11 We included 16 cohorts of Caucasian sporadic ALS patients (n = 4,638) and/or unaffected controls (n = 14,038) from six European countries and the USA for whom genome-wide genotype data were available. Previous replication cohorts with selected SNP sets (for example obtained by TaqMan genotyping) could not be included. For all cohorts, the diagnosis of probable or definite ALS was made according to the revised El Escorial criteria.21
We obtained raw genotype data for 658 individuals that were originally genotyped for the FTD-TDP GWAS, and were recruited from 11 countries in Europe, USA, Canada and Australia.16 In the original publication, 598 cases with FTD-TDP pathology matched the inclusion criteria, of which 515 were used for analysis. For the present study, we only included cases with FTD-TDP confirmed by TDP-43 immunohistochemistry, a single proband per pedigree, and only individuals of European descent. We excluded cases with mutations in GRN or VCP, resulting in 453 FTD-TDP cases that remained for further quality control. Clinical data on the presence of motor neuron symptoms were recorded. The Wellcome Trust Case Control Consortium (WTCCC) 1958 Birth Cohort was used for population controls.16
For replication purposes, we collected genomic DNA from a total of 2,218 sporadic ALS patients and 2,261 unaffected controls from The Netherlands, Germany, Sweden and Switzerland. In addition, in silico genotypes were obtained from Illumina beadchip data for 1,838 sporadic ALS patients and 1,697 controls from Italy.
Dutch patients were recruited by neuromuscular centers at the University Medical Center Utrecht, the Radboud University Nijmegen Medical Center, and the Academic Medical Center Amsterdam, as part of an ongoing population-based study of ALS in The Netherlands. Unrelated control samples without any neuromuscular disease were matched for age and gender. Swedish samples were included from the Umeå University ALS Clinic that had not yet participated in previous GWAS included in the present meta-analysis. For the Swiss stratum, patients were recruited at Kantonsspital St. Gallen. German ALS patients were recruited through Ulm University Hospital and Charité University Hospital, Berlin. Control samples were unrelated individuals, free of any neuromuscular disease. Italian ALS patients were included through different Italians ALS centers as part of the Italian SLAGEN Consortium. Controls consisted of Italian healthy individuals who did not have a personal or family history for neurodegenerative diseases.
For all strata, patients with ALS fulfilled the revised El Escorial Criteria for possible, probable, or definite ALS. Cases with a family history of ALS or non-Caucasian descent were excluded. As the discovery ALS and FTD GWAS samples include individuals with C9orf72 repeat expansions (no complete data available), we did not exclude C9orf72 repeat expansion carriers from the replication sets.
All participants gave written informed consent and approval was obtained from the local institutional review boards. More detailed information on ALS or FTD subject selection methods has been published previously and can be found in Supplementary Table 1.10,11,16
Genotypes and quality control
For each cohort, raw Illumina beadchip genotype data were obtained. The following quality control measures were applied to each cohort separately. We removed A/T and C/G SNPs in order to prevent allele swaps, SNPs with a minor allele frequency (MAF) < 5%, a genotyping call rate < 95%, or with deviation from Hardy-Weinberg equilibrium (HWE) in controls (p < 0.001). Samples with missing phenotype data, a genotyping call rate < 95%, high (inbreeding coefficient F > 0.05) or low (F < −0.025) heterozygosity rates, or where the clinically reported gender did not match the genotypic gender (based on chromosome X markers), were removed. SNP identifiers and positions were mapped to dbSNP 126 and NCBI genome build 36. For the WTCCC 2 cohort, markers and samples listed for exclusion (as provided by the WTCCC), and samples previously included in the WTCCC 1 cohort, were removed. Cohorts consisting of control samples only were merged with cohorts that included affected patients from the same country, ultimately forming twelve balanced strata for ALS and one for FTD. Per stratum, we excluded SNPs with differing missing rates between cases and controls (p < 1×10−3), or SNPs that, as determined by flanking SNPs, are missing higher for one allele (p < 1×10−10). For population stratification analysis purposes, strata were merged into a separate dataset containing only SNPs common to all strata. Population substructure was determined by using principal components analysis in EIGENSTRAT, also incorporating HapMap 3 release 2 samples.22 After the removal of population outliers (based on deviation from the CEU+TSI population cluster in a plot of the first two principal components, duplicate (PI_HAT value > 0.9), and related samples (PI_HAT > 0.2), new principal components were calculated. For the ALS and FTD meta-analysis, we removed duplicate and related individuals across disease datasets. See Supplementary Table 2 for Hardy-Weinberg equilibrium (HWE) p values and call rates for significantly associated SNPs per stratum.
For the replication of association with the chromosome 8q24.13 locus, we used KASPar (KBioscience) and TaqMan (Applied Biosystems) assays to determine rs13268726 and rs12546767 genotypes in the replication set, according to the manufacturer’s protocols. We used an ABI Prism 7900HT analyzer (Applied Biosystems) and SDS v2.3 software (Applied Biosystems) to determine genotype clusters, and outliers were excluded from further analyses. For the Italian SLAGEN cohort, in silico genotypes were obtained from Illumina Human-660W Quad Beadchips for rs12546767, and rs13268726 genotypes were imputed using IMPUTE v2 and HapMap3 release 2 and 1000 Genomes pilot reference panels.
Statistical analysis
Because of the use of many different genotyping platforms, and a relatively small number of markers with genotypes in all strata, we used genome-wide SNP imputation to extend genome-wide coverage and to increase comparability.23 Imputation was carried out using IMPUTE2 v2.1.2 in genomic chunks of 5 Mb, leaving all options at the program’s defaults. We preferred the HapMap3r2 CEU+TSI reference (~1.4M markers) as a reference panel, because of the relatively large number of reference haplotypes (n = 410). We, additionally, imputed using the HapMap2 reference (120 haplotypes, ~2.5M markers), to determine if we would not miss important associations compared to the HapMap3r2 reference. Imputation using the HapMap2 reference did not yield additional significant results (data not shown).
Imputed genotypes were stored as continuous allele dosage data, which are continuous numerical values indicating the estimated number of minor alleles (ranging from 0 to 2), thus incorporating a measure of imputation uncertainty.
