Abstract
The responses of high-level neurons tend to be mixtures of many different types of signals. While this diversity is thought to allow for flexible neural processing, it presents a challenge for understanding how neural responses relate to task performance and to neural computation. To address these challenges, we have developed a new method to parse the responses of individual neurons into weighted sums of intuitive signal components. Our method computes the weights by projecting a neuron's responses onto a predefined orthonormal basis. Once determined, these weights can be combined into measures of signal modulation; however, in their raw form these signal modulation measures are biased by noise. Here we introduce and evaluate two methods for correcting this bias, and we report that an analytically derived approach produces performance that is robust and superior to a bootstrap procedure. Using neural data recorded from inferotemporal cortex and perirhinal cortex as monkeys performed a delayed-match-to-sample target search task, we demonstrate how the method can be used to quantify the amounts of task-relevant signals in heterogeneous neural populations. We also demonstrate how these intuitive quantifications of signal modulation can be related to single-neuron measures of task performance (d′).
Keywords: orthonormal basis, signal modulation, bias correction
the responses of neurons at higher stages of neural processing in the brain tend to reflect heterogeneous mixtures of many different types of task-relevant signals (e.g., Bennur and Gold 2011; Brody et al. 2003; Buckley et al. 2009; Miller and Desimone 1994; Rigotti et al. 2013). This diversity is thought to be advantageous insofar as a population that contains a diversity of neural responses is capable of performing a diversity of tasks (Rigotti et al. 2013). However, response heterogeneity also makes these high-level brain areas difficult to understand with classical single-neuron approaches, which inherently rely on identifying regularities in the response properties of individual neurons across a population (e.g., discovering that the majority of V1 neurons are tuned for orientation).
Here we present a method to deconstruct the responses of heterogeneous neurons as weighted sums of intuitive signals. Our method is useful when applied to experimental designs that involve changing multiple experimental parameters, which is of course a prerequisite for uncovering signal “mixtures.” Examples include tasks that require finding a “match” to a target, which involves changing the identities of the “stimuli” and the “target” (e.g., Maunsell et al. 1991; Miller and Desimone 1994; Pagan et al. 2013). Likewise, tasks that require flexible rule-based mappings of sensory stimuli onto behavioral responses involve manipulating the sensory stimulus and the rule (e.g., Bennur and Gold 2011; Mansouri et al. 2007). A slightly less obvious example is a task that requires a subject to remember the specific sequence with which objects appear; the different conditions in such a task can be envisioned as combinations of object identity and time (Naya and Suzuki 2011).
To address the challenges associated with understanding how the responses of a heterogeneous neural population reflect different task-relevant components, we have developed a method to parse the responses of individual neurons into weighted sums of intuitive components. Our method computes the weights by projecting a neuron's responses onto a predefined orthonormal basis. Once determined, these weights can then be combined to quantify different types of signal modulation in a manner that does not depend on sign (e.g., firing rate increases or decreases). From a neural coding perspective, both firing rate increases and decreases convey information and thus unsigned modulation measures more accurately reflect signal magnitude. Additionally, because firing rate increases and decreases tend to be balanced in many high-level brain areas (see, e.g., Maunsell et al. 1991; Miller and Desimone 1994; Romo et al. 1999), the “average” signed modulation across a population is not a useful quantity (i.e., because it takes on a value near zero) whereas the “average” unsigned (absolute valued or squared) modulation is meaningful.
As we describe in detail below, our method is related to other approaches, including the analysis of variance (ANOVA), the multiple linear regression (MLR), the principal components analysis (PCA), and a recent PCA extension [demixed PCA (dPCA); Machens 2010]. While these methods have advantages over our method for some applications, one advantage of our method over the others is that it produces unsigned and unbiased estimates of signal modulation magnitudes. Unbiased signal estimates are important when one wants to compare signals across brain areas, across different points in time, or across different types of signals. However, we note that our method is not ideal for describing exactly “how” neurons are tuned for a particular parameter (e.g., for describing tuning curves).
In addition to introducing a new way to measure neural signals, we demonstrate how these measures can be related to task performance. Quantifying task performance for individual neurons by performing a receiver operating characteristic (ROC) analysis or by calculating the related discriminability measure d′ is a common way to compare neural signals—between different brain areas, between different points in time within the same brain area, or with behavior (e.g., Adret et al. 2012; Bennur and Gold 2011; Gu et al. 2012; Liebe et al. 2011; Newsome et al. 1989; Swaminathan and Freedman 2012). Understanding the underlying sources of neural task performance differences (e.g., overall firing rate changes vs. changes in different types of tuning modulation) is crucial for accurate interpretation of what these differences mean for neural coding. Here we show how our method can be used to derive a precise understanding of how task performance depends on different types of signal modulation.
METHODS
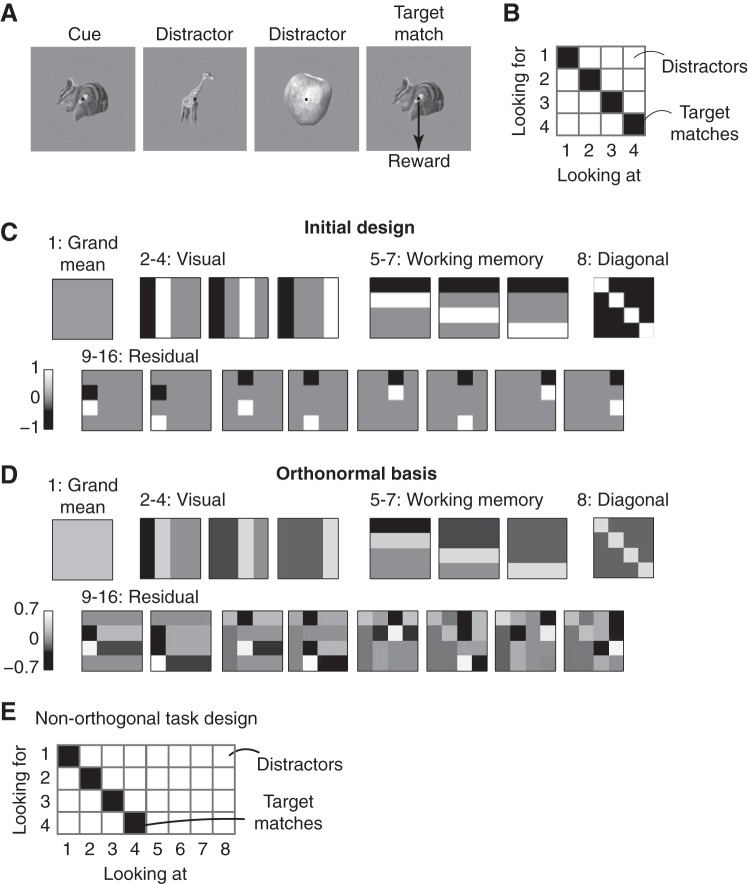
The data we use to describe our method have been reported previously (Pagan et al. 2013). All procedures were reviewed and approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Briefly, we recorded neural responses in inferotemporal cortex (IT) and perirhinal cortex (PRH) as monkeys performed a delayed-match-to-sample (DMS), sequential target search task that required treating the same images as targets and as distractors on different trials (Fig. 1A). Monkeys initiated a trial by fixating a small dot, and after a short delay a cue indicating the target for that trial was presented, followed by a random number (0–3) of distractors and then the target match. Monkeys indicated the presence of the target match by making a saccade to a specific location on the screen before the onset of the next stimulus and were rewarded for correct responses. Altogether, four images were presented in all possible combinations as a visual stimulus (“looking at”) and as a target (“looking for”), resulting in a four by four matrix, and at least 20 repeated trials of each condition were collected (Fig. 1B).
Fig. 1.
Constructing an orthonormal basis for a delayed-match-to-sample (DMS) task. A: each trial of the DMS task began with the presentation of a cue indicating the target for that trial, followed by the presentation of 0–3 distractors and then the target match. Images were presented for 400 ms, followed by a 400-ms blank. Monkeys were required to maintain fixation throughout the distractors and saccade to a response dot after the target match appeared (within 800 ms) to receive a reward. B: the experimental design included 4 images, each presented as a visual stimulus (“looking at”) in the context of every other image as a target (“looking for”), thus defining a 4 × 4 matrix. In this matrix, target matches fall along the diagonal and distractors fall off the diagonal. C: the matrices produced by the first stage of the orthonormal basis design process (see text). D: the matrices produced by applying the Gram-Schmidt process to the matrices described in C. E: an experimental design in which the “target match” and “visual” conditions cannot be orthogonalized (see text).