Associations between genotypes and disease susceptibility, were tested in logistic regression models for each of the strata in PLINK. Gender and principal components (PCs) that were strongly (p < 1×10−5) associated with disease status were included as covariates in the logistic regression analyses (seven PCs for ALS, two PCs for FTD). Results from each of the strata were joined in a fixed effect inverse variance meta-analysis in PLINK, both per disease (ALS or FTD) separately, and combined.24,25 See Supplementary Table 2 for allele and genotype frequencies per stratum for the topmost associated SNPs. We calculated genotypic odds ratios (ORgeno) for top-associated markers in heterozygotes (1 minor allele vs. 2 major allele carriers) and homozygotes (2 minor allele vs. 2 major allele carriers) using logistic regression, and incorporating the same covariates as were used in the main disease susceptibility analyses.
We used the GCTA software package to calculate trait heritability estimates for ALS and FTD-TDP cohorts, as well as to estimate the cross-traits heritability, using a bivariate restricted maximum likelihood analysis, as has been described previously.26,27 Imputed dosage data were used as input, excluding SNPs with a poor imputation quality score (R2 < 0.3) and minor allele frequency < 0.01. The first seven principal components calculated from a combined dataset of ALS and FTD were included as covariates. Additionally, we performed a conditional and joint multiple-SNP analysis of the ALS and FTD meta-analysis results in order to determine possible independent association signals within a genomic locus reaching genome-wide significance.28 The conditional and joint analyses were carried out after converting imputed dosage data to hard-called genotypes (as required by the GCTA software).
As an additional approach, in order to determine SNPs with shared susceptibility to both ALS and FTD, we selected SNPs with p < 1×10−4 from the above ALS meta-analysis and used the FTD-TDP data to replicate the associations with these SNPs. We used linkage disequilibrium (LD)-based clumping of SNPs in PLINK in order to cluster multiple genotyped and imputed SNPs within a region of strong LD, thus determining independent loci.25 Per clump, we looked up p values of SNPs from the above logistic regression results in the FTD analysis. We applied a Bonferroni multiple-testing correction for the number of independent loci (clumps) tested. Subsequently, SNPs with p values below the threshold of 1×10−3 in the FTD analysis were selected for replication in the ALS data. For the selection of SNPs from the FTD analysis, we used a less stringent p value threshold (p < 1×10−3) in order to avoid the omission of false-negative associations due to limited statistical power of the FTD-TDP data.
Furthermore, as sample sizes differ substantially between the joined ALS strata and the FTD-TDP stratum, we would be almost exclusively measuring the GWAS signal from the ALS patients. We, therefore, used a conservative rank products (RP) analysis to compare results from both ALS and FTD.29 The rank products method originates from the analysis of multiple expression micro-array experiments, and provides a non-parametric test that is independent of sample size. For the RP analysis, we ranked SNPs by increasing p value for each disease (ALS or FTD). Only SNPs with the same direction of effect were included. For each SNP we calculated the RP. Ultimately, SNPs were sorted by increasing rank product, and a permutation test was used to determine statistical significance for each RP. In order to obtain empirical p values, we permuted the ranks of each SNP 100,000,000 times and counted the number of times the permuted RP was equal to or higher than the observed RP. SNPs with an empirical p value < 5×10−8 were considered to be significantly top-ranked for both diseases.
Replication of association with locus 8q24.13
In the replication set of sporadic ALS patients and unaffected controls, for each stratum, association between SNP and disease status was tested in a logistic regression model corrected for gender. Subsequently, results were analyzed in a weighted inverse variance meta-analysis in PLINK.
Results
After quality control, there were twelve ALS strata (4,377 cases, 13,017 controls), and one FTD stratum (435 cases, 1,414 controls). For some cohorts, the total number of cases and controls differed from the original publications, due to different quality control methods. Genomic inflation factors (λGC) ranged from 1.01 – 1.07, indicating adequate quality control and correction for population stratification (Supplementary Table 1). Associations did not reach genome-wide significance (p < 5×10−8) in any of the 13 strata separately.
Analysis of ALS and FTD separately
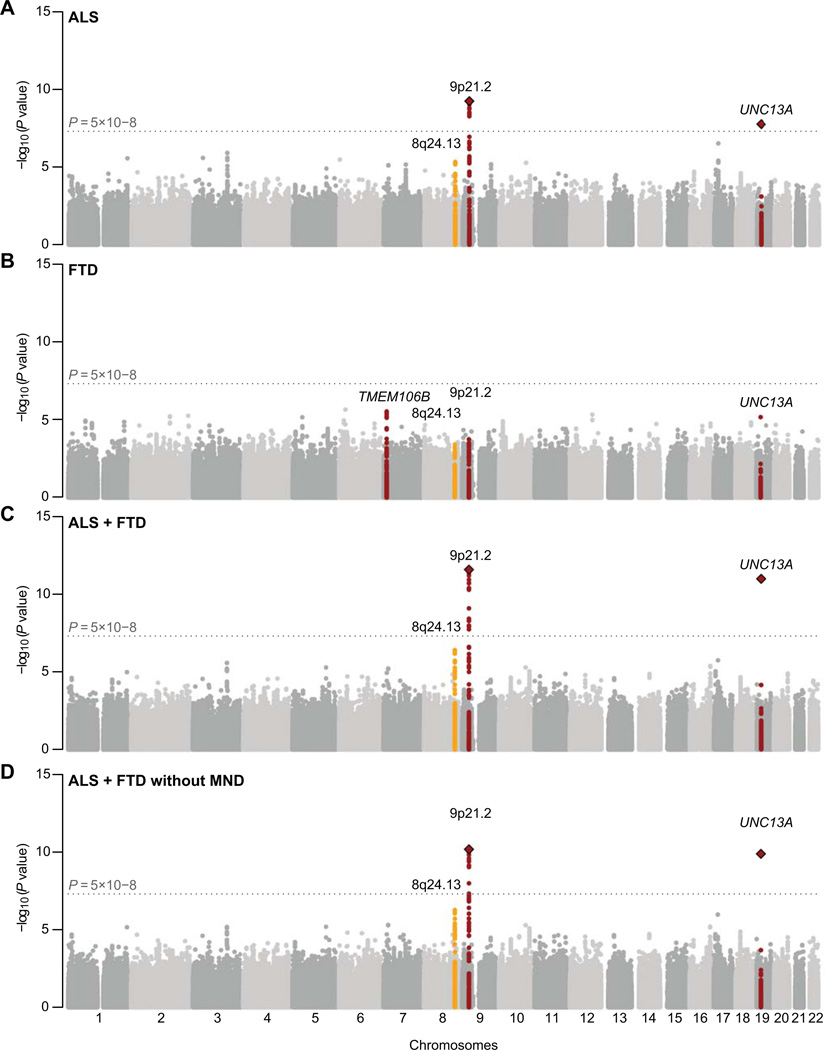
First, we inspected a meta-analysis of the ALS strata. Consistent with previous findings, we found genome-wide significant hits near C9orf72 on chromosome 9p21.2 (top SNP rs3849943) and in gene UNC13A (top SNP rs12608932) on chromosome 19p13.11 (Fig 1 and 2; Table 1).10,11
Figure 1.
Figure 2.
TABLE 1.
Top association results for independent loci containing SNPs, per disease and in the joint meta-analysis
| Chr | Locus | n SNPs | Top SNP | Minor allele | MAF | ALS analysis | FTD analysis | Meta-analysis | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | p | OR | p | OR | p | ||||||
| 9 | 9p21.2 | 29 | rs3849943 | C | 0.24 | 1.22 | 5.48×10−10 | 1.38 | 5.53×10−4 | 1.24 | 2.60×10−12 |
| 19 | UNC13A | 1 | rs12608932 | C | 0.35 | 1.18 | 1.70×10−8 | 1.46 | 6.57×10−6 | 1.21 | 1.02×10−11 |
| 8 | 8q24.13 | 11 | rs13268726 | G | 0.10 | 0.80 | 4.69×10−6 | 0.70 | 0.020 | 0.79 | 3.91×10−7 |
| 17 | CENPV | 1 | rs7477 | A | 0.49 | 1.16 | 2.91×10−7 | 0.98 | 0.785 | 1.14 | 1.82×10−6 |
| 3 | TXNDC6 | 8 | rs7638688 | A | 0.29 | 0.85 | 1.20×10−6 | 0.97 | 0.757 | 0.87 | 2.66×10−6 |
| 16 | KIAA0513 | 3 | rs16975170 | T | 0.12 | 1.17 | 2.27×10−4 | 1.53 | 5.99×10−4 | 1.20 | 4.20×10−6 |
| 5 | FAM13B | 1 | rs9327807 | C | 0.18 | 0.85 | 1.24×10−5 | 0.87 | 0.196 | 0.85 | 5.25×10−6 |
| 7 | 7p21.1 | 2 | rs10233425 | C | 0.01 | 1.88 | 7.46×10−6 | 1.44 | 0.391 | 1.83 | 6.15×10−6 |
| 10 | BUB3 | 1 | rs11248416 | G | 0.11 | 1.23 | 3.64×10−5 | 1.27 | 0.103 | 1.24 | 9.19×10−6 |
| 17 | 17p13.2 | 1 | rs12950017 | C | 0.24 | 1.16 | 1.47×10−5 | 1.11 | 0.293 | 1.15 | 9.19×10−6 |
Per locus, the number of SNPs with p < 1×10−5 is indicated, and association results for the SNP with the most significant p value (top SNP) are presented. The ALS analysis was based on 4,377 sporadic ALS patients and 13,017 controls. The FTD analysis was based on 435 sporadic FTD-TDP cases and 1,414 population controls. The meta-analysis comprises a total of 4,811 patients with either ALS or FTD, and 14,428 controls. Chr = chromosome; MAF = weighted minor allele frequency across all datasets; OR = odds ratio.
Subsequently, we analyzed the separate FTD-TDP stratum. We found no genome-wide significant associations, which was consistent with the results for patients without progranulin (GRN) mutations in the original publication (Fig 1).16 The exact association results differed minimally from the original publication, most probably due to the use of a partly different control cohort, and different methods for quality control and statistical analysis.
Combined ALS and FTD analysis
To examine common genetic variants that are shared in ALS and FTD-TDP we applied three complementary methods to avoid only picking up ALS effects, considering the imbalance in cohort size. First, all strata were joined into a single meta-analysis. Not only was the signal at 9p21.2 (C9orf72) greatly enhanced, but also at UNC13A (Fig 1). For rs3849943 at 9p21.2, the genotypic odds ratio (ORgeno) in heterozygotes is 1.25 (p = 3.19×10−7), in homozygotes the ORgeno is 1.19 (p = 6.76×10−5); while for rs12608932 in UNC13A, the heterozygote ORgeno and homozygote ORgeno are 1.12 (p = 0.014) and 1.29 (p = 3.33×10−15), respectively. Results for markers previously associated with sporadic ALS or FTD-TDP can be found in Supplementary Table 3. We did not identify any new genome-wide significant associations in the joint meta-analysis, although one new locus nearing the genome-wide significance threshold emerged at chromosome 8q24.13 (Fig 1).
Using a restricted maximum likelihood analysis, we estimated trait heritability for both ALS and FTD cohorts separately, as well as the cross-traits heritability between ALS and FTD-TDP. We estimated a SNP heritability for ALS of 0.21 (standard error 0.02), while for FTD-TDP no reliable heritability estimate could be calculated due to lack of statistical power. The genetic correlation between ALS and FTD-TDP was modest, but significant (0.20, standard error 0.098, p = 0.012). The SNP-based coheritability was estimated at 0.02 (standard error 0.007).
To further demonstrate shared susceptibility to both ALS and FTD-TDP for the C9orf72 and UNC13A loci, we selected top-associated SNPs from one disease and tried to replicate their association in the other disease. We selected 191 SNPs with p < 1×10−4 in 75 independent loci from the ALS meta-analysis, and looked up association results for these SNPs in the FTD analysis, applying a Bonferroni multiple-testing correction for the number of independent loci tested. We thus identified six SNPs in two loci (UNC13A and C9orf72) that were significantly replicated in the FTD-TDP data (Table 2). Conversely, we selected 1,450 SNPs with p < 1×10−3 in 658 independent loci from the FTD analysis and replicated these SNPs in the ALS data. We, again, identified two loci (seven SNPs near C9orf72 and one in UNC13A) that were significantly replicated in the ALS data (Table 2).
TABLE 2.