Most of our methods are described in results. Here we describe the statistical procedures we used to evaluate the statistical significance of the observed differences in the mean values of various indices between IT and PRH (see Fig. 5). Because many of these measures were not normally distributed, we calculated these P values via a bootstrap procedure. On each iteration of the bootstrap, we randomly sampled the true values from each population, with replacement, and we computed the difference between the means of the two newly created populations. We computed the P value as the fraction of 1,000 iterations on which the difference was flipped in sign relative to the actual difference between the means of the full data set (e.g., if the mean for PRH was larger than the mean for IT, the fraction of bootstrap iterations in which the IT mean was larger than the PRH mean; Efron and Tibshirani 1994).
Fig. 5.
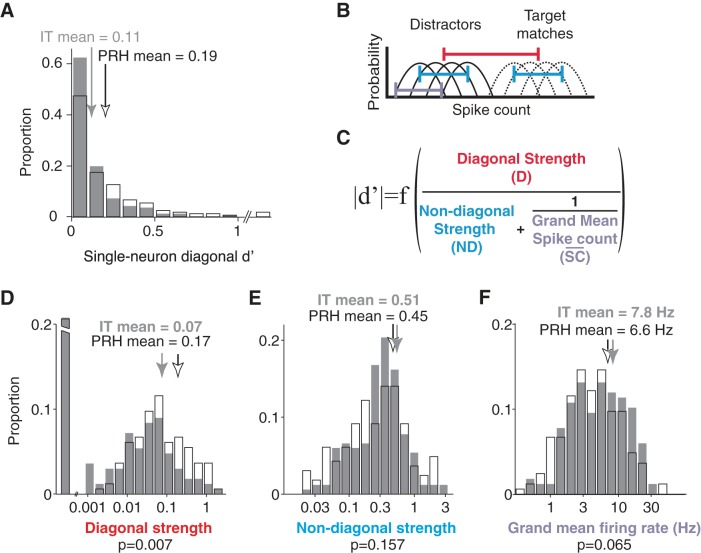
Relating signal modulation magnitudes with task performance (d′). A: diagonal d′, computed as the absolute value of d′, followed by bias correction (Eq. 13). Histograms are shown for 167 neurons recorded in IT and 164 neurons recorded in PRH. PRH neurons with neuron diagonal d′ of 1.1, 1.3, and 2.1 are included in the last bin. Arrows indicate means. B: the diagonal d′ calculation was based on the distributions of responses over trials to the target matches (dashed lines) and to the distractors (solid lines). C: diagonal d′ can be expressed as a function of 3 intuitive components. B and C: diagonal strength (red) is computed as a function of the normalized diagonal weight (Eq. 15), and this component determines the distance between the average response to all target matches and the average response to all distractors (red line); diagonal d′ is proportional to this component (see text). The nondiagonal strength (cyan) is computed as a function of the combined normalized nondiagonal weights (Eq. 16), and this component determines the spread within the target matches and within the distractors (cyan line); diagonal d′ is inversely related to this component (see text). The final component (lavender) captures the trial-by-trial variability of the neuron (lavender line). Within a Poisson process and after normalization, this term can be estimated by the inverse of the grand mean spike count across all conditions (Eq. 40), and thus diagonal d′ increases monotonically with the grand mean spike count (see text). D–F: diagonal strength (D), nondiagonal strength (E), and grand mean firing rate (F), in IT (gray) and PRH (white). Arrows indicate means. In D, the 1st bin includes neurons with a diagonal strength <0.001 and the broken axis extends to a proportion of 0.46 in IT and 0.34 in PRH.
RESULTS
The methods we describe here are useful for analyzing the neural data from experiments in which experimental conditions are combinations of multiple stimulus parameters (e.g., sensory stimuli combined with different task instructions). Additionally, they can be applied to both parametric variation (e.g., systematic changes in motion direction) and nonparametric variation (e.g., changes in object identity where the relationships between different identities are not well defined). The ultimate goal of our method is to measure the magnitude by which a neuron's responses are modulated by different experimental parameters, and below we refer to these modulation magnitudes as “signals.” Our method involves parsing a neuron's firing responses to N different combinations of the stimulus parameters (i.e., experimental conditions), which we refer to as a “response matrix,” into a weighted sum of N intuitively defined signals. This process begins by constructing an orthonormal basis of N vectors. “Ortho” refers to the fact that the vectors are “orthogonal,” and this allows the original matrix to be deconstructed into a weighted sum (i.e., none of the neural responses is counted twice). “Norm” refers to the fact that all the vectors have the same length (i.e., the “norm” of each vector, computed as the square root of the summed squared values, is equal to 1). “Basis” refers to the fact that, together, the vectors capture all possible types of response modulation that could occur given the specific experimental design. As described in more detail below, once the orthonormal basis is determined, the weights are calculated for each neuron by taking the projection (i.e., the dot product) of the neuron's average firing rate responses and each basis vector and the “signals” are determined by combining weights of the same type.
Constructing an orthonormal basis.
To construct the basis, we begin by constructing a set of N vectors that capture the types of modulation we are interested in. Next we apply the Gram-Schmidt process to convert the set of vectors into an orthonormal basis. To describe the method, we apply this procedure to an example experimental design taken from our previous work: a DMS target search task (Pagan et al. 2013). In these experiments, monkeys viewed a series of sequentially presented images and indicated when a “target match” appeared within a sequence of “distractors” (Fig. 1A). Altogether, monkeys viewed each of four visual images in the context of each image as a target, resulting in a four-by-four matrix of experimental conditions (Fig. 1B). In this matrix, target matches fall along the diagonal and distractors fall off the diagonal. This “response matrix” R is computed as the average spike count response across 20 repeated trials for each of the 16 experimental conditions. Below, we treat R as a 16-entry vector to perform our calculations.
To design an orthonormal basis for this task, we began by constructing a first vector that corresponds to the grand mean spike count response across all conditions; all entries in this vector take on the same, constant value (e.g., 1/16; Fig. 1C). The remaining vectors are designed to capture the types of modulation that neural responses might reflect, which follow from the task design. In the case of our experiment, this included three vectors to describe the visual modulation, reflected by columns in the response matrix (Fig. 1C). Notably, while there are four different visual images, only three are required to capture the visual modulation once the mean firing rate response has also been defined (i.e., degrees of freedom for the visual conditions = 4 − 1). The second type of modulation is reflected by rows in this matrix and corresponds to response modulations that can be attributed to changing the identity of the target; because target identity must be held in working memory during this task, we refer to this as “working memory” modulation. The third type of modulation differentiates whether a condition was a target match or a distractor, and this corresponds to modulation along the diagonal. The final type of modulation is that which is required to describe responses that are “peppered” across the matrix, such as differential responses to the same visual image under two different distractor conditions, and we refer to this modulation as “residual.” More technically, residual modulations reflect all nonlinear combinations of visual and working memory signals that are not diagonal.
Once this initial set of vectors is defined, we apply the Gram-Schmidt procedure to convert it into an orthonormal basis. Specifically, we define each of the N original vectors as vi and each of the vectors of the resulting orthonormal basis as bi. The Gram-Schmidt process is applied iteratively to each initially defined vector, and consists of two stages: first, the vector is orthogonalized relative to all the vectors already incorporated into the final, orthonormal basis, and second, the resulting vector is normalized by its norm ||bi||:
| (1) |
| (2) |
where bij indicates the jth element of the ith vector bi.
The final orthonormal basis obtained for our experiment is shown in Fig. 1D. A crucial requirement is that the originally defined vectors v1 . . . vN span the full space; if this is not the case, the Gram-Schmidt process will fail to produce a valid orthonormal basis. It is possible to verify this simply by measuring the rank of the matrix obtained by juxtaposing the original vectors [v1 . . . vN] and checking that it is equal to N.