Replication of top SNPs from ALS analysis in FTD data and vice versa
| Top SNPs from ALS replicated in FTD | |||||||||
| Chr | Locus | SNP | Minor allele | ALS analysis | FTD analysis | ||||
| OR | p | OR | p | p Bonferroni | |||||
| 19 | UNC13A | rs12608932 | C | 1.18 | 1.70×10−8 | 1.46 | 6.57×10−6 | 4.93×10−4 | |
| 9 | 9p21.2 | rs2453554 | T | 1.21 | 3.06×10−9 | 1.39 | 3.35×10−4 | 0.025 | |
| 9 | 9p21.2 | rs700791 | A | 1.22 | 1.51×10−9 | 1.39 | 4.21×10−4 | 0.032 | |
| 9 | 9p21.2 | rs3849942 | T | 1.22 | 9.12×10−10 | 1.38 | 5.41×10−4 | 0.041 | |
| 9 | 9p21.2 | rs3849943 | C | 1.22 | 5.48×10−10 | 1.38 | 5.53×10−4 | 0.041 | |
| 9 | 9p21.2 | rs774359 | C | 1.18 | 1.09×10−7 | 1.37 | 5.67×10−4 | 0.042 | |
| Top SNPs from FTD replicated in ALS | |||||||||
| FTD analysis | ALS analysis | ||||||||
| 9 | 9p21.2 | rs3849943 | C | 1.38 | 5.53×10−4 | 1.22 | 5.48×10−10 | 3.16×10−7 | |
| 9 | 9p21.2 | rs10967965 | T | 1.39 | 8.41×10−4 | 1.24 | 5.80×10−10 | 3.34×10−7 | |
| 9 | 9p21.2 | rs3849942 | T | 1.38 | 5.41×10−4 | 1.22 | 9.12×10−10 | 5.25×10−7 | |
| 9 | 9p21.2 | rs700791 | A | 1.39 | 4.21×10−4 | 1.22 | 1.51×10−9 | 8.71×10−7 | |
| 9 | 9p21.2 | rs17779457 | G | 1.36 | 8.35×10−4 | 1.21 | 2.93×10−9 | 1.69×10−6 | |
| 9 | 9p21.2 | rs2453554 | T | 1.39 | 3.35×10−4 | 1.21 | 3.06×10−9 | 1.76×10−6 | |
| 19 | UNC13A | rs12608932 | C | 1.46 | 6.57×10−6 | 1.18 | 1.70×10−8 | 9.77×10−6 | |
| 9 | 9p21.2 | rs774359 | C | 1.37 | 5.67×10−4 | 1.18 | 1.09×10−7 | 6.30×10−5 | |
| Top SNPs from ALS replicated in FTD without MND signs | |||||||||
| Chr | Locus | SNP | Minor allele | ALS analysis | FTD without MND signs analysis | ||||
| OR | p | OR | p | p Bonferroni | |||||
| 19 | UNC13A | rs12608932 | C | 1.18 | 1.70×10−8 | 1.39 | 4.52×10−4 | 0.034 | |
| Top SNPs from FTD without MND signs replicated in ALS | |||||||||
| FTD without MND signs analysis | ALS analysis | ||||||||
| 19 | UNC13A | rs12608932 | C | 1.39 | 4.52×10−4 | 1.18 | 1.70×10−8 | 8.63×10−6 | |
The upper part of the table shows SNPs with p < 1×10−4 in the ALS analysis and with p < 0.05 after Bonferroni correction for the number of independent loci (n = 191) tested in the FTD analysis. Conversely, the second part of the table shows SNPs with p < 1×10−3 in the FTD analysis and with p < 0.05 after Bonferroni correction for the number of independent loci (n = 658) tested in the ALS analysis. Subsequently, SNPs are shown with p < 1×10−4 in the ALS analysis, and with p < 0.05 after Bonferroni correction for the number of independent loci (n = 191) tested in the FTD-TDP without MND signs analysis. The lower part of the table shows SNPs with p < 1×10−3 in the FTD-TDP without MND signs analysis and with p < 0.05 after Bonferroni correction for the number of independent loci (n = 1,374) tested in the ALS analysis. Results are sorted by increasing Bonferroni-corrected p value. Chr = chromosome; OR = odds ratio; MND = motor neuron disease.
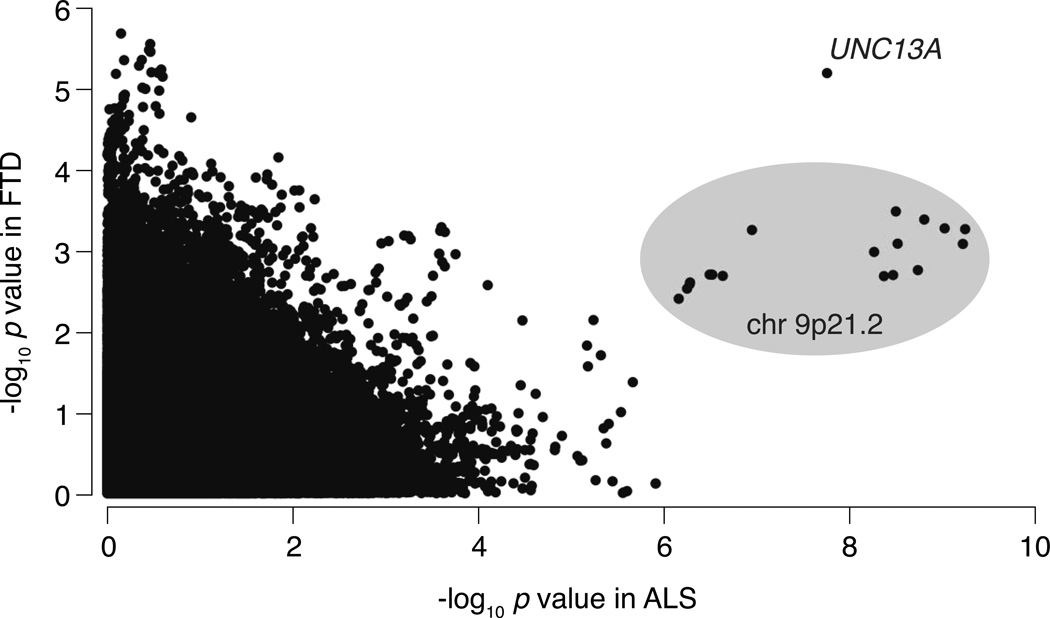
Subsequently, we used a third approach to investigate whether the associations near C9orf72 and UNC13A were not exclusively driven by one of the disease cohorts. Because of the large sample size of the ALS strata, the combined meta-analysis signals might be largely driven by ALS patients. To take this into account, we used a conservative, sample size-independent, rank products (RP) analysis. This analysis identifies SNP associations whose p values that are ranked highest in both diseases, thus allocating equal weight to both ALS and FTD results. Supplementary Table 4 shows the top ten SNPs from this analysis, arranged according to increasing RP. The −log10 p values of SNPs with the lowest RPs are ranked highest in both diseases. Empirical p values for the rank products, as determined by 100,000,000 permutations, were highly significant for the top six SNPs (p < 5×10−8). SNP rs12608932 in UNC13A had the strongest signal in both diseases. The following nine most significant SNPs were all in the C9orf72 locus. Fig 3 shows a more visual representation of the results from both studies; associations present in both ALS and FTD strata clearly stand out from the disease-specific and non-significant associations.