There is no unique way to parse a set of N vectors into an orthonormal basis. For example, one might consider the “standard basis” as the set of vectors that define each experimental condition (e.g., 10000, 01000, 00100, etc.). While this basis is orthonormal, it is not very useful because a projection of a neuron's responses R onto this basis would simply return the mean firing rate response to each experimental condition (i.e., each entry in R). Decisions about how to create the initial vectors when designing the basis depend on what one is trying to achieve. We often find it useful to begin by considering the task “inputs” and whether the task “output” (i.e., the solution) can be expressed as a linear or nonlinear combination of the inputs, because this approach formalizes the mapping between the computational goals of the task and the neural signals. For the DMS task described above, the task inputs include “visual” and “working memory” signals (i.e., the monkey is presented with the identity of the target, which he holds in working memory, and the identity of the visual image). These are equivalent to the “linear terms” of a two-factor ANOVA analysis. The solution for this task—differentiating whether each condition is a target match or a distractor (i.e., the diagonal matrix)—cannot be expressed by any linear combination of inputs but instead requires a nonlinear computation. However, it is only one of many possible nonlinear vectors, and it is thus essential to parse it from the “residual” vectors, which also reflect nonlinear combinations of visual and working memory signals. We note that “diagonal” and “residual” signals would be combined into a single “nonlinear interaction term” in a two-factor ANOVA (for a more extensive description of the relationship between the orthonormal basis and the ANOVA, see discussion).
Not all experimental designs allow for orthogonalization, or equivalently, not all experimental parameters can be orthogonalized. For example, Fig. 1E depicts a modified experimental design in which some visual images are always presented as distractors and never as targets. In this case, there is no way to produce a component that captures “target match” signals (e.g., one that reflects tuning for whether a condition is a target match or a distractor) that can be orthogonalized with the “visual” components. This is because the experimental design introduces a correlation between image identity and whether the condition is a target match: once you know that the identity of an image is 5, 6, 7, or 8, you know with certainty that the image is a distractor. Stated differently, in this experimental design “target match” and “visual” signals are confounded. Thus an additional advantage of our method is that it introduces a means to evaluate and improve a candidate experimental design through the attempted construction of a useful orthonormal basis.
Computing and interpreting signal modulation magnitudes.
Once the orthonormal basis has been defined, we can compute the corresponding signal modulation magnitudes. A neuron's response matrix R can always be decomposed into a weighted sum of the orthonormal components:
| (3) |
where bi indicates the ith component and wi indicates the weight associated with the ith component. The weights wi are thus determined by computing the projection (i.e., the dot product) of the vector R and each basis component bi:
| (4) |
Ultimately, we are interested in quantifying how much of a neuron's firing rate modulation can be attributed to changes in specific type of experimental manipulation (e.g., the amount of firing rate modulation that can be attributed to changes in the visual stimulus), and thus we need to group together weights that correspond to the same type (e.g., the 3 visual weights). When doing so, it is important to consider that these weights can be negative as well as positive. Positive weights correspond to neural responses that directly resemble an orthonormal basis component, whereas negative weights correspond to neural responses that are simply flipped in sign and thus they also reflect relevant firing rate modulations. To convert a set of weights into a measure of a particular type of modulation, we square the weights, sum across the set, and then take the square root. For the DMS task:
| (5) |
where Mvis is the amount of visual modulation, Mwm is working memory modulation, Mdiag is diagonal modulation, and Mresidual is residual modulation. When computed this way, each type of signal modulation measures the standard deviation (i.e., the spread) of the responses, averaged across repeated trials, and has units of spike count. For example, a “visual signal equal to 2” means that the trial-averaged spike count was spread two standard deviations around the grand mean firing rate as a result of changes in the visual stimulus.
Next we introduce three different ways to normalize these signal modulation magnitudes, each designed to highlight a different aspect of signal modulation. First, one might wish to produce signal modulation measures that are not “raw” (Eq. 5) but instead are compared to the amount of noise. This type of “signal-to-noise” modulation measure can be obtained by simply normalizing by the average trial-by-trial variability of a neuron:
| (6) |
where σ̄noise is computed as
| (7) |
and σi,noise2 indicates the trial-by-trial variability (variance) associated with the ith condition. In this formulation, modulations are unitless and they measure the ratio between the signal and noise modulations. For example, a “visual signal equal to 2” now means that changes in the visual signal produced a spread in the trial average spike counts with a standard deviation twofold larger than the standard deviation of the noise. To anticipate and prevent confusion, we note that the issue of whether a signal modulation estimate is biased by noise is distinct from the issue of normalizing the size of the signal relative to the size of the noise; the former is related to the issue of getting an accurate estimate of signal size (discussed below), whereas the latter informs how much a given amount of signal will be actually “useful” at conveying information. In other words, a fixed amount of signal can provide perfect information in the absence of noise, or it can be almost impossible to detect within a very large amount of noise.
As a second consideration, we note that in some situations, including the DMS task, different types of signals have different numbers of components and it may be desirable to normalize by the number of components to arrive at a measure of modulation “per degree of freedom”:
| (8) |
where Nvis indicates the number of visual components (= 3), Nwm indicates the number of working memory components (= 3), and Nres indicates the number of residual components (= 8). For example, a “visual signal equal to 2” now means that each visual component was (on average) responsible for spreading the trial-averaged spike count two standard deviations around the grand mean. This normalization can also be combined with the noise normalization in Eq. 6 to produce a measurement of the signal-to-noise ratio per component.
Finally, one might wish to produce a measure of signal modulation that is affected by changes in the “pattern” of the response matrix but not by an overall rescaling of the firing rates, whereas in their raw form signal modulations (Eq. 5) are directly proportional to the overall grand mean firing rate. Scale-invariant modulation measures can be computed by normalizing each type of signal modulation by the grand mean response to produce quantities that we refer to as “signal strengths.” This normalization is described in more detail in Relating signal modulations and task performance.
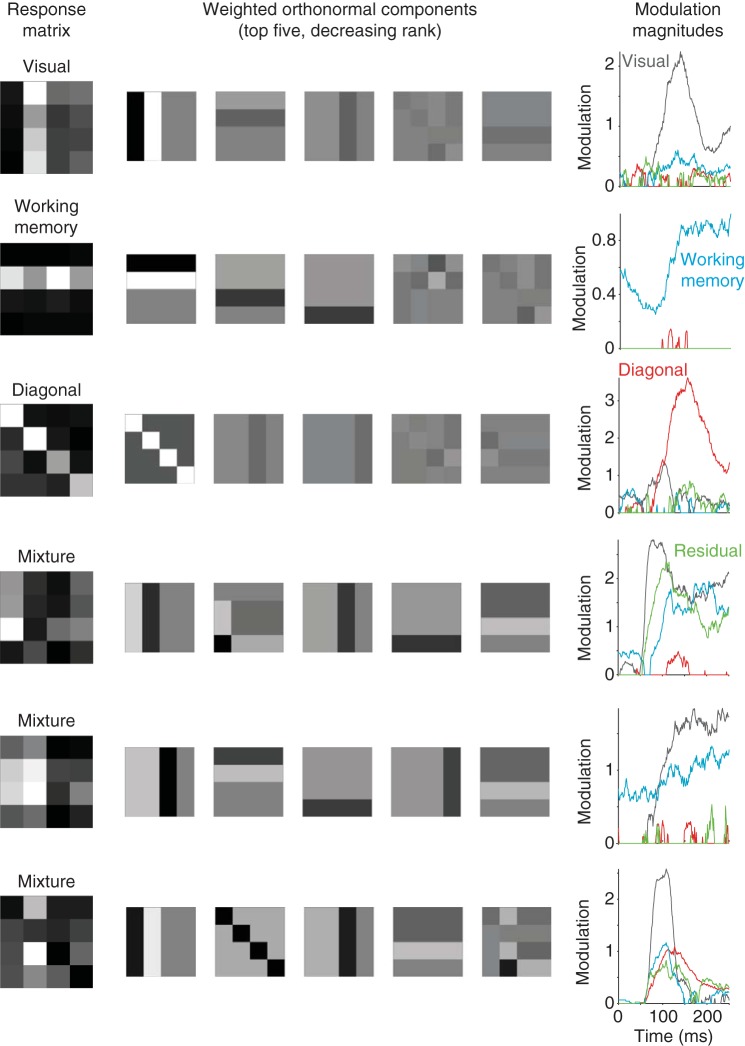
To illustrate an example of signal modulations, Fig. 2 shows the result of our method applied to six neurons collected during the DMS task, including three neurons whose responses reflect relatively pure selectivity for signals of a single type (Fig. 2, top) and three neurons whose responses reflect mixtures of different types of signals (Fig. 2, bottom). Shown in Fig. 2 are the response matrices for each neuron (Fig. 2, left) and the top five orthonormal components rescaled by their weights (Fig. 2, center). As described above, weights can be positive or negative and negative weights invert the polarity of the orthonormal component (e.g., compare the diagonal matrices in the 3rd and 6th rows of Fig. 2). Also shown in Fig. 2 are the signal modulations computed as the square root of the summed, squared weights for each type of signal, normalized by the number of components for each signal type (Fig. 2, right; Eq. 8). To produce these plots, response matrices were computed by counting spikes in 50-ms windows systematically shifted relative to response onset. Signal modulations are computed by squaring the weights, so that both positive and negative weights contribute equally to measured modulations. Signal modulations thus provide an intuitive quantification of “how much” of a particular type of signal is reflected in the responses of a particular neuron, regardless of the “sign” of that weight (i.e., responses increases or decreases) and regardless of “how” that modulation is distributed across the different components (i.e., tuning). Importantly, computing modulations in this way is biased (Figs. 3 and 4), and this bias must be corrected for to get an accurate measure of modulation (as described below).