Figure 3.
Our study most likely includes a substantial number of C9orf72 repeat expansion carriers, however, we are not able to retrieve repeat expansion status for all patients included. As a proxy for C9orf72 repeat expansion status, we adjusted association tests for rs3849943 genotype (the top SNP at the 9p21.2 locus). The association at UNC13A was not dependent on rs3849943 SNP genotype in ALS (OR 1.18, p = 8.64×10−9), FTD (OR 1.45, p = 9.09×10−6), and the ALS-FTD meta-analysis (OR 1.21, p = 5.68×10−12). Instead, the statistical significance of the association with UNC13A increased. Similarly, for the 8q24.13 locus (top SNP rs13268726), association results that were corrected for rs3849943 genotype in ALS (OR 0.80, p = 6.04×10−6), FTD (OR 0.71, p = 0.029), and the ALS-FTD meta-analysis (OR 0.79, p = 6.47×10−7) were very similar to the unadjusted results (Table 1). Furthermore, we performed a systematic, genome-wide conditional analysis to identify possible additional, independent association signals in our meta-analysis of ALS and FTD-TDP. The conditional analysis did not yield any additional independent association signals at the chromosome 9p21.2 locus, gene UNC13A and chromosome 8q24.13. For each locus, association signals were driven by the SNP with the lowest p value. Adjusted, joint p values, did not differ notably from the original p values, indeed indicating these three SNPs represent true independent signals (data not shown).
Analyses including only FTD-TDP cases without motor neuron symptoms
As noted previously, a substantial proportion of FTD-TDP patients also have motor neuron symptoms. In order to determine if association signals from the FTD-TDP cohort were not solely driven by patients with motor neuron symptoms, we repeated the ALS and FTD meta-analysis after removing all FTD-TDP patients with signs of motor neuron disease (n = 99) from the FTD stratum. Of course, statistical power was attenuated for the set of FTD-only patients. However, signals at C9orf72 and UNC13A were still enhanced by adding the FTD-only cases into the meta-analysis (Fig 1). Also, the rank products analysis showed consistent ranking of the top 6 markers (Supplementary Table 4).
New signal on chromosome 8q24.13
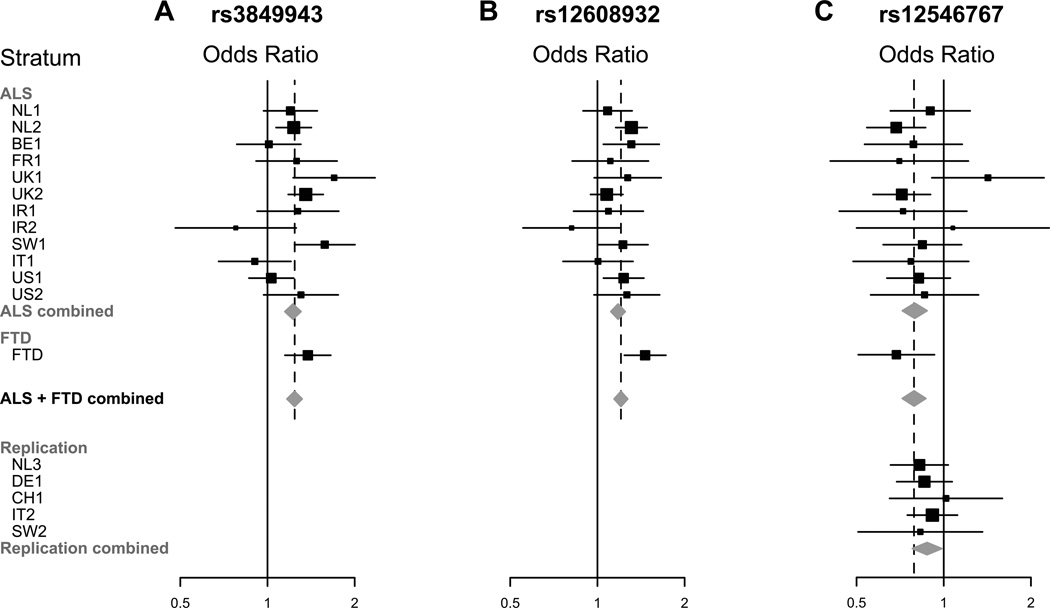
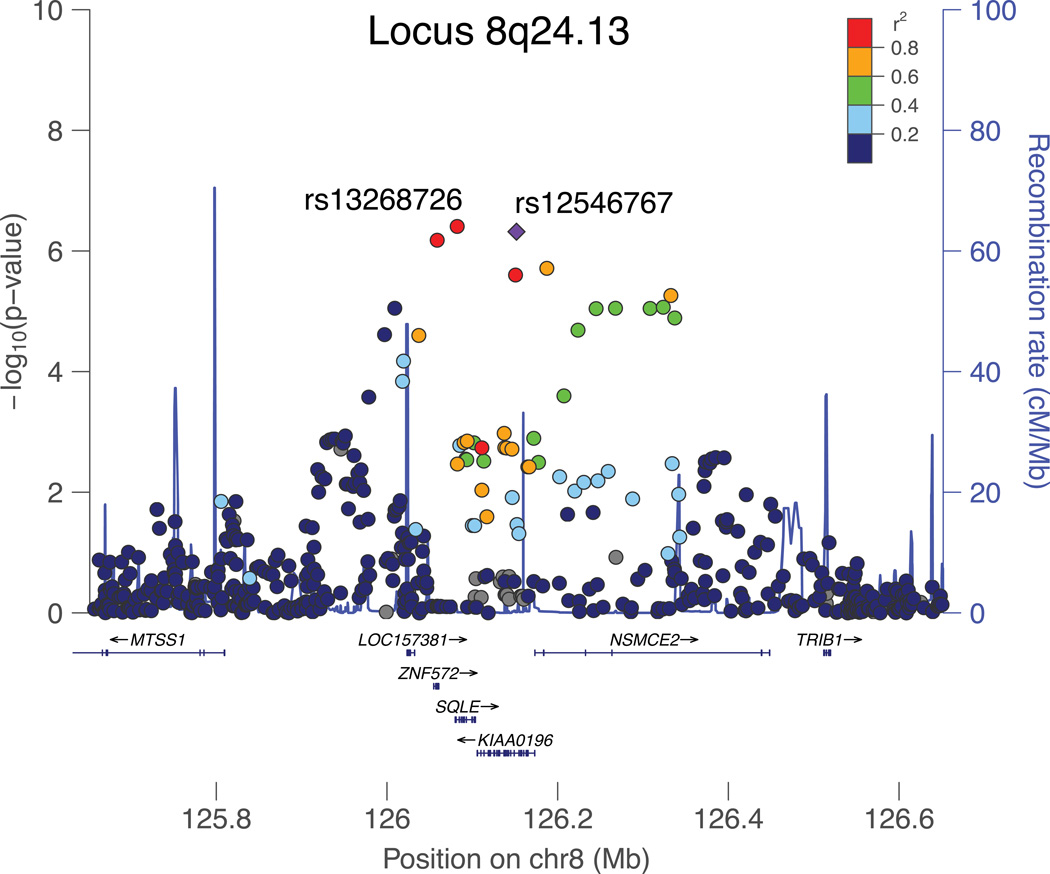
In the combined ALS and FTD meta-analysis, there was one new signal on chromosome 8q24.13 with top SNPs rs13268726 (imputed, OR 0.79, p = 3.91×10−7), and rs12546767 (genotyped, OR 0.80, p = 4.68×10−7) and mapping to a region with strong LD comprising genes SQLE, KIAA0196, and NSMCE2 (Fig 4). Both top SNPs are in strong LD (r2 = 0.95, D' = 0.975). Additionally, in the analysis of FTD-TDP cases without motor neuron symptoms, we found consistent associations for rs13268726 (OR 0.69, p = 0.029) and rs12546767 (OR 0.67, p = 0.022) compared to the full FTD stratum (Table 3).
Figure 4.
TABLE 3.