Fig. 2.
Example neurons. Each row depicts a single example neuron, where the responses of the top 3 neurons reflect relatively pure selectivity and the responses of the bottom 3 neurons reflect mixtures of different selectivity types. Left: the mean spike count responses computed within a window 50–250 ms after stimulus onset to each of the 16 conditions (the “response matrix”), averaged over 20 repeated trials, normalized to range from the minimum (black) to the maximum (white). Center: the orthonormal components with the 5 largest weights, plotted as shown in Fig. 1D but with intensity scaled by the weight applied to each component. The response matrix can be reconstructed as a weighted sum of these matrices (once the grand mean spike count is also factored in, which is not shown). Right: the temporal evolution of the closed-form bias-corrected signal modulation magnitudes for each type of signal, computed as the square root of the sum of squares of the bias-corrected weights normalized by the number of components for each signal type (Eq. 8). To perform this analysis, spikes were counted in 50-ms sliding windows shifted 1 ms for each successive time bin. The example neuron depicted in the 4th row was recorded in inferotemporal cortex (IT); the other neurons were recorded in perirhinal cortex (PRH).
Fig. 3.
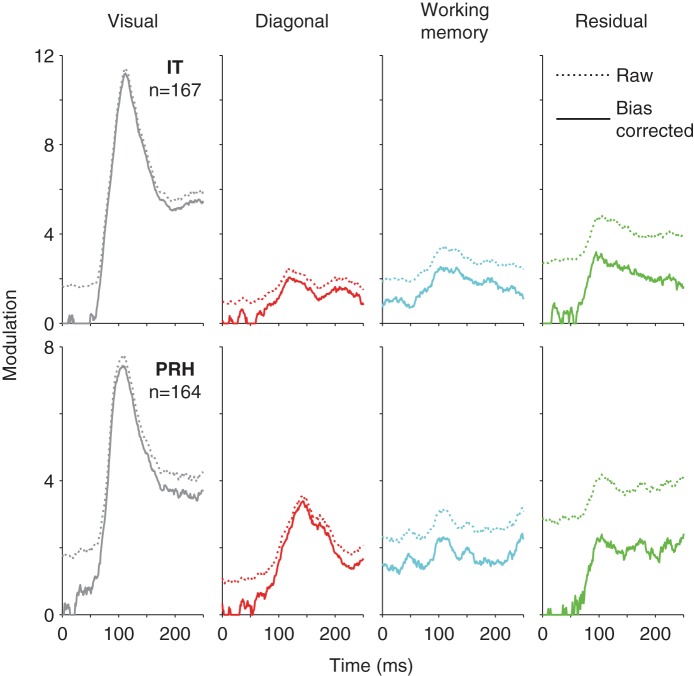
Empirical demonstration of bias. Raw (dotted) and closed-form bias-corrected (solid) measures of signal modulation summed across the IT (top) and PRH (bottom) populations plotted with the same conventions as Fig 2, right.
Fig. 4.
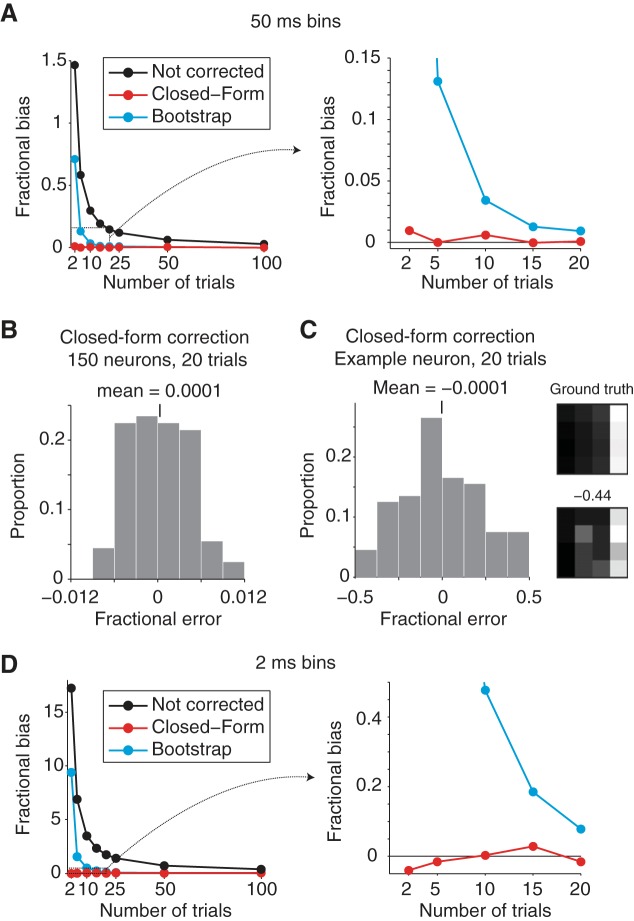
Evaluation of bias-correction procedures. To evaluate the accuracy of our signal modulation measures, a population of 150 simulated neurons with known amounts of signal modulation were created from measured responses in IT and PRH. A: fractional bias, calculated as the ratio of the total bias (summed across all signal modulations) divided by the total signal (summed across all signal modulations), plotted for the uncorrected simulated population (black), the closed-form bias correction (red), and the bootstrap bias correction (cyan) as a function of the number of Poisson trials collected in each simulated experiment when spikes were counted in 50-ms bins centered 125 ms after stimulus onset. Shown are the averages over 100 simulated experiments. Plot on right shows an enlargement of the boxed region indicated on left. B: a histogram of the average (across the 150 simulated neurons) fractional error (total error/total signal) remaining after the closed-form correction for each of 100 simulated experiments with 20 Poisson trials to show that the fractional bias measured per experiment is always near 0. C, left: histogram of the fractional bias remaining for 1 representative simulated neuron to show that fractional error measured per neuron can be large. Right: the “ground truth” response matrix for this neuron plotted along with 1 example matrix measured from a simulated experiment that produced an extreme fractional error. D: results of the same analysis presented in A, but performed from responses counted in 2-ms windows. As in A, plot on right shows an enlargement of the boxed region indicated on left.
As highlighted above, an orthonormal basis is not uniquely defined for a given experimental design. This statement also applies to subsets of different types of components—for example, one could define an orthonormal basis with “visual” vectors that are different from those presented in Fig. 1D but capture the visual modulations equally well because the three new visual vectors will define the same linear subspace as the original vectors. Thus the combined projection of a neuron's response vector onto the three visual components uniquely captures the amount of modulation that can be attributed to changes in the identity of the visual stimulus even if the specific visual vectors themselves are not uniquely defined. Under what situations is a particular type of signal modulation uniquely defined? In our experiment, the uniquely defined parsing of different signal types follows from the two-dimensional “looking at”/“looking for” matrix structure of this task, in which the “visual” and “working memory” conditions are presented in all possible combinations and are thus independent from one another (Fig. 1B). Similarly, because the diagonal matrix is a single dimension, it is also uniquely defined. Finally, the “residual” subspace is uniquely defined because it describes everything that remains after the other uniquely defined subspaces have been accounted for. In contrast, if we were to, for example, combine the first visual, the first working memory, and the first residual dimension into a measure of signal modulation, we would obtain a subspace that is strictly dependent on the particular choice of basis, i.e., a different orthonormal basis would produce a different linear subspace when the same three components are considered.
Bias and bias correction.
When estimating the amount of modulation in a signal, noise and limiting sampling size are known to introduce a positive bias (Panzeri et al. 2007). For example, consider a hypothetical neuron that responds with the exact same average firing rate response to each of a set of experimental conditions. Because neurons are noisy, if we were to estimate these mean rates on the basis of a limited number of repeated trials, we would get different values for different conditions and this could lead to the erroneous impression that the neural responses are in fact modulated by the stimuli. Similarly, applying the orthonormal basis method to this data would produce weights shifted away from zero as a result of noise for at least a subset of the basis vectors. While the mean of the weights themselves would be unbiased (because noise would shift the weights to both more positive and more negative values), the process of converting the weights into signal modulation magnitudes by squaring (Eq. 5) would result in a positive mean bias.
To illustrate this bias, Fig. 3 includes plots of summed raw and bias-corrected signal modulation magnitudes plotted as a function of time for the IT and PRH populations (using the “closed form” bias correction described below). These results reveal that under physiologically relevant conditions these biases can be considerable when signals are small or absent (e.g., at stimulus onset, biased estimates of visual modulation are ∼1.7 standard deviations in IT and PRH compared with bias-corrected measures of ∼0) and that these biases become smaller when signals are larger (e.g., at the peak of the visual signal, the bias is ∼0.25 standard deviations in IT and PRH, which is only ∼3% of the bias-corrected value). This is because the bias is additive in the domain of the squared weights but to compute signal modulations we take the square root. The square root operation has the effect of enhancing the effect of the bias when the modulation is small and shrinking it when the modulation is larger. The reason why we prefer to take the square root rather than operating on the squared modulations is that we find that measures of signal modulations in units of “spike counts” are preferable to units of “squared spike counts” in that they more clearly map onto our intuitive definitions of signals (e.g., signals double when firing rates double).
To estimate bias, we compared two methods: an analytical solution and a bootstrap technique. Under the assumption that trial-by-trial variability is Gaussian distributed, which is a reasonable approximation of Poisson distributions when the mean spike counts are sufficiently large, the amount of bias can be derived and unbiased measures of the squared weights ŵi2 can be computed as (see appendix)
| (9) |
where T equals the number of repeated trials for each experimental condition.
Because the analytical solution assumes that spike counts are Gaussian distributed whereas spike count distributions are known to deviate from this assumption, particularly at low firing rates, we also introduce a bootstrap procedure. The first step in estimating the bias for a given weight involves resampling with replacement T responses to each condition and recomputing the squared weight for these bootstrapped responses w̃i2 with Eq. 4. Next, the bias can be estimated by subtracting the modulation computed from the actual responses from the bootstrapped modulation estimates, and finally a corrected estimate for each squared weight ŵi2 can be computed simply by subtracting the bias (Efron and Tibshirani 1994):
| (10) |
In practice, we find that the bias estimated on any one resampling can be noisy, and thus we find it useful to calculate the bias a number of times (e.g., 100) and average the bias across those calculations.
To test our bias correction procedures, we performed simulations in which we created “ground truth” neurons with known amounts of underlying modulation, simulated their trial-by-trial variability as Poisson, and compared the ground truth and estimated modulation magnitudes. To test the bias correction in a relevant regime, we performed these simulations by creating a population of 150 “ground truth” neurons that were inspired by actual neurons we recorded in IT and PRH (examples include the neurons shown in Fig. 2). Specifically, we computed each simulated neuron's underlying responses by applying a bias correction to 150 randomly selected raw response matrices measured in our experiments, and we then used these mean values to generate N Poisson simulated trials. Figure 4A shows the fractional bias (total bias/total signal), computed for a population of 150 neurons, averaged over 100 simulated experiments. This plot reveals that, as expected, total bias decreases as a function of the number of repeated trials (Fig. 4A). With only 2 trials the magnitude of the bias exceeded the magnitude of the signal (fractional bias ∼1.5), and fractional bias dropped to ∼0.15 for 20 trials and ∼0.025 for 100 trials. However, at all numbers of trials, the closed-form bias correction did a very good job at correcting bias (maximal fractional bias remaining after correction = 0.01 for 2 trials; Fig. 4A). In contrast, fractional bias remained high after the bootstrap correction for small numbers of trials (∼0.7 for 2 trials) but converged to the closed-form correction for more than 25 trials (Fig. 4A). Poor bootstrap performance with small sample size is a well-known phenomenon (Chernick 2007).
For a closer look at the closed-form bias correction, Fig. 4B displays the distribution of fractional error (total error/total signal) after correction across the 100 simulated experiments when 20 trials for each condition were collected. This distribution is centered around 0, thus confirming that the bias has been successfully removed, and it shows that the remaining average fractional error is small (<0.012 in magnitude) for individual experiments. These results support the validity of our procedure for estimating signal modulation magnitudes averaged across a population. However, we caution the reader that while the average signal modulation estimates are very accurate, no method can correct for the specific “noise” patterns within the data for a particular neuron. To illustrate this, Fig. 4C, left, displays the distribution of fractional error remaining after correction for a representative simulated neuron across the 100 simulated experiments. On average, the error was zero (thus showing no bias); however, on individual simulated experiments, the fractional error ranged from −0.45 to 0.46. Figure 4C, right, shows the “ground truth” response matrix for this neuron as well as one response matrix collected during a simulated experiment. As depicted by the “ground truth” matrix, this neuron was largely visual modulated and responsive to the visual presentation of image 4 but the response matrices collected in this simulated experiment reflect other types of modulation as a result of trial-by-trial variability; even after bias correction, these translate to signal modulation estimates that deviate from the true underlying value. These results demonstrate that the signal modulation magnitudes computed for any individual neuron need to be interpreted cautiously.
The simulations reported above were performed with spike counting windows of 50 ms, and with that size counting window we found that the closed-form bias correction was better at estimating bias than the bootstrap bias correction (Fig. 4A). We wondered whether the bootstrap might perform better than the closed-form correction for smaller counting windows where spike count distributions deviate more from the Gaussian assumption (e.g., are Poisson), and thus we compared both types of correction for spike count windows of 2 ms. In these narrow windows, the total fractional bias increased dramatically relative to the broader windows (bias was 16-fold larger than signal for 2 trials; Fig. 4D) but the closed-form bias correction continued to perform well (maximal magnitude fractional bias remaining after correction = −0.04 for 2 trials; Fig. 4D). In contrast, the bootstrap correction performed considerably worse at all numbers of trials, with the most discrepant differences for small numbers of trials; even with 10 trials, the average fractional bias remaining after bootstrap correction was 47% the magnitude of the signal (Fig. 4D). These results suggest that for small spike count windows and the numbers of trials typically collected in these types of experiments (n = 5–20), the bootstrap correction is highly inaccurate. In contrast, the closed-form bias correction is highly accurate within this regime despite its assumption of Gaussian-distributed trial-by-trial variability.
Relating signal modulations and task performance.
Quantifying the performance of individual neurons on a task by calculating the discriminability measure d′ is a commonly used approach to compare neurons within or between brain areas. For tasks that involve multiple experimental parameters or require the combination of multiple information sources to compute a solution, arriving at a quantitative understanding of how different signal types relate to task performance can be challenging. Here we derive this relationship for the DMS task (Fig. 1A). We then go on to demonstrate how this type of quantitative understanding can be used to, for example, determine which of many possible accounts can explain why two populations have different average d′, by applying the analysis to data collected in IT and PRH.