Association results for top SNPs at the chromosome 8q24.13 locus
| SNP | Gene | Minor allele |
MAF | ALS analysis | FTD analysis | ALS + FTD meta-analysis |
Replication ALS |
Meta-analysis + replication combined |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | p | OR | p | OR | p | OR | p | OR | p | ||||
| rs13268726 | SQLE | G | 0.10 | 0.80 | 4.69×10−6 | 0.79 | 0.020 | 0.79 | 3.91×10−7 | 0.89 | 0.044 | 0.82 | 1.85×10−7 |
| rs12546767 | KIAA0196 | C | 0.10 | 0.80 | 6.63×10−6 | 0.69 | 0.015 | 0.79 | 4.78×10−7 | 0.88 | 0.026 | 0.82 | 1.01×10−7 |
Association results are shown for a weighted inverse-variance meta-analysis of 12 sporadic ALS cohorts (4377 ALS cases, 13017 controls), for logistic regression analysis in the FTD-TDP cohort (435 FTD-TDP cases, 1414 controls), for the combined ALS-FTD meta-analysis (4811 cases, 14428 controls), and for an independent replication cohort of sporadic ALS (4056 ALS, 3958 controls). In the righthand column, results for a combined analysis of both the ALS-FTD meta-analysis data and replication cohort are shown (8867 cases, 18386 controls). MAF = weighted minor allele frequency across all datasets; OR = odds ratio.
We followed up rs13268726 (in SQLE) and rs12546767 (in KIAA0196) in a replication set of 4,056 sporadic ALS patients and 3,958 controls. We replicated the associations between the two SNPs in this locus and ALS susceptibility, with the lowest p value for rs12546767 (OR 0.88, p = 0.026) in gene KIAA0196 (Fig 2). Joint analysis of both genome-wide and replication data reached a p value of 1.01×10−7 for this locus, but did not reach genome-wide significance (p < 5×10−8). Detailed results are shown in Table 3.
Discussion
In the present study, we conducted a large genome-wide meta-analysis in a combined cohort of nearly 5,000 patients and over 14,000 controls. We aimed at finding new genetic variants that would affect susceptibility to both sporadic ALS and TDP-43 positive FTD. With three complementary methods, to avoid only picking up effects of the larger ALS cohort, we identified not only the known 9p.21 locus including C9orf72, but also the UNC13A locus as shared between both neurodegenerative diseases. By combining results from ALS and FTD datasets in a joint meta-analysis, we found a strong increase of association signals at both loci. Furthermore, by replication of top-associated SNPs from one disease in the other and vice versa, we found both the C9orf72 and UNC13A loci to be significantly associated in the ALS as well as in the FTD analysis. We also showed that signals at both loci are not solely driven by the relatively large number of ALS patients. Furthermore, by repeating the meta-analysis selecting only FTD patients without motor neuron disease, we demonstrated that the signals from the meta-analysis are not unique to individuals with motor neuron symptoms in both groups. Lastly, we found a modest, but significant genetic correlation between ALS and FTD-TDP.