The DMS task described in Fig. 1A requires a subject to determine whether each test image is a target match or a distractor, and thus can be envisioned as a two-way discrimination between the set of all target matches versus the set of all distractors. Because target matches and distractors correspond to conditions on versus off the diagonal of the response matrix, respectively, we refer to this as “diagonal d′.” Diagonal d′ is calculated as the absolute value of the difference between the mean response to all target matches and the mean response to all distractors, divided by their pooled standard deviation:
| (11) |
Because target match modulations in IT and PRH result from both increases and decreases in the firing rates (e.g., Fig. 2, 3rd vs. 6th rows), the absolute value of diagonal d′ best quantifies the linearly discriminable match/distractor information in each neuron. Similar to the signal modulation bias described above, merely taking the absolute value of d′ produces a biased estimate of performance in which any modulations, including noise, translate into positive d′. In particular, note that this bias is directly dependent on the numerator of the d′ (i.e., the estimated absolute difference can be larger than 0 even if the true difference was 0), while the denominator corresponds to the classic estimator of the standard deviation and it does not have a direct impact on the bias (see appendix). To correct for the bias of the d′ we can thus focus on correcting for the bias of the numerator, which requires a calculation analogous to that described above for the case of signal modulations (Eq. 9). In particular, it is possible to show (see appendix) that the bias of the squared numerator is equal to
| (12) |
where mi indicates the response to the ith match, di indicates the response to the ith distractor, and T indicates the number of trials. Therefore, a corrected estimate of the absolute d′ can be obtained as
| (13) |
where is set to 0 if the numerator takes on a negative value. Below, we also apply the bias correction to estimate the orthonormal weights (Eq. 9).
To derive the relationship between the signal modulations of a neuron (Fig. 2) and its diagonal d′, we begin by computing the orthonormal basis weights (Eq. 4). As described above, rescaling a neuron's firing rate responses (e.g., multiplying a neuron's response matrix by 2) will result in a rescaling (i.e., a doubling) of all its weights, and, consequently, its signal modulations will also increase. To describe the signal modulations in a manner that does not depend on the overall scaling of firing, we normalized the weights by the grand mean spike count . We then considered the grand mean spike count as a separate term.
Using the orthonormal basis, the diagonal d′ of a neuron can be deconstructed as a function of three intuitive “signal strengths” (see appendix for the derivation):
| (14) |
The first signal strength, “D,” which we call the “diagonal strength” (Fig. 5) is computed as
| (15) |
where wdiag corresponds to the weight applied to the diagonal basis component (Fig. 1D). This signal strength determines the distance between the average of the diagonal responses (the target matches), and the average of the off-diagonal responses (the distractors), averaged across all images and all trials (Fig. 5B), and this term is proportional to diagonal d′ (Fig. 5C).
The second signal strength, “ND,” which we call the “nondiagonal strength” (Fig. 5), is computed as
| (16) |
where the weights used are those corresponding to the visual, working memory, and residual components (Fig. 1D). This term determines the spread of the firing rate responses within the target matches and within the distractors (Fig. 5B), and it is inversely related to diagonal d′ (Fig. 5C).
The final term (Fig. 5) is designed to capture the trial-by-trial variability of a neuron. When trial-by-trial variability is generated by a Poisson process, the grand mean spike count can be used as a good approximation of the variance across trials within each condition, averaged across the 16 conditions, and this term can be described by the inverse of the grand mean spike count (see appendix). This term is also inversely related to diagonal d′, as an increase in the spread within each condition will produce an overall increase in the spread across the set of all target matches and the set of all distractors.
We now demonstrate how understanding the relationship between different signal types and single-neuron task performance can be used to gain insight into neural processing by applying these analyses to our data from IT and PRH. We begin with the observation that diagonal d′ was significantly higher in PRH compared with IT (mean IT = 0.11, PRH = 0.19, P < 0.001; Fig. 5A). We can use the derivation of diagonal d′ presented above to discriminate between different possible explanations of why diagonal d′ is higher in PRH. Our decomposition suggests three possible factors that might account for this result (which are not mutually exclusive): 1) the diagonal strength (Fig. 5C) could be higher in PRH than in IT; 2) the nondiagonal strength (Fig. 5C) could be lower in PRH than in IT; and/or 3) grand mean firing responses (Fig. 5C) could be higher in PRH than in IT. First and foremost, the diagonal strength was significantly higher in PRH than in IT (Fig. 5D), suggesting that this factor contributed to higher average neuron diagonal d′ in PRH. Second, the nondiagonal strength was not significantly different between IT and PRH (Fig. 5E), suggesting that this factor could not account for the difference in neuron diagonal d′. Finally, the grand mean firing rates were slightly lower in PRH compared with IT but not significantly so (Fig. 5F), and, notably, lower firing rates in PRH are the opposite of what would be required to account for higher average PRH neuron diagonal d′ (Fig. 5C). Taken together, these results suggest that higher neuron diagonal d′ results from a twofold increase in diagonal structure within the response matrices of PRH neurons compared with IT neurons, as opposed to alternative explanations (such as increases in firing rate in PRH or more nondiagonal modulation in IT).
DISCUSSION
In our own work, we have found this method of estimating signal modulation magnitudes to be useful for a variety of applications. For example, we have used these signal quantifications as a benchmark to assess model performance (Pagan et al. 2013). We have also used these methods to compare the latencies with which specific types of signals arrive in different brain areas to infer the direction of information flow between them (Pagan et al. 2013). As described above, these methods can also be used to uncover the underlying source of differences in single-neuron performance measures between brain areas to gain insights into neural coding. These are but a few examples of the potential uses of this method.
Relationship to other analyses.
The method we describe here is similar to a multi-way ANOVA, but it incorporates two important extensions: it parses the signal into more terms, and it produces a bias-corrected estimate of signal modulation. For the DMS task described above, a two-way ANOVA would parse the total response variance into two linear terms, a nonlinear interaction term, and an error term. The two ANOVA linear terms map directly onto the summed squared projections onto the visual and working memory orthonormal basis vectors (e.g., in Eq. 5, the computation of Mvis and Mwm before taking the square root). Similarly, the ANOVA nonlinear interaction term maps onto the summed squared projections onto the “diagonal” and “residual” terms in our analysis. We note that parsing the diagonal signals from the other nonlinear terms is crucial in our analysis because this signal reflects the task solution, whereas the other types of nonlinear terms do not. The final term in the ANOVA analysis, the error term, is equal to the square of the σ̄noise term described in Eq. 6. We remind the reader that in this raw form the values of the orthonormal basis, as well as the ANOVA, are biased because of trial-by-trial variability (i.e., response matrix structure that arises from noise). The ANOVA deals with this bias by computing the probability (the P value) that each term is significantly higher than expected by chance, given the trial-by-trial variability, by considering the ratio between each term and the error term (the “F statistic”), based on the assumption that the noise is Gaussian distributed. However, the ANOVA does not produce bias-corrected estimates of signal modulation, whereas here we describe two ways to estimate and correct for this bias.
Our method also has similarities with an approach related to the ANOVA, MLR. Similar to our procedure, MLR seeks to describe a neuron's responses as a weighted sum of multiple terms. In practice, it is most often applied to continuous variables (e.g., motion direction or color), and often in cases in which one has an specific underlying model of how different stimulus parameters combine to determine a neuron's response (e.g., knowledge that neurons have Gaussian-shaped tuning functions for motion direction). MLR can also be applied in nonparametric cases, and when used in this way multiple terms are required to capture response modulation of a single variable type (e.g., for object identity, response = baseline + weight_1 × identity_1 + weight_2 × identity_2 + . . .) and, in fact, our method could be described as an MLR with the regressors specified by an intuitive orthonormal basis. When viewed from this perspective, our method can provide multiple insights for those wishing to perform this type of MLR. First, a crucial consideration with MLR is the degree to which the different regressors are correlated with one another, because the values of the weights (i.e., the “beta coefficients”) can be misleading in case of strongly correlated regressors. One solution to this problem is to orthogonalize the variables of interest, although we note that for some data sets the experimental variables simply cannot be orthogonalized (e.g., Fig. 1E). Our method provides a straightforward way to evaluate the degree to which different candidate experimental designs can be orthogonalized for MLR. Second, determining the weights for a complete orthonormal basis guarantees a full account of a neuron's spike count modulation, whereas an MLR against a few (e.g., linear) terms might provide only a partial account. Finally, if one desires to convert MLR “beta coefficients” into positive-valued measures of modulation, these measures will be biased in the exact same manner we describe above, and here we introduce a way to correct for that bias.
Our method also has similarities with PCA and related techniques (Machens 2010). A PCA applied to the mean firing rate responses of a population of neurons to N experimental conditions returns an orthonormal basis of N “eigenvectors,” and each neuron's mean firing rate response to the N conditions can be decomposed into a weighted sum of these vectors by projecting the neuron's responses onto the basis, as described above. PCA differs from our method in that the eigenvectors are produced via a procedure that iteratively determines the stimulus dimensions that account for the most response variance across the population with the constraint that each successive vector must be orthogonal to all the others. Consequently, PCA dimensions are not guaranteed to be intuitive. As an illustration, Fig. 6A shows the results of a PCA applied to our IT and PRH populations. While the two largest eigenvectors for each population are primarily visual, they are not purely so, and eigenvectors of rank three and lower capture mixtures of different types of modulation. Thus PCA is not very useful in providing an intuitive description of the types of signals reflected in these populations. Rather, PCA is most often used as a “dimensionality reduction” technique. For example, in the case of the reverse correlation method “spike-triggered covariance” (Schwartz et al. 2006) one applies a PCA to the spike-triggered stimuli in an attempt to find a small number of stimulus dimensions that can account for individual neuron's responses within a linear-nonlinear model framework.
Fig. 6.

Results of principal components analysis (PCA) and demixed PCA (dPCA). A: illustration of the orthonormal components corresponding to the 8 largest eigenvectors obtained by applying PCA to our IT and PRH data. B: 8 largest orthonormal components resulting from the application of dPCA.
One extension of the PCA framework, demixed PCA (dPCA; Brendel et al. 2011; Machens 2010) has recently been introduced as a solution to the “mixing” issues described above for PCA. dPCA allows one to specify the experimental parameters that should not be mixed and thus to perform dimensionality reduction within specific linear subspaces. It is advantageous over our method in scenarios in which, for example, one wants to determine whether the responses to a specific type of stimulus parameter can be captured with a simple (i.e., low dimensional) description or, equivalently, to uncover specific types of “tuning.” For example, dPCA has provided important insights into how the working memory delay period activity of neurons in prefrontal cortex depends on time (Machens et al. 2010). The results of a dPCA applied to our data in IT and PRH are shown in Fig. 6B. The input to dPCA included the neural responses to all conditions as well as the task parameters (i.e., the visual and target identities) associated with each condition. This information, which is not provided to traditional PCA, allows dPCA to search for a set of components that capture most of the modulation while avoiding mixing different types of signals (e.g., visual and working memory). In contrast to a regular PCA (Fig. 6A), the first three components in each area are almost exclusively visual and the fourth component for PRH corresponds to the “diagonal” component of the orthonormal basis. However, one can also see from this analysis that if the desired outcome is a characterization of “how much” of specific, predefined signal types are present in a population, the orthonormal basis provides a better approach for two reasons: 1) the components retrieved by dPCA still present some degree of “noise,” and thus if the relevant axes are known in advance it is better to measure their modulations directly, and 2) in situations in which one wants to make a quantitative comparison between two populations, some compromise has to be established when different dPCA components are retrieved for each population (e.g., compare IT and PRH in Fig. 6B).
Finally, a complementary approach for quantifying signals is to measure single-neuron performance either by a ROC analysis (e.g., Bennur and Gold 2011; Newsome et al. 1989; Swaminathan and Freedman 2012) or by the related (boundless) discriminability measure d′ (e.g., Adret et al. 2012; Gu et al. 2012; Liebe et al. 2011). Under the assumption that trial-by-trial variability is Gaussian distributed, one can convert between the two measures with a simple nonlinear function (i.e., the complementary error function; Dayan and Abbott 2001). As our results show, in a multiparameter task like DMS, single-neuron task performance does not necessarily depend on a single type of signal but instead can reflect the combination of multiple signal types. Additionally, it is important to note that if one wishes to compute a measure of task performance that is unsigned (i.e., by taking the absolute value or squaring), these task performance measures will be biased. However, this bias can be estimated and corrected with the approaches we describe here.
GRANTS
This work was supported by National Eye Institute Grant R01 EY-020851, an Alfred P. Sloan Research Fellowship to N. C. Rust, and a McKnight Foundation Scholar award to N. C. Rust.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.P. and N.C.R. conception and design of research; M.P. and N.C.R. performed experiments; M.P. and N.C.R. analyzed data; M.P. and N.C.R. interpreted results of experiments; M.P. and N.C.R. prepared figures; M.P. and N.C.R. drafted manuscript; M.P. and N.C.R. edited and revised manuscript; M.P. and N.C.R. approved final version of manuscript.
Appendix
Derivation of the bias correction for signal modulations.
When estimating the amount of modulation (or information) in a signal, noise and limited sample size are known to introduce a positive bias (see, e.g., Treves and Panzeri 1995). Here we quantify the magnitude of this bias for the weights associated to the different signal components (Eq. 4), which are used to produce the estimated modulation components (Eq. 5) of a single neuron.
We begin by making the simplifying assumption that responses to each condition j are normally distributed with mean μj and variance σj2 and that for each condition, μj is approximately equal to σj2. We indicate the estimate of the mean response μj to each condition as rj defined as the average of the responses sampled over T trials. The value of the estimate rj will itself be normally distributed, with mean equal to the true mean μj and variance equal to σj2/T.
By expanding Eq. 4, the estimated weight associated to each component i can also be written as
| (17) |
where rj indicates the neuron's average response to the jth condition and bij indicates the jth entry of the ith basis component. Since wi is a linear combination of normally distributed variables, it will also be normally distributed. The mean of wi will thus be equal to the linear combination of the means of the estimated rj (i.e., the true mean responses μj) and the entries bij, while the variance will be equal to the linear combination of the variances of rj (i.e., σj2/T) and the squared entries bij2:
| (18) |
Note that is the value of the “true” weight, and this estimate of wi is unbiased. However, a bias is introduced by squaring the estimated weight wi. The square of a normally distributed variable with nonzero mean takes the form of a noncentral χ2 distribution, whose mean is equal to the sum of the squared mean of the original normally distributed variable plus its variance. In our case:
| (19) |
Under the assumption that the variance for each condition is equal to its mean:
| (20) |
where the first term corresponds to the “true” squared weight and the second term represents the additive bias. If we substitute the true mean responses with their estimates, we obtain an estimator of the bias:
| (21) |
where R indicates the neuron's response matrix (“flattened” into a vector) and (biT)2 indicates the ith basis function, squared element by element. Finally, an unbiased estimator of wi2 is given by
| (22) |
Derivation of the bias correction for the diagonal d′.
The equation for the absolute value of the diagonal d′ is presented in Eq. 11. As in the case of signal modulations, for simplicity we proceed by estimating the bias for the squared diagonal d′. As above, we make the simplifying assumption that responses to each condition j are normally distributed with mean μj and variance σj2 and that for each condition, μj is approximately equal to σj2.
The numerator of the squared diagonal d′ is given by the square of the difference between the mean match response and the mean distractor response. Since the response to each condition is assumed to be normally distributed, the difference between mean match and mean distractor is a linear combination of normal random variables and is also normally distributed. The numerator is equal to the square of this value, and it thus follows a noncentral χ2 distribution, whose mean is equal to the sum of the squared mean of the original normally distributed variable plus its variance:
| (23) |
where mi indicates the mean response to the ith match and σmi,noise2 is its corresponding trial-by-trial variance, di indicates the mean response to the ith distractor and σdi,noise2 is its corresponding trial-by-trial variance, and we used the assumption that the trial-by-trial variance for a given condition is equal to its corresponding mean response. The bias of the numerator of the squared d′ is then equal to
| (24) |
The denominator of the squared diagonal d′ is equal to the pooled variance of the noise across matches and distractors. In the general case in which different conditions elicit different amounts of trial-by-trial variability, the denominator results in a linear combination of χ2 variables, and its parameters can only be estimated in an approximated form (Satterthwaite 1946). However, one can note that the estimate of each individual trial-by-trial variance is unbiased, and therefore a linear combination of unbiased quantities is unbiased itself; thus no bias is introduced by the denominator. Consequently, the bias of the diagonal d′ can be corrected by subtracting the bias of the squared numerator according to Eq. 24, dividing it by the estimate of the pooled variance, and taking the square root (Eq. 13).