The observation that the UNC13A association signal is shared between ALS and FTD-TDP cohorts, highlights UNC13A not only as susceptibility gene in ALS, but also as a susceptibility factor for FTD-TDP, further corroborating the role of UNC13A in neuronal degeneration. Previously we have shown that rs12608932 acts as a modifier of survival in ALS, which was recently replicated in an Italian cohort of ALS patients.30,31 UNC13A, therefore, poses an interesting therapeutic target. The protein encoded by UNC13A is a member of a family of presynaptic proteins present throughout the nervous system and involved in the priming of presynaptic vesicles containing neurotransmitters before their release.32 Aberrant function of UNC13A disrupts the exocytosis of excitatory and inhibitory neurotransmitters. This not only affects the biochemical communication between neurons, but also triggers structural changes in existing neuronal networks.32 Furthermore, changes in the release of neurotransmitters as a consequence of defects in UNC13A are in line with the glutamate excitotoxicity hypothesis, previously implicated in motor neuron degeneration. Thus, our findings implicate changes in neurotransmitter release and synaptic function as a converging mechanism in the pathogenesis of ALS and FTD.33
The highly significant signal generated by 19 SNPs at the chromosome 9p21.2 locus is most likely linked to C9orf72 repeat expansion status. As we have C9orf72 repeat expansion status available for only a few of the strata included in the present meta-analysis, we were not able to perform an analysis with C9orf72 wild-type carriers only to search for a residual effect of this locus in ALS-FTD. As a proxy for C9orf72 repeat expansion status, we conditioned on rs3849943 genotype and showed that the association signal at UNC13A increased. Moreover, it is interesting to note that C9orf72 is structurally related to DENN RabGEFs and that Rabs cooperate with UNC13A to regulate synaptic functions.34–36 Therefore, these observations hint at the exciting possibility that defects in UNC13A and C9orf72 in ALS and FTD converge upon the same synaptic mechanisms.
The present study identified one locus with increased disease association signal in the combined ALS and FTD meta-analysis on chromosome 8q24.13, nearing the genome-wide significance threshold. We successfully replicated disease association for the two top SNPs in this locus (rs13268726 and rs12546767) in an independent cohort of sporadic ALS patients. The 8q24.13 locus maps to a region of high LD encompassing SQLE, KIAA0196 and NSMCE2 (Fig 4). Of these genes KIAA0196 appears to be of particular interest. First, mutations in KIAA0196 cause hereditary spastic paraplegia, which shares clinical upper motor neuron dysfunction with ALS.37 Second, KIAA0196 (alias SPG8) encodes for strumpellin, a valosin-containing protein (VCP) binding partner in the human central nervous system.38 Mutations in VCP have previously been identified in both ALS and FTD patients.13 Third, protein aggregates containing strumpellin have been found in patients with inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia (IBMPFD).38 Nevertheless, since the combined analysis of the discovery and replication samples did not show a genome-wide significant association (p=1.01×10−7), further work is needed to establish the role of KIAA0196 and other genes in the associated locus in ALS-FTD pathogenesis.
Our study is the largest GWAS including sporadic ALS and TDP-43 positive FTD patients in search of new loci for neurodegeneration. With over 4,300 ALS patients and over 14,000 controls the study was well powered to detect associations of common variants with modest effect size. For example, we estimated 97% power for the detection of an association similar to rs3849943 on chromosome 9p.21 at α = 5×10−8. In terms of sample sizes required for GWAS, the FTD-TDP cohort was relatively small, but unique due to the homogenous TDP-43 pathology. The relatively small increase in statistical power obtained by adding 435 TDP-43 positive FTD cases might provide an important explanation for why we did not identify new shared susceptibility loci with genome-wide significance.
Also, in order to replicate our findings in TDP-43 positive FTD cohorts, one would require a minimum of 1,000–8 1,950 TDP-43 pathology-proven FTD cases and controls to achieve 80% power for detecting an effect with OR 0.8 and 1.2 (with minor allele frequency 0.1 – 0.35 at α=0.05), which is clearly challenging and requires further international collaborations. In addition, future studies implementing a combined analysis with large cohorts of patients with neurodegenerative disease including FTD, late-onset Alzheimer’s disease or Parkinson’s disease might add more statistical power and improve chances of finding new shared susceptibility loci for neurodegeneration.
For the FTD-TDP cohort, clinical information was available allowing us to identify patients with and without motor neuron signs, although we cannot definitively rule out the possibility that a small proportion of FTD cases classified as ‘without motor neuron signs’ still developed motor neuron disease after the last clinical follow-up. For the ALS strata, however, data on frontotemporal dementia or cognitive impairment in ALS patients were not available. Therefore, the extent to which signals from the meta-analysis are driven by signs of frontotemporal dementia or cognitive impairment is not known exactly, although previous studies have estimated a proportion of 5–10% of ALS patients also having FTD.1,2 Furthermore, previously, an association was reported of variants in TMEM106B with susceptibility to FTD and with cognitive impairment in ALS patients.18,19 In the present meta-analysis we did find this locus in FTD, but we did not find a significant association of variants in TMEM106B with ALS. It is possible that an association with TMEM106B exists within a subset of ALS patients with cognitive impairment. Careful deep phenotyping of samples in future GWAS studies will help to shed light on the genetic determinants of motor neuron dysfunction versus cognitive impairment.
In conclusion, our meta-analysis identifies UNC13A as a novel link between ALS and TDP-43 positive FTD, which identifies synaptic defects as a shared disease mechanism and further corroborates the role of UNC13A and synaptic mechanisms in neuronal degeneration. Our results provide a novel starting point for further dissection of shared pathogenic pathways underlying ALS and FTD.
Supplementary Material
Acknowledgement
We would like to thank all patients and healthy volunteers who participated in this project; the study staff, general practitioners and pharmacists. The authors are very grateful to the participants and staff of the Rotterdam Study. We thank the Motor Neurone Disease Association UK, the Medical Research Council (UK), the Wellcome Trust and the Psychiatry Research Trust (Tim Perkins Fund and Charcot Fund).