Derivation of diagonal d′ as a function of the orthonormal basis.
Here we demonstrate that a neuron's diagonal d′ can be deconstructed into a function of three “signal strengths” defined in terms of the orthonormal basis presented in Fig. 1D. Diagonal d′ is defined as the absolute value of the difference between the mean response to all target matches and the mean response to all distractors, divided by the pooled standard deviation of the noise (Eq. 11).
The numerator of diagonal d′ can thus be expressed as the absolute value of the dot product between the flattened response matrix R and a similarly formatted vector c, in which the target matches are scaled by 1/4 and the distractors are scaled by −1/12:
| (25) |
where mi denotes the mean response to the ith match and di denotes the mean response to the ith distractor. The orthonormal basis function corresponding to the diagonal modulation bdiag is equal to c multiplied by √3 to impose unitary norm. As a result, the numerator of a neuron's diagonal d′ can be rewritten as
| (26) |
The denominator of the diagonal d′ is equal to the pooled standard deviation, i.e., the square root of the pooled variance (Eq. 11). Our goal is to arrive at a formulation of the pooled standard deviation as a function of the orthonormal basis weights.
We begin by expanding the terms for the variance of spike count responses to target matches σMatch2 and to distractors σDistractor2. If we indicate with mit the response to the ith match on the tth trial, σMatch2 can be rewritten as
| (27) |
where σ̄noise,Match2 indicates the average trial-by-trial variability across the four matches and mi denotes the mean response to the ith match. Similarly, if we indicate with dit the response to the ith distractor on the tth trial, σDistractor2 can be written as
| (28) |
where σ̄noise,Distractor2 is the average trial-by-trial variability across the 12 distractors and di is the mean response to the ith distractor. Now we substitute σMatch2 and σDistractor2 from Eqs. 27 and 28 into Eq. 11 and express the pooled standard deviation as
| (29) |
where σ̄noise2 indicates the average trial-by-trial variability across all conditions (as defined in Eq. 7) and σMD2 indicates the sum of the variance across matches and the variance across distractors:
| (30) |
We now wish to express σMD2 as a function of the orthonormal basis components. Here we indicate the average response to the ith condition as ri and the grand mean spike count across all conditions as and we derive an expansion of the sum of the squared responses by substituting :
| (31) |
Equation 31 can be rearranged as
| (32) |
The diagonal basis function bdiag and the grand mean basis function bmean are defined such that their weights wdiag and wmean take the following values:
| (33) |
Because b1 . . . b16 form an orthonormal basis,
| (34) |
Substituting Eqs. 33 and 34 into Eq. 32 allows us to derive
| (35) |
We now substitute Eq. 35 into Eq. 29:
| (36) |
Diagonal d′ can thus be written as
| (37) |
In the raw formulation of the weights, rescaling a neuron's firing rate responses (e.g., multiplying a neuron's response matrix by 2) results in a rescaling (i.e., a doubling) of all its deconstructed matrix weights, and, consequently, modulations due to changes in the pattern of responses within the matrix and overall firing rates are entangled. To capture matrix structure in a manner that does not depend on the overall scaling of firing, we compute the “normalized weights” by dividing each weight by the grand mean spike count . Dividing both numerator and denominator of Eq. 37 by allows us to express diagonal d′ as a function of the normalized weights:
| (38) |
Finally, we express diagonal d′ as a function of three components:
| (39) |
where
| (40) |
using the assumption that is equal to σ̄noise2.
REFERENCES
- Adret P, Meliza CD, Margoliash D. Song tutoring in presinging zebra finch juveniles biases a small population of higher-order song-selective neurons toward the tutor song. J Neurophysiol 108: 1977–1987, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennur S, Gold JI. Distinct representations of a perceptual decision and the associated oculomotor plan in the monkey lateral intraparietal area. J Neurosci 31: 913–921, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel W, Romo R, Machens CK. Demixed principal component analysis. Adv Neural Inform Process Syst 24: 222–223, 2011 [Google Scholar]
- Brody CD, Hernandez A, Zainos A, Romo R. Timing and neural encoding of somatosensory parametric working memory in macaque prefrontal cortex. Cereb Cortex 13: 1196–1207, 2003 [DOI] [PubMed] [Google Scholar]
- Buckley MJ, Mansouri FA, Hoda H, Mahboubi M, Browning PG, Kwok SC, Phillips A, Tanaka K. Dissociable components of rule-guided behavior depend on distinct medial and prefrontal regions. Science 325: 52–58, 2009 [DOI] [PubMed] [Google Scholar]
- Chernick MR. Bootstrap Methods: A Guide for Practitioners and Researchers (2nd ed.) New York: Wiley, 2007 [Google Scholar]
- Dayan P, Abbott LF. Theoretical Neuroscience. Cambridge, MA: MIT Press, 2001 [Google Scholar]
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Boca Raton, FL: CRC, 1994 [Google Scholar]
- Gu Y, Deangelis GC, Angelaki DE. Causal links between dorsal medial superior temporal area neurons and multisensory heading perception. J Neurosci 32: 2299–2313, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe S, Logothetis NK, Rainer G. Dissociable effects of natural image structure and color on LFP and spiking activity in the lateral prefrontal cortex and extrastriate visual area V4. J Neurosci 31: 10215–10227, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machens CK. Demixing population activity in higher cortical areas. Front Comput Neurosci 4: 126, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machens CK, Romo R, Brody CD. Functional, but not anatomical, separation of “what” and “when” in prefrontal cortex. J Neurosci 30: 350–360, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri FA, Buckley MJ, Tanaka K. Mnemonic function of the dorsolateral prefrontal cortex in conflict-induced behavioral adjustment. Science 318: 987–990, 2007 [DOI] [PubMed] [Google Scholar]
- Maunsell JH, Sclar G, Nealey TA, Depriest DD. Extraretinal representations in Area-V4 in the macaque monkey. Vis Neurosci 7: 561–573, 1991 [DOI] [PubMed] [Google Scholar]
- Miller EK, Desimone R. Parallel neuronal mechanisms for short-term memory. Science 263: 520–522, 1994 [DOI] [PubMed] [Google Scholar]
- Naya Y, Suzuki WA. Integrating what and when across the primate medial temporal lobe. Science 333: 773–776, 2011 [DOI] [PubMed] [Google Scholar]
- Newsome WT, Britten KH, Movshon JA. Neuronal correlates of a perceptual decision. Nature 341: 52–54, 1989 [DOI] [PubMed] [Google Scholar]
- Pagan M, Urban LS, Wohl MP, Rust NC. Signals in inferotemporal cortex and perirhinal cortex suggest an untangling of visual target information. Nat Neurosci 16: 1132–1139, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri S, Senatore R, Montemurro MA, Petersen RS. Correcting for the sampling bias problem in spike train information measures. J Neurophysiol 98: 1064–1072, 2007 [DOI] [PubMed] [Google Scholar]
- Rigotti M, Barak O, Warden MR, Wang XJ, Daw ND, Miller EK, Fusi S. The importance of mixed selectivity in complex cognitive tasks. Nature 497: 585–590, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romo R, Brody CD, Hernandez A, Lemus L. Neuronal correlates of parametric working memory in the prefrontal cortex. Nature 399: 470–473, 1999 [DOI] [PubMed] [Google Scholar]
- Satterthwaite FE. An approximate distribution of estimates of variance components. Biometrics Bull 2: 110–114, 1946 [PubMed] [Google Scholar]
- Schwartz O, Pillow JW, Rust NC, Simoncelli EP. Spike-triggered neural characterization. J Vis 6: 484–507, 2006 [DOI] [PubMed] [Google Scholar]
- Swaminathan SK, Freedman DJ. Preferential encoding of visual categories in parietal cortex compared with prefrontal cortex. Nat Neurosci 15: 315–320, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves A, Panzeri S. The upward bias in measures of information derived from limited data samples. Neural Comput 7: 399–407, 1995 [Google Scholar]