This work was supported by the Prinses Beatrix Fonds (Kersten Foundation); VSB fonds; The Netherlands ALS Foundation and J.R. van Dijk; and the Adessium Foundation to LHB. JHV is supported by the Brain Foundation of The Netherlands; and the Thierry Latran Foundation. RJP is supported by the Prinses Beatrix Fonds and the Thierry Latran Foundation. The GWA study was funded by the Netherlands Organization of Scientific Research NWO Investments [grant numbers 175.010.2005.011, 911-03-012]; the Research Institute for Diseases in the Elderly (014-93-015; RIDE); and the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) [project number 050-060-810]. In addition, the research leading to these results has received funding from the European Community's Health Seventh Framework Programme (FP7/2007-2013) [grant agreement number 259867]. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam; Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); and the Municipality of Rotterdam. The work was also supported by grants from the German Federal Ministry of Education and Research (01GI0704; network for ALS research (MND-NET)), the Charcot Foundation for ALS Research (ACL, JHW), and the Swabian ALS Registry. The Irish study was funded by The Muscular Dystrophy Association (USA); The Health Research Board of Ireland; The Irish Neurological Association Travel Award; and The Irish Motor Neuron Disease Research Foundation. In Sweden, this project was supported by the Swedish Brain Research Foundation; the Hållstens Research Foundation; the Swedish Medical Society; the Björklund Foundation for ALS Research; and the Swedish Association for the Neurologically Disabled to PMA. In Switzerland, this study was supported by the Swiss ALS Association. WR was supported by grants from the University of Leuven (Methusalem); and the Interuniversity Attraction Poles program P6/43 of the Belgian Federal Science Policy Office. In Italy, this study has been funded in part by a grant of the “Associazione Centro Dino Ferrari”, of the Italian Ministry of Health [GWA-SLA, Ricerca Finalizzata 2007, grant number 31 and ALS-FTD, Ricerca Finalizzata 2009, grant number 276] (IF, VS), and of the Fondazione Istituto Auxologico Italiano. Also in Italy, the study was financed by the Italian Health Ministry (Ricerca Sanitaria Finalizzata 2007), Fondazione Vialli e Mauro ONLUS, and Federazione Italiana Giuoco Calcio (AC). In France, this study was funded by the Association pour la recherché sur la sclérose latérale amyotrophique (ARSla); and the Association Réseau SLA Ile de France. Support was also provided by the ALS Therapy Alliance; Project ALS; the Angel Fund; the Pierre L. de Bourgknecht ALS Research Foundation; the Al-Athel ALS Research Foundation; the ALS Family Charitable Foundation and the National Institute of Neurological Disorders and Stroke [NS050557]. JEL received funding from the National Institutes of Health/National Institute of Neurological Disorders and Stroke [1R01NS073873]. VMV was supported by NIH grant AG017586.
References
- 1.Murphy J, Henry R, Lomen-Hoerth C. Establishing subtypes of the continuum of frontal lobe impairment in amyotrophic lateral sclerosis. Arch Neurol. 2007;64:330–334. doi: 10.1001/archneur.64.3.330. [DOI] [PubMed] [Google Scholar]
- 2.Ringholz GM, Appel SH, Bradshaw M, et al. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 2005;65:586–590. doi: 10.1212/01.wnl.0000172911.39167.b6. [DOI] [PubMed] [Google Scholar]
- 3.Brettschneider J, Del Tredici K, Toledo JB, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol. 2013;74:20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gijselinck I, Engelborghs S, Maes G, et al. Identification of 2 Loci at chromosomes 9 and 14 in a multiplex family with frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Arch Neurol. 2010;67:606–616. doi: 10.1001/archneurol.2010.82. [DOI] [PubMed] [Google Scholar]
- 5.Le Ber I, Camuzat A, Berger E, et al. Chromosome 9p-linked families with frontotemporal dementia associated with motor neuron disease. Neurology. 2009;72:1669–1676. doi: 10.1212/WNL.0b013e3181a55f1c. [DOI] [PubMed] [Google Scholar]
- 6.Vance C, Al-Chalabi A, Ruddy D, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 7.Dejesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laaksovirta H, Peuralinna T, Schymick JC, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shatunov A, Mok K, Newhouse S, et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Es MA, Veldink JH, Saris CGJ, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 12.Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borroni B, Bonvicini C, Alberici A, et al. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum Mutat. 2009;30:E974–E983. doi: 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- 15.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 16.Van Deerlin VM, Sleiman PMA, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patsopoulos NA, de Bakker PIW the Bayer Pharma MS Genetics Working Group tSCoSEI-baaC-A, ANZgene Consortium, GeneMSA, International Multiple Sclerosis Genetics Consortium. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol. 2011;70:897–912. doi: 10.1002/ana.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Zee J, Van Langenhove T, Kleinberger G, et al. TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain. 2011;134:808–815. doi: 10.1093/brain/awr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vass R, Ashbridge E, Geser F, et al. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:373–380. doi: 10.1007/s00401-010-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strong MJ, Lomen-Hoerth C, Caselli RJ, et al. Cognitive impairment, frontotemporal dementia, and the motor neuron diseases. Ann Neurol. 2003;54:S20–S23. doi: 10.1002/ana.10574. [DOI] [PubMed] [Google Scholar]
- 21.Brooks BR, Miller RG, Swash M, et al. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bakker PIW, Ferreira MAR, Jia X, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SH, Yang J, Goddard ME, et al. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 30.Chiò A, Mora G, Restagno G, et al. UNC13A influences survival in Italian amyotrophic lateral sclerosis patients: a population-based study. Neurobiol Aging. 2013;34:357.e351–357.e355. doi: 10.1016/j.neurobiolaging.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diekstra FP, Van Vught PWJ, van Rheenen W, et al. UNC13A is a modifier of survival in amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:630.e633–630.e638. doi: 10.1016/j.neurobiolaging.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 32.Varoqueaux F, Sons MS, Plomp JJ, Brose N. Aberrant morphology and residual transmitter release at the Munc13-deficient mouse neuromuscular synapse. Mol Cell Biol. 2005;25:5973–5984. doi: 10.1128/MCB.25.14.5973-5984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65(suppl 1):S3–S9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 34.Huang C-C, Yang D-M, Lin C-C, Kao L-S. Involvement of Rab3A in Vesicle Priming During Exocytosis: Interaction with Munc13-1 and Munc18-1. Traffic. 2011;12:1356–1370. doi: 10.1111/j.1600-0854.2011.01237.x. [DOI] [PubMed] [Google Scholar]
- 35.Levine TP, Daniels RD, Gatta AT, et al. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdmanis PN, Meijer IA, Reynolds A, et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet. 2007;80:152–161. doi: 10.1086/510782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clemen CS, Tangavelou K, Strucksberg K-H, et al. Strumpellin is a novel valosin-containing protein binding partner linking hereditary spastic paraplegia to protein aggregation diseases. Brain. 2010;133:2920–2941. doi: 10.1093/brain/awq222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